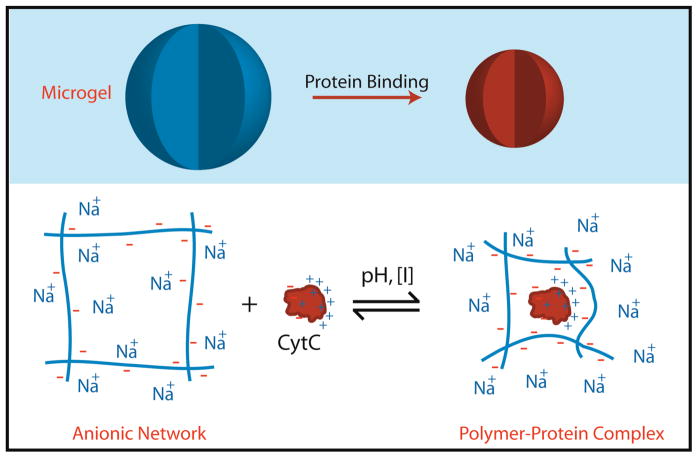
Abstract
The binding of cytochrome c to pH and thermoresponsive colloidal hydrogels was investigated using multiangle light scattering, measuring loading through changes in particle molar mass and root mean square radius. Loosely cross-linked microgels [composed of a random copolymer of N-isopropylacrylamide (NIPAm) and acrylic acid (AAc)] demonstrated a high loading capacity for protein. Encapsulation was dependent on both the charge characteristics of the network and the salinity of the medium. Under favorable binding conditions (neutral pH, low ionic strength), microgels containing the highest studied charge density (30 mol% AAc) were capable of encapsulating greater than 9.7 × 105 cytochrome c molecules per particle. Binding resulted in the formation of a polymer-protein complex and condensation of the polymer. Anionic microgels demonstrated a change in density ~20-fold in the presence of oppositely charged proteins. These studies of cytochrome c encapsulation represent a significant step towards direct measurement of encapsulation efficiency in complex media as we pursue responsive nanogels and microgels for the delivery of macromolecular therapeutic agents.
Introduction
The development of nanoparticles as competent drug delivery vehicles remains as a challenging yet promising pursuit within materials science. In principle, nanoparticles are capable of improving the pharmacological properties of existing and new therapeutic agents. Dozens of nanoparticle-based drug products have already been approved for clinical use,1 and several new nanoparticle delivery platforms are expected to emerge in the coming years. Particular enthusiasm has arisen in the delivery of new classes of drug agents, such as therapeutic proteins or small interfering RNA (siRNA).2
Effective delivery systems are greatly needed for therapeutic proteins, which are characterized by several undesirable physicochemical properties that limit their widespread medical use. For example, the bioavailability of proteins is generally poor since they are unable to cross many biological membranes. Unlike more stable compounds, protein drugs are highly susceptible to loss of their pharmacologically-active structure (e.g. by proteolysis, oxidation, deamidation), decreasing their therapeutic activity.3 Proteins are also sensitive to their local environment, being prone to aggregation, adsorption, or denaturation.4 To overcome the shortcomings of protein drugs, hydrogel particles (e.g. micro- or nanogels) may be used to encapsulate, protect, and subsequently release the agents at disease sites.5–6 Hydrogels are composed of loosely cross-linked hydrophilic polymers, yielding a low density structure that can be used to encapsulate proteins. Importantly, using microgels and nanogels scales the favorable encapsulation properties down to the sub-cellular level, suggesting the potential for cellular targeting of therapeutics. Microgels are also mechanically flexible,7 a property that modulates cellular uptake and biodistribution.8–10 In addition to those features, stimuli-responsive microgels may be synthesized, yielding particles that undergo rapid changes in swelling in response to an external stimulus (e.g. pH, ionic strength, temperature).11 Hydrogel deswelling due to environmental triggers has been proposed as a mechanism to drive the release of internalized drug compounds.3, 12–13
Several researchers have examined the encapsulation of therapeutic peptides and proteins within hydrogels.3, 13–20 In previous studies, protein loading was shown to depend on the physicochemical properties of the gel (e.g. cross-link density, charge, hydrophilicity) and the strength of protein-polymer interactions within the polymer network. However, the majority of reports have focused on encapsulation within gels of macroscopic dimensions. Whereas macroscopic gels show promise for certain delivery routes (e.g. implantable drug depots, topical application, oral administration), the smaller dimensions of colloidal particles enables access to regions of the body inaccessible to their larger counterparts. For example, we and others are pursuing microgels and nanogels as drug delivery vehicles for siRNA and therapeutic proteins.18, 21–24 In work described below, we systematically investigated the loading of cationic proteins within anionic microgels (<0.7 μm in diameter), assessing the relationships between microgel charge and binding stoichiometry. Encapsulation was measured by monitoring the changes in microgel molar mass and radius upon protein loading via multiangle light scattering (MALS).
Methods
Materials
All reagents were purchased from Sigma-Aldrich (St Louis, MO) and used as received, unless otherwise noted. The monomer N-isopropylacrylamide (NIPAm) was recrystallized from hexanes (VWR international, West Chester, PA) and dried in vacuo prior to use. Reagents N,N′-methylenebisacrylamide (BIS), sodium dodecyl sulfate (SDS), and ammonium persulfate (APS) were all used as received. Water used in all reactions and particle purifications was purified to a resistance of 18 MΩ (Barnstead E-Pure system), and filtered through a 0.2-μm filter to remove particulate matter.
Synthesis
Particle syntheses were performed via surfactant-stabilized precipitation polymerization, using a modified approach to that previously reported.25 The total monomer concentration of all reactions was maintained at 100 mM with a total reaction volume of 100 mL. A series of microgels were synthesized with varying AAc content (30%, 20%, 10%, and 0% of the total monomer concentration) with identical BIS content (2%). The concentrations of NIPAm monomer were adjusted accordingly to achieve the desired total monomer content. Reactions were performed by first dissolving NIPAm, AAc, and BIS in distilled, deionized water. A small amount of SDS was added to each suspension to yield a total surfactant concentration of 1 mM. Monomer solutions were filtered through 0.2 μm Acrodisc syringe filters and subsequently added to 200 mL three-neck round-bottom flasks. Once equilibrated at 70 °C, the reaction mixtures were purged with N2 for 1 hr while stirring (400 RPM). The polymerizations were initiated by delivering a 1.0 mL aliquot of 0.100 M APS solution by pipette. All reactions were allowed to proceed for 24 hours under an N2 blanket while continuously stirring. Once cooled, reaction products were filtered through 0.8 μm Acrodisc syringe filters and purified via repeated ultracentrifugation and resuspension in distilled, deionized water.
Microgel Characterization
The weight-average molar mass (Mw) and refractive index increment (dn/dc) values of all microgels were determined via the Calypso syringe pump system (Wyatt Technology Corporation, Santa Barbara, CA), equipped with MALS and dRI detection. A diagram of the instrument configuration is shown in Scheme S1. The flow system consisted of a computer-controlled triplet syringe pump and a multichannel degasser, equipped with in-line mixers and valves to allow rapid and automated batch measurements. Multiangle light scattering was performed using the DAWN-EOS (Wyatt Technology Corporation, Santa Barbara, CA) equipped with a temperature-regulated K5 flow cell with a GaAs laser light source (λ = 685 nm). Data collection and subsequent light scattering analysis was performed using the Astra software Version 5.3.4.14 (Wyatt Technology Corporation, Santa Barbara, CA). Accurate measurements of microgel molar mass require the characterization of the particle refractive increment (dn/dc). Differential refractive index analysis was performed via composition-gradient static light scattering using the Calypso and Optilab rEX system, equipped with an LED light source (λ = 690 nm). The diffusion coefficient values of particles were measured via dynamic light scattering (DLS). DLS experiments were performed at a 90° scattering angle using a Protein Solutions DLS (Wyatt Technology Corporation, Santa Barbara, CA). Hydrodynamic radii (rh) were calculated from measured diffusion coefficients through the Stokes-Einstein relation (rh = kT/6πηD, where D is the diffusion coefficient, k is Boltzmann’s constant, η and T are the solution viscosity and temperature). Microgel electrophoretic mobility values (vE) were determined using a Malvern Zetasizer Nano. The ζ-potential (ζ) values were derived from the Smoluchowski relation (ζ = vEη/ε0ε, where η is the solution viscosity, ε0 is the relative dielectric constant, and ε is the electrical permittivity). It should be noted, however, that the ζ-potential is only applicable to microgels in a semi-quantitative manner, since no well-defined slipping plane exists between the microgel surface and the medium. Additionally, most of the charges in the microgels are buried, and contribute only fractionally to the calculated ζ-potential.
Protein Binding Analysis
Protein loading was quantified in this work via MALS. MALS is frequently employed to measure the root-mean-square radius (rrms) and weight-average molar mass (Mw) of polymers. The principles employed in MALS measurements have been described in detail elsewhere.26 At low concentration, suspensions of polymers scatter light with an intensity governed by the molar mass of the polymer and the concentration. For large particles ( ), angular dependence to the scattered intensity is observed, requiring extrapolation to 0° scattering angle to accurately characterize the polymer mass. It is at low scattering angles where a robust relationship exists between concentration, molar mass, and scattering intensity according to the following equation,
| (1) |
where , c is the polymer concentration, R(θ) is the Rayleigh ratio, and λ is the wavelength of light in vacuo. The bracketed component of Eq. 1 is often referred to as the scattering function of the polymer, P(θ). For spheres, extrapolations of the scattering data are most accurate with the Debye method for constructing the Debye plot.26 A Debye plot is constructed with R(θ)/Kc as the ordinate and sin2(θ/2) as the abscissa (Figure S5). Using Eq. 1, the Mw is derived from the intercept (0° scattering angle), whereas the rrms is derived from the slope at the intercept, ∂(R(θ)/Kc)/∂(sin2(ϑ/2). For large spheres (rrms > 50 nm) the angular dependence of scattering becomes more complex, requiring higher order polynomial fitting to extrapolate to low angles with accuracy. For accurate measurement of Mw, the differential refractive index increment (dn/dc) was measured for all microgels in pH 7 phosphate buffer. The dn/dc value is an important factor in accurate molar mass determination, being a component of the optical constant K in Eq. 1, and is dependent on solvent conditions. Thus, the dn/dc values of microgels were determined in several different ionic strength environments and in the presence of cytochrome c.
For estimations of protein loading via light scattering, the overall change in Mw must be approximated for microgels in the presence or absence of protein. However, it is important to note that changes in particle swelling from loading introduces error into the approximation of Mw, where an increase in polymer segment density causes an increase in the intensity of scattering and thus the calculated change in molar mass. In order to compensate for this error, one may simply account for the overall change in particle volume in the loaded and unloaded states to more accurately estimate an overall change in Mw from binding via Equation 2
| (2) |
where V0 is the volume of the microgel in the absence of protein and Vloaded is the volume of the microgel in the presence of the macromolecule. The validity of this approach was investigated in this work by comparing the measured loading via light scattering with offline methods (i.e traditional centrifugation/supernatant assay methods) and by comparing the measured values with reported loading capacities for similar hydrogels in the literature (described below in the Discussion section).
Solutions of microgels were prepared by diluting a purified stock of particles to a concentration of 1.50 μg/mL, 0.86 μg/mL, 0.663 μg/mL, or 1.82 μg/mL for 30 mol%, 20 mol%, 10 mol% and 0 mol% AAc microgels, respectively in pH 7.0 phosphate (10 mM, ionic strength = 20 mM). All buffers were filtered through 0.1 μm syringe filters prior to use. In a typical measurement, a baseline was first measured by delivering buffer alone through syringe S1 of the delivery system (from reservoir R1) (Scheme S1). Microgel solutions (loaded in reservoir R2) were subsequently introduced using the computer-controlled tri-syringe dispenser. The Calypso software enables programmable control over reactant concentrations through the volume and rate of syringe flow. For reported microgel Mw and rrms values in this work, particles were diluted to 50% concentration by dispensing and mixing the microgel solutions (S2) with buffer (S1). Scattering data was collected at 15 angles (0.5 s collection interval) and the resultant scattering curves were interpreted using the ASTRA software.
Measurement of microgel-protein interactions at a single concentration of cyt c was performed by dissolving the protein to a concentration of 48 μM in pH 7.0 phosphate buffer (10 mM, ionic strength = 20 mM). Protein solutions were filtered through 0.1 μM syringe filters to remove any particulate matter. The final concentration of protein stock was verified through differential refractometry. A baseline scattering signal was first measured by delivering cyt c (loaded in syringe S3) with buffer (loaded in syringe S1), resulting in 50% dilution ([cyt c] = 24 μM). After baseline measurement, the previously described microgel stock solutions (loaded in syringe S2) were administered with cytochrome c (syringe S3), diluting both reactants to 50% stock concentration. The Mw and rrms of each particle type was measured at equilibrium via MALS, subtracting the baseline light scattering contribution from the protein alone. In an investigation of the effect of ionic strength on binding, the buffer used in microgel and protein stock preparation was replaced with pH 7.0 phosphate (10 mM) with varying salinity (ionic strength range of 20 – 150 mM).
Traditional centrifugation/supernatant assay methods were used to validate the light scattering approach for binding measurements. Solutions were prepared at 24 μm cyt c and varying 30 mol% AAc microgel concentrations (3.7 × 10−4 – 1.7 × 10−5 g/mL polymer) in pH 7.0 phosphate buffer (10 mM, ionic strength = 20 mM). Microgels were sedimented at 50,000 RPM (RCF = 136,000 × g) for 15 min using an Optilab Preparative Ultracentrifuge (Beckman Coulter, USA). Supernatant solutions were collected and assayed for protein concentration via standard curve using a UV-1601 spectophotometer (Shimadzu Corporation, Kyoto, Japan). Protein concentrations in the supernatant were used to calculate the mass of bound cyt c per gram of polymer, enabling estimation of the polyelectrolyte capacity for the microgel (PC = g protein/1 g polymer). It is important to note that the polymer concentrations employed in this assay are much greater than those employed in light scattering analysis. Thus, the PC value for microgels was estimated using the concentration series to extrapolate the PC value to lower polymer concentrations (~0.7 μg/mL).
Microgel Size Analysis
Investigations of microgel size by DLS were performed offline at particle concentrations of 0.04 mg/mL, 0.04 mg/mL, 0.01 mg/mL, and 0.005 mg/mL for 30 mol%, 20 mol%, 10 mol %, and 0 mol% AAc microgels, respectively. Samples were prepared by diluting a concentrated, purified stock of particles to the indicated concentration in pH 7.0 phosphate buffer. Microgel rh was measured at 25 °C using the Dynapro DLS. The ζ-potential of microgels was measured at the same stock concentrations and in identical buffers. In an investigation of microgel binding, 30 mol% AAc microgels were diluted in a concentrated protein stock ([cyt c] = 24 μM). The diffusion coefficients for the microgels at varying ionic strength were measured, enabling calculation of particle rh in response to protein binding.
Titrations
Potentiometric titrations were performed to measure acid incorporation into microgels using a Corning pH Meter 430 (Corning Incorporated, NY). Microgels were diluted to a total concentration of 1 mg/mL and a volume of 20.00 mL in distilled, deionized water. Samples were titrated with 0.11 M NaOH under an N2 purge while stirring at 22 °C. Measurements of solution pH were made after 300 s equilibration after each addition of titrant.
Results and Discussion
To systematically investigate the role of hydrogel charge in protein encapsulation, we synthesized a series of poly(N-isopropylacrylamide-co-acrylic acid) microgels, loosely cross-linked with N,N′-methylenebisacrylamide (BIS, 2 mol%). Acrylic acid (AAc) was chosen as a comonomer since it incorporates quantitatively into pNIPAm microgels during precipitation polymerization, as reported previously.27 In order to tune the acid content, microgels were synthesized by varying the mol% AAc added during precipitation polymerization. The set of synthesized microgel particles had similar weight-average molar mass (Mw) values, densities (ϱ), and topological features (Table 1). The topology of the spheres may be inferred through the ratio of the rrms and the rh values.28 Ratios observed in this work (rrms/rh < 0.7) are indicative of a radially heterogeneous network topology. Heterogeneity in the microgel structure is imparted through non-uniform cross-linker incorporation during precipitation polymerization; particles prepared in this fashion typically have greater cross-linking within their interior than the periphery.29 Additionally, the microgels have a low density, a common characteristic for superabsorbent hydrogels. The density was calculated from the mass of a single microgel per the measured volume (from DLS analysis). Acid incorporation was quantified via potentiometric titration and was found to scale quantitatively with the moles of AAc included in synthesis for 30 mol% (2.9 × 10−3 mol COOH/g polymer), 20 mol% (1.9 × 10−3 mol COOH/g polymer), and 10 mol% AAc (1.1 × 10−3 mol COOH/g polymer) particles (Figure S1). The ζ-potential for all spheres was measured to determine the influence of the AAc content on particle ionization at neutral pH. The magnitude of ζ-potential was found to increase with increasing amounts of AAc included during synthesis, as reported in Table 1.
Table 1.
Physical properties of microgels synthesized via precipitation polymerization.1
| AAc Content (mol%) | rh, DLS (nm) | rrms, MALS (nm) | Mw (g/mol) | ρ (g/mL) | ζ-potential (mV) |
|---|---|---|---|---|---|
| 30 | 358 ± 3 | 230.5 ± 0.5 | 2.60 ± 0.02 × 109 | 2.26 ± 0.01 × 10−2 | −32.9 ± 0.8 |
| 20 | 327 ± 4 | 187.5 ± 0.2 | 1.62 ± 0.01 × 109 | 1.89 ± 0.01 × 10−2 | −26.6 ± 0.5 |
| 10 | 229 ± 4 | 138.0 ± 0.3 | 9.41 ± 0.01 × 108 | 3.10 ± 0.01 × 10−2 | −16.4 ± 0.7 |
| 0 | 121 ± 1 | 76 ± 2 | 2.91 ± 0.01 × 108 | 6.4 ± 0.1 × 10−2 | −0.7 ± 0.7 |
Error represents ± one standard deviation in the error of measurements in replicate (n = 4).
Protein binding was measured for the charged microgels in the presence of cytochrome c (cyt c). The structure and charge distribution of cyt c have been explored in-depth,30 whereas the binding characteristics of the protein to macroscopic hydrogels has also been reported.15–16 The protein is a small, highly water soluble, heme protein with a hydrodynamic diameter of 3.5 nm (1.75 nm Rh, by DLS).30 The heme ligand is located in a lysine-rich region of the protein, imparting a positive charge to the macromolecule at pH 7.31 It has been hypothesized that this front face electrostatically guides the cytochrome complex towards negatively charged proteins and lipids in vivo. The protein has a net charge of +9.3 (pI = 10.1, Mw = 12,327 Da) at neutral pH.31
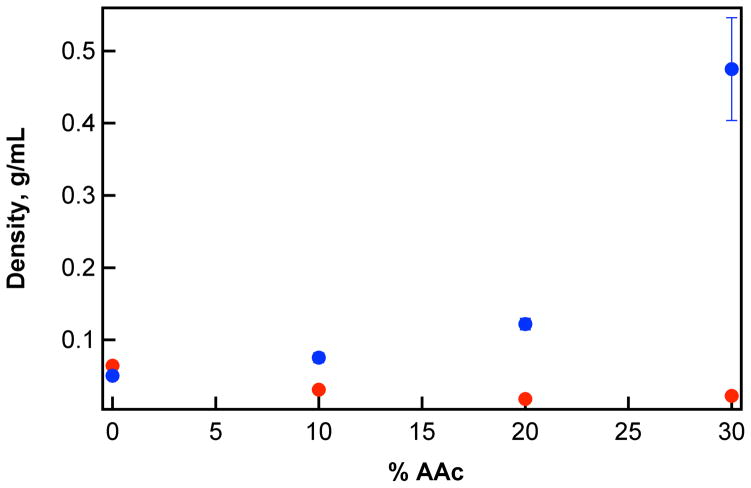
We employed multiangle light scattering (MALS) to quantitatively determine the relative cyt c binding to microgels of varying AAc content. The Mw values of dilute microgels in buffer alone (0.332 – 0.910 μg/mL) were compared to the Mw measured in the presence of 24 μM cyt c (~300 μg/mL) (Figure 1).
Figure 1.
The AAc content of microgels influences binding to cyt c, resulting in an increase in apparent microgel Mw in the presence (blue, 24 μM protein) versus absence (red) of cyt c. Inset: calculated polyelectrolyte capacity (PC) for microgels. Error bars represent one standard deviation about the mean of measurements performed in triplicate.
A significant increase in Mw was observed for microgels containing AAc in the presence of cyt c (Figure 1). Notably, the Mw of 30 mol% AAc microgels increased by approximately an order of magnitude, indicating a large amount of cyt c loading within the hydrogel network. The loading results in a decrease in particle radius (rh and rrms). The rrms of microgels containing 30 mol% AAc decreased from 229 nm to 174 nm from the empty to loaded states, respectively (pH 7.0, I = 20 mM, [cyt c] = 24 μM). This size change correlates to a volume decrease of ~55% during the binding event. A similar volume decrease was observed in the measurement of rh by DLS under the same solution conditions (~53%) (Figure S3). Microgels interact with oppositely charged cyt c via polyelectrolyte interactions. Coulombic forces between the microgel and cyt c results the formation of a polymer-protein complex, releasing counterions associated with the polymer and protein. Entropy is gained through counterion release, providing a net gain in free energy which drives the formation of a cross-linked protein-polyelectrolyte complex (Scheme 1).16
Scheme 1.
Proposed interaction of microgels with oppositely charged cyt c.
Using the scattering data, the polyelectrolyte capacity (PC) of the polymer may be calculated (Figure 1). The PC is a frequently used metric for the loading capacity of macroscopic hydrogels reported in the literature.16 Conventionally, the amount of loaded material is measured by weighing the dehydrated bulk gels (equation 2).
| (3) |
Using MALS, the PC is instead determined through the ratio of the mass of loaded protein per the mass of a single sphere. The PC was derived through the simplified relationship (see Supporting Information for a derivation):
| (4) |
To validate the MALS method for the measurement of PC in this fashion, traditional centrifugation/supernatant recovery assay methods were used to measure the fraction of protein bound under similar conditions (pH 7.0, [cytc] = 24 μm). As described in the Methods, microgels at varying concentration were mixed with the protein and subsequently separated by centrifugation. The concentration of bound cyt c was quantified by analysis of the supernatant by UV-vis (Figure S2). The PC was found to converge to a value of ~4.4 for the particles in the presence of excess cyt c. Using the MALS approach, the PC was found to be ~4.5 for the 30 mol% AAc microgels, which is in good agreement with traditional centrifugation/supernatant assay methods for measuring encapsulation. To further validate the MALS method, the calculated PC for 30 mol% AAc microgels was compared with the PC values for macroscopic anionic hydrogels reported in the literature. The PC value varies significantly with the pH and the ionic strength of the medium. However, Kabanov et al. have reported PC values ranging from 13.2 – 16.5 for poly(acrylic acid) macroscopic gels under similar binding conditions to those reported in this work (pH 7, low ionic strength).16 From the PC range reported by Kabanov (for AAc homopolymer gels), one would anticipate a PC value for 30 mol% AAc microgel to range from ~4.0 – 5.0. Our measured PC value for 30 mol% AAc microgels (~4.5) is thus in good agreement with the expected capacity of the hydrogels for protein.
Using MALS, loading was quantified for each particle in solutions of high protein concentration ([cytc] = 24 μM). For instance, the 30 mol% AAc microgels bind ~9.7 × 105 molecules of cyt c per sphere. Using the number of bound proteins and the volume of the microgel, we approximated the effective internal protein concentration to be ~17 mM (~210 g/L), which is a ~700-fold increase in the protein concentration relative to the 24 μM solution used for loading. The loaded concentration exceeds the solubility of cyt c in water (100 g/L at 25 °C), which suggests that the microgel network facilitates solubilization of the protein. Microgels with lower AAc content loaded less cyt c: 20 mol% AAc microgels bound ~4.0 × 105 cyt c and 10 mol% AAc microgels bound ~1.0 × 104 cyt c per sphere. Those loading stoichiometries correspond to effective internal protein concentrations of ~4.0 mM and ~0.6 mM cyt c for 20 mol% and 10 mol% AAc particles, respectively. For particles synthesized without AAc, the MALS method was unable to detect a significant increase in light scattering in the presence of cyt c, which suggests minimal interaction with the protein.
Additional differences between particles were found by comparing the mole fraction (θ) of acids in the microgel per the total moles of cytochrome c loaded (θ= moles AAc available/moles cyt c loaded). Whereas the PC value reflects the total capacity of the polymer for protein, the value of θ reflects the capacity of the microgel to interact with the protein on a per site basis. For example, particles with 30 mol% AAc bind a significantly greater fraction of charged groups (θ = 0.13) than either the 20 mol% (θ = 0.05) or 10 mol% AAc microgels (θ = 0.01). In other words, the proteins are able to access a much larger number of the available charges within the high acid content microgels. The higher loading capacities for 30 mol% AAc microgels likely result from the increased network swelling, caused by the Donnan effect.27, 32 The higher charge density in these microgels increases swelling pressure in the network. Gel swelling yields a lower polymer segment density in the network and greater gel porosity, yielding a higher loading capacity for cyt c. In the case of the lower acid content particles, it appears that the decrease in charge density dramatically decreases the accessibility of those charges to protein binding. This effect was especially apparent for 10 mol% AAc microgels (θ = 0.01), where the amount of protein binding (Figure 1) is commensurate with a sub-monolayer of the cyt c on the particle surface. The low binding stoichiometry suggests limited access of the protein to the interior polymer network in the case of low acid-content microgels. It may be the case that the initial protein binding events are surface localized, causing a condensation of the particle periphery, which limits subsequent protein binding within the microgels. Conversely, for the higher acid content microgels, the initial protein binding events may occur deeper within the microgel or may cause less network condensation, thereby presenting less of a steric hindrance to subsequent binding events.
Previous reports have described an anti-cooperative mechanism for binding between microgels and cationic molecules, being strongly dependent on the physicochemical properties of the loaded macromolecule (e.g. charge, hydrophobicity, molar mass).15–16, 20 For example, the uptake of lysozyme (a small, cationic protein similar to cyt c) results in condensation of the polymer network in the periphery of particles, forming a collapsed shell and a semi-swollen particle interior.20 Protein-protein interactions cause lysozyme to aggregate in the shell, resulting in a biphasic distribution of loaded constituents at the final stages of loading (with the greatest protein concentration in the particle periphery). Malmsten and coworkers noticed a similar effect in loading cationic peptides into charge microgels, where hydrophobic interactions between peptides greatly influences the resulting distribution of macromolecules in the microgels.33 Although cytochrome c has been reported to form a collapsed polymer shell during loading, a recent study has shown that the high stability of the molecule (by protein-protein repulsion) enables diffusion through the collapsed shell during loading, eventually resulting in a uniform distribution of protein throughout the microgel at equilibrium.15 Guided by the literature precedent, we anticipate that the highly acidic microgels (30 mol% AAc) likely load cytochrome c with a similar mechanism as their macroscopic counterparts, resulting in a uniform distribution of proteins within the structure (Scheme 1). However, the decreased accessibility of the charge groups within 20 mol% and 10 mol% AAc microgels (θ = 0.05 and 0.01, respectively) may suggest a different loading mechanism, where the decreased swelling of the particles limits loading within the microgel exterior. However, additional work is needed to verify this hypothesis.
The interaction of cyt c with polyanionic microgels results in a highly condensed polymer structure presumably due to the release of counterions and solvent from the network, causing an overall decrease in particle volume. The observed decrease in volume, coupled with the increase in particle molar mass, results in a density increase (Figure 2). For example, the density of 30 mol% AAc microgels increases ~20-fold in the presence of cyt c. For reference, the density of globular proteins typically ranges from 0.8–1.2 g/mL (density of cyt c = 0.9 g/mL). The microgel density increase is representative of a transition from of a porous, highly-swollen network (~0.02 g/mL), to a condensed, protein-rich material (~0.5 g/mL). This result may suggest that protein loading affects the modulus of the microgel particles. Others have described a strong relationship between particle shear modulus with the ionization of the network.34 Protein loading and the occupancy of available charged groups may decrease network ionization, and thus decrease particle softness. This outcome may influence the deformability of particles in response to applied forces.
Figure 2.
Influence of cyt c binding on density for loaded (blue, 24 μM) and unloaded (red) microgels. Density values for microgels were calculated through measurement of microgel volume and molar mass via DLS and MALS, respectively (pH 7.0, I = 20 mM, [CytC] = 24 μM).
Under neutral pH conditions, AAc-functionalized microgels are ionized, and in the case of AAc containing pNIPAm microgels, have a uniform distribution of charges throughout the particle structure.27 Conversely, cyt c is positively charged due to the presence of a lysine-rich region of the macromolecule (adjacent to the heme group). The mixing of microgels and cyt c under low ionic strength conditions results in microgel-protein association (Figures 1 and 2). However, the extent and strength of Coulombic interactions between polyanionic gels and cyt c is dependent on the solution composition, as reported for macroscopic hydrogels.15–16, 20 For example, changes in pH or ionic strength modulate the swelling of charged hydrogels. These volume changes are primarily driven by the distribution of ions between the internal gel network and the external environment by the Donnan potential.27, 32 This additional swelling pressure is thus dependent on the AAc content, pH, and the salt concentration in the medium. Additionally, as the ionic strength is increased, the strength of Coulombic interactions between ionized gel and the loaded protein is diminished as the charges become more screened.
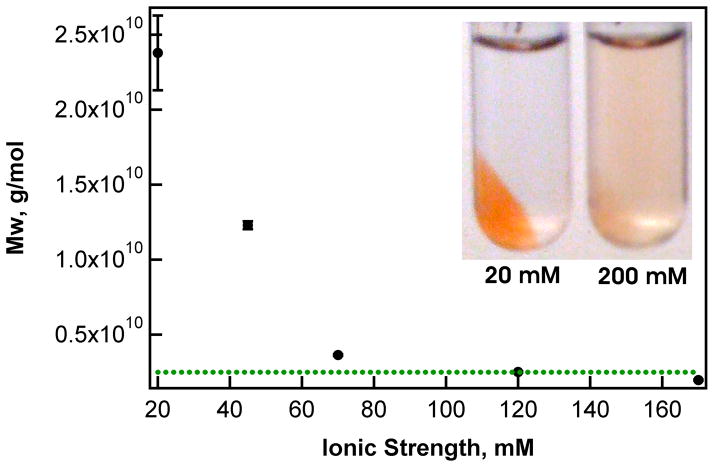
To look more closely at this effect, the impact of solution ionic strength on protein loading was investigated for 30 mol% AAc microgels, which have the highest capacity for cyt c of the microgels studied here. For microgels in the absence of protein, a decrease in particle rh was measured with increasing ionic strength (20 – 100 mM) at neutral pH, as expected for polyanionic networks (Figure S3 and S4). The stoichiometry of protein binding to microgel is also affected by ionic strength; as the salt concentration is increased, the measured Mw of the complex decreases. At the highest ionic strengths studied (170 mM), the microgel Mw approaches a value that is similar to that measured for microgels in the absence of protein (Figure 3, green dotted line). The loading of microgels was also qualitatively observed by mixing 30 mol% microgels with cyt c and separating the bound and unbound fractions by centrifugation. Photographs were taken after centrifugation (Figure 3, inset). At low ionic strength, where charge screening is limited, cyt c is co-localized with particles in the pellet.
Figure 3.
Influence of ionic strength on microgel-cyt c interactions. Increasing salinity results in a decrease in the measured Mw of 30 mol% AAc microgels. For reference, the Mw of unloaded microgels is shown (green dotted line). All measurements were made at neutral pH and at identical microgel and protein concentrations. Inset: visual observation of loading via mixing and separating microgels from free protein by centrifugation. Cyt c (red color) is localized with microgels in the pellet at low ionic strength (I = 20 mM).
The decrease in protein loading at high ionic strength is likely caused by two factors, the change in swelling of the microgel network and charge screening between binding partners. As the salt concentration is increased, the overall volume of the microgels diminishes (Figure S3 and S4). Decreased microgel swelling results in a decrease in the porosity, limiting the diffusion of cyt c into the polymer network. Meanwhile, the Debye screening length is reduced at high ionic strength, decreasing the effective strength of Coulombic interactions. For microgels composed of 30 mol% AAc, binding was not detected above 120 mM ionic strength. A similar trend has recently been reported for the interaction between polyanionic macrogels and oppositely charged peptides, where increasing ionic strength screened the Coulombic interactions between peptides bound to the gel.33
Conclusions
The physicochemical properties of microgels is altered through binding with various compounds, including polyelectrolytes,35–36 polymers,37 surfactants,38 and small molecule compounds.39 Namely, the adsorption or absorption of macromolecules affects swelling, density, charge and the stability of the polymer. In this work, we assessed cyt c loading into colloidal microgels by monitoring the properties of the particles (e.g. molar mass, radius) directly during encapsulation. Similar to their macroscopic counterparts,15–16, 20 colloidal microgels load oppositely charged proteins via Coulombic interactions. The increase in particle Mw in the presence of cyt c was used to quantify the number and concentration of macromolecules within the particles. Microgels containing 30 mol% AAc were capable of loading extremely high concentrations of cyt c (~17 mM), beyond the solubility limit of the protein in aqueous media (8 mM, 25 °C). Modulating AAc content was an effective means to tune binding stoichiometry; particles with greater AAc content showed the greatest capacity for loading. Microgels with low charge content (e.g. 10 mol% AAc) encapsulated a smaller fraction of proteins due to the decreased swelling and low porosity of those networks.
Our detailed assessment of encapsulation revealed import design parameters to consider as we pursue microgels and nanogels as peptide and protein delivery vehicles. For example, considerable changes in microgel morphology resulted from loading, where the density of the highly charged spheres (30 mol% AAc) increased ~20-fold. Decreased swelling during loading is likely to influence the modulus or “softness” of the particles, where previous reports have shown a strong relationship between polyelectrolyte shear modulus and the ionization of the network.34 The microgel softness is a critical property for the material in biomedical applications, affecting their behavior in confined environments.7–8 The investigation also elucidated the effects of gel network structure on macromolecule encapsulation. Using network swelling and ionization as a tunable variable, future delivery vehicles may be designed with specific encapsulation and release properties for biomedical applications.
Supplementary Material
Acknowledgments
This work was partially supported by the National Institutes of Health (1 R01 GM088291-01). Additional funding for MHS was provided by U.S. Department of Education GAANN awards, the Georgia Tech Center for Drug Design, Development and Delivery, and the Georgia Tech TI:GER® program.
Footnotes
Supporting Information Available: Experimental procedures, supporting data, and additional discussion. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wagner V, Dullaart A, Bock AK, Zweck A. Nat Biotechnol. 2006;24(10):1211–1217. doi: 10.1038/nbt1006-1211. [DOI] [PubMed] [Google Scholar]
- 2.Farokhzad OC, Langer R. ACS Nano. 2009;3(1):16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 3.Bromberg LE, Ron ES. Adv Drug Delivery Rev. 1998;31(3):197–221. doi: 10.1016/s0169-409x(97)00121-x. [DOI] [PubMed] [Google Scholar]
- 4.Antosova Z, Mackova M, Kral V, Macek T. Trends Biotechnol. 2009;27(11):628–635. doi: 10.1016/j.tibtech.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Raemdonck K, Demeester J, De Smedt S. Soft Matter. 2009;5(4):707–715. [Google Scholar]
- 6.Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Adv Mater. 2006;18(11):1345–1360. [Google Scholar]
- 7.Hendrickson GR, Lyon LA. Angew Chem, Int Ed. 2010;49(12):2193–2197. doi: 10.1002/anie.200906606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merkel TJ, Jones SW, Herlihy KP, Kersey FR, Shields AR, Napier M, Luft JC, Wu HL, Zamboni WC, Wang AZ, Bear JE, DeSimone JM. Proc Natl Acad Sci U S A. 2011;108(2):586–591. doi: 10.1073/pnas.1010013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banquy X, Suarez F, Argaw A, Rabanel JM, Grutter P, Bouchard JF, Hildgen P, Giasson S. Soft Matter. 2009;5(20):3984–3991. [Google Scholar]
- 10.Beningo KA, Wang YL. J Cell Sci. 2002;115(4):849–856. doi: 10.1242/jcs.115.4.849. [DOI] [PubMed] [Google Scholar]
- 11.Nayak S, Lyon LA. Angew Chem, Int Ed. 2005;44(47):7686–7708. doi: 10.1002/anie.200501321. [DOI] [PubMed] [Google Scholar]
- 12.Malmsten M. Soft Matter. 2006;2(9):760–769. doi: 10.1039/b608348j. [DOI] [PubMed] [Google Scholar]
- 13.Wu JY, Liu SQ, Heng PWS, Yang YY. J Control Release. 2005;102(2):361–372. doi: 10.1016/j.jconrel.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Bysell H, Malmsten M. Langmuir. 2006;22(12):5476–5484. doi: 10.1021/la060452a. [DOI] [PubMed] [Google Scholar]
- 15.Johansson C, Hansson P. Soft Matter. 2010;6(16):3970–3978. [Google Scholar]
- 16.Kabanov VA, Skobeleva VB, Rogacheva VB, Zezin AB. J Phys Chem B. 2003;108(4):1485–1490. [Google Scholar]
- 17.Eichenbaum GM, Kiser PF, Dobrynin AV, Simon SA, Needham D. Macromolecules. 1999;32(15):4867–4878. doi: 10.1021/ma970897t. [DOI] [PubMed] [Google Scholar]
- 18.Huo D, Li Y, Qian Q, Kobayashi T. Colloids Surf, B. 2006;50(1):36–42. doi: 10.1016/j.colsurfb.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Zhu W, Wang B, Ding J. J Control Release. 2005;105(3):260–268. doi: 10.1016/j.jconrel.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Johansson C, Gernandt J, Bradley M, Vincent B, Hansson P. J Colloid Interface Sci. 2010;347(2):241–251. doi: 10.1016/j.jcis.2010.03.072. [DOI] [PubMed] [Google Scholar]
- 21.Hoare T, Pelton R. Biomacromolecules. 2008;9(2):733–740. doi: 10.1021/bm701203r. [DOI] [PubMed] [Google Scholar]
- 22.Blackburn WH, Dickerson EB, Smith MH, McDonald JF, Lyon LA. Bioconjug Chem. 2009;20(5):960–968. doi: 10.1021/bc800547c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickerson E, Blackburn W, Smith M, Kapa L, Lyon LA, McDonald J. BMC Cancer. 2010;10(1):10. doi: 10.1186/1471-2407-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murthy N, Xu MC, Schuck S, Kunisawa J, Shastri N, Frechet JMJ. Proc Natl Acad Sci U S A. 2003;100(9):4995–5000. doi: 10.1073/pnas.0930644100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nayak S, Gan D, Serpe M, Lyon L. Small. 2005;1(4):416–421. doi: 10.1002/smll.200400089. [DOI] [PubMed] [Google Scholar]
- 26.Andersson M, Wittgren B, Wahlund KG. Anal Chem. 2003;75(16):4279–4291. doi: 10.1021/ac030128+. [DOI] [PubMed] [Google Scholar]
- 27.Hoare T, Pelton R. Langmuir. 2006;22(17):7342–7350. doi: 10.1021/la0608718. [DOI] [PubMed] [Google Scholar]
- 28.Stieger M, Richtering W, Pedersen JS, Lindner P. J Chem Phys. 2004;120(13):6197–6206. doi: 10.1063/1.1665752. [DOI] [PubMed] [Google Scholar]
- 29.Baselga J, Llorente MA, Nieto JL, Hernández-Fuentes I, Piérola IF. Eur Polym J. 1988;24(2):161–165. [Google Scholar]
- 30.Mirkin N, Jaconcic J, Stojanoff V, Moreno A. Proteins: Struct, Funct, Bioinf. 2008;70(1):83–92. doi: 10.1002/prot.21452. [DOI] [PubMed] [Google Scholar]
- 31.Dumetz AC, Snellinger-O’Brien AM, Kaler EW, Lenhoff AM. Protein Sci. 2007;16(9):1867–1877. doi: 10.1110/ps.072957907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ricka J, Tanaka T. Macromolecules. 1984;17(12):2916–2921. [Google Scholar]
- 33.Bysell H, Hansson P, Malmsten M. J Phys Chem B. 2010;114(21):7207–7215. doi: 10.1021/jp1016664. [DOI] [PubMed] [Google Scholar]
- 34.Skouri R, Schosseler F, Munch JP, Candau SJ. Macromolecules. 1995;28(1):197–210. [Google Scholar]
- 35.Kleinen J, Klee A, Richtering W. Langmuir. 2010;26(13):11258–11265. doi: 10.1021/la100579b. [DOI] [PubMed] [Google Scholar]
- 36.Wong JE, Díez-Pascual AM, Richtering W. Macromolecules. 2008;42(4):1229–1238. [Google Scholar]
- 37.Bradley M, Ramos J, Vincent B. Langmuir. 2005;21(4):1209–1215. doi: 10.1021/la047966z. [DOI] [PubMed] [Google Scholar]
- 38.Mears SJ, Deng Y, Cosgrove T, Pelton R. Langmuir. 1997;13(7):1901–1906. [Google Scholar]
- 39.Hoare T, Pelton R. Langmuir. 2008;24(3):1005–1012. doi: 10.1021/la7024507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.