Genome organizer Satb1 is regulated by p63 and contributes to epidermal morphogenesis by remodeling chromatin structure and gene expression at the epidermal differentiation complex locus.
Abstract
During development, multipotent progenitor cells establish tissue-specific programs of gene expression. In this paper, we show that p63 transcription factor, a master regulator of epidermal morphogenesis, executes its function in part by directly regulating expression of the genome organizer Satb1 in progenitor cells. p63 binds to a proximal regulatory region of the Satb1 gene, and p63 ablation results in marked reduction in the Satb1 expression levels in the epidermis. Satb1−/− mice show impaired epidermal morphology. In Satb1-null epidermis, chromatin architecture of the epidermal differentiation complex locus containing genes associated with epidermal differentiation is altered primarily at its central domain, where Satb1 binding was confirmed by chromatin immunoprecipitation–on-chip analysis. Furthermore, genes within this domain fail to be properly activated upon terminal differentiation. Satb1 expression in p63+/− skin explants treated with p63 small interfering ribonucleic acid partially restored the epidermal phenotype of p63-deficient mice. These data provide a novel mechanism by which Satb1, a direct downstream target of p63, contributes in epidermal morphogenesis via establishing tissue-specific chromatin organization and gene expression in epidermal progenitor cells.
Introduction
During the execution of developmental programs, the components of the chromatin-remodeling and transcription machinery are expressed in a highly organized manner, resulting in the establishment of tissue-specific patterns of gene activation and silencing (Hemberger et al., 2009). However, the mechanisms underlying these series of events to achieve terminal differentiation of specific cell types are still unclear.
The program of epidermal differentiation and barrier formation in mice begins at about embryonic day 14.5 (E14.5) followed by the appearance of the spinous and granular layers at E16.5 and establishment of a functional epidermal barrier at E18.5 (Fuchs, 2007; Koster and Roop, 2007; Blanpain and Fuchs, 2009). This program is tightly controlled by several transcription regulators, including the p63 transcription factor (Koster and Roop, 2007; Truong and Khavari, 2007; Crum and McKeon, 2010), as well as by the DNA- and chromatin-remodeling factors (DNA methyltransferase DNMT1, histone demethylase JMJD3, histone deacetylases 1/2, ATP-dependent chromatin-modifying enzymes Brg1 and Mi-2β, and the polycomb group protein Ezh2; Indra et al., 2005; Kashiwagi et al., 2007; Sen et al., 2008, 2010; Ezhkova et al., 2009; LeBoeuf et al., 2010). It remains to be explored how transcription factors and chromatin-remodeling enzymes are coordinately regulated to establish tissue-specific gene expression programs during terminal differentiation of epidermal progenitor cells.
The p63 transcription factor serves as a master regulator of epidermal development, and p63 knockout (−/−) mice fail to form stratified epithelium and to express several epidermis-specific genes (Mills et al., 1999; Yang et al., 1999). In keratinocytes, p63 regulates a large number of genes that encode distinct adhesion/signaling molecules, transcription factors, and cell cycle–associated proteins as well as tissue-specific proteins, such as keratins, involucrin, and loricrin (Viganò and Mantovani, 2007).
Eukaryotic genomes appear to be organized for gene regulation with a tendency of linear clustering of coexpressed genes, often related in function (Kosak and Groudine, 2004). In the mammalian epidermis, differentiation of the multipotent progenitor cells in the basal epidermal layer into keratinocytes of the suprabasal layer is accompanied by coordinated activation of the specific sets of genes; e.g., a cluster of such genes is found in the epidermal differentiation complex (EDC) located in the gene-rich region of mouse chromosome 3 (Fig. S1). Genes in the EDC encode the components of the cornified cell envelope (involucrin, loricrin, filaggrin, small proline-rich and late-cornified envelope proteins, etc.) essential for epidermal barrier function (Marshall et al., 2001; Martin et al., 2004; Bazzi et al., 2007; Brown et al., 2007).
It is well accepted now that higher-order chromatin organization and spatial arrangement of genes within the nuclear space, as well as nuclear compartmentalization of chromatin-remodeling complexes and transcription machinery, all play an important role in controlling gene expression (Fraser and Bickmore, 2007; Lanctôt et al., 2007; Takizawa et al., 2008; Rando and Chang, 2009; Zhao et al., 2009; Joffe et al., 2010; Naumova and Dekker, 2010; Schoenfelder et al., 2010). It has been previously shown that the differentiation process in cultured epidermal keratinocytes is also associated with changes in intranuclear positioning of the EDC and distinct chromosomes (Elder and Zhao, 2002; Marella et al., 2009). siRNA knockdown of p63 in keratinocytes results in marked alterations of gene expression in the EDC (Truong et al., 2006). However, it remains to be determined whether the p63 protein itself directly regulates some of these events during epidermal differentiation or whether the proteins encoded by p63 downstream target genes play a role in some of these processes.
In several other tissue-specific gene loci (e.g., TH2 cytokine locus and β-globin locus), higher-order chromatin remodeling for establishing specific 3D conformations is regulated by the specialized adenine and thymine–rich binding protein Satb1, which functions as the genome organizer (Dickinson et al., 1992; Alvarez et al., 2000; Cai et al., 2003, 2006; Kumar et al., 2007; Han et al., 2008; Wang et al., 2009). Satb1 targets chromatin-remodeling enzymes and transcription factors to specific genomic regions, establishes region-specific epigenomic modification status, and plays a fundamental role in the execution of tissue-specific gene expression programs (Yasui et al., 2002; Cai et al., 2006; Pavan Kumar et al., 2006). However, the upstream mechanisms controlling Satb1 expression in distinct cell types are unknown.
Here, we show that during epidermal morphogenesis, p63 transcription factor directly regulates expression of the genome organizer Satb1. Satb1 remodels chromatin architecture at the tissue-specific EDC gene locus, as indicated by compression of the central domain of this locus where terminal differentiation–associated genes are clustered. Upon Satb1 ablation, these genes cannot be properly activated, and the epidermal morphology becomes impaired, thus demonstrating an essential role for Satb1 as a part of the p63-dependent developmental program, through higher-order chromatin remodeling and gene regulation in epidermal progenitor cells.
Results
Expression of genes encoding chromatin-remodeling factors is markedly altered in the p63-null epidermis
Because p63 transcription factor is a master regulator for epidermal development, we hypothesized that it may regulate expression of a multitude of genes encoding chromatin-remodeling factors to establish specific chromatin architecture in epidermal progenitor cells. Global gene expression profiles were compared between the epidermis isolated by laser capture microdissection (LCM) from E16.5 p63−/− and wild-type (WT) mice, as previously described (Fig. 1 A; Mammucari et al., 2005; Sharov et al., 2006). This approach allowed us to compare gene expression profiles between the epidermal cells of p63−/− and WT mice in situ and to exclude alterations in gene expression induced by the proteolysis-associated cell separation procedures (Zhou et al., 2006). Global microarray analysis showed ≥1.8-fold changes in expression of 2,522 up-regulated and 998 down-regulated genes between the epidermis of p63−/− and WT mice (Fig. 1 B and Tables S1 and S2).
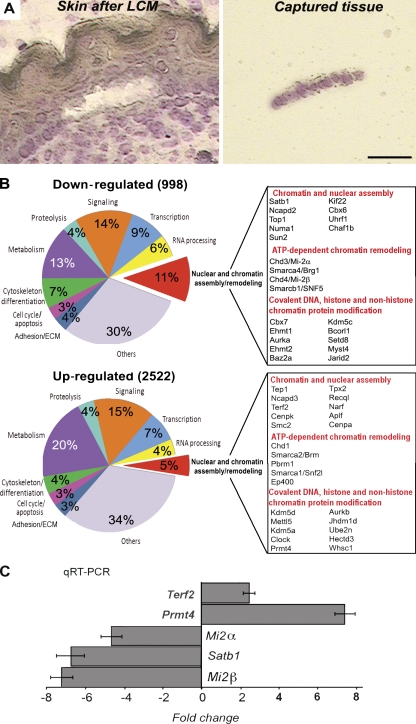
Figure 1.
Changes in global transcription profile and expression of the genes encoding chromatin-remodeling factors in the epidermis of p63−/− mice. The skin of E16.5 p63−/− and WT mice was processed for LCM to isolate fragments of the epidermis. RNA was isolated from captured tissue, amplified, and processed for microarray and RT-PCR analyses. (A) Cryosection of the skin after LCM including a fragment of the captured epidermis. Bar, 25 µm. (B) Agilent microarray analysis of the laser-captured epidermis of p63−/− and WT mice. Diagrams showing the ontology of the down- and up-regulated genes in p63-null versus WT epidermis. Selected genes involved in the control of nuclear structure and chromatin remodeling, the expression of which was changed in the epidermis of p63−/− versus WT mice, are listed (a full list of the genes is shown in Tables S1 and S2). (C) Real-time PCR for Satb1, Mi-2α, Mi-2β, Terf2, and Prmt4 in the E16.5 epidermis of p63−/− mice normalized to the corresponding levels in age-matched WT mice. Error bars represent SEM.
Among the distinct groups of genes, the expression of which was either down- or up-regulated in p63−/− mice versus WT mice, gene ontology analysis revealed a significant enrichment of the genes (P < 0.05) involved in the control of nuclear and chromatin assembly and remodeling (Fig. 1 B). These genes constituted ∼11 and 5% of the total number of down-regulated or up-regulated genes, respectively. Interestingly, in the epidermis of p63−/− mice versus WT controls, the expression of the genes encoding several key regulators of the higher-order chromatin structure and ATP-dependent chromatin remodeling (Satb1, Mi-2α, and Mi-2β) was down-regulated, whereas the expression of some other ATP-dependent chromatin-remodeling factors (Chd1 and Brm) was up-regulated. The top list of most affected genes is shown in Fig. 1 B (see the entire list in Tables S1 and S2). The relative expression levels of some of the most down-regulated genes between the WT and p63−/− mice were further validated by RT-PCR (Fig. 1 C). These data suggest that the genes encoding chromatin-remodeling factors represent a novel class of the p63 targets, raising a possibility that some of these factors may be in charge of the remodeling of the higher-order and/or local chromatin structures and tissue-specific gene expression during differentiation of the epidermal progenitor cells.
p63 controls the expression of Satb1 in epidermal progenitor cells
We studied whether p63 controls tissue-specific chromatin organization and gene expression during development of the epidermis through regulation of some of the chromatin-remodeling factors. For this purpose, we focused on the genome organizer Satb1 as a potential p63 target because of its known function in regulating large-scale chromatin remodeling in several cell type–specific gene loci (e.g., TH2 cytokine locus and β-globin locus; Cai et al., 2003; Wang et al., 2009) as well as cancer-related gene loci in aggressive breast cancer (Han et al., 2008). We found that Satb1 was coexpressed with p63 in basal cells of the epidermis of embryonic (E16.5) and postnatal skin (Fig. 2 A and not depicted). Skin epithelium of p63−/− mice expressing Keratin 18 as a marker of undifferentiated epithelial cells showed strongly reduced Satb1 protein levels (Fig. 2, B and C), which was consistent with greatly reduced Satb1 transcript expression in the epidermis of p63−/− mice versus WT controls (Fig. 1 C).
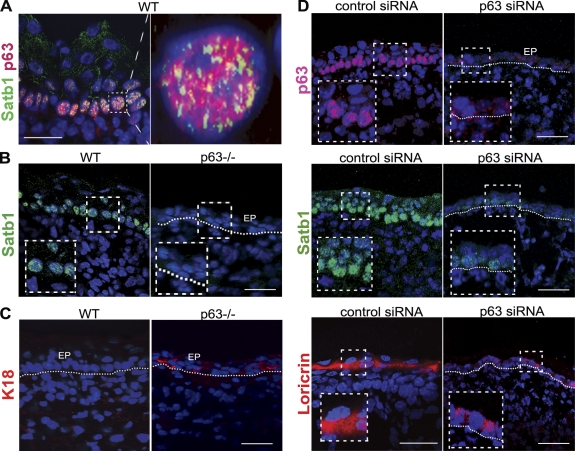
Figure 2.
p63 controls the expression of Satb1 in epidermal progenitor cells. The skin of p63-deficient and WT embryos was processed for immunofluorescent analyses as well as for modulation of gene expression in organ culture experiments with p63 siRNA. (A) Double immunofluorescence showing coexpression of p63 and Satb1 in the nuclei of basal epidermal cells in E16.5 WT mice. (B and C) Immunostaining for Satb1 (B) and K18 (C) in the E16.5 skin of WT and p63−/− mice. Marked decrease of Satb1 expression in the epidermis (EP) of p63−/− mice is magnified in the dashed boxes. (D) Immunostaining for p63, Satb1, and Loricrin in the E13.5 skin samples of heterozygous p63 knockout (+/−) mice cultured in the presence of p63 siRNA or control siRNA for 48 h. In epidermis, the expression of p63, Satb1, and Loricrin proteins in the p63 siRNA–treated samples was decreased compared with the controls. (B and D) The epidermal–dermal junction is shown by dotted lines. Bars, 25 µm.
In addition, the expression of the Satb1 protein was strongly reduced in the E13.5 skin explants of heterozygous p63 knockout (+/−) mice treated with p63 siRNA when compared with age-matched skin samples treated by the control siRNA (Figs. 2 D and S2 [A and B]). The expression of Loricrin, a marker of differentiated keratinocytes, was also decreased in the epidermis of embryonic skin explants after treatment with p63 siRNA compared with controls (Fig. 2 D).
Furthermore, chromatin immunoprecipitation (ChIP) assays using primary epidermal keratinocytes isolated from E16.5 embryos or newborn mice revealed binding of the p63 to the consensus sequence identified in the upstream region at −1662 to −1738 bp from the Satb1 transcription start site (Fig. 3, A and B). p63 binding was also seen at the consensus sequence in the Cldn1 promoter region, an established p63 target gene (Fig. 3 B; Lopardo et al., 2008). However, no binding was seen at the predicted p63 site located in the first intron of the Satb1 gene, serving as a negative control for this assay (Fig. 3 B).
Figure 3.
Satb1 is a direct target of p63 in keratinocytes. Primary keratinocytes isolated from E16.5 embryos or newborn mice were processed for ChIP analysis with an antibody against the p63 protein or purified rabbit IgGs. HaCaT cells were used in cotransfection experiments with Satb1 promoter–driven reporter construct. (A and B) Quantitative RT-PCR analysis of two distinct regions of the Satb1 (a predicted high-affinity p63-binding site and a negative control site) showing a specific high-affinity p63-binding site at the Satb1 promoter. In A, the position 1 refers to 5′ of Satb1 transcript (AK037740). The uppercase letters of the sequence show the p63 putative binding sites in the promoter region of the Satb1 gene chosen for the quantitative PCR analysis after ChIP. The promoter region of the Cldn1 was used as a positive control. The input levels of unprecipitated chromatin DNA were used as loading controls. Error bars represent SEM, and three independent experiments were run in triplicate. (C) HaCaT keratinocytes were cotransfected with the luciferase reporter plasmid containing a mouse Satb1 promoter fragment and pΔNp63 expression plasmid or control pcDNA3 plasmid. The increase in the pSatb1-luc activities by cotransfection with pΔNp63 compared with the control pcDNA3 was ∼3.3 fold (mean ± SEM, n = 3; *, P < 0.05, Student’s t test).
To assess whether p63 controls the activity of the Satb1 promoter, HaCaT keratinocytes were cotransfected with a luciferase reporter plasmid containing a mouse Satb1 promoter (pSatb1luc; unpublished data) and either the ΔNp63 expression vector (pΔNp63) or a control vector (pcDNA3). Transfection of cells with pΔNp63, but not with pcDNA3, resulted in an increase of the Satb1 promoter–driven luciferase activity by threefold compared with control (Fig. 3 C). These data strongly suggest that Satb1 is indeed a direct target gene of p63 in keratinocytes.
p63 or Satb1 ablation results in altered conformation of the chromatin domain containing the EDC locus
We further investigated whether there is any evidence for changes as a result of either p63 or Satb1 ablation in the higher-order chromatin organization in epidermal progenitor cells during development. Using 3D FISH, we analyzed the conformation of the 5 Mbp chromatin domain of mouse chromosome 3 that contains the tissue-specific EDC gene locus in the epidermis of E16.5 p63−/−, Satb1−/−, and corresponding WT embryos. The EDC occupies ∼3.1 Mbp in this domain located in the gene-rich region of mouse chromosome 3 and is flanked by several genes that are expressed in the epidermis, including Rps27 and Gabpb2, encoding ribosomal protein S27 and guanine and adenine–binding protein subunit 2, respectively (Fig. 4 A).
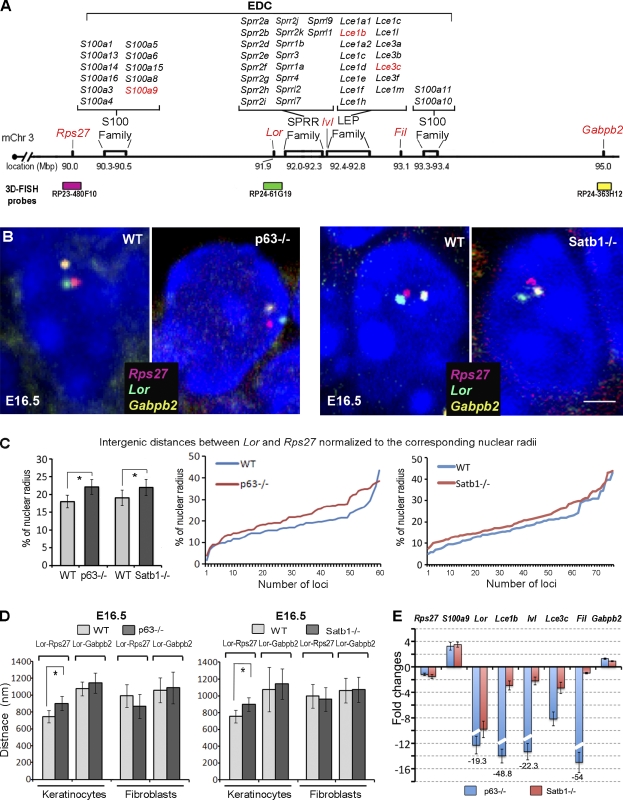
Figure 4.
Alterations in the conformation of the 5 Mb chromatin domain containing EDC in the epidermis of p63−/− and Satb1−/− mice. The skin of E16.5 p63−/−, Satb1−/−, and corresponding WT mice was processed for 3D FISH analyses, which were correlated with changes in gene expression determined by quantitative RT-PCR. (A) A schematic structure of the 5 Mb domain on mouse chromosome 3 (mChr3) containing the EDC locus, Rps27, and Gabpb2 genes. 3D FISH DNA probes detecting the corresponding domains are shown in green (Loricrin), pink (Rps27), and yellow (Gabpb2). Genes selected for quantitative RT-PCR analysis (shown in E) are shown in red. LEP, late-cornified envelope protein. (B) Multicolor 3D FISH with BACs covering the Rps27, Lor, and Gabpb2 in the epidermal cells of p63−/−, Satb1−/−, and corresponding WT mice at E16.5 (representative single Z sections). Bar, 2 µm. (C) 3D FISH distances between the Rps27and Lor normalized to the radius of each nuclei in basal epidermal cells of p63−/−, Satb1−/−, and corresponding WT mice. Pairwise comparisons represent a significant increase in the Lor-Rps27 distances between the E16.5 WT versus p63−/− or Satb1−/− mice (mean ± SEM, n = 60; *, P < 0.01, Newman–Keuls test after a one-way ANOVA test). (D) 3D FISH distances between the Rps27and Lor and the Lor and Gabpb2 in basal epidermal cells and dermal cells of p63−/−, Satb1−/−, and corresponding WT mice. Pairwise comparisons represent a significant increase in the Lor-Rps27 distances between the E16.5 WT versus p63−/− or Satb1−/− mice (mean ± SEM, n = 60; P < 0.01, Newman–Keuls test after a one-way ANOVA test). (E) Quantitative RT-PCR analyses of gene expression in the 5 Mb domain containing EDC in the epidermis of E16.5 p63−/− and Satb1−/− mice normalized to the expression levels in the corresponding WT mice. White slashes represent the interruption of these bars to indicate that they show the fold changes in gene expression levels higher than the maximum value in the y axis. The exact fold changes are indicated under each bar. SEM is shown in the appropriate scale (relative to the y axis). Three independent experiments were run in triplicate.
Despite the fact that the mean radius of the nuclei of basal epidermal cells was quite similar in p63−/−, Satb1−/−, and corresponding WT embryos, the distances between the Rps27 and Lor genes in the E16.5 epidermis of both p63−/− and Satb1−/− mice (without or after normalization to the corresponding nuclear radius) were significantly increased (P < 0.05) versus the corresponding controls, whereas distances between the Lor and Gabpb2 genes remained unchanged (Figs. 4 [B–D] and S2 C). Distribution of the distances between the Rps27 and Lor in individual nuclei showed a lack of peaks in the corresponding histograms, thus suggesting similarities in the positioning of both alleles of each gene relative to other genes in the keratinocyte nuclei (Fig. S2 D). However, there was no significant difference observed in the distances between any of those genes in dermal cells of the p63−/− or Satb1−/− mice and WT controls, indicating that the effects of p63 and Satb1 on 3D chromatin structure in keratinocytes are tissue specific (Fig. 4 D).
Next, we examined whether there is any correlation of the changes in chromatin organization and gene expression at the EDC locus for E16.5 epidermis of p63−/− and Satb1−/− embryos. The EDC contains several genes, including those activated during terminal keratinocyte differentiation (Fig. 4 A). Using quantitative RT-PCR, we observed that genes within the EDC and its flanking regions (Rps27 and Gabpb2) showed quite similar changes in expression in the epidermis of E16.5 p63−/− and Satb1−/− embryos versus the WT controls (Fig. 4 E).
In both p63−/− and Satb1−/− mice, the expression of the genes located in the central EDC domain and involved in the epidermal barrier formation, such as Lor, Inv, Lce1b, and Lce3c, was decreased, whereas the expression of the S100a9 located at the EDC 5′ end increased versus the corresponding WT controls (Fig. 4 E). In Satb1−/− mice, similar changes in gene expression in the EDC were observed in both dorsal skin and foot pads (Figs. 4 E and S2 E). The higher levels of reduction in expression for some of the genes tested in the epidermis of p63−/− mice compared with those of the Satb1−/− mice at E16.5 suggest the requirement of additional factors downstream of p63 for regulation of their expression. Expression of Fil, located at the EDC 3′ end, is solely dependent on p63 and not on Satb1, indicating that Satb1 is insufficient to replace global gene regulation by p63 (Fig. 4 E). Nevertheless, a similarity in altered expression patterns of the EDC genes seen in the p63−/− and Satb1−/− mice, paralleled by similar changes in the conformation of the 5 Mbp chromatin domain containing EDC, suggests a role for Satb1 in mediating, at least in part, the p63 functions in the developing epidermis.
Satb1−/− mice have impaired epidermal morphology and altered gene expression in keratinocyte-specific genomic loci
If Satb1 is a critical downstream target gene of p63 in epidermal progenitor cells, it is expected that the skin of Satb1−/− mice will exhibit altered epidermal morphogenesis. In fact, Satb1−/− mice at postnatal day 0.5 (P0.5) show a significant decrease (P < 0.01) of the epidermal thickness, as well as a thinning of the granular epidermal layer compared with WT mice, more pronounced in the foot pads versus dorsal skin (Fig. 5, A and B). In addition, Satb1−/− mice showed significantly (P < 0.05) decreased epidermal proliferation compared with WT controls (Fig. 5 C).
Figure 5.
Satb1 knockout mice show alterations in the epidermal structure and Loricrin expression. Cryosections of the footpad skin of newborn Satb1−/− and WT mice were processed for morphometric and immunofluorescent analyses. (A) Alterations in the structure and thinning of granular layer (arrows) in the epidermis of Satb1−/− mice compared with WT mice. Bars, 25 µm. (B) Significant (*, P < 0.05) decrease of the epidermal thickness in the epidermis of Satb1−/− mice compared with WT mice. Measurements of the epidermal thickness were performed in three Satb1−/− and three WT mice. 20 measurements were performed in each mouse. (C) Significant (*, P < 0.05) decrease of cell proliferation in the epidermis of Satb1−/− mice versus WT controls. The percentages of the Ki-67+ nuclei were determined in the epidermis of two Satb1−/− and two WT mice; 40–50 nuclei were analyzed in each animal. (B and C) Error bars represent SEM. (D) Decrease of Loricrin expression in the epidermis of Satb1−/− versus WT mice. Bars, 25 µm. (E) Agilent microarray data demonstrating changes in gene expression in the EDC between P0.5 Satb1−/− and WT mice. Asterisks indicate the fold changes in gene expression levels validated by quantitative RT-PCR.
To gain mechanistic insight into the role of Satb1 in the regulation of the keratinocyte terminal differentiation, samples of total RNA isolated from the skin of newborn Satb1−/− and WT mice were processed for microarray analysis (Sharov et al., 2006). In addition, the formaldehyde–cross-linked chromatin fragments from primary epidermal keratinocytes were immunoprecipitated with anti-Satb1 antibody, and a NimbleGen MM8 Deluxe Promoter HX1 array analysis was performed. ChIP-on-chip analysis showed that Satb1 does not always bind in the vicinity of the transcription start sites of the genes. In some cases, it binds sites remote from the gene proximal regulatory regions (Tables S4–S6), which is consistent with its role as a genome organizer important for the proper higher-order chromatin folding previously shown (Cai et al., 2003, 2006; Wang et al., 2009). Microarray and ChIP-on-chip data were merged, and 2,503 genes whose expression was significantly changed in Satb1−/− mice (≥1.8-fold) and showed Satb1 binding within 200 kb from the corresponding transcription start sites were selected as targets for Satb1 in keratinocytes (Fig. S3 A and Tables S4 and S5).
Interestingly, bioinformatic analyses on the Satb1 target genes involved in many biological processes (viz., regulation in cell adhesion/ECM remodeling, metabolism, cytoskeleton, signaling, and transcription) revealed that a large number of those genes are grouped into large genomic loci (>0.5 Mbp each) of gene clusters, containing >20 functionally relevant genes per locus, and that such genomic loci include epidermis-specific loci (Fig. S3 B and Tables S4 and S5). In fact, analyses of 33 such loci that are distributed genome wide showed significant preferential binding (P < 0.05) of Satb1 and Satb1-dependent changes in gene expression in the keratinocyte-specific loci (EDC, keratin type I/II loci, and keratin-associated protein locus) located on mouse chromosomes 3, 11, 15, and 16 (Fig. S3 B). Many of those genes found in the keratinocyte-specific loci were altered in expression upon p63 ablation (Fig. 5 E and Tables S1 and S2).
Microarray analysis revealed a profound decrease in gene expression in all keratinocyte-specific loci in Satb1−/− mice compared with WT controls (Fig. 3 B and Tables S4 and S5). Similarly to p63−/− mice, the expression of Krt10, Krt23, and Krt78 decreased in Satb1−/− mice compared with WT controls (Tables S1 and S4). In the EDC locus, Satb1 ablation resulted in decreased expression of a large number of genes that belong to the small proline-rich and late-cornified envelope families involved in the control of terminal keratinocyte differentiation, whereas an increase in expression was detected for selected members of the S100 family (Fig. 5 E). Consistent with microarray and quantitative RT-PCR data (Fig. 4 E), immunohistochemical analysis revealed markedly reduced expression of the Loricrin in Satb1-null epidermis versus the controls (Fig. 5 D). Thus, alterations of epidermal morphology in Satb1−/− mice and marked similarity in the Satb1- and p63-dependent changes in gene expression in four major keratinocyte-specific loci suggest that Satb1 is indeed a significant component of p63-regulated molecular network in the epidermis.
Gain of Satb1 expression partially rescues the epidermal phenotype in embryonic skin explants of p63-deficient mice
To assess whether the restoration of Satb1 expression is capable of partially rescuing the skin phenotype of p63−/− mice, E13.5 skin explants isolated from heterozygous (+/−) p63 knockout mice were cultured ex vivo and treated with p63 siRNA in combination with either Satb1-expressing or control lentiviruses (Fig. 6). Treatment of p63+/− skin explants with p63 siRNA resulted in an ∼80–90% decrease of the p63 mRNA levels compared with WT skin (Fig. S2 A). p63 siRNA treatment also caused marked decrease of the Satb1 mRNA levels, which, however, were restored by cotreatment with Satb1-expressing but not the control lentiviruses (Fig. 6 E).
Figure 6.
Treatment with Satb1-expressing lentivirus partially rescues alterations of epidermal phenotype in skin explants caused by p63 deficiency. The skin of p63+/− and WT E13.5 embryos was processed for modulation of gene expression in organ culture with p63 siRNA and Satb1-expressing lentivirus. (A and B) Morphology of E13.5 skin explants of p63+/− mice treated with combinations of p63 or control siRNAs and Satb1-expressing or control lentiviruses (LV) for 48 h. The decrease of the epidermal thickness induced by p63 siRNA is significantly (**, P < 0.01) rescued by the cotreatment with Satb1-expressing lentivirus compared with corresponding controls. Epidermal thickness was measured in three samples for each experimental group; 60 measurements were conducted in each sample. (C and D) Cotreatment with Satb1-expressing lentivirus prevents the decrease of cell proliferation in the epidermis of skin explants induced by p63 siRNA compared with controls (*, P < 0.05). The percentages of Ki-67+ cells were analyzed in three samples for each experimental group; 30 cells were analyzed in each sample. (A and D) The dotted lines show the position of the basement membrane separating the epidermis and dermis. (E and F) Quantitative RT-PCR for Loricrin and Satb1 (E) and immunostaining for Loricrin (F). Levels of Loricrin transcripts and the expression of protein decreased after p63 siRNA treatment restored in the epidermis after cotreatment with Satb1-expressing lentivirus compared with the controls (F, arrows). Treatment with Satb1-expressing lentivirus resulted in a marked increase of Satb1 transcript levels in the epidermis after decrease induced by p63 siRNA (E). (A, D, and F) The epidermal–dermal junction is shown by dotted lines. (B, C, and E) Error bars represent SEM. Bars: (A and F) 50 µm; (D) 25 µm.
In contrast to the p63+/− skin explants treated by control siRNA, treatment with p63 siRNA alone or in combination with control lentivirus caused a marked decrease of the epidermal thickness, cell proliferation, and Loricrin expression, thus reproducing in part the skin phenotype of p63−/− mice (Fig. 6, A–F). However, cotreatment of p63+/− skin samples with p63 siRNA and Satb1-expressing lentivirus resulted in a significant increase in epidermal thickness, cell proliferation, and Loricrin expression compared with the samples treated with p63 siRNA and control lentivirus (Fig. 6 A–F). Restoration of epidermal proliferation after treatment with Satb1-expressing lentivirus was consistent with data showing the reduced cell proliferation in the epidermis of Satb1−/− mice compared with WT controls (Fig. 5 C). Therefore, it is likely that Satb1 plays a role in regulating both keratinocyte proliferation and differentiation and is capable of partially restoring the epidermal phenotype of p63-deficient mice.
Satb1 binds to the distinct genomic regions within the EDC locus in keratinocytes
Because the distance between the Lor and Rps27 in the 5 Mbp chromatin domain is increased in Satb1−/− epidermis in comparison with WT mice (Fig. 4, B–D), we hypothesized that Satb1 may bind to specific sites in the EDC locus to regulate the EDC chromatin conformation. To determine whether any regions within the EDC locus are direct binding targets of Satb1, the formaldehyde–cross-linked chromatin fragments from keratinocytes isolated from E16.5 embryos or newborn skin were immunoprecipitated with anti-Satb1 antibody, and ChIP-on-chip analysis was performed (Fig. 7 A).
Figure 7.
Satb1 binds the central EDC domain and regulates its conformation in epidermal cells. Primary mouse keratinocytes were processed for ChIP-on-chip analyses with Satb1 antibody, and skin cryosections of E16.5 and P0.5 Satb1−/− and WT mice were processed for 3D FISH analyses of the EDC chromatin structure. (A) ChIP-on-chip analysis of the Satb1 binding to the distinct genomic regions of the 5 Mb chromatin domain on mouse chromosome 3 (chr3) containing EDC in primary keratinocytes. Enrichment of the Satb1-binding sites at the distinct EDC regions including the central domain. Lack of Satb1 binding to the gene desert region between the 5′ end and the central domain of the locus. LEP, late-cornified envelope protein. (B) 3D FISH analyses of the volume of central EDC domain as well as the EDC length with probes detecting the 5′ and 3′ ends of the EDC (S100a6 and S100a10 genes, respectively) and the central domain (Lor-Lce3c) in the basal epidermal cells of Satb1−/− and WT mice. Representative single Z optical sections of newborn WT and Satb1 knockout keratinocyte nuclei are shown. Consecutive Z sections of the same nuclei are shown in Fig. S5 E. Arrows indicate the FISH signals. Bar, 2 µm. (C) The volume of the EDC central domain and the length of the total EDC locus were measured and compared between nuclei of basal epidermal cells of Satb1−/− and WT mice. The distribution of the data after measuring the total EDC length with or without normalization to the corresponding nuclear radii (left and middle, respectively) and the volume of the central domain (right) in 60–70 loci of basal epidermal cells at P0.5 Satb1−/− and WT mice is shown. Individual loci are shown in the x axis. (D) Statistical analysis shows a significant increase of the volume of central EDC domain in Satb1−/− mice compared with that of WT mice. The length of the total EDC increases in P0.5 Satb1−/− mice compared with WT mice.
Analyses of the ChIP-on-chip data revealed Satb1 binding to many genomic sites, and these sites are found enriched in three distinct regions within the EDC locus (at the 5′ end domain adjacent to the gene desert region, the central domain, and the 3′ end domain). Importantly, Satb1-binding sites in the central domain correspond to the small proline-rich protein (Sprr) and late-cornified envelope (Lce) gene families (Fig. 7 A and Table S6), which are activated during terminal keratinocyte differentiation (Marshall et al., 2001; Martin et al., 2004; Bazzi et al., 2007; Brown et al., 2007). ChIP-on-chip data were further validated by ChIP–quantitative PCR, and the results confirmed that, in both E16.5 and newborn mice, Satb1 binds to the proximal regulatory regions of the Sprr1b and Lce3b genes located at the central EDC domain (Fig. S4).
Satb1 compresses chromatin conformation of the EDC locus and its central domain
To further examine the potential roles of Satb1 in chromatin remodeling and the execution of gene expression programs in epidermal progenitor cells, 3D FISH analysis of the chromatin conformation was performed in the epidermis of E16.5 and P0.5 Satb1−/− and age-matched WT mice, focusing on total EDC and its central domain containing a large number of the keratinocyte-specific genes (such as genes belonging to the Sprr and Lce families as well as Lor and Inv). Using DNA probes corresponding to S100a6 (5′ end), Lor-Lce3c (the central domain), and S100a10 (3′ end), we analyzed the 3D structure of the EDC locus by measuring the volume of its central domain as well as distances between its 5′ end, central domain, and 3′ end. We found that in E16.5 or P0.5 Satb1−/− and WT mice, the EDC locus was localized in the nuclear interior of epidermal progenitor cells, and Satb1 ablation did not significantly alter its radial positions compared with WT mice (Fig. S5, A and B).
However, the most striking feature of the EDC conformation in the E16.5 and P0.5 Satb1−/− epidermis, in comparison with age-matched WT mice, was the significantly (P < 0.001) expanded central domain, determined as the volume of the Lor-Lce3c signal (representative Z optical sections of the corresponding DNA FISH images are shown in Fig. 7 B, and continuous Z section images are shown in Fig. S5 E). As evident from the distribution of the data, we consistently observed the increased volume of the central domain in each individual nuclei of epidermal cells in P0.5 Satb1−/− mice compared with aged-matched WT mice (Fig. 7 C). In contrast to the epidermal cells, a lack of changes in the volume of the central EDC domain was seen between dermal cells of Satb1−/− and WT mice (Fig. S5 D), thus suggesting that the effects of the Satb1 ablation on its conformation are indeed keratinocyte specific.
In addition, we found an overall expansion in the length of the whole EDC in P0.5 Satb1−/− epidermis (P < 0.01) compared with age-matched WT mice (Figs. 7 [C and D] and S5 C), which is likely a result of elongation of its 5′ domain, determined as a distance between the S100a6 and the center of the Lor-Lce3c signal (Table S7). These results are consistent with data demonstrating the significant elongation of the distance between Lor and Rps27 genes in Satb1−/− embryos versus WT mice (Fig. 4, B–D) and suggest that Satb1 ablation leads to significant decompression of the locus.
Interestingly, Satb1 binding at specific sites within the EDC locus was not always detected at the proximal promoter regions of the genes whose expression was markedly altered in the Satb−/− epidermis (e.g., Lor; Fig. 7 A and Table S6). Our 3D FISH data indicate that Satb1 compresses the chromatin conformation at the EDC locus, including the regions enriched in genes activated during terminal keratinocyte differentiation (Fig. 7, B–D). We observed marked differences in the expression of the genes located in the EDC central domain upon either Satb1 or p63 ablation (Figs. 4 E and 5 E). Therefore, it is likely that Satb1 regulates gene expression in the EDC locus in epidermal progenitor cells by establishing its tissue-specific chromatin conformations and potentially by forming a densely looped chromatin structure, rather than by directly targeting proximal cis-regulatory elements of individual genes.
Discussion
Here, we show that p63 transcription factor governs a program of the development of stratified epithelia (Koster and Roop, 2007; Truong and Khavari, 2007; Crum and McKeon, 2010) via regulating expression of the genes that control higher-order chromatin remodeling, covalent histone modifications, and nuclear assembly. Among the genes that regulate chromatin structure and gene expression, we focused on Satb1, which is known to regulate large-scale chromatin remodeling in selected cell types and progenitor cells (Cai et al., 2003, 2006; Kumar et al., 2007; Han et al., 2008; Agrelo et al., 2009). Satb1 was identified among the genes involved in chromatin remodeling that were down-regulated in p63−/− epidermis, consistent with data previously reported using cultured epithelial cells (Carroll et al., 2006; Truong et al., 2006; Pozzi et al., 2009). Our data unveil a novel mechanism by which p63 functions in epidermal development, at least in part, via direct regulation of Satb1.
We demonstrate that in keratinocytes, Satb1 shows preferential binding to four major tissue-specific gene loci (EDC, keratin type I/II loci, and keratin-associated protein locus). Satb1 ablation, in striking similarity to p63 deficiency, results in profound alterations of expression of a large number of epidermis-specific genes. Furthermore, we show that the restoration of Satb1 levels partially rescues the epidermal phenotype of embryonic skin explants from p63-deficient mice, further suggesting Satb1 as an important part of the p63-regulated network in the skin.
Among many chromatin-remodeling factors (such as Smarca4/Brg1, Mi-2α, Mi-2β, etc.) that are altered in p63−/− mice and other p63 targets previously identified (Koster and Roop, 2007; Viganò and Mantovani, 2007), Satb1 represents an important functional component of the p63-regulated genetic network because of its ability to reorganize chromatin structure and regulate higher-order chromatin remodeling in epidermal progenitor cells. In fact, our expression data together with ChIP-on-chip data for Satb1-binding sites indicate that once Satb1 is induced by p63, it establishes its own regulatory network, binding to the target gene loci and controlling the expression of ∼2,500 genes, including those not associated with the epidermis-specific gene loci. Among these genes, cell cycle–associated genes are of interest because we found that Satb1 has an effect in promoting cell proliferation in the developing epidermis, which is consistent with data obtained from the studies on aggressive breast cancer cells (Han et al., 2008). The mechanisms underlying Satb1-induced cell proliferation need to be studied in the future.
Differences in gene expression profiles in keratinocytes observed after either p63 or Satb1 ablation in many gene groups may suggest that the roles of p63 and Satb1 in the control of expression of the genes located outside of the epidermis-specific loci are quite distinct. However, we must be aware that p63 ablation, which deregulates many key transcriptional repressors/activators and chromatin-remodeling factors, in addition to Satb1, could alter expression of many Satb1 target genes beyond the effect of Satb1 ablation alone. This could explain, at least for some gene groups, why their expression profiles are quite distinct between p63−/− and Satb1−/− mice. Nevertheless, we provide compelling evidence that within a spiderweb of a gene regulatory network governed by p63, Satb1, as its direct downstream target gene, constitutes an important component driving epidermal development via regulating the epidermis-specific gene loci. It still remains to be investigated whether Satb1 acts together with p63 or whether it acts independently from p63 in controlling the establishment of specific conformation of the EDC and other genomic loci in keratinocytes.
We show here that Satb1 binds to distinct genomic regions including the central domain of the EDC locus, which consists of a large number of genes activated during terminal keratinocyte differentiation (Martin et al., 2004; Brown et al., 2007) and which regulates the establishment of its specific conformations in keratinocytes. It has been previously shown that intranuclear EDC positioning in cultured keratinocytes is cell type specific, whereas their treatment with 5-azacytidine and sodium butyrate results in an increase of gene expression within the EDC (Elder and Zhao, 2002; Williams et al., 2002). These and other studies suggest that changes in local epigenomic modification of the EDC gene locus are required for efficient expression of keratinocyte-specific genes (Frye et al., 2007; Ezhkova et al., 2009; Sen et al., 2010).
Furthermore, we demonstrate that Satb1 deficiency results in decompression of the EDC locus, specifically its central domain. These alterations are associated with a marked decrease in expression of terminal differentiation–associated genes and are accompanied by morphological changes in the epidermis (thinning of the granular layer and decrease of the epidermal thickness). Lack of similar dynamics in the nuclei of dermal cells suggests that the developmentally regulated remodeling of the higher-order chromatin structure in the EDC associated with increase of its transcription activity is indeed a tissue-specific event and that other factors may control EDC condensation in dermal cells.
During the execution of cell differentiation programs, tissue-specific genes organized into large genomic loci are expressed in a tightly coordinated manner to achieve the proper balance of the distinct proteins in differentiated cells (Kosak and Groudine, 2004). Increased evidence of data suggests that for such coordinated regulation, distinct spatial organization of tissue-specific gene loci is required, and Satb1 was shown to be an essential factor in controlling formation of the specific loop structures in many tissue-specific gene loci (Cai et al., 2003, 2006). It has been shown that during TH2 lymphocyte activation, Satb1 promotes the formation of a specific 3D structure of the cytokine locus, in which chromatin is folded into numerous small loops that bring proximal and distal gene regulatory elements together to activate the gene expression (Cai et al., 2006). More recent data demonstrate that Satb1 also controls the formation of the chromatin loops in the β-globin locus (Wang et al., 2009). We hypothesize that Satb1 may form a similar chromatin structure, characterized by dense chromatin loops, in the EDC central domain and the flanking regions, which may result in reduction of the total physical volume of chromatin in these regions. Such a higher-order transcriptionally active chromatin structure may facilitate the accessibility of protein complexes to target gene loci as well as interactions between proximal and distal regulatory gene regions.
If this hypothesis is correct, in the absence of Satb1, the EDC locus cannot be properly folded, and, therefore, this region on mouse chromosome 3 will remain more decompressed (or elongated) compared with WT cells and will not be able to support efficient expression of the terminal differentiation–associated genes. To test this hypothesis, additional studies using the chromatin conformation capture techniques will be required to identify the role of Satb1 in the formation of such chromatin loop–like structures within the EDC. However, significant changes in the volume of the central EDC domain and total EDC length upon Satb1 ablation provide strong support for this hypothesis.
Importantly, these data are also consistent with previous observations that strong Satb1 binding may not be necessarily detected at the proximal promoter regions of its target genes. This is because the main binding targets of Satb1 are the specialized DNA domains called base unpairing regions (Kohwi-Shigematsu and Kohwi, 1990; Bode et al., 1992), which are enriched in the gene-rich regions throughout the genome (Cai et al., 2006). For instance, Satb1 binding was undetected at the proximal regulatory region of the Lor gene per se, and yet Lor appears to be regulated in the context of Satb1-dependent 3D chromatin folding.
The data presented in this paper link, for the first time, the functions of a master regulator of tissue development (viz., p63) with a protein that controls higher-order chromatin remodeling (viz., Satb1), and this link provides an important foundation for further studies of the role of p63 as a potential upstream regulator of Satb1 activity in other cell types. Because the Satb1-dependent step in the p63-mediated program of higher-order chromatin remodeling in the EDC locus is highly important for the proper execution of differentiation programs in epidermal progenitor cells, it remains to be determined whether similar mechanisms underlie the involvement of p63 and Satb1 during the developmentally regulated activation of gene expression in other keratinocyte-specific loci as well as during activation of the epidermis-specific gene expression programs in epithelial stem cells in adult skin (Ito et al., 2007; Watt et al., 2008; Blanpain and Fuchs, 2009). Importantly, recent data indeed revealed that Satb1 has a critical role in regulating the differentiation potential of embryonic stem cells (Savarese et al., 2009).
Collectively, our data demonstrate that Satb1 is an important hitherto unrecognized component of the global regulatory network of p63, a master regulator of epidermal morphogenesis. These data provide a novel fundamental mechanism showing how master regulators of tissue morphogenesis establish tissue- and developmental stage–specific patterns of chromatin organization and remodeling to control the fate of multipotent progenitor cells.
Materials and methods
Experimental animals and tissue collection
All animal studies were performed under protocols approved by the Boston University and University of California Berkeley Institutional Animal Care and Use Committee and the Home Office Project License. C57Bl/6 mice were purchased from Charles River. Skin samples were collected from mice at distinct days of embryonic and postnatal development (E16.5 and P0.5). p63−/− and p63+/+ control embryos at E16.5 were obtained by breeding p63+/− animals purchased from The Jackson Laboratory. Genotyping of mice was performed using PCR, as recommended by the supplier. For each developmental stage, six to seven samples were collected. Tissue samples were frozen in liquid nitrogen and processed for immunofluorescent or microarray analyses as previously described (Sharov et al., 2003, 2005b).
In brief, for FISH and immunofluorescent analysis of 3D preserved nuclei, skin samples were processed through the fixation protocol with formaldehyde and gradual equilibration in buffered sucrose solutions according to previously published recommendations (Solovei et al., 2009). The embryos or tissues were fixed in 4% formaldehyde at 4°C overnight and incubated in 50 mM ammonium chloride solution at room temperature for 5 min followed by washes with 0.1 M phosphate buffer (pH 7.0) and by incubations in 5 and 12.5% sucrose solution in the phosphate buffer (1 h at room temperature) and in 20% sucrose solution (overnight at 4°C). Finally, the samples were frozen and embedded into optimal cutting temperature medium.
LCM, microarray, and quantitative RT-PCR analysis
LCM of whole mouse epidermis at selected stages of development was performed followed by RNA isolation, amplification, and microarray analysis as previously described (Sharov et al., 2006). In brief, 8-µm-thick frozen skin cryosections were dehydrated to preserve RNA integrity, and LCM was performed from the epidermis of WT and p63−/− mice at E16.5 using an LCM microscope (Life Technologies). Total RNAs were isolated using the PicoPure RNA isolation kit (Life Technolgoies) followed by two rounds of linear RNA amplification using the RiboAmp RNA amplification kit (Life Technologies). Equal amounts of RNA from each sample were labeled by Cy3 using a fluorescent labeling kit (Agilent Technologies). Quality and size distribution of targets was determined by RNA 6000 Nano Laboratory–on-chip assay (Agilent Technologies), and quantification was determined using a microscale spectrophotometer (NanoDrop). After one round of linear amplification in all analyses, Universal Mouse Reference RNA (Agilent Technologies) was used as a control. All microarray analyses were performed by MOgene, LC using the 41K Whole Mouse Genome 60-mer Oligo Microarray kit (Agilent Technologies). Real-time PCR of unamplified reference RNA and reference RNA obtained after two rounds of amplification was used for validation of possible alterations in gene expression caused by the amplification procedure. All microarray data on gene expression were normalized to the corresponding data obtained from the reference RNA. Two independent datasets were obtained from WT and transgenic mice, and p values were calculated by Feature Extraction software (version 7.5; Agilent Technologies) using distribution of the background intensity values to signal intensity and using a Student’s t test. For quantitative RT-PCR analysis of RNA isolated and amplified after LCM, PCR primers were designed on Beacon Designer software (Table S6; PREMIER Biosoft International). Real-time PCR was performed using iQ SYBR green Supermix and the MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad Laboratories). Differences between samples and controls were calculated using the Gene Expression Macro program (Bio-Rad Laboratories) based on the ΔΔCt equitation method with glyceraldehyde 3-phosphate dehydrogenase and peptidylprolyl isomerase A genes as internal housekeeping controls.
3D FISH and immunofluorescent analyses
Labeled DNA probes were prepared, and 3D FISH analysis was performed on 20-µm tissue sections as previously described (Solovei et al., 2009). Individual or pooled bacterial artificial chromosome (BAC) DNA for the genetic loci of interest was used for the probe synthesis (Table S3). The probes were synthesized by nick translation using Biotin-dUTP, FITC-dUTP, or Cy3-dUTP. Whole chromosome paints were labeled with Biotin-dUTP by degenerate oligonucleotide–primed PCR. The frozen sections were briefly dried, heated in 10 mM sodium citrate (pH 6.0) to partially reverse cross-linking, and equilibrated in 50% formamide/2× SSC. Labeled DNA probe mixtures were dissolved in hybridization buffer and applied on the tissue under glass chambers. Genomic and probe DNA were denatured by incubating slides at 85°C for 5 min, and hybridization was performed at 37°C for 2 d. After hybridization, slides were washed three times at 2× SSC for 10 min at 37°C followed by one wash in 0.1× SSC for 10 min at 60°C. When biotinylated probes were used, the slides were incubated with avidin–Alexa Fluor 488 (Invitrogen), streptavidin-Cy3, or streptavidin-Cy5 (Rockland Immunochemicals). DNA was stained with DAPI (Sigma-Aldrich), and slides were embedded using VectaShield medium (Vector Laboratories). Immunofluorescent analysis was performed on 10–20-µm quickly frozen or 3D preserved tissue with antibodies (Table S10) using standard protocols (Fessing et al., 2006).
Microscopy and image analyses
For analysis of epidermal morphology, images of the tissue after staining with hemotoxylin were acquired at room temperature using a microscope (Eclipse 50i; Nikon) equipped with a Plan Fluor 20×/0.50 or 40×/0.75 objective lens (Nikon), VisiCam 3.0 camera (VWR International), and VisiCam Image Analyzer software. The morphometric analysis was performed using ImageJ software (National Institutes of Health). Immunofluorescent images were acquired at room temperature using a microscope (Eclipse 50i), EXi Aqua camera (QImaging), and Image-Pro Express software (version 6.3; Media Cybernetics).
3D FISH images were collected using a laser-scanning confocal microscope (510 META; Carl Zeiss), Plan Apochromat 63×/1.4 oil differential interference contrast objective lens, and laser-scanning microscopy software (510 META). Imaging was performed at room temperature using Immersol immersion oil (Carl Zeiss). For 3D image analysis, the nuclei were scanned with a Z axial distance of 200 nm yielding separate stacks of 8-bit grayscale images for each fluorescent channel with a pixel size of 100–200 nm. For each optical section, images were collected sequentially for all fluorophores, and axial chromatic shift was corrected for each channel using 500-nm TetraSpeck beads (Invitrogen; Solovei et al., 2009). The images were processed and analyzed using ImageJ software. For the illustration purposes, 3D FISH and immunofluorescent images of the skin cryosections were adjusted using the levels and brightness/contrast tools in Photoshop (CS4; Adobe); separate adjustments were applied to every pixel in each RGB channel.
To determine radial positions of the loci, coordinates of at least 200 random points from the nuclear surface depicted by DAPI counterstain were acquired and exported to Excel (Microsoft), where the nuclear geometric center was calculated by coordinate averaging (Höfers et al., 1993). The mean nuclear radius for each nucleus was calculated as a mean of distances from the nuclear geometric center to all surface points. Distances between the centers of the nucleus and distinct loci were calculated on the basis of their spatial coordinates and were normalized to the mean nuclear radius using Excel (Ronneberger et al., 2008). Distances between individual gene loci were calculated after determination of the spatial coordinates of the corresponding centers of the signals using ImageJ software. The distances were normalized to the mean radii of the corresponding nuclei, determined as described above.
The volume of the central EDC domain was calculated using the approach previously described (Rauch et al., 2008). First, the threshold of the fluorescent signal was determined via selection of 1% of the brightest voxels from 3D intensity histograms using ImageJ software. Second, using this threshold, the number of voxels within each fluorescent signal was calculated. Next, the physical volume indicated by the fluorescent signal was determined as a sum of the volumes from all voxels after correction for optical aberrations by normalizing the data to the volume of beads of distinct size (100 nm, 500 nm, and 4 µm; TetraSpeck).
Embryonic tissue culture
Whole dorsal skin culture from E13.5 p63+/− embryos were prepared and maintained as previously described (Botchkarev et al., 1999). In brief, skin samples were dissected from embryos and cultured in the 6-well plates containing Williams’ E culture medium for 48 h. Within 2 h after the culture preparation, tissue samples were transfected with Stealth siRNA, recognizing all isoforms of p63 (Antonini et al., 2006), or medium containing negative control Stealth siRNA (Invitrogen) at 100 nM final concentration using the RNAiMax reagent (Invitrogen). Samples were also incubated with combinations of p63 siRNA or control siRNA and Satb1-expressing lentivirus or control lentivirus (10 µg/ml) for 48 h. The tissue was frozen and embedded into optimal cutting temperature medium as described in the Experimental animals and tissue collection section for subsequent immunofluorescent analyses. The penetration of the RNAi into the tissue under the used experimental conditions was demonstrated using BLOCK-iT Fluorescent Oligo (Invitrogen) in parallel experiments.
Western blot analysis
Proteins were extracted from snap-frozen skin samples or cultured keratinocytes with lysis buffer as previously described (Antonini et al., 2006). 5 µg of protein was processed for Western blot analysis followed by incubation with primary antibodies against p63 or Erk (Table S10) overnight at 4°C. HRP-tagged IgG antibody was used as a secondary antibody (1:5,000; Thermo Fisher Scientific). Antibody binding was visualized using an ECL system (SuperSignal West Pico kit; Thermo Fisher Scientific) followed by autoradiography with x-ray film (Thermo Fisher Scientific).
ChIP and ChIP-on-chip assays
ChIP assay was performed using epidermal keratinocytes isolated from E16.5 or newborn mouse skin with anti-p63 or anti-Satb1 antibodies (Table S8) as previously described (Antonini et al., 2006; Nguyen et al., 2006). PCR analysis for enrichment in the promoter sequences of Satb1, Cldn1, Lor, Sprr1b, Lce3b, or Inv genes in precipitated DNA was performed using the PCR primers listed in Table S7. For ChIP-on-chip analysis, precipitated DNA was analyzed by quantitative PCR or after one round of amplification (WGA2; Sigma-Aldrich) and applied to a general extended NimbleGen MM8 Deluxe Promoter HX1 array (MOgene, LC).
Reporter assay
Plasmids expressing pSatb1-luc construct bearing a promoter region containing the far-upstream CpG island and the p63 consensus sequence of mouse Satb1 were generated. For reporter assay, HaCaT cells were seeded into 96-well plates (104 cells per well) 1 d before transfection. Cells were cotransfected with pΔNp63 (Antonini et al., 2006) or pcDNA3 plasmids (120 ng/well; Invitrogen) along with the reporter pSatb1-Luc construct using Lipofectamine 2000 (0.5 µl/well; Invitrogen). After 48 h, reporter activity was detected using the Dual-Glo Luciferase Assay System (Promega). Two independent assays were performed in triplicate, and results were normalized to the pNull-Renilla construct activity, the data were pooled, a mean ± SEM was calculated, and statistical analysis was performed using a Student’s t test.
Statistical analysis and gene ontology analysis
For statistical evaluation of differences in radial positions, intergene distances, and 3D FISH signal volumes, normality of data distributions was evaluated by plotting them on histograms. Equality of their variances was tested by the Levene’s test. Significance of the differences for the pairwise comparisons was analyzed by a two-tailed t test (α = 0.05). For more than two-sample comparisons, a one-way analysis of variance (ANOVA) followed by the Newman–Keuls test (α = 0.05) was performed. Enrichment of the Satb1 target genes in the distinct genomic loci was assessed by using the hypergeometric or Fisher’s exact tests. Functional annotation of the overrepresented and underrepresented genes was performed as previously described (Sharov et al., 2009; Fessing et al., 2010) using the National Institute on Aging Array Analysis software (National Institutes of Health) according to previously published recommendations (Sharov et al., 2005a).
Online supplemental material
Fig. S1 shows localization of the EDC in gene-dense regions of mouse chromosome 3. Fig. S2 shows validation of the effects of p63 knockdown after siRNA treatment in skin explants and keratinocytes as well as the data on the analyses of 3D FISH distances and gene expression in the epidermis of p63−/−, Satb1−/−, and corresponding WT mice. Fig. S3 shows the results of the microarray, ChIP-on-chip, and bioinformatic analyses of the genes whose expression was altered in the skin of Satb1−/− mice versus WT controls and which showed association of Satb1 with the genomic regions within 200 kb from the transcription start sites in the ChIP-on-chip assay. Fig. S4 shows the results of ChIP–quantitative PCR analyses of keratinocytes demonstrating binding of Satb1 to the promoter regions of the distinct genes within the EDC. Fig. S5 shows the results of 3D FISH analyses of radial positions of Rps27 and Lor genes and differences in the intergene distances in the EDC locus as well as the confocal stacks demonstrating intranuclear localization of the distinct EDC domains in the epidermis of Satb1−/− and WT mice. Tables S1 and S2 show the lists of the genes down- and up-regulated in the skin epithelium of the E16.5 p63−/− versus WT mice, respectively. Table S3 shows the list of 3D FISH BAC probes generated to the distinct regions of mouse chromosome 3. Tables S4 and S5 show the list of genes that demonstrate Satb1 binding to their regulatory regions and down- and up-regulated in the skin of the P0.5 Satb1−/− versus WT mice, respectively. Table S6 shows SATB1 binding sites in the EDC locus and its 5′ and 3′ flanking regions based on the ChIP-on-chip analysis. Table S7 shows the length of the distinct EDC domains in Satb1−/− and WT mice. Tables S8 and S9 show the list of the primers used for quantitative RT-PCR and ChIP–quantitative PCR analyses, respectively. Table S10 shows the list of primary antibodies used for immunohistochemical analyses. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201101148/DC1.
Acknowledgments
The authors greatly appreciate the invaluable help and support of Prof. T. Cremer, Dr. I. Solovei, and Dr. B. Joffe (University of Munich, Munich, Germany) in establishing 3D FISH technique and analyzing the data.
This study was supported in part by the grants from the Medical Research Council (G0901666) to V.A. Botchkarev, from the National Institutes of Health (R37CA039681) to T. Kohwi-Shigematsu (under the Department of Energy Contract no. DE-AC02-05CH11231), and from the Fondazione Telethon (GGP09230) to C. Missero.
Footnotes
Abbreviations used in this paper:
- BAC
- bacterial artificial chromosome
- ChIP
- chromatin immunoprecipitation
- EDC
- epidermal differentiation complex
- LCM
- laser capture microdissection
- WT
- wild type
References
- Agrelo R., Souabni A., Novatchkova M., Haslinger C., Leeb M., Komnenovic V., Kishimoto H., Gresh L., Kohwi-Shigematsu T., Kenner L., Wutz A. 2009. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev. Cell. 16:507–516 10.1016/j.devcel.2009.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez J.D., Yasui D.H., Niida H., Joh T., Loh D.Y., Kohwi-Shigematsu T. 2000. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 14:521–535 [PMC free article] [PubMed] [Google Scholar]
- Antonini D., Rossi B., Han R., Minichiello A., Di Palma T., Corrado M., Banfi S., Zannini M., Brissette J.L., Missero C. 2006. An autoregulatory loop directs the tissue-specific expression of p63 through a long-range evolutionarily conserved enhancer. Mol. Cell. Biol. 26:3308–3318 10.1128/MCB.26.8.3308-3318.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi H., Fantauzzo K.A., Richardson G.D., Jahoda C.A., Christiano A.M. 2007. Transcriptional profiling of developing mouse epidermis reveals novel patterns of coordinated gene expression. Dev. Dyn. 236:961–970 10.1002/dvdy.21099 [DOI] [PubMed] [Google Scholar]
- Blanpain C., Fuchs E. 2009. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 10:207–217 10.1038/nrm2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode J., Kohwi Y., Dickinson L., Joh T., Klehr D., Mielke C., Kohwi-Shigematsu T. 1992. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 255:195–197 10.1126/science.1553545 [DOI] [PubMed] [Google Scholar]
- Botchkarev V.A., Botchkareva N.V., Roth W., Nakamura M., Chen L.-H., Herzog W., Lindner G., McMahon J.A., Peters C., Lauster R., et al. 1999. Noggin is a mesenchymally derived stimulator of hair-follicle induction. Nat. Cell Biol. 1:158–164 10.1038/11078 [DOI] [PubMed] [Google Scholar]
- Brown S.J., Tilli C.M., Jackson B., Avilion A.A., MacLeod M.C., Maltais L.J., Lovering R.C., Byrne C. 2007. Rodent Lce gene clusters; new nomenclature, gene organization, and divergence of human and rodent genes. J. Invest. Dermatol. 127:1782–1786 [DOI] [PubMed] [Google Scholar]
- Cai S., Han H.J., Kohwi-Shigematsu T. 2003. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34:42–51 10.1038/ng1146 [DOI] [PubMed] [Google Scholar]
- Cai S., Lee C.C., Kohwi-Shigematsu T. 2006. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 38:1278–1288 10.1038/ng1913 [DOI] [PubMed] [Google Scholar]
- Carroll D.K., Carroll J.S., Leong C.O., Cheng F., Brown M., Mills A.A., Brugge J.S., Ellisen L.W. 2006. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 8:551–561 10.1038/ncb1420 [DOI] [PubMed] [Google Scholar]
- Crum C.P., McKeon F.D. 2010. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu. Rev. Pathol. 5:349–371 10.1146/annurev-pathol-121808-102117 [DOI] [PubMed] [Google Scholar]
- Dickinson L.A., Joh T., Kohwi Y., Kohwi-Shigematsu T. 1992. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 70:631–645 10.1016/0092-8674(92)90432-C [DOI] [PubMed] [Google Scholar]
- Elder J.T., Zhao X. 2002. Evidence for local control of gene expression in the epidermal differentiation complex. Exp. Dermatol. 11:406–412 10.1034/j.1600-0625.2002.110503.x [DOI] [PubMed] [Google Scholar]
- Ezhkova E., Pasolli H.A., Parker J.S., Stokes N., Su I.H., Hannon G., Tarakhovsky A., Fuchs E. 2009. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 136:1122–1135 10.1016/j.cell.2008.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessing M.Y., Sharova T.Y., Sharov A.A., Atoyan R., Botchkarev V.A. 2006. Involvement of the Edar signaling in the control of hair follicle involution (catagen). Am. J. Pathol. 169:2075–2084 10.2353/ajpath.2006.060227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessing M.Y., Atoyan R., Shander B., Mardaryev A.N., Botchkarev V.V.J., Jr, Poterlowicz K., Peng Y., Efimova T., Botchkarev V.A. 2010. BMP signaling induces cell-type-specific changes in gene expression programs of human keratinocytes and fibroblasts. J. Invest. Dermatol. 130:398–404 10.1038/jid.2009.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P., Bickmore W. 2007. Nuclear organization of the genome and the potential for gene regulation. Nature. 447:413–417 10.1038/nature05916 [DOI] [PubMed] [Google Scholar]
- Frye M., Fisher A.G., Watt F.M. 2007. Epidermal stem cells are defined by global histone modifications that are altered by Myc-induced differentiation. PLoS ONE. 2:e763 10.1371/journal.pone.0000763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. 2007. Scratching the surface of skin development. Nature. 445:834–842 10.1038/nature05659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H.J., Russo J., Kohwi Y., Kohwi-Shigematsu T. 2008. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 452:187–193 10.1038/nature06781 [DOI] [PubMed] [Google Scholar]
- Hemberger M., Dean W., Reik W. 2009. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat. Rev. Mol. Cell Biol. 10:526–537 10.1038/nrm2727 [DOI] [PubMed] [Google Scholar]
- Höfers C., Baumann P., Hummer G., Jovin T.M., Arndt-Jovin D.J. 1993. The localization of chromosome domains in human interphase nuclei. Three-dimensional distance determinations of fluorescence in situ hybridization signals from confocal laser scanning microscopy. Bioimaging. 1:96–106 [DOI] [Google Scholar]
- Indra A.K., Dupé V., Bornert J.M., Messaddeq N., Yaniv M., Mark M., Chambon P., Metzger D. 2005. Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development. 132:4533–4544 10.1242/dev.02019 [DOI] [PubMed] [Google Scholar]
- Ito M., Yang Z., Andl T., Cui C., Kim N., Millar S.E., Cotsarelis G. 2007. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 447:316–320 10.1038/nature05766 [DOI] [PubMed] [Google Scholar]
- Joffe B., Leonhardt H., Solovei I. 2010. Differentiation and large scale spatial organization of the genome. Curr. Opin. Genet. Dev. 20:562–569 10.1016/j.gde.2010.05.009 [DOI] [PubMed] [Google Scholar]
- Kashiwagi M., Morgan B.A., Georgopoulos K. 2007. The chromatin remodeler Mi-2beta is required for establishment of the basal epidermis and normal differentiation of its progeny. Development. 134:1571–1582 10.1242/dev.001750 [DOI] [PubMed] [Google Scholar]
- Kohwi-Shigematsu T., Kohwi Y. 1990. Torsional stress stabilizes extended base unpairing in suppressor sites flanking immunoglobulin heavy chain enhancer. Biochemistry. 29:9551–9560 10.1021/bi00493a009 [DOI] [PubMed] [Google Scholar]
- Kosak S.T., Groudine M. 2004. Gene order and dynamic domains. Science. 306:644–647 10.1126/science.1103864 [DOI] [PubMed] [Google Scholar]
- Koster M.I., Roop D.R. 2007. Mechanisms regulating epithelial stratification. Annu. Rev. Cell Dev. Biol. 23:93–113 10.1146/annurev.cellbio.23.090506.123357 [DOI] [PubMed] [Google Scholar]
- Kumar P.P., Bischof O., Purbey P.K., Notani D., Urlaub H., Dejean A., Galande S. 2007. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 9:45–56 10.1038/ncb1516 [DOI] [PubMed] [Google Scholar]
- Lanctôt C., Cheutin T., Cremer M., Cavalli G., Cremer T. 2007. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 8:104–115 10.1038/nrg2041 [DOI] [PubMed] [Google Scholar]
- LeBoeuf M., Terrell A., Trivedi S., Sinha S., Epstein J.A., Olson E.N., Morrisey E.E., Millar S.E. 2010. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev. Cell. 19:807–818 10.1016/j.devcel.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopardo T., Lo Iacono N., Marinari B., Giustizieri M.L., Cyr D.G., Merlo G., Crosti F., Costanzo A., Guerrini L. 2008. Claudin-1 is a p63 target gene with a crucial role in epithelial development. PLoS ONE. 3:e2715 10.1371/journal.pone.0002715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C., Tommasi di Vignano A., Sharov A.A., Neilson J., Havrda M.C., Roop D.R., Botchkarev V.A., Crabtree G.R., Dotto G.P. 2005. Integration of Notch 1 and calcineurin/NFAT signaling pathways in keratinocyte growth and differentiation control. Dev. Cell. 8:665–676 10.1016/j.devcel.2005.02.016 [DOI] [PubMed] [Google Scholar]
- Marella N.V., Seifert B., Nagarajan P., Sinha S., Berezney R. 2009. Chromosomal rearrangements during human epidermal keratinocyte differentiation. J. Cell. Physiol. 221:139–146 10.1002/jcp.21855 [DOI] [PubMed] [Google Scholar]
- Marshall D., Hardman M.J., Nield K.M., Byrne C. 2001. Differentially expressed late constituents of the epidermal cornified envelope. Proc. Natl. Acad. Sci. USA. 98:13031–13036 10.1073/pnas.231489198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N., Patel S., Segre J.A. 2004. Long-range comparison of human and mouse Sprr loci to identify conserved noncoding sequences involved in coordinate regulation. Genome Res. 14:2430–2438 10.1101/gr.2709404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills A.A., Zheng B., Wang X.J., Vogel H., Roop D.R., Bradley A. 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 398:708–713 10.1038/19531 [DOI] [PubMed] [Google Scholar]
- Naumova N., Dekker J. 2010. Integrating one-dimensional and three-dimensional maps of genomes. J. Cell Sci. 123:1979–1988 10.1242/jcs.051631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen B.C., Lefort K., Mandinova A., Antonini D., Devgan V., Della Gatta G., Koster M.I., Zhang Z., Wang J., Tommasi di Vignano A., et al. 2006. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 20:1028–1042 10.1101/gad.1406006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan Kumar P., Purbey P.K., Sinha C.K., Notani D., Limaye A., Jayani R.S., Galande S. 2006. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol. Cell. 22:231–243 10.1016/j.molcel.2006.03.010 [DOI] [PubMed] [Google Scholar]
- Pozzi S., Zambelli F., Merico D., Pavesi G., Robert A., Maltère P., Gidrol X., Mantovani R., Vigano M.A. 2009. Transcriptional network of p63 in human keratinocytes. PLoS ONE. 4:e5008 10.1371/journal.pone.0005008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando O.J., Chang H.Y. 2009. Genome-wide views of chromatin structure. Annu. Rev. Biochem. 78:245–271 10.1146/annurev.biochem.78.071107.134639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch J., Knoch T.A., Solovei I., Teller K., Stein S., Buiting K., Horsthemke B., Langowski J., Cremer T., Hausmann M., Cremer C. 2008. Light optical precision measurements of the active and inactive Prader-Willi syndrome imprinted regions in human cell nuclei. Differentiation. 76:66–82 [DOI] [PubMed] [Google Scholar]
- Ronneberger O., Baddeley D., Scheipl F., Verveer P.J., Burkhardt H., Cremer C., Fahrmeir L., Cremer T., Joffe B. 2008. Spatial quantitative analysis of fluorescently labeled nuclear structures: problems, methods, pitfalls. Chromosome Res. 16:523–562 10.1007/s10577-008-1236-4 [DOI] [PubMed] [Google Scholar]
- Savarese F., Dávila A., Nechanitzky R., De La Rosa-Velazquez I., Pereira C.F., Engelke R., Takahashi K., Jenuwein T., Kohwi-Shigematsu T., Fisher A.G., Grosschedl R. 2009. Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 23:2625–2638 10.1101/gad.1815709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S., Clay I., Fraser P. 2010. The transcriptional interactome: gene expression in 3D. Curr. Opin. Genet. Dev. 20:127–133 10.1016/j.gde.2010.02.002 [DOI] [PubMed] [Google Scholar]
- Sen G.L., Webster D.E., Barragan D.I., Chang H.Y., Khavari P.A. 2008. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 22:1865–1870 10.1101/gad.1673508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G.L., Reuter J.A., Webster D.E., Zhu L., Khavari P.A. 2010. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 463:563–567 10.1038/nature08683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov A.A., Weiner L., Sharova T.Y., Siebenhaar F., Atoyan R., Reginato A.M., McNamara C.A., Funa K., Gilchrest B.A., Brissette J.L., Botchkarev V.A. 2003. Noggin overexpression inhibits eyelid opening by altering epidermal apoptosis and differentiation. EMBO J. 22:2992–3003 10.1093/emboj/cdg291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov A.A., Dudekula D.B., Ko M.S. 2005a. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 21:2548–2549 10.1093/bioinformatics/bti343 [DOI] [PubMed] [Google Scholar]
- Sharov A.A., Fessing M., Atoyan R., Sharova T.Y., Haskell-Luevano C., Weiner L., Funa K., Brissette J.L., Gilchrest B.A., Botchkarev V.A. 2005b. Bone morphogenetic protein (BMP) signaling controls hair pigmentation by means of cross-talk with the melanocortin receptor-1 pathway. Proc. Natl. Acad. Sci. USA. 102:93–98 10.1073/pnas.0408455102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov A.A., Sharova T.Y., Mardaryev A.N., Tommasi di Vignano A., Atoyan R., Weiner L., Yang S., Brissette J.L., Dotto G.P., Botchkarev V.A. 2006. Bone morphogenetic protein signaling regulates the size of hair follicles and modulates the expression of cell cycle-associated genes. Proc. Natl. Acad. Sci. USA. 103:18166–18171 10.1073/pnas.0608899103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov A.A., Mardaryev A.N., Sharova T.Y., Grachtchouk M., Atoyan R., Byers H.R., Seykora J.T., Overbeek P., Dlugosz A., Botchkarev V.A. 2009. Bone morphogenetic protein antagonist noggin promotes skin tumorigenesis via stimulation of the Wnt and Shh signaling pathways. Am. J. Pathol. 175:1303–1314 10.2353/ajpath.2009.090163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I., Kreysing M., Lanctôt C., Kösem S., Peichl L., Cremer T., Guck J., Joffe B. 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 137:356–368 10.1016/j.cell.2009.01.052 [DOI] [PubMed] [Google Scholar]
- Takizawa T., Meaburn K.J., Misteli T. 2008. The meaning of gene positioning. Cell. 135:9–13 10.1016/j.cell.2008.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong A.B., Khavari P.A. 2007. Control of keratinocyte proliferation and differentiation by p63. Cell Cycle. 6:295–299 10.4161/cc.6.3.3753 [DOI] [PubMed] [Google Scholar]
- Truong A.B., Kretz M., Ridky T.W., Kimmel R., Khavari P.A. 2006. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 20:3185–3197 10.1101/gad.1463206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viganò M.A., Mantovani R. 2007. Hitting the numbers: the emerging network of p63 targets. Cell Cycle. 6:233–239 10.4161/cc.6.3.3802 [DOI] [PubMed] [Google Scholar]
- Wang L., Di L.J., Lv X., Zheng W., Xue Z., Guo Z.C., Liu D.P., Liang C.C. 2009. Inter-MAR association contributes to transcriptionally active looping events in human beta-globin gene cluster. PLoS ONE. 4:e4629 10.1371/journal.pone.0004629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F.M., Frye M., Benitah S.A. 2008. MYC in mammalian epidermis: how can an oncogene stimulate differentiation? Nat. Rev. Cancer. 8:234–242 10.1038/nrc2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.R., Broad S., Sheer D., Ragoussis J. 2002. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 272:163–175 10.1006/excr.2001.5400 [DOI] [PubMed] [Google Scholar]
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. 1999. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 398:714–718 10.1038/19539 [DOI] [PubMed] [Google Scholar]
- Yasui D., Miyano M., Cai S., Varga-Weisz P., Kohwi-Shigematsu T. 2002. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 419:641–645 10.1038/nature01084 [DOI] [PubMed] [Google Scholar]
- Zhao R., Bodnar M.S., Spector D.L. 2009. Nuclear neighborhoods and gene expression. Curr. Opin. Genet. Dev. 19:172–179 10.1016/j.gde.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Li X.M., Lavker R.M. 2006. Transcriptional profiling of enriched populations of stem cells versus transient amplifying cells. A comparison of limbal and corneal epithelial basal cells. J. Biol. Chem. 281:19600–19609 10.1074/jbc.M600777200 [DOI] [PubMed] [Google Scholar]