Abstract
This study investigated the effect of reactive carbonyl species (RCS)-trapping agents on the formation of protein carbonyls during depletion of brain glutathione (GSH). To this end, rat brain slices were incubated with the GSH-depletor diethyl maleate in the absence or presence of chemically different RCS scavengers (hydralazine, methoxylamine, aminoguanidine, pyridoxamine, carnosine, taurine and z-histidine hydrazide). Despite their strong reactivity towards the most common RCS, none of the scavengers tested, with the exception of hydralazine, prevented protein carbonylation. These findings suggest that the majority of protein-associated carbonyl groups in this oxidative stress paradigm do not derive from stable lipid peroxidation products like malondialdehyde (MDA), acrolein and 4-hydroxynonenal (4-HNE). This conclusion was confirmed by the observation that the amount of MDA-, acrolein- and 4-HNE-protein adducts does not increase upon GSH depletion. Additional studies revealed that the efficacy of hydralazine at preventing carbonylation was due to its ability to reduce oxidative stress, most likely by inhibiting mitochondrial production of superoxide and/or by scavenging lipid free radicals.
Keywords: Carbonyl scavengers, glutathione depletion, hydralazine, oxidative stress, protein carbonylation, reactive oxygen species
Introduction
Carbonylation constitutes the major and most common oxidative modification of proteins [1]. Protein carbonyls (PCOs) have been shown to affect the function and/or metabolic stability of the modified proteins [2] and thus they are likely to play an important role in the pathophysiology of disorders with considerable oxidative stress. Carbonylation of brain proteins has been implicated in the aetiology and/or progression of several neurodegenerative disorders including Alzheimer’s disease [3], Parkinson’s disease [4], amyotrophic lateral sclerosis [5] and multiple sclerosis [6].
Carbonyl groups are introduced into proteins by two distinct mechanisms: oxidative (direct) and non-oxidative (indirect). Oxidative mechanisms, which are metal ion-catalysed, involve the direct reaction of certain reactive oxygen species (e.g. hydrogen peroxide, lipid hydroperoxides, etc.) with protein side-chains. Common amino acid targets of direct oxidative processes leading to carbonylation are Thr, Lys, Arg and Pro. Threonine side-chains are oxidized to α-amino-β-ketobutyric acid while oxidative deamination of Lys, Arg and Pro generates α-aminoadipic semi-aldehyde and glutamic semi-aldehyde [7]. Non-oxidative carbonylation of proteins involves the reaction of the nucleophilic centres in Cys, His or Lys residues with reactive carbonyl species (RCS). RCS are carbonyl-containing molecules derived from the oxidation of lipids (e.g. 4-hydroxynonenal (4-HNE), malondialdehyde (MDA), acrolein) and carbohydrates (e.g. glyoxal, methylglyoxal) [7].
To understand the process underlying the formation and accumulation of PCOs during oxidative stress, we recently utilized a brain slices system where carbonyls are induced by acute depletion of cellular glutathione (GSH) with diethyl maleate (DEM) or with 1,2-bis(2-chloroethyl)-1-nitrosourea [8]. We found that under these conditions there is an increased mitochondrial production of reactive oxygen species (ROS), which leads to extensive lipid peroxidation (LPO) and protein carbonylation by a metal ion-catalysed process likely involving the formation of hydroxyl radical. More recently we demonstrated that LPO is required for the carbonylation of cytoskeletal and membrane proteins [9]. Furthermore, evidence was presented suggesting that the mechanism underlying LPO-mediated protein carbonylation is the direct oxidation of amino acid side-chains by lipid hydroperoxides (or their immediate decomposition products lipid peroxyl and lipid alkoxyl radicals) rather than the attachment of RCS like 4-HNE and MDA.
The present study was designed to determine if carbonylation of brain proteins that takes place during acute GSH depletion indeed occurs by an oxidative mechanism. To achieve this objective we have (1) investigated the possible presence of protein-bound lipid aldehydes using antibodies against MDA, 4-HNE and acrolein and (2) tested the efficacy of several RCS-trapping agents (hydralazine, methoxylamine, aminoguanidine, pyridoxamine, carnosine, taurine and histidine hydrazide) at preventing protein carbonylation in rat brain slices incubated with DEM. The results show that GSH depletion does not generate protein carbonyls by the indirect (non-oxidative) mechanism. This was confirmed by the finding that none of the RCS scavengers tested, with the exception of hydralazine, prevented protein carbonylation. Additional studies revealed that the efficacy of hydralazine is mostly due to its antioxidant properties. A preliminary account of these results has been presented in abstract form [10].
Materials and methods
Chemicals
Aminoguanidine, apocynin, L-carnosine, clorgyline, deprenyl, DEM, hydralazine, hydroxylamine, methoxylamine and taurine were purchased from Sigma-Aldrich (St. Louis, MO). Pyridoxamine was from Fluka (Ronkonkoma, NY). z-Histidine hydrazide was from Peninsula Laboratories (San Carlos, CA). 15-Hydroper oxy-5,8,11,13-eicoanotetraenoic acid (15-HpETE) was obtained from Cayman Chemicals (Ann Arbor, MI). Anti-4-HNE (AB46544), anti-MDA (AB27642) and anti-acrolein (AB37110) rabbit polyclonal antibodies were from Abcam (Cambridge, MA). All other reagents were of the highest purity available.
Incubation of rat brain slices
Forty-day old Sprague-Dawley male rats were used throughout. Housing and handling of the animals as well as the euthanasia procedure were in strict accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee. Animals were killed by decapitation and the brains were rapidly removed and sliced in two directions at right angle in sections 400 µm-thick using surgical grade, carbon steel razor blades. Slices corresponding to ~ 80 mg of tissue were transferred to flasks containing 3 ml of Hank’s balanced salt solution supplemented with 10 mM D-glucose and were incubated at 37°C under 95% O2/5% CO2. Drugs were added at the beginning of the incubation period as indicated in the figure legends. After 2 h, aliquots were taken from the supernatant for H2O2 determination using the Fe/xylenol orange (FOX) assay [11]. Tissue sections were then collected by low-speed centrifugation and rinsed twice with 2 ml of ice-cold saline solution. Slices were homogenized by sonication in PEN buffer (10 mM sodium phosphate pH 7.0, 1 mM EDTA and 0.1 mM neocuproine) containing 1 mM 4,5-dihydroxy-1,3-benzene sulphonate and 0.5 mM dithiothreitol to prevent further protein oxidation. Homogenates were kept at −20°C until use. Protein concentration was assessed with the Bio-Rad DCT protein assay using bovine serum albumin (BSA) as standard.
Determination of non-protein thiols (NPSHs)
NPSHs, which are made mostly of GSH and small amounts of cysteine and homocysteine, were determined spectrophotometrically with 5,5’-dithiobis(2-nitrobenzoic) acid [12].
Measurement of lipid peroxidation
Lipid peroxidation was assessed by measuring the amount of thiobarbituric acid reactive substances (TBARS) in the tissue homogenates as described previously [13].
Measurement of protein carbonylation by western blotting
Protein carbonyl groups were measured by western blot analysis using the OxyBlot™ protein oxidation detection kit (Intergen Co., Purchase, NY) as we described earlier [9]. In brief, proteins (5 µg) were incubated with 2,4-dinitrophenyl-hydrazine to form the 2,4-dinitrophenyl (DNP) hydrazone derivatives. Proteins were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted to polyvinylidene difluoride (PVDF) membranes. DNP-containing proteins were then immunostained using rabbit anti-DNP antiserum (1:500) and goat anti-rabbit IgG conjugated to horseradish peroxidase (HRP) (1:2000). Blots were developed by enhanced chemioluminescence (ECL) using the Western Lightning ECL™ kit from Perkin-Elmer (Boston, MA). The developed films were scanned in a Hewlett Packard Scanjet 4890 and the images quantified using the NIH image analysis program version 1.63.
Reaction of oxidized brain proteins with carbonyl scavengers
Proteins from DEM-treated brain slices were precipitated with 1% sulphosalicylic acid and suspended 20 mM sodium phosphate buffer pH 7.5 containing 2 mM EDTA. Aliquots (50 µg of protein) were incubated at 20°C in the absence or presence of various carbonyl scavengers. After 2 h, proteins were derivatized with DNPH and analysed by western blotting as described above.
Assessment of MDA-, 4-HNE- and acrolein-protein adducts by western blotting
Proteins from control and GSH-depleted brain slices were separated by SDS-PAGE and blotted against PVDF membranes. RCS-protein adducts were detected using polyclonal rabbit antibodies against MDA (1:1000), 4-HNE (1:1000) and acrolein (1:2000) and HRP-conjugated goat anti-rabbit IgG (1:2000). Blots were developed by ECL as described above.
Additional assays
The effect of hydralazine (50 µM–50 mM) on the rate of pyrogallol auto-oxidation was carried out as described by Semsei et al. [14]. The effectiveness of RCS-scavengers at removing MDA was assessed by titrating the unreacted dialdehyde with thiobarbituric acid [13]. The efficacy of RCS-trapping drugs at scavenging 4-HNE was measured by titrating the unreacted unsaturated alkenal with N-methyl-2-phenylindole in the presence of methanesulphonic acid [6]. The ability of RCS-trapping agents at scavenging acrolein was determined with treating unreacted acrolein with cysteine ethyl ester and then titrating the excess thiols with 5,5’-dithiobis(2-nitrobenzoic) acid.
Statistical analysis
Results were analysed for statistical significance with Student’s unpaired t-test utilizing the GraphPad Prism® (version 4) program (GraphPad Software Inc., San Diego, CA).
Results
DEM-induced protein carbonylation in brain slices
DEM is an α,β-unsaturated dicarboxylic acid that conjugates to GSH via a reaction catalysed by glutathione-S-transferase [15]. Incubation of brain slices with 10 mM DEM for 2 h diminishes NPSHs by 85% and increased TBARS and PCO levels by 70% and 100%, respectively [8]. As described in our previous study [9], the majority of the carbonylated proteins from both control and DEM-treated slices have molecular weights between 40–120 K. Among these species are the major cytoskeleton proteins including α/β-tubulin, β-actin, the neuronal intermediate filament proteins and glial fibrillary acidic protein.
Effect of carbonyl scavengers on DEM-induced protein carbonylation in brain slices
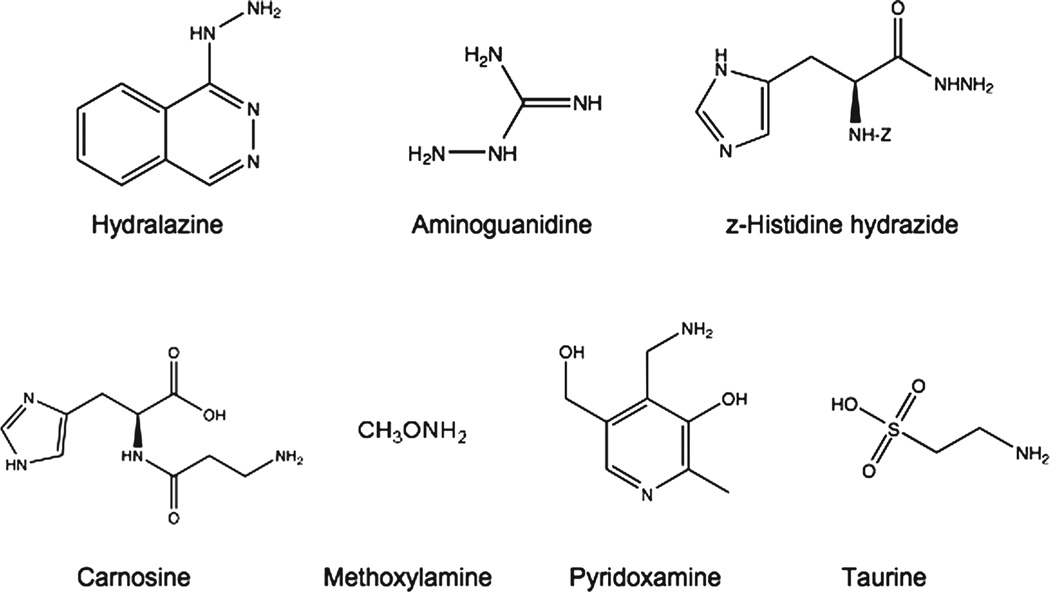
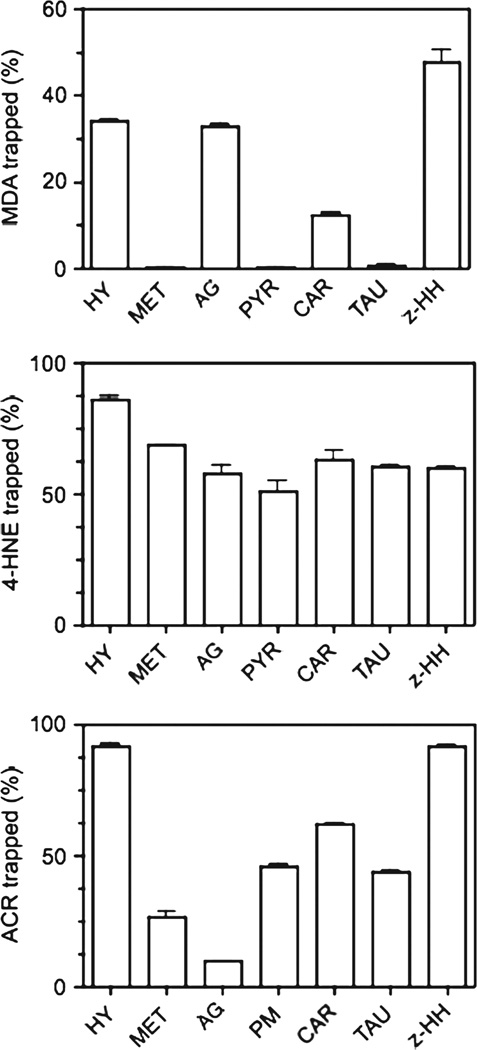
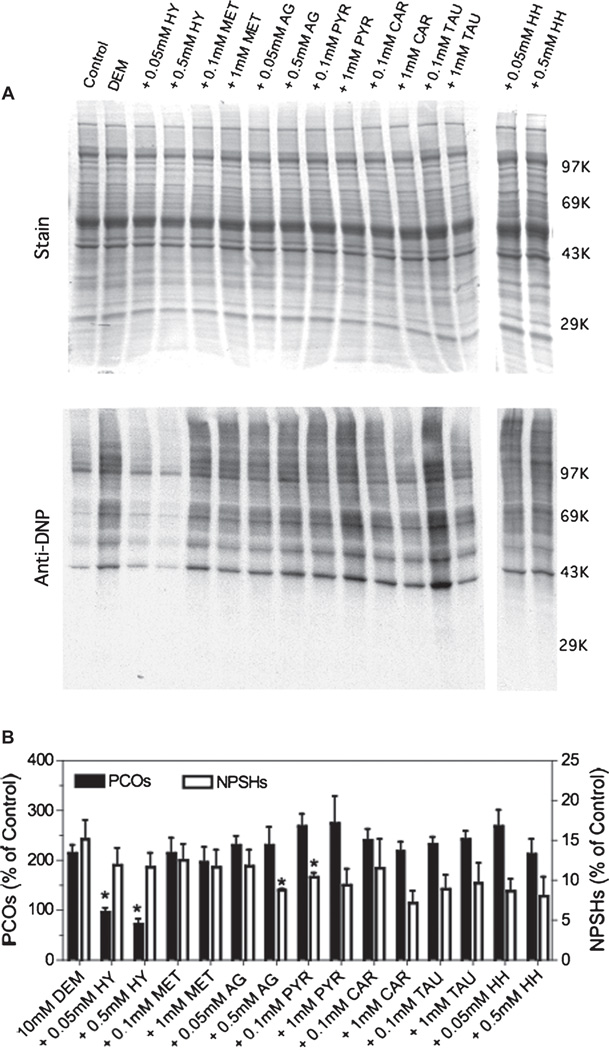
The detailed chemical structure of the carbonyl trapping agents used in this study is shown in Figure 1. These agents include three hydrazines (hydralazine [16], aminoguanidine [17] and z-histidine hydrazide [18]) and four primary amines (methoxylamine [19], pyridoxamine [20], carnosine [21] and taurine [22]). We chose several, chemically different RCS scavengers because their reactivity toward various RCS differs considerably [23]. In cell-free systems we found that only the three hydrazines and to a lesser extent carnosine, effectively trap MDA (Figure 2A), while all the scavengers showed high reactivity towards 4-HNE (Figure 2B). Except for aminoguanidine, most drugs also adducted the highly reactive acrolein (Figure 2C). It is important to note that these scavengers were also able to prevent the formation protein-RCS adducts in tissue slices incubated with MDA or 4-HNE, indicating that they are cell-permeable (data not shown). We then tested the ability of these RCS scavengers to prevent the appearance of protein carbonyls in brain sections incubated with DEM. As shown in Figure 3, none of the carbonyl scavengers prevented DEM-induced glutathione depletion and, among the seven drugs tested, only hydralazine (50–500 µM) inhibited the formation of protein carbonyls. Aminoguanidine shows an effect only at concentrations ≥ 1 mM [9]. The observation that just hydralazine prevents PCO formation was surprising and suggests that either protein carbonylation takes place through an indirect mechanism and that only hydralazine effectively traps RCS in intact cells or that the efficacy of this drug is due to some property that is unrelated to RCS adduction.
Figure 1.
Chemical structure of the various carbonyl-trapping agents used in this study. z-, carbobenzoxy-.
Figure 2.
Ability of carbonyl scavengers to trap various RCS in a cell-free system. MDA (36 µM), 4-HNE (25 µM) and acrolein (100 µM) were incubated for 2 h at room temperature in the presence or absence of various carbonyl scavengers (1 mM). After incubation, the amount of unreacted RCS was determined as described in Materials and methods. Values are expressed as the percentage of RCS trapped by the scavenger and represent the mean ± SEM of three separate incubations. The concentration of acrolein in these experiments was 4-times higher than that of 4-HNE solely because the sensitivity of the assays for measuring each unsaturated alkenal is different. Abbreviations: HY, hydralazine; MET, methoxylamine; AG, aminoguanidine; PYR, pyridoxamine; CAR, carnosine; TAU, taurine; z-HH, z-histidine hydrazide.
Figure 3.
Effect of carbonyl scavengers on DEM-induced protein carbonylation. Rat brain slices were incubated with 10 mM DEM in the absence or presence of two different concentrations of various carbonyl scavengers. After 2 h, slices were homogenized in PEN buffer and aliquots of the homogenate were used to determine the PCOs by western blot as described in Materials and methods. (A) Representative OxyBlot. The molecular weight markers are: phosphorylase b (97K), bovine serum albumin (69K), ovalbumin (43K) and carbonic anhydrase (29K). Other abbreviations are as in Figure 2. (B) Protein carbonylation levels obtained from western blots and NPSH levels determined spectrophotometrically with 5,5′-dithiobis(2-nitrobenzoic) acid. Values are expressed as percentage of control and represent the mean ± SEM of 3–4 experiments. Control values for carbonyls and NPSHs are 0.21 ± 0.02 nmol/mg protein and 10.8 ± 0.8 nmol/mg protein, respectively. Asterisks denote those numbers that are significantly different (p < 0.05) from DEM-treated slices.
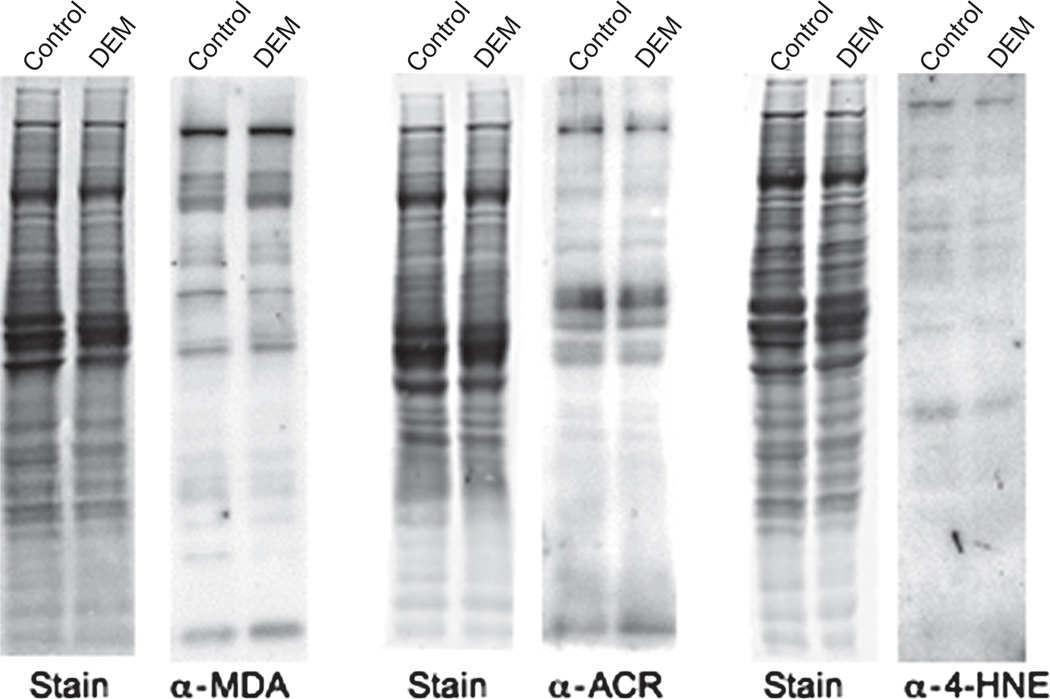
To distinguish between the above possibilities, we determined by western blot analysis whether or not RCS-protein adducts are formed in GSH-depleted brain slices. As depicted in Figure 4, antibodies against MDA, acrolein and 4-HNE labelled a number of distinct bands and the intensity of neither the whole lane not the individual modified proteins was changed in DEM-treated sections. This indicates that MDA-protein, ACR-protein and 4-HNE-protein adducts are not formed to any appreciable degree in this model of oxidative stress and that the effect of hydralazine on protein carbonylation is not due to its ability to remove RCS.
Figure 4.
Detection of RCS-protein adducts in control and GSH-depleted brain slices. Rat brain slices were incubated in the absence (control) or presence of 10 mM DEM. After 2 h, slices were homogenized in PEN buffer and aliquots of the homogenate were analysed by western blotting using antibodies against the various RCS as described in Materials and methods.
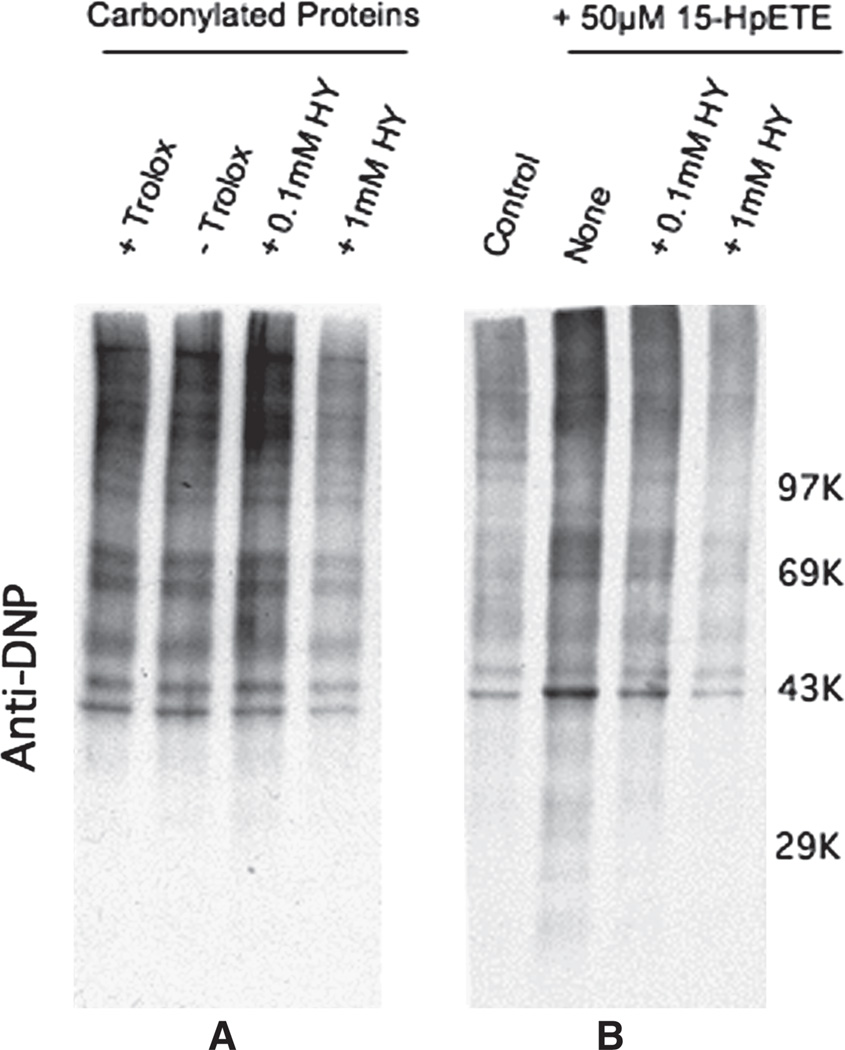
The possibility that hydralazine reacts directly with protein carbonyls was examined by incubating carbonylated proteins derived from DEM-treated brain slices with hydralazine (0.1–1 mM). Only 1 mM hydralazine efficiently reduced the amount of protein bound carbonyls, suggesting that the effectiveness of 50 µM hydralazine at preventing protein carbonylation in intact cells is unlikely caused by a direct reaction between the drug and protein-bound carbonyl groups (Figure 5A). Interestingly, 100 µM hydralazine was found to partially inhibit the 15-HpETE-induced oxidation of proteins (Figure 5B) and this could be due to scavenging of alkoxyl and peroxyl radicals produced during the metal ion-catalysed decomposition of the lipid hydroperoxide and/or to the sequestration of iron and copper ions. In any case, the concentration of hydralazine needed to cause this effect in a cell-free system is still higher than that required to suppress PCO formation in GSH-depleted brain sections (see below).
Figure 5.
(A) Effect of hydralazine on protein-bound carbonyl groups. Oxidized proteins prepared from DEM-treated brain slices were incubated for 2 h in the absence or presence of two different concentration of hydralazine as described in Materials and methods. PCOs were derivatized with DNPH and analysed by OxyBlot. The antioxidant trolox (1 mM) was included in some samples to ascertain that further protein oxidation does not occur during the incubation period. (B) Effect of hydralazine on lipid hydroperoxide-induced protein carbonylation in a cell-free system. Proteins prepared from control brain slices were incubated for 2 h with 15-HpETE in the absence or presence of two different concentration of hydralazine. After incubation PCOs were derivatized with DNPH and analysed by OxyBlot.
Antioxidant properties of hydralazine
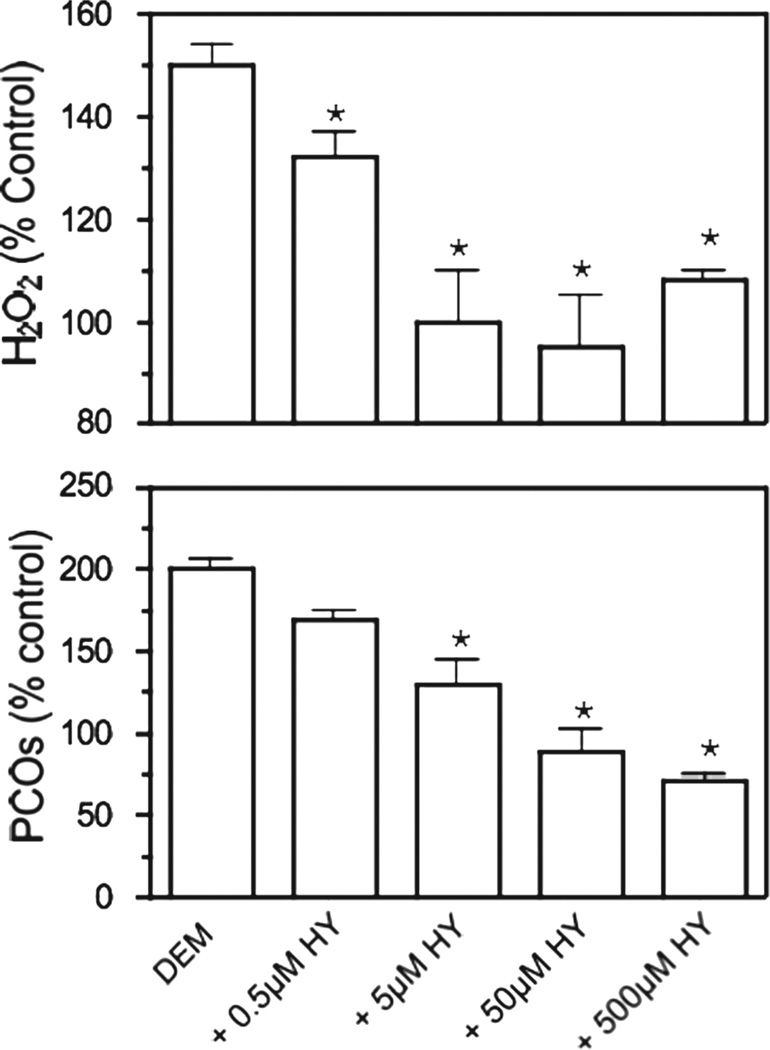
Because hydralazine is known to have antioxidant properties in several oxidative stress settings [24] and based on the observation that it also lowers the levels of TBARS in GSH-depleted brain sections (data not shown), we hypothesized that this drug may just be reducing oxidative stress and indirectly carbonyl formation. To address this issue, we determined hydrogen peroxide and PCO levels in DEM-treated brain slices incubated with different concentrations of hydralazine. As shown in Figure 6, both H2O2 levels and the amount of protein carbonyls diminished with increasing concentrations of hydralazine (0.5–500 µM), indicating that the inhibitor is acting mostly as an antioxidant.
Figure 6.
Effect of increasing concentrations of hydralazine on DEM-induced protein carbonylation and hydrogen peroxide production. Rat brain slices were incubated with 10 mM DEM in the absence or presence of increasing concentrations (0.5–500 µM) of hydralazine. After 2 h, aliquots from the incubation media were removed and used to determine H2O2 levels using the FOX assay. Slices were homogenized in PEN buffer and aliquots of the homogenate were used to determine PCOs by OxyBlot. Values are expressed as percentage of control and represent the mean ± SEM of 3–4 experiments. Asterisks denote values that are significantly different (p < 0.05) from DEM-treated slices.
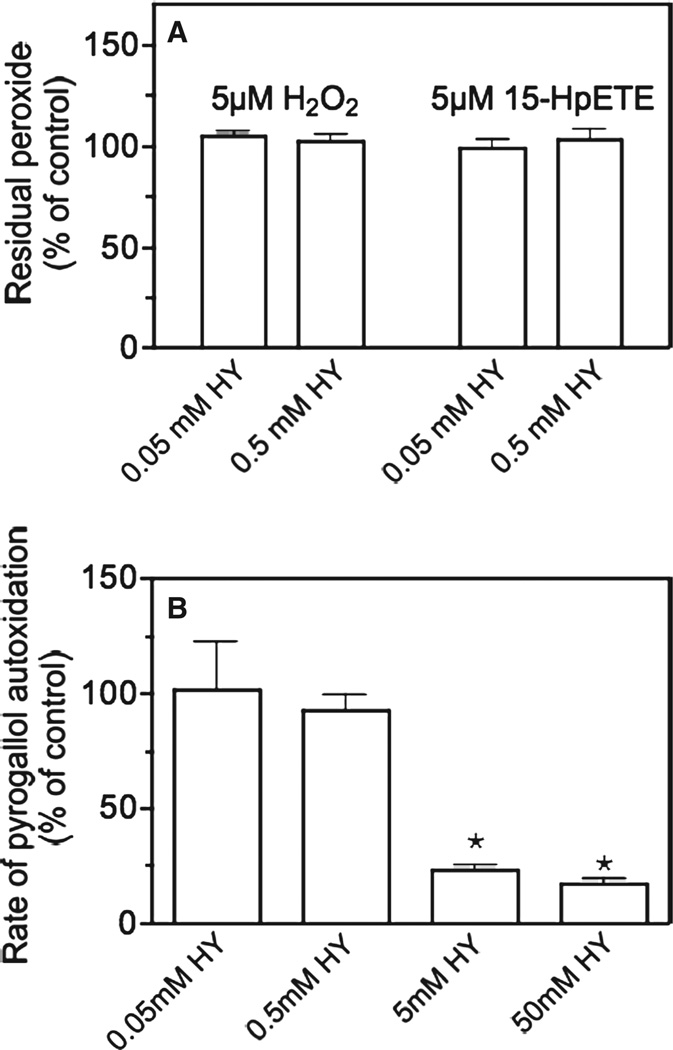
A series of additional studies were conducted to ascertain the mechanism underlying the antioxidant properties of hydralazine. We first investigated the possibility that hydralazine could be scavenging peroxides (H2O2, lipid hydroperoxides) or superoxide radicals. Figure 7A shows that hydralazine does not react with either H2O2 or 15-HpETE, even at high concentrations (0.5 mM). The ability of hydralazine to scavenge superoxide was tested with pyrogallol. In the pyrogallol system, superoxide radicals are formed from molecular oxygen and the detecting system is the pyrogallol itself. Hydralazine reduced superoxide levels only at a concentration ≥ 5 mM, which is 1000-times higher than that required to abolish H2O2 production (Figure 7B). Thus, the most likely mechanism underlying the antioxidant effects of hydralazine is the inhibition of processes responsible for ROS production.
Figure 7.
(A) Effect of hydralazine on hydrogen peroxide and lipid hydroperoxide stability. Hydrogen peroxide and the lipid hydroperoxide 15-HpETE were incubated in the absence or presence of two different concentrations of hydralazine. After 2 h, residual peroxide levels were determined with the FOX assay as described in Materials and methods. (B) Effect of hydralazine on the stability of pyrogallol-generated superoxide. Pyrogallol (0.2 mM) was incubated in the absence or presence of two different concentrations of hydralazine and the rate of auto-oxidation was determined as described in Materials and methods. Values are expressed as percentage of control (i.e. without hydralazine) and represent the mean ± SEM of three experiments. Asterisks denote values that are significantly different (p < 0.05) from controls.
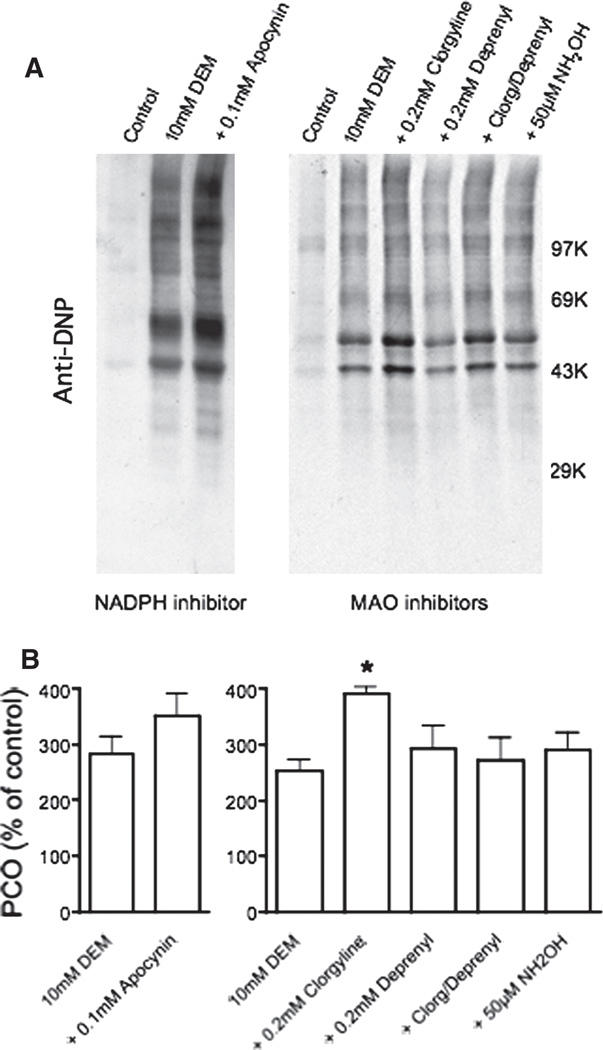
In a previous study we showed that most of the superoxide generated during GSH depletion comes from mitochondria (inhibited by carbonyl cyanide 3-chlorophenylhydrazone) and to a lesser extent from cytochrome P-450 (inhibited with proadifen), but not from xanthine oxidase (unaltered by oxypurinol) [8]. However, other possible sources of superoxide that are targeted by hydralazine, like NADPH oxidase [24] and monoamine oxidase (MAO) [25], were not investigated in that study. To address this issue, brain slices were incubated with DEM in the presence or absence of apocynin (NADPH oxidase inhibitor [26]), clorgyline (MAO-A inhibitor [25]), deprenyl (MAO-B inhibitor [25]) or hydroxylamine (MAO-A/B inhibitor [27]). As shown in Figure 8, protein carbonylation was not decreased by any of these drugs, indicating that these enzymes are not involved in the generation of superoxide during GSH depletion. In sum, the above results are in agreement with previous findings suggesting that hydralazine reduces superoxide production from mitochondria [28,29]. At this time, however, we cannot rule out the possibility that hydralazine acts also by scavenging of lipid alkoxyl and peroxyl radicals, mainly because the concentration of these radicals and hydralazine in the cell membranes may be quite different than those used in our cell-free experiment (Figure 5B).
Figure 8.
Effect of NADPH oxidase and MAO inhibitors on DEM-induced protein carbonylation. Rat brain slices were incubated with 10 mM DEM in the absence or presence of a NADPH oxidase inhibitor (apocynin) or MAO inhibitors (clorgyine, deprenyl and hydroxylamine). After 2 h, slices were homogenized in PEN buffer and aliquots of the homogenate were used to determine the PCOs by OxyBlot as described in Materials and methods. (A) Representative OxyBlots. (B) Protein carbonylation levels obtained from the western blots. Values are expressed as percentage of control and represent the mean ± SEM of three experiments. Asterisk denotes the value that is significantly different (p < 0.05) from DEM-treated slices.
Discussion
In this study, we present evidence that RCS-trapping drugs do not prevent the carbonylation of brain proteins during depletion of the antioxidant glutathione. Commonly used RCS-scavengers like aminoguanidine, pyridoxamine, methoxylamine, carnosine, taurine and z-histidine hydrazide were unable to reduce the appearance of protein carbonyls during DEM-induced oxidative stress. Only hydralazine prevented PCO accumulation at relatively low concentrations, but the effect is due to its antioxidant properties rather than to its ability to trap RCS or to react with PCOs directly. This suggests that the majority of protein-associated carbonyl groups in this oxidative stress paradigm do not derive from stable LPO products like MDA, acrolein and 4-HNE. This conclusion was confirmed by the observation that the amount of MDA-, acrolein- and 4-HNE-protein adducts does not increase in GSH-depleted sections. Thus, this system is best suited to test the effect of antioxidants on protein carbonylation rather than to explore the efficacy of new RCS-trapping agents.
Oxidation of polyunsaturated fatty acids gives rise to three major products, all of which are know to introduce carbonyl groups into proteins. These compounds include (i) dialdehydes (e.g. MDA), which react with lysine residues to form carbonyl derivatives; (ii) α,β-unsaturated aldehydes (e.g. 4-HNE, 4-hydroxy-2-hexenal, acrolein), which undergo a Michael addition reaction with the ε-amino group of lysine residues, the thiol group of cysteine residues and the imidazole group of histidine residues; and (iii) lipid hydroperoxides, which can undergo metal ion-catalysed decomposition to produce alkoxyl and peroxyl radicals that can react directly with amino acid residues. Previous work from our laboratory suggested that lipid hydroperoxide-mediated oxidation is the major mechanism by which brain proteins (particularly cytoskeletal and membrane proteins) are carbonylated during acute GSH depletion [9]. In the present study we have strengthened this conclusion by the finding that none of the classical RCS scavengers prevent the carbonylation of proteins but more importantly by the absence of MDA-, 4-HNE- and ACR-protein adducts as measured on western blots. Lipid hydroperoxide-induced protein carbonylation was initially proposed by Refsgaard et al. [30], who discovered that metal-catalysed oxidation of proteins is greatly enhanced by addition of polyunsaturated fatty acids. These investigators speculated that alkoxyl radicals, derived from metal-catalysed heterolytic cleavage of lipid hydroperoxides, are responsible for the introduction of carbonyls into proteins by a mechanism that might involve site-specific interaction with lysine residues. Lipid-derived alkoxyl radicals were found to be involved also in the oxidation of retinal proteins in diabetes [31], suggesting that this mechanism is more common than previously thought.
Accumulation of PCOs has been implicated in the aetiology and/or progression of several neurodegenerative disorders such as Alzheimer’s disease [3], Parkinson’s disease [4] and amyotrophic lateral sclerosis [5]. Our recent discovery that carbonylation of CNS proteins is augmented in multiple sclerosis [6,32] and its animal model experimental allergic encephalomyelitis [33] suggests that this type of protein modification may play a critical pathophysiological role in inflammatory demyelinating diseases as well. Therefore, approaches to reduce the extent of protein carbonylation may be beneficial for treating these disorders. In recent years carbonyl trapping has received considerable attention as a specific treatment for conditions with severe carbonyl stress. Carbonyl trapping agents like those tested in this study react with RCS at a faster rate than do cell macromolecules, thereby ensuring the safe excretion of drug-carbonyl conjugates. However, this approach is effective only if protein carbonylation takes place by an indirect mechanism, which does not seem to be the case during acute depletion of glutathione. Furthermore, α-aminoadipic semi-aldehyde and glutamic semi-aldehyde, which result from direct oxidation of lysine, arginine and proline residues, are the major carbonylated amino acids in CNS proteins during ageing and in neurodegenerative disorders [34,35]. Thus, in most cases, prevention of protein carbonylation will have to be achieved with agents that reduce oxidative stress, limit LPO and/or interfere with the direct oxidation of amino acids. The anti-hypertensive hydralazine seems to possess all of these properties since it reduced oxidative stress in intact cells (Figure 6) and interfered with lipid hydroperoxide-induced protein carbonylation in a cell-free system (Figure 5B) and there is also evidence that it strongly inhibits LPO [36]. Preliminary studies in our laboratory have shown that administration of hydralazine reduces CNS damage and decrease neurological symptoms of rats with acute experimental autoimmune encephalomyelitis, suggesting that this drug could be potentially useful for treating neuroinflammatory disorders.
The antioxidant properties of hydralazine have been reported in several studies and have been attributed to: (1) inhibition of ROS-generating enzymes such as NADPH oxidase [24], monoamine oxidase [25] and xanthine oxidase [37], (2) scavenging of superoxide and peroxynitrite [29] and (3) decreased mitochondrial superoxide production [28,29]. In this study we ruled out the participation of NADPH oxidase and monoamine oxidases in the generation of ROS during GSH depletion and that of xanthine oxidase was excluded in our previous study [8]. In addition, we have eliminated the possibility of a direct reaction of superoxide, hydrogen peroxide and lipid hydroperoxides with hydralazine. The possibility that hydralazine reduces protein carbonylation by scavenging peroxynitrite is also unlikely since this oxidant is not produced in this oxidative stress paradigm as demonstrated by the lack of nitrotyrosine [9]. Furthermore, addition of two peroxynitrite-scavengers (uric acid and dimethylthiourea) [8] and a nitric oxide synthetase inhibitor aminoguanidine (this study) has no effect on protein oxidation in this system. All of these findings, along with recent observations that hydralazine does not inhibit cytochrome P-450 [38], point to mitochondria as the most likely target of hydralazine in the DEM-treated brain sections. There is some evidence that hydralazine reduces the mitochondrial production of superoxide and consequently hydrogen peroxide [28,29] and we are currently investigating the molecular mechanism underlying this effect.
Acknowledgements
The authors thank Ms. Savanna Reyes for her technical help in some experiments.
This work was supported by PHHS grant NS057755 from the National Institutes of Health.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chem Res Toxicol. 1997;10:485–494. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- 2.Levine RL. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic Biol Med. 2002;32:790–796. doi: 10.1016/s0891-5849(02)00765-7. [DOI] [PubMed] [Google Scholar]
- 3.Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, Markesbery WR. Protein oxidation in the brain in Alzheimer’s disease. Neuroscience. 2001;103:373–383. doi: 10.1016/s0306-4522(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 4.Floor E, Wetzel MG. Increased protein oxidation in human sustantia nigra pars compacta in comparison with basal ganglia and prefrontal cortex measured with an improved dinitrophenylhydrazine assay. J Neurochem. 1998;70:268–275. doi: 10.1046/j.1471-4159.1998.70010268.x. [DOI] [PubMed] [Google Scholar]
- 5.Ferrante RJ, Browne SE, Shinobu LA, Bowling AC, Baik MJ, MacGarvey U, Kowall NW, Brown RH, Beal MF. Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis. J Neurochem. 1997;69:2064–2074. doi: 10.1046/j.1471-4159.1997.69052064.x. [DOI] [PubMed] [Google Scholar]
- 6.Bizzozero OA, Dejesus G, Callahan K, Pastuszyn A. Elevated protein carbonylation in the brain white matter and gray matter of patients with multiple sclerosis. J Neurosci Res. 2005;81:687–695. doi: 10.1002/jnr.20587. [DOI] [PubMed] [Google Scholar]
- 7.Adams S, Green P, Claxton R, Simcox S, Williams MV, Walsh K, Leeuwenburgh C. Reactive carbonyl formation by oxidative and non-oxidative pathways. Front Biosci. 2001;6:17–24. doi: 10.2741/adams. [DOI] [PubMed] [Google Scholar]
- 8.Bizzozero OA, Ziegler JL, DeJesus G, Bolognani F. Acute depletion of reduced glutathione causes extensive carbonylation of rat brain proteins. J Neurosci Res. 2006;83:656–667. doi: 10.1002/jnr.20771. [DOI] [PubMed] [Google Scholar]
- 9.Bizzozero OA, Reyes S, Ziegler JL, Smerjac S. Lipid peroxidation scavengers prevent the carbonylation of cytoskeletal brain proteins induced by glutathione depletion. Neurochem Res. 2007;32:2114–2122. doi: 10.1007/s11064-007-9377-y. [DOI] [PubMed] [Google Scholar]
- 10.Zheng J, Reyes S, Bizzozero OA. Effect of hydralazine and other carbonyl scavengers at preventing the carbonylation of brain proteins induced by GSH depletion. J Neurochem. 2008;104 Suppl 1:PTW06–PTW15. [Google Scholar]
- 11.Nourooz-Zadeh A, Tajaddini-Sarmadi J, Ling KL, Wolff SP. Low-density lipoprotein is the major carrier of lipid hydroperoxides in plasma. Biochem J. 1996;313:781–786. doi: 10.1042/bj3130781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riddles PW, Blakely RL, Zerner B. Ellman’s reagent: 5,5’-dithiobis (2-nitrobenzoic acid)—a reexamination. Anal Biochem. 1979;94:75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- 13.Ohkawa H, Ohishi N, Yagi K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal Biochem. 1979;95:351–358. doi: 10.1016/0003-2697(79)90738-3. [DOI] [PubMed] [Google Scholar]
- 14.Semsei I, Nagy K, Zs-Nagy I. In vitro studies on the OH and O2-free radical scavenger properties of idebenone in chemical systems. Arch Gerontol Geriatr. 1990;11:187–197. doi: 10.1016/0167-4943(90)90064-d. [DOI] [PubMed] [Google Scholar]
- 15.Buchmuller-Rouiller Y, Corrandin SB, Smith J, Schneider P, Ransijn A, Jongeneel CV, Mauel J. Role of glutathione in macrophage activation: effect of cellular glutathione depletion on nitrite production and leishmanicidal activity. Cell Immunol. 1995;164:73–80. doi: 10.1006/cimm.1995.1144. [DOI] [PubMed] [Google Scholar]
- 16.Kaminskas LM, Pyke SM, Burcham PC. Strong protein adduct trapping accompanies abolition of acrolein-mediated hepatotoxicity by hydralazine in mice. J Pharmacol Exp Ther. 2004;310:1003–1010. doi: 10.1124/jpet.104.067330. [DOI] [PubMed] [Google Scholar]
- 17.Al-Abed Y, Bucala R. Efficient scavenging of fatty acid oxidation products by aminoguanidine. Chem Res Toxicol. 1997;10:875–879. doi: 10.1021/tx970035l. [DOI] [PubMed] [Google Scholar]
- 18.Tang SC, Arumugam TV, Cutler RG, Jo DG, Magnus T, Chan SL, Mughal MR, Telljohann RS, Nassar M, Ouyang X, Calderan A, Ruzza P, Guiotto A, Mattson MP. Neuroprotective actions of a histidine analogue in models of ischemic stroke. J Neurochem. 2007;101:729–736. doi: 10.1111/j.1471-4159.2006.04412.x. [DOI] [PubMed] [Google Scholar]
- 19.Burcham PC, Fontaine FR, Kaminskas LM, Petersen DR, Pyke SM. Protein adduct-trapping by hydrazinophthalazine drugs: mechanisms of cytoprotection against acrolein-mediated toxicity. Mol Pharmacol. 2004;65:655–664. doi: 10.1124/mol.65.3.655. [DOI] [PubMed] [Google Scholar]
- 20.Voziyan PA, Metz TO, Baynes JW, Hudson BG. A post-amadori inhibitor pyridoxamine also inhibits chemical modification of proteins by scavenging carbonyl intermediates of carbohydrate and lipid degradation. J Biol Chem. 2002;277:3397–3403. doi: 10.1074/jbc.M109935200. [DOI] [PubMed] [Google Scholar]
- 21.Hipkiss AR, Brownson C. A possible new role for the anti-aging peptide carnosine. Cell Mol Life Sci. 2000;57:747–753. doi: 10.1007/s000180050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devamanoharan PS, Ali AH, Varma SD. Prevention of lens protein glycation by taurine. J Mol and Cell Biochem. 1997;177:245–250. doi: 10.1023/a:1006863322454. [DOI] [PubMed] [Google Scholar]
- 23.Negre-Salvayre A, Coatrieux C, Ingueneau C, Salvayre R. Advanced lipid peroxidation end oxidative damage to proteins. Potential diseases and therapeutic prospects inhibitors. Br J Pharmacol. 2008;153:6–20. doi: 10.1038/sj.bjp.0707395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Münzel T, Kurz S, Rajagopalan S, Thoenes M, Berrington WR, Thompson JA, Freeman BA, Harrison DG. Hydralazine prevents nitroglycerin tolerance by inhibiting activation of a membrane-bound NADH oxidase: a new action for an old drug. J Clin Invest. 1996;98:1465–1470. doi: 10.1172/JCI118935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maher P, Davis JB. The role of monoamine metabolism in oxidative glutamate toxicity. J Neurosci. 1996;16:6394–6401. doi: 10.1523/JNEUROSCI.16-20-06394.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.’t Hart BA, Simons JM, Knaan-Shanzer S, Bakker NP, Labadie RP. Antiarthritic activity of the newly developed neutrophil oxidative burst antagonist apocynin. Free Radic Biol Med. 1990;9:127–131. doi: 10.1016/0891-5849(90)90115-y. [DOI] [PubMed] [Google Scholar]
- 27.Roh JH, Suzuki H, Azakami H, Yamashita M, Murooka Y, Kumagai H. Purification, characterization, and crystallization of monoamine oxidase from Escherichia coli. Biosci Biotechnol Biochem. 1994;58:1652–1656. doi: 10.1271/bbb.58.1652. [DOI] [PubMed] [Google Scholar]
- 28.Kishi H, Kishi T, Folkers K. Bioenergetics in clinical medicine. III. Inhibition of conezyme Q10-enzymes by clinically used antihypertensive agents. Res Commun Chem Pathol Pharmacol. 1975;12:533–540. [PubMed] [Google Scholar]
- 29.Daiber A, Oelze M, Coldewey M, Kaiser K, Huth C, Schildknecht S, Bachschmid M, Nazirisadeh Y, Ullrich V, Mülsch A, Münzel T, Tsilimingas N. Hydralazine is a powerful inhibitor of peroxynitrite formation as a possible explanation for its beneficial effects on prognosis in patients with congestive heart failure. Biochem Biophys Res Commun. 2005;338:1865–1874. doi: 10.1016/j.bbrc.2005.10.106. [DOI] [PubMed] [Google Scholar]
- 30.Refsgaard HF, Tsai L, Stadman ER. Modification of proteins by polyunsaturated fatty acid peroxidation products. Proc Natl Acad Sci USA. 2000;97:611–691. doi: 10.1073/pnas.97.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pennathur S, Ido Y, Heller JI, Byun J, Danda R, Pergola P, Williamson JR, Heinecke JW. Reactive carbonyls and polyunsaturated fatty acids produce hydroxyl radical-like species. J Biol Chem. 2005;280:22706–22714. doi: 10.1074/jbc.M500839200. [DOI] [PubMed] [Google Scholar]
- 32.Bizzozero OA. Chapter 23 Protein carbonylation in neurodegenerative and demyelinating CNS diseases. In: Lajtha, Banik, Ray, editors. Handbook of neurochemistry and molecular neurobiology—Brain and spinal cord trauma. Springer; 2008. pp. 543–562. [Google Scholar]
- 33.Smerjac S, Bizzozero OA. Cytoskeletal protein carbonylation and degradation in experimental autoimmune encephalomyelitis. J Neurochem. 2008;105:763–772. doi: 10.1111/j.1471-4159.2007.05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Requena JS, Chao CC, Levine R, Stadtman ER. Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc Natl Acad Sci USA. 2001;98:69–74. doi: 10.1073/pnas.011526698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pamplona R, Dalfo E, Ayala V, Bellmunt MJ, Prat J, Ferrer I, Portero-Otin M. Proteins in human brain cortex are modified by oxidation, glycoxidation, and lipoxidation. J Biol Chem. 2005;280:21522–21530. doi: 10.1074/jbc.M502255200. [DOI] [PubMed] [Google Scholar]
- 36.Metha R, Wong L, O’Brien PJ. Cytoprotective mechanisms of carbonyl scavenging drugs in isolated rat hepatocytes. Chem-Biol Interact. 2009;178:317–323. doi: 10.1016/j.cbi.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 37.Leiro JM, Alvarez E, Arranz JA, Cano E, Orallo F. Antioxidant activity and inhibitory effects of hydralazine on inducible NOS/COX-2 gene and protein expression in peritoneal macrophages. Int Immunopharmacol. 2004;4:163–177. doi: 10.1016/j.intimp.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Svensson CK, Knowlton PW, Ware JA. Effect of hydralazine on the elimination of antipyrine in the rat. Pharm Res. 1987;4:515–518. doi: 10.1023/a:1016487824200. [DOI] [PubMed] [Google Scholar]