Regulation of Arpc3 by miRNA alters dendritic spine morphology.
Abstract
Previous studies have demonstrated that microribonucleic acids (miRs) are key regulators of protein expression in the brain and modulate dendritic spine morphology and synaptic activity. To identify novel miRs involved in neuronal plasticity, we exposed adult mice to chronic treatments with nicotine, cocaine, or amphetamine, which are psychoactive drugs that induce well-documented neuroadaptations. We observed brain region– and drug-specific changes in miR expression levels and identified miR-29a/b as regulators of synaptic morphology. In vitro imaging experiments indicated that miR-29a/b reduce mushroom-shaped dendritic spines on hippocampal neurons with a concomitant increase in filopodial-like outgrowths, suggesting an effect on synapse formation via actin cytoskeleton remodeling. We identified Arpc3, a component of the ARP2/3 actin nucleation complex, as a bona fide target for down-regulation by miR-29a/b. This work provides evidence that targeting of Arpc3 by miR-29a/b fine tunes structural plasticity by regulating actin network branching in mature and developing spines.
Introduction
Structural plasticity and reorganization of neuronal connections underpin many brain functions including memory, learning, and addiction. In neurons, polymerized actin is the major structural constituent of dendritic spines, specialized structures involved in synaptic communication. Regulation of the actin cytoskeleton influences filopodial outgrowth, novel spine production, and changes in spine morphology as well as spine loss (Fischer et al., 2000; Dillon and Goda, 2005). Structural reorganization of spine morphology is an ongoing dynamic process sensitive to Ca2+ influx through glutamatergic N-methyl-d-aspartate (NMDA) receptors and voltage-operated Ca2+ channels (Fischer et al., 2000; Saneyoshi et al., 2010). Longer-term consolidation of structural changes occurs through altered gene expression and insertion of novel proteins into the synaptic spine (West et al., 2002; Lamprecht and LeDoux, 2004; Greer and Greenberg, 2008; McClung and Nestler, 2008).
microRNAs (miRs) are endogenous noncoding RNA molecules (Lee at al., 1993; Wightman et al., 1993; Bartel, 2004) that mediate posttranscriptional regulation of gene expression by targeting the 3′ untranslated region (UTR; 3′-UTR) of specific mRNA sequences and by repressing their translation into proteins (Bartel, 2004; Winter et al., 2009). miRs exhibit developmental regulation, tissue specificity, and differential expression in specialized subcellular compartments (Miska et al., 2004; Sempere et al., 2004; Kosik, 2006; Christensen and Schratt, 2009; Fineberg et al., 2009). In neurons, several miRs are specifically enriched in dendritic spines where they fine tune localized protein synthesis (Schratt et al., 2006; Schratt, 2009; Siegel et al., 2009). Here, miRs appear to regulate dendritic spine morphogenesis (Wayman et al., 2008; Siegel et al., 2009; Edbauer et al., 2010), spine structure (Schratt et al., 2006; Siegel et al., 2009), and synaptic physiology (Edbauer et al., 2010). miR-134 reduces spine size by targeting the synaptic protein LIMPK1, a kinase that enhances actin polymerization via inactivation of the actin depolymerization factor cofilin (Schratt et al., 2006). Similarly, miR-138 modulates spine size by targeting APT1 and activating the RhoA pathway (Schratt, 2009; Siegel et al., 2009). Importantly, levels of both miR-134 and miR-138 are regulated by neuronal activity, thereby demonstrating a feedback mechanism between synaptic signaling, miR expression, localized regulation of translation, and spine morphology.
Despite these examples, the role of miRs in the regulation of synaptic structure and function, and hence synaptic plasticity, remains poorly understood. In this study, we have used chronic treatments with psychostimulants (nicotine, cocaine, and amphetamine) to induce widespread neuroplastic changes in adult mouse brain. We have then screened for individual miRs and miR clusters regulated during these treatments, as we postulated that miRs targeted during periods of drug-induced neuroplasticity were also likely to have a physiological role in the maintenance of synaptic connections. Via cross-referencing with a second, more focused, screen of miR regulation during periods of synaptogenesis in cultured neurons, we now identify the miR-29a/b cluster as an important regulator of dendritic spine morphology and synaptic connectivity. Furthermore, we demonstrate that miR-29a/b directly target the mRNA encoding for Arpc3, a subunit of the ARP2/3 actin nucleation complex (Goley and Welch, 2006). This complex is required for the production of branched actin filaments and is involved in dendritic spine morphogenesis (Hotulainen et al., 2009). We suggest that miR-29a/b fine tune structural plasticity via regulating the sensitivity of ARP2/3 to remodeling cues in the spine.
Results
Regulation of miRs during neuroplasticity in vivo
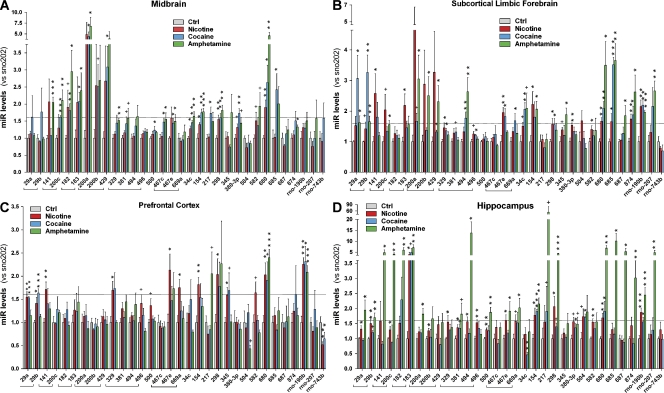
Chronic treatments with psychostimulants produce substantial neuroplastic changes in the brain (Robinson and Kolb, 2004; Tang and Dani, 2009). Recent data now suggest that altered miR levels contribute to the plastic changes underlying addictive behaviors (Chandrasekar and Dreyer, 2009; Hollander et al., 2010; Im et al., 2010). To identify miRs involved in psychostimulant-induced neuroplasticity, C57BL6 mice (four to five per group) were exposed to saline, 1 mg/kg nicotine (tartrate), 10 mg/kg cocaine (hydrochloride), or 5 mg/kg amphetamine (sulfate) for 5 d (one injection/day) and sacrificed 4 h after the last injection. For each animal, the hippocampus, prefrontal cortex, subcortical limbic forebrain, and ventral midbrain were dissected and total RNA (including small RNAs) extracted. Our initial screen, performed by pooling samples between animals that were exposed to the same treatment (but keeping areas separated), demonstrated that chronic drug exposure affected the miR transcriptome in a drug- and region-specific manner (Fig. S1). Potential candidates for further investigation were identified according to the following criteria: (a) miRs that were consistently up-regulated by different psychoactive drugs, (b) miRs that were consistently up-regulated across different brain areas, and (c) genetically clustered up-regulated miRs that were likely to be expressed as a single transcriptional unit (Lee et al., 2002; Kim et al., 2009). According to these criteria, 32 miRs were selected, and changes in expression levels between brain areas and drug treatments were quantified using custom-made real-time PCR plates (Fig. 1). The drug and brain region specificity of miR up-regulation is outlined in Fig. S2, with several miRs being targeted by two or more drug treatments. In particular, we identified consistent up-regulation of the miR-29a/b and miR-182/183 clusters in most brain regions. In contrast, the miR-200/429 cluster appeared to be more heavily targeted in the ventral midbrain and subcortical limbic forebrain (Figs. 1 and S2). Of the single transcripts, miR-680 and miR-190b also appeared to be consistently up-regulated in most of the areas assessed. Therefore, these 32 miRs were selected for further study.
Figure 1.
In vivo screening for miRs involved in neuroplasticity. (A–D) Adult C57BL6 mice (four to five per group) were treated with a single i.p. injection of saline, 1 mg/kg nicotine, 10 mg/kg cocaine, or 5 mg/kg amphetamine once a day for 5 d. 4 h after the final injection, brain regions were dissected, and miRs were extracted. Changes in expression levels of a subset of 32 miRs were analyzed in custom-made 96-well miR arrays (TaqMan). Four areas that receive dopaminergic inputs from the ventral tegmental area were assessed, namely the ventral midbrain (A), subcortical limbic forebrain (B), prefrontal cortex (C), and hippocampus (D). Dotted lines represent the threshold of physiological relevance with a >1.6-fold up-regulation. Data represent the mean value ± SEM from four to five animals/group, and comparisons were performed using an unpaired t test. Ctrl, control; +, 0.1 < P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Activity-dependent up-regulation of miRs during synaptogenesis
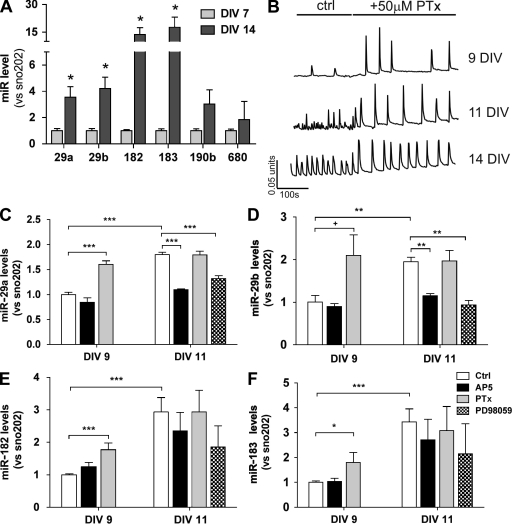
The formation of synaptic contacts during development demonstrates a high level of plasticity, as new spines are formed and then either removed or consolidated according to the strength of connections made (Dunaevsky et al., 1999; Yang et al., 2009). As we had postulated that miRs targeted during periods of drug-induced neuroplasticity were also likely to be regulators of more general neuroplastic changes, we next examined the expression levels of the selected 32 miRs in an in vitro model of synaptogenesis (Grabrucker et al., 2009). Expression levels of miRs were therefore measured in primary cultures of mouse hippocampal neurons at 7 d in vitro (DIV), a point at which the neurons are relatively immature. miR levels were then measured in the same cultures at 14 DIV, when mature synapses are being formed, and synaptic connectivity has been consolidated (Fig. 2 B; Ogura et al., 1987; Bacci et al., 1999). An initial screen demonstrated that a small portion of the previously identified 32 miRs was also up-regulated in this in vitro model system (unpublished data). Quantification, via real-time PCR, demonstrated that the miR cluster containing miR-29a/b was up-regulated around 3.5–4.2 fold, whereas the miR-182/183 cluster was up-regulated 13.6–17.6 fold (Fig. 2 A). In contrast, miR-680 and miR-190b were not significantly up-regulated over this time period and so appeared less likely to be actively involved in the process of synaptogenesis.
Figure 2.
Activity-dependent miR up-regulation. Primary cultures of C57BL6 mouse hippocampal neurons were grown for up to 14 DIV. (A) miRs were extracted from primary neuronal cultures at 7 and 14 DIV. Changes in the levels of 32 selected miRs were then examined via miR assay (TaqMan; data represent the mean ± SEM from three experiments; P = 0.0359 for 29a, 0.0224 for 29b, 0.0273 for 182, and 0.0388 for 183 using an unpaired t test, whereas none of the other selected miRs demonstrated significant up-regulation). (B) Primary neuronal cultures at 9, 11, and 14 DIV were loaded with fura-4f, and intracellular Ca2+ transients were imaged. Data are representative traces of network Ca2+ activity either in control (ctrl; untreated) neurons or where 50 µM PTx had been added to the same neurons. (C–F) Clusters containing either miR-29a/b or miR-182/183 demonstrated activity-dependent regulation between 9 and 11 DIV. Enhancement of neuronal activity via treatment with 50 µM PTx from 7–9 DIV induced up-regulation of miR-29a by 60 ± 7.1% (P < 0.0001), 29b by 109 ± 48% (P = 0.0073), 182 by 74 ± 20%, (P = 0.0008), and 183 by 80 ± 39% (P = 0.0234). Addition of PTx from 9 and 11 DIV did not enhance miRs levels compared with control. Treatment with 10 µM AP5 between 9 and 11 DIV reduced the miR-29a levels by 61.8 ± 0.5% (P < 0.0001 vs. control) and 29b levels by 58.4 ± 2.6% (P = 0.0025 vs. control). The reduction is present also in miR-182/183 although not significant. 10 µM PD98059 also reduces the spontaneous increase of miR-29a (53 ± 5.1% of the control; P = 0.0004) and 29b (27 ± 2.7% of the control; P = 0.0025), but, again, the effect is not significant on miR-182/183. All miR levels are relative to the housekeeping sno202. Data in panels C–F represent the mean ± SEM from three to four independent experiments, and comparisons were performed using an unpaired t test. (A and C–F) +, 0.1 < P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Neuronal activity regulates gene transcription via frequency- and amplitude-encoded Ca2+ signals (Greer and Greenberg, 2008). To determine whether up-regulation of miR-29a/b and miR-182/183 was similarly activity dependent, neuronal activity in hippocampal neurons was stimulated via the addition of picrotoxin (PTx; a GABAA receptor antagonist that enhances activity by blocking the inhibitory influence of GABAergic interneurons present in the culture; Bacci et al., 1999). Drug additions were made at 7 or 9 DIV, and miR levels were examined after a further 48 h. The ability of PTx to enhance Ca2+ activity in the mouse hippocampal cultures was first confirmed using single-cell imaging of fura-4f–loaded neurons (Fig. 2 B). Treatment with 50 µM PTx induced up-regulation of both sets of miR clusters at 9 DIV. The magnitude of responses was broadly similar in both sets of clusters, ranging from increases of 60 ± 7 to 109 ± 48% for miR-29a and miR-29b (P = 0.0001 and 0.073, respectively; n = 3) and 74 ± 20% for miR-182 and 80 ± 39% for miR-183 (P = 0.0008 and 0.0234, respectively; n = 3). In contrast, PTx treatment did not enhance expression at 11 DIV above the already raised levels. At this point, basal neuronal activity (Fig. 2 B) in the absence of PTx appeared capable of maintaining miR levels. To investigate this further, basal Ca2+ activity in the cultures was inhibited via the addition of the NMDA receptor antagonist AP5. This produced a marked reduction in miR-29a and -29b as measured at 11 DIV (61.8 ± 0.5 and 58.4 ± 2.6% reductions, P = 0.0001 and 0.0025, respectively; n = 3; Fig. 2, C and D). In comparison, expression levels of miR-182 and -183 were only partially inhibited by AP5, and, as a result of the greater degree of variability between experiments, these decreases were not significant (48 ± 19 and 20 ± 24% reductions; n = 4; Fig. 2, E and F). Expression of both sets of miR clusters was also reduced by the addition of 10 µM PD98059, suggesting an involvement of MEK1/extracellular signal-regulated kinase, which was most probably acting downstream of NMDA receptor–mediated Ca2+ entry (53 ± 5% reduction for miR-29a, P = 0.0004; 27 ± 3% reduction for miR-29b, P = 0.0025; 37 ± 22% reduction for miR-182, P = 0.19; and 38 ± 35% reduction for miR-183, P = 0.28; n = 4; Fig. 2, C–F).
Effect of candidate miRs on synaptic structure and function
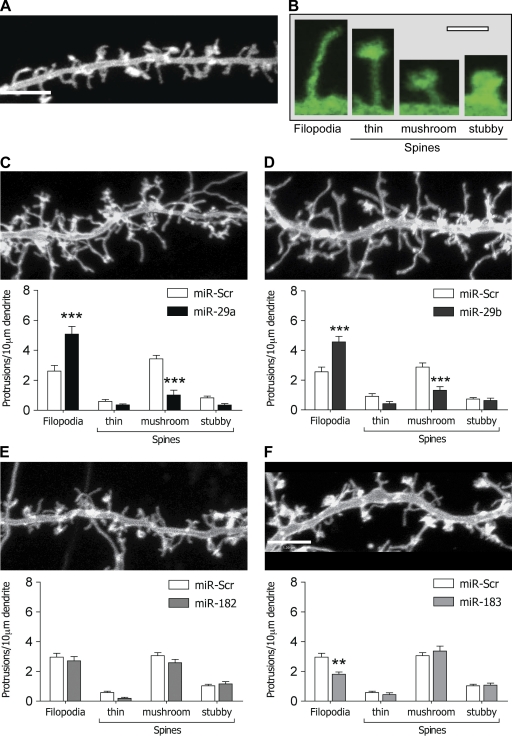
Activity-dependent remodeling of synaptic structure plays an import role in neuronal plasticity (Lamprecht and LeDoux, 2004; McClung and Nestler, 2008). To determine whether the identified miRs altered synaptic spine structure, primary hippocampal neurons were cotransfected with a plasma membrane–targeted GFP construct (GFP-MEM) plus oligonucleotides for miR-29a, -29b, -182, or -183, and spine morphology was examined using confocal microscopy. This approach was validated using a dsRed-based reporter construct with miR seed regions cloned into the 3′-UTR (Fig. S4). Hippocampal neurons, which were transfected at 9 DIV and imaged at 14 DIV, demonstrated a mixed phenotype of dendritic protrusions (Fig. 3, A and B). These were characterized as either filopodia (which lacked any spine head enlargement) or subclassed into long, mushroom, or stubby spine categories (Fig. 3 B; Hotulainen et al., 2009). Mushroom-shaped spines were, by far, the most prevalent spine shape observed, and, broadly speaking, at 14 DIV, these and filopodia were present in approximately equal numbers. Cotransfection of neurons with 10 nM miR-29a or -29b altered the dendritic phenotype measure at 14 DIV. miR-29a reduced the number of mushroom spines from 3.4 ± 0.2 to 1.0 ± 0.3 per 10-µm dendritic length (P < 0.001 compared with scrambled control; n = 26), with a concomitant increase in the number of filopodia (2.6 ± 0.4 to 5.1 ± 0.5 per 10-µm dendritic length; P < 0.001; n = 26; Fig. 3 C). However, the total number of protrusions remained unchanged (Fig. 2, C and D). miR-29b produced similar results, with a 2.9 ± 1.3 to 1.3 ± 0.2 decrease in the number of mushroom spines, and a 2.6 ± 0.3 to 4.6 ± 0.4 increase in filopodia (P < 0.001, n = 32). In contrast, neither miR-182 nor -183 affected spine number (Fig. 3, E and F), although miR-183 did cause a modest decrease in filopodia (3.0 ± 0.3 to 1.8 ± 0.2, n = 48 and 23, respectively; P < 0.001; Fig. 3 F). Finally, the effect of the miR-29b oligonucleotide could be mimicked by transfection of a miR-29b expression plasmid (pEZX; GeneCopoeia). Transfected neurons were identified via the expression of a GFP reporter integral to the plasmid. However, as GFP expression was low compared with the EGFP-MEM, neuronal morphology was examined via coexpression with tdTomato (Fig. S3). Overall, these data suggested that miR-29a/b expression prevented the conversion of dendritic filopodia to a mature spine phenotype.
Figure 3.
Effect of miRs on spine morphology. Primary cultures of hippocampal neurons were cotransfected with EGFP-MEM plus miR oligonucleotides at 9 DIV, and effects on neuronal structure were investigated at 14–15 DIV using confocal microscopy. (A) A representative image of a hippocampal dendrite at 14 DIV transfected with EGFP-MEM. Bar, 5 µm. (B) Sample images demonstrating the classification of dendritic filopodia and thin, mushroom, or stubby spines in EGFP-MEM–transfected neurons. Bar, 1 µm. (C–F) Representative images of dendritic regions of neurons transfected with miR-29a (C), miR-29b (D), miR-182 (E), or miR-183 (F). Data represent the mean ± SEM number of protrusions per 10-µm dendritic length for 23–48 transfected neurons. Both miR-29a and -29b enhanced the number of filopodia and reduced the number of mushroom spines (P < 0.001). **, P < 0.01; ***, P < 0.001. Bar, 5 µm.
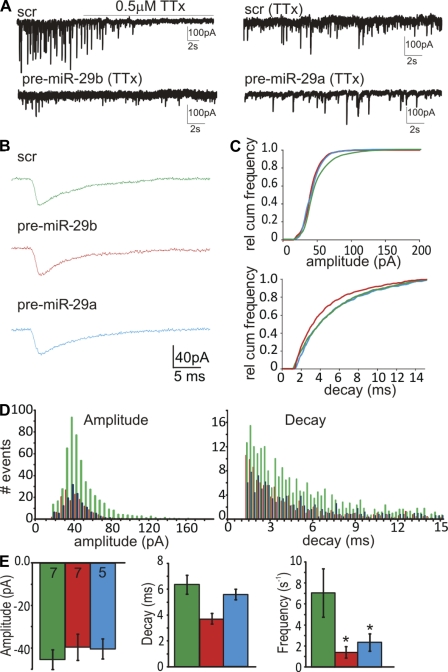
To determine the effects on synaptic transmission, miniature excitatory postsynaptic currents (mEPSCs) were measured in miR-29a– and -29b–transfected primary hippocampal neurons (Fig. 4 A). Neuronal cultures were first treated with 0.5 µM tetrodotoxin (TTx) to block spontaneous evoked network glutamatergic transmission (Fig. 4 B, top trace). Under these conditions, miR-29a and -29b expression reduced the frequency of mEPSCs from 7.05 ± 2.3 to 1.4 ± 0.4 and 2.4 ± 0.8 s−1, respectively (P < 0.05; n = 5–7), without altering mEPSC amplitude or decay kinetics (Fig. 4, C–E). The mEPSC amplitude was −45.4 ± 4.7 pA under control conditions and −39.6 ± 6.1 and −40.4 ± 4.7 pA in miR-29a and miR-29b cells, respectively. Decay rates were 6.4 ± 0.7 ms for control cells, 3.7 ± 0.4 for miR-29a cells, and 5.6 ± 0.4 ms for miR-29b cells (n = 5–7). This supports the notion that miR-29a/b expression reduces the number of synaptic connections via a decrease in the number of mushroom-shaped dendritic spines. However, the lack of effect on mEPSC amplitudes and kinetics suggests that the postsynaptic machinery, where present, was unaffected by miR-29a or -29b (Edbauer et al., 2010). In support of this, network Ca2+ transients, induced by the addition of 50 µM PTx, were unaffected by miR-29a/b expression (Fig. S3).
Figure 4.
Expression of miR-29a/b reduces the frequency of mEPSCs. Primary cultures of hippocampal neurons were cotransfected with EGFP-MEM plus miR oligonucleotides at 9 DIV, and effects on synaptic activity were investigated at 14–15 DIV. (A, top left) Application of 0.5 µM TTx suppresses spontaneous activity leaving only mEPSCs. (top right) Basal level of mEPSC activity in neuron-expressing miR-Scr. (bottom) Neurons expressing miR-29a or -29b show reduced mEPSC activity (all recordings were performed in the presence of 0.5 µM TTx). (B) Representative averaged mEPSCs from neurons containing either miR-Scr, miR-29a, or miR-29b. (C) Relative cumulative frequency (cum rel) histograms for amplitude and decay distributions. There is no significant difference between miR-Scr and miR expression using the Kolmogorov–Smirnov test. (D) Histograms for amplitude and decay distributions. (E) Mean ± SEM values for amplitude, decay, and frequency from neurons expressing miR-Scr (green), miR-29b (red), or miR-29a (blue) for 5–7 cells (the actual numbers of cells are indicated within the bars on left graph). *, P < 0.05 using an analysis of variance with a post-hoc test.
Down-regulation of Arpc3 mRNA mimics the miR-29a/b phenotype
The miR-29a/b expression phenotype suggested potential gene targets associated with remodeling of the actin cytoskeleton (e.g., Rho family–associated proteins or actin nucleation factors). Using two different target prediction software packages (TargetScan and miRBase; Lewis et al., 2005), we selected 24 putative mRNAs that encoded proteins capable of regulating neuronal morphology and that demonstrated 3′-UTR complementarity to miR-29a/b (Fig. S4 A). miRs can inhibit protein expression by repressing mRNA translation or promoting mRNA destabilization/degradation (Lim et al., 2005; Bartel, 2009), and recent work suggests that the predominant mechanism of action involves a reduction in levels of target mRNA (Guo et al., 2010). Using BACE1 mRNA, a previously identified miR-29 target, as a positive control (Hébert et al., 2008), we observed that transfection of miR-29b into primary hippocampal neurons also reduced mRNA levels for the following: ArhGAP28, Arpc3, Camk2b, Coro6, Daam1, Dnm3, Fez2, Gprin1, Lasp1, Ophn1, Pcdha1, RapGEFL1, and Rhobtb1 (Fig. S4 A).
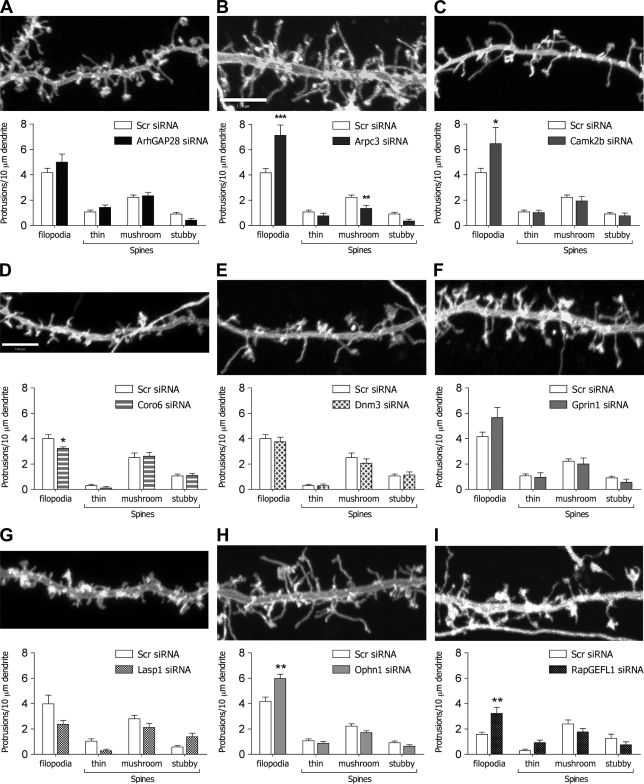
Next, candidate proteins were subjected to siRNA-mediated knockdown. ArhGAP28, Arpc3, Camk2b, Coro6, Dnm3, Gprin1, Lasp1, Ophn1, and RapGEFL1 were selected, as these appeared most likely to regulate synaptic morphology. Validated siRNA oligonucleotides (Fig. S4 B) were cotransfected with GFP-MEM into primary hippocampal neurons at 9 DIV, and images of dendritic morphology were captured at 14 DIV. Dendritic protrusions were then scored as described under the previous subheading. Of the selected proteins, only knockdown of Arpc3 produced a phenotype similar to miR-29a/b expression (Fig. 5). Thus, in Arpc3 siRNA–treated neurons, there was a reduction of 2.2 ± 0.2 to 1.4 ± 0.2 per 10-µm dendritic length in the number of mushroom spines (n = 35 and 15, respectively; P = 0.008) and an increase of 4.2 ± 0.4 to 7.1 ± 0.8 per 10-µm dendritic length in filopodia (P < 0.001; Fig. 5 B). Knockdown of Camk2b, Ophn1, and RapGEFL1 also increased the number of filopodia but had no significant effect on the number of mushroom spines (Fig. 5, C, H, and I).
Figure 5.
Effect of siRNA on spine morphology. (A–I) Primary cultures of hippocampal neurons were cotransfected with EGFP-MEM plus siRNA oligonucleotides at 9 DIV, and effects on neuronal structure were investigated at 14–15 DIV using confocal microscopy. Data are representative images plus graphs demonstrating the mean ± SEM number of protrusions for Scr control siRNA and siRNA targeted against ArhGAP28 (n = 27; A), Arpc3 (**, P = 0.008; ***, P < 0.001; n = 15; B), Camk2b (*, P = 0.02; n = 12; C), Coro6 (*, P = 0.048; n = 13; D), Dnm3 (n = 10; E), Gprin1 (n = 11; F), Lasp1 (n = 8; G), Ophn1 (**, P = 0.001; n = 21; H), and RapGEFL1 (**, P = 0.001; n = 12; I). Bars, 5 µm.
Arpc3 mRNA and protein expression are targeted by miR-29a/b
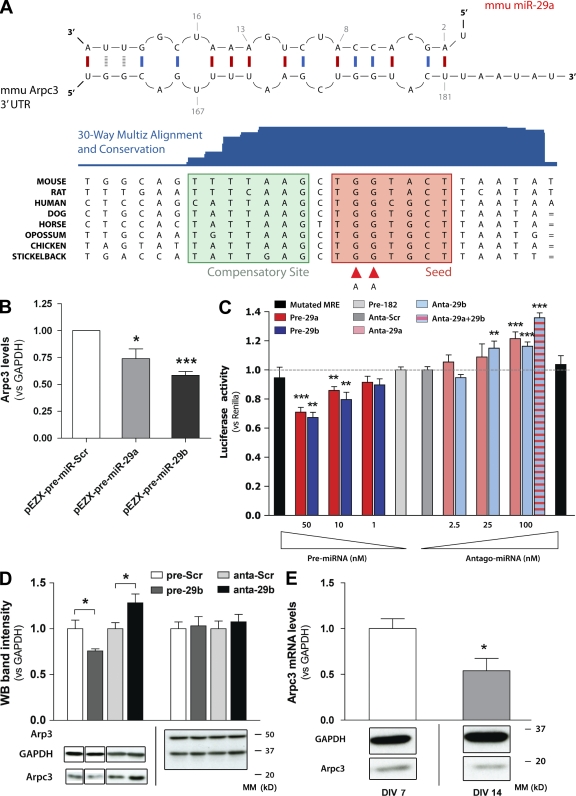
To determine whether Arpc3 was a bona fide target for miR-29a/b–mediated down-regulation, we first investigated the predicted interaction between miR-29a/b with the 3′-UTR of Arpc3. The alignment for mouse Arpc3, according to the MicroCosm Targets database, is illustrated in Fig. 6 A. The miR-responsive element (MRE) is highly conserved throughout species, as shown by the 30-way Multiz Alignment software (University of California, Santa Cruz genome browser; Fig. 6 A). To confirm that Arpc3 is a genuine target of miR-29a/b, mouse N2a cells were transfected with pEZX plasmid expression clones for miR-29a or -29b (see Effect of candidate miRs on synaptic structure and function). The coexpression of GFP from the pEZX vector enabled selection of miR-29a– or -29b–overexpressing cells only. As demonstrated in Fig. 6 B, the Arpc3 transcript was significantly down-regulated by both miR-29a and -29b (27 ± 9%, P = 0.048; 42 ± 4%, P = 0.0003; n = 3; Fig. 6 B). In these experiments, selected cells demonstrated an increase in miR levels of around 10–25 fold, which appeared comparable with the method whereby the miR oligonucleotide was coexpressed with EGFP-MEM (Fig. S5 B).
Figure 6.
Targeting of Arpc3 by miR-29a/b. (A, top) Predicted interaction between the Arpc3 3′-UTR MRE and the miR-29a microseed using MicroCosm analysis. (bottom) A comparison of the Arpc3 MRE in different species demonstrates a high degree of conservation both in the seed region and in the compensatory site. mmu, Mus Musculus. (B) Plasmid overexpression of miR-29a and -29b in N2a cells results in significant down-regulation of Arpc3 mRNA levels (27 ± 9%, P = 0.048 and 42 ± 3.5%, P = 0.0003, respectively; data represent the mean ± SEM from three independent experiments, and comparisons were performed using an unpaired t test). Levels were assessed by Universal ProbeLibrary assay (TaqMan) after FACS sorting for GFP-expressing cells. (C) Concentration response effect of miR-29a and -29b oligonucleotides and miR-29a and -29b antagonists on pGL3-mArpc3 in HEK-293 cells. 10 and 50 nM miR-29a and -29b oligonucleotides increasingly inhibited luciferase activity (P = 0.005 and 0.004, respectively, at 10 nM, and P = 0.002 and P = 0.0004, respectively, at 50 nM; n = 10). In contrast, 1 nM oligonucleotide did not produce any significant reduction. 50 nM miR-182 was used as a control (light gray bar). Transfection of 25 or 100 nM miR-29a and -29b antagonists increased luciferase activity (P = 0.0004 and 0.0002, respectively, at 100 nM, and P = 0.008 for 29b at 25 nM; n = 10). A double point mutation in two nucleotides of the MRE in the Arpc3 3′-UTR (A, red arrowheads) abolishes the regulatory effect of both miR oligonucleotides (50 nM) and miR antagonists (100 nM; black bars) without altering basal luciferase activity. The dotted lines represent the control value (1) to help visualize changes in luciferase activity upon up-regulation or inhibition of miR-29a/b. Data are mean ± SEM, and analysis was performed using an unpaired t test. (D) Protein levels of Arpc3 are down-regulated by miR-29b in primary mouse hippocampal neurons. Transfection of 50 nM miR-29b oligonucleotides results in a significant down-regulation of Arpc3 protein levels, as measured via Western Blotting (WB; 25 ± 2% decrease; P = 0.04; n = 3). In contrast, 100 nM miR-29b antagonist increased expression levels by 28 ± 9.6 (P = 0.04; n = 3). In comparison, Arp3, another component of the Arp2/3 complex, was unaffected by pre–miR-29b or miR-29b antagonist. Data represent the mean ± SEM, and comparisons were performed using an unpaired t test. MM, molecular mass. (E) Arpc3 levels in primary hippocampal neurons decrease between 7 (P = 0.05) and 14 (P = 0.005) DIV (47 ± 13 and 67 ± 4% decrease for mRNA and protein, P = 0.05 aand 0.005, respectively; n = 3 each; data represent the mean ± SEM). (B–E) *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, we cloned the 3′-UTR of Arpc3 from either human (pGL3-hsArpc3) or mouse (pGL3-mmArpc3) downstream of a luciferase-coding sequence (pGL3–multiple cloning site). The constructs were overexpressed in HEK-293 cells, and luciferase chemiluminescence was detected in control (transfected with the miR-Scr oligonucleotide) or miR-29a or -29b oligonucleotide–transfected cells (50 nM). In addition, miR-182 was selected as a further control condition. miR-29a and -29b selectively inhibited pGL3-mmArpc3 and pGL3-hsArpc3 expression, thereby demonstrating a direct targeting effect of these miRs on Arpc3 expression (pGL3-mmArpc3 29a: 29 ± 3.2%, P = 0.0018; 29b: 33 ± 3.5%, P = 0.0004; pGL3-hsArpc3 29a: 22 ± 3.4%, P = 0.0012; 29b: 30 ± 3.4%, P < 0.0001; n = 16; Fig. S5 C).
To further characterize the posttranscriptional regulation of the pGL3-mArpc3 construct, we examined the concentration relationship of miR-29a and -29b oligonucleotides on luciferase activity in HEK-293 cells. The greatest level of inhibition was observed with 50 nM oligonucleotide, with 10 nM also producing a significant inhibition (Fig. 6 C). In contrast, the effect of 1 nM oligonucleotide was not significantly different from control. Quantitative real-time PCR experiments suggested that HEK-293 cells expressed significant endogenous levels of miR-29a/b. To determine whether these endogenous miRs were suppressing luciferase activity, HEK-293 cells were transfected with increasing concentrations of miR antagonists against either miR-29a or -29b. As demonstrated in Fig. 6 C, increasing the miR antagonist concentration (ranging from 2.5 to 100 nM) produced a progressive increase in luciferase activity (maximum effect obtained with miR-29a/b antagonist at 100 nM; 35.8 ± 3.4% increase, P < 0.0001; n = 8; Fig. 6 C). The specificity of this effect was demonstrated by inducing point mutations in two nucleotides in the mArpc3 seed region (Fig. 6 A, red arrowheads). This mutated luciferase construct was insensitive to miR-29a or -29b or miR-29a or -29b antagonist oligonucleotides (Fig. 6 C).
The aforementioned data demonstrate that Arpc3 levels are directly regulated by the targeting of miR-29a or -29b to the identified 3′-UTR MRE. To demonstrate regulation of Arpc3 levels in situ, 50 nM miR-29b or 100 nM miR-29b antagonist was transfected into hippocampal neurons, and protein expression levels were examined (Fig. 6 D). Transfection with miR-29b oligonucleotides reduced Arpc3 protein levels by 25 ± 2% (P = 0.041; n = 3). In contrast, transfection with the miR-29b antagonist increased expression levels by 28 ± 10% (P = 0.040; n = 3). Protein levels of Arp3, a different subunit of the Arp2/3 actin nucleation complex, were unchanged by these treatments (Fig. 6 D). Of note, down-regulation of Arpc3 protein levels by miR-29b was only moderately less efficient than down-regulation by Arpc3-specific siRNA (Fig. S5 D). Finally, as data in Fig. 2 indicated that miR-29a/b levels increased between 7 and 14 DIV, we investigated whether this increase correlated with a decrease in Arpc3 in these cultures. As demonstrated in Fig. 6 E, both Arpc3 mRNA and protein levels decreased between 7 and 14 DIV (47 ± 13 and 67 ± 4% decrease for mRNA and protein, respectively; P = 0.05 and 0.0053) relative to the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
Rescue of the miR phenotype via mutated Arpc3
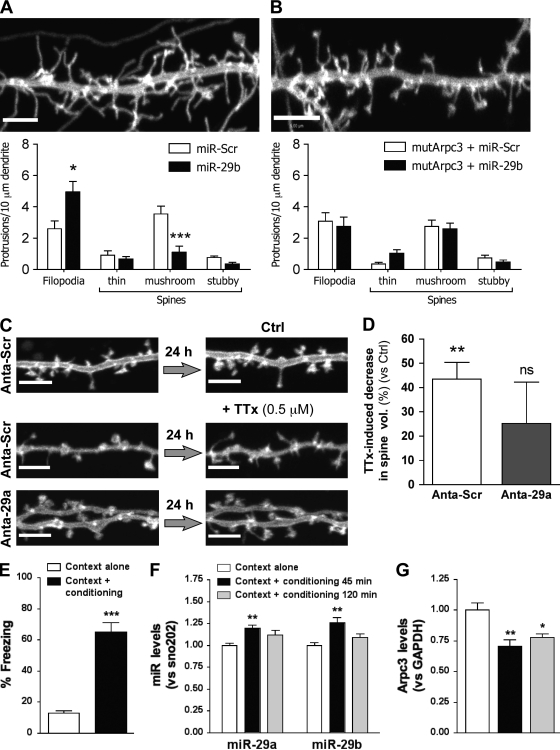
Arpc3 is a component of the seven-protein ARP2/3 actin nucleation complex, responsible for initiating branching in actin filaments (Goley and Welch, 2006). Previous work has demonstrated the importance of this complex for the conversion of dendritic filopodia to a mature spine phenotype (Hotulainen et al., 2009). Our data suggest that down-regulation of Arpc3 may be responsible for the increased filopodia to spine ratio observed in response to miR-29a/b. To examine this further, we created a construct encoding for the full mRNA of mouse Arpc3 containing the same two point mutations in the MRE as used in the luciferase assay mentioned in the previous section (Fig. 6 A). This mutated construct (mutArpc3) was cotransfected with GFP-MEM and either miR-Scr or miR-29b into primary hippocampal neurons, and effects on dendritic morphology were measured. As described in Fig. 3 D, miR-29b increased the number of filopodia present along a 10-µm dendritic length, with a concomitant decrease in the number of mushroom spines (Fig. 7 A). In this set of experiments, the number of filopodia per 10 µm increased from 2.6 ± 0.5 to 5.0 ± 0.7 (P = 0.01; n = 12). The number of mushroom spines decreased from 3.5 ± 0.5 to 1.1 ± 0.4 (P < 0.001; n = 12). In contrast, in parallel experiments, the effect of miR-29b was prevented by the expression of mutArpc3 (Fig. 7 B). This phenotypic rescue via overexpression of an Arpc3 construct lacking in miR-29a/b target sites strongly suggests that down-regulation of Arpc3 is central to the mechanism via which these miRs modulate neuronal structure. Expression of mutArpc3 alone did not notably alter the dendritic phenotype (3.1 ± 0.6 filopodia and 2.7 ± 0.4 mushroom spines per 10 µm for mutArpc3 plus miR-Scr; n = 12).
Figure 7.
miR-29a and -29b regulate sensitivity to neuroplastic changes. (A and B) Primary cultures of hippocampal neurons at 9 DIV were transfected with EGFP-MEM ± Arpc3 in which the miR-29 seed region contained two point mutations (mutArpc3) plus siRNA oligonucleotides. Effects on neuronal structure were investigated at 14–15 DIV using confocal microscopy. Data are representative images plus graphs demonstrating the mean ± SEM number of protrusions for miR-Scr and miR-29b for EGFP-MEM alone (A) or EGFP-MEM plus mutArpc3 (B). (A) As demonstrated previously, miR-29b enhances the number of filopodia (P = 0.01) and reduces the number of mushroom spines (P < 0.001; n = 12). (B) miR-29b did not alter dendritic morphology in neurons expressing mutArpc3 (n = 12). (C–E) miR-29a antagonist reduces spine volume changes induced by TTx. Dendritic regions of hippocampal neurons cotransfected with EGFP-MEM plus 50 nM miR-Scr or miR-29a antagonist were imaged via confocal microscopy at 14 DIV and then again after 24 h in the presence or absence of 0.5 µM TTx. Changes in spine volume were calculated from 3D volume–rendered images. (C) Representative dendritic images for Scr antagonist and miR-29a antagonist neurons before and after a 24-h period under control (Ctrl) conditions or in neurons treated with TTx. Note the loss of spine structure in Scr antagonist after 24 h in the presence of TTx (middle). (D) Expression of miR-29a antagonist did not alter spine volume measured at 14 DIV (data from 200 and 170 spines; not depicted). Treatment with TTx for 24 h reduced spine volume in Scr antagonist neurons by 43.5 ± 6.9% (P = 0.002 from untreated control). Treatment with TTx for 24 h on miR-29a antagonist neurons did not significantly reduce spine volume (25.2 ± 16.9% reduction; ns from untreated control). Error bars represent the mean ± SEM. (E and F) Contextual fear conditioning induces up-regulation of miR-29a/b. (E) Percentage of freezes measured in the conditioning context 24 h after fear conditioning. Data are expressed as the mean (±SEM) percentage of freezes. Statistical analysis was performed by an independent sample Student’s t test in which t = 8.10 and degrees of freedom = 10 (P < 0.001 vs. context alone). (F and G) Levels of miR-29a and -29b in hippocampal tissue extracted from mice that underwent contextual fear conditioning or control context exposure (F) and levels of Arpc3 in the same samples as F (G). Data are expressed as the mean ± SEM. Statistical analysis was performed by a one-way analysis of variance followed by Bonferroni’s correction for multiple comparisons. miR-29a: F(2,17) = 6.51 (P = 0.008) using a post-hoc Bonferroni’s test versus context alone with 45 min of conditioning (P = 0.007); miR-29b: F(2,17) = 7.33 (P = 0.005) using a post-hoc Bonferroni’s test versus context alone with 45 min of conditioning (P = 0.005); Arpc3: F(2,18) = 9.76 (P = 0.0013) using a post-hoc Bonferroni’s test versus context alone with 45 min of conditioning (P < 0.01) and 2 h of conditioning (P < 0.05). (A and D–G) *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars: (A and C) 3.5 µm; (B) 5 µm.
miR-29a regulates sensitivity of synaptic spines to neuroplastic changes
Mature and developing spines demonstrate structural plasticity in response to alterations in synaptic transmission. Inherent to this process is an initial alteration of the actin cytoskeleton (Engert and Bonhoeffer, 1999), which is then stabilized through the introduction of novel expression of synaptic proteins (Lamprecht and LeDoux, 2004). Thus, short-term loss of synaptic transmission enhances spine motility (Korkotian and Segal, 2001; Richards et al., 2005); however, longer-term inhibition of synaptic transmission alters spine morphology (Papa and Segal, 1996). Our data suggest that miR-29a/b may fine tune the sensitivity of mature synaptic spines to neuroplastic cues by regulating the expression of Arpc3 and hence the functioning of the ARP2/3 actin nucleation complex (Gournier et al., 2001). To investigate this hypothesis in more detail, we set up a protocol to measure changes in the volume of spines over a 24-h period. 3D volume–rendered images were created from GFP-MEM–expressing primary hippocampal neurons at 14 DIV. Changes in spine volume were then calculated by imaging the same dendritic regions after 24 h in culture medium alone or in culture medium in which neurotransmission was inhibited by the addition of 0.5 µM TTx (Fig. 7 C). Under control conditions, spines increased in volume by 1.4 ± 0.1 fold over the 24-h period, indicating that the process of spine morphogenesis was ongoing. In these cultures, average spine volume was not altered by the presence of 50 nM miR-29a antagonist (0.95 ± 0.07 µm3 for miR-Scr and 0.84 ± 0.06 µm3 for miR-29a antagonist, with data from 200 and 170 spines, respectively, and measured at 14 DIV in non-TTx–treated cultures; Siegel et al., 2009). In control neurons containing 50 nM Scr antagonist, treatment with TTx for 24 h reduced spine volume by 43.5 ± 6.9% (89–104 spines from four experiments; P = 0.002). In contrast, neurons containing miR-29a antagonist were much less sensitive to the loss of synaptic transmission. Thus, TTx treatment reduced spine volume only by 25.2 ± 16.9% over the same 24-h period (55–111 spines from four experiments; NS from control miR-29a antagonist neurons; Fig. 7, C and D). These data suggest that, in the absence of miR-29a, mature neurons are less sensitive to morphological changes induced by loss of synaptic connectivity. The ability of the miR-29a antagonist to inhibit miR-29a in primary hippocampal neurons was validated using a dsRed sensor (Siegel et al., 2009) containing the miR-29a seed region in the 3′-UTR (see Materials and methods; Fig. S4).
Contextual fear conditioning induces up-regulation of miR-29a/b in the hippocampus
A physiological form of hippocampal spine remodeling is the observed increase in spine density that is rapidly induced by several forms of learning (Leuner et al., 2003; Knafo et al., 2004; Restivo et al., 2009). Contextual fear conditioning is a hippocampus-dependent learning test that consists in the association of a brief electrical shock with the context in which the shock is administered. This type of learning depends on both the formation of dendritic spines (Restivo et al., 2009) and actin remodeling (Fischer et al., 2004) in hippocampal circuits. Adult mice were first exposed to a conditioning procedure that consisted in the administration of a tone for 30 s with the administration of a 0.5 mA electrical foot shock during the last 2 s of the tone. This was then repeated three times in a new cage to create the conditioning context. When reexposed to the same context 24 h after the conditioning procedure, mice exposed to the foot shocks showed significantly increased fear-conditioned freezing responses (P < 0.001) relative to mice exposed to the context in the absence of the conditioning procedure (Fig. 7 E). Alterations in levels of miR-29a/b in the hippocampus were then measured in control and conditioned animals after the contextual fear conditioning. A highly significant increase in both miR-29a and miR-29b (P = 0.007 and 0.005, respectively) was detected 45 min after contextual conditioning with respect to mice sacrificed 45 min after a simple exposure to the context. This appeared to be a transient response, as the enhancement was no longer significant 2 h after conditioning (Fig. 7 F). In contrast, miR-182, miR-183, miR-190b, and miR-680 were not altered by this treatment (unpublished data). Furthermore, levels of Arpc3 mRNA were significantly decreased at both 45 min (0.7 ± 0.05, P < 0.01) and 2 h (0.77 ± 0.02, P < 0.05) after contextual fear conditioning, suggesting a close temporal regulation of Arpc3 levels by miR-29a/b (Fig. 7 G).
Discussion
Dendritic spines are discrete compartments specialized in synaptic communication and are equipped with the essential machinery for protein synthesis. Local control of mRNA translation allows neurons to respond to extracellular cues at a subcellular resolution, and this accounts for the tight spatial regulation of synaptic strength that occurs during plasticity (Schratt, 2009). Recent evidence suggests that miRs are involved in the regulation of gene expression at the synapse. Several miRs are enriched in the synaptodendritic compartment, where they fine tune the production of proteins that determine spine head size (Schratt et al., 2006; Siegel et al., 2009), regulate dendritic arborization (Fiore et al., 2009), or mediate synaptic plasticity (Rajasethupathy et al., 2009; Gao et al., 2010). In this study, we identify a novel miR cluster involved in the regulation of spine morphology and provide evidence that this may be involved in activity-dependent structural plasticity. miR-29a/b target Arpc3 of the ARP2/3 actin nucleation complex, thereby reducing the probability of actin branch formation, a fundamental step in the development of a fully mature spine (Hotulainen et al., 2009). Interestingly, miR-29a had already been identified in a screen for miRs enriched in rat forebrain synaptosomes (Siegel et al., 2009), suggesting a correlation between this miR and localized repression of protein translation in synaptic spines.
Psychostimulants induce activity-dependent rewiring of neuronal circuits and long-term neuroadaptations via synapse-specific Hebbian plasticity, structural plasticity, and modulation of gene expression (Russo et al., 2010). Transcription factors like CREB (Walters et al., 2005), ΔFosB (McClung and Nestler, 2003), and NF-kB (Russo et al., 2007) are persistently activated after chronic treatments with psychostimulants. Notably, drugs such as cocaine can target genes that encode for cytoskeleton regulatory proteins, therefore inducing spine restructuring (Toda et al., 2006). Recent observations demonstrate regulation of miR levels in response to chronic cocaine exposure (Hollander et al., 2010), suggesting that the miR transcriptome may be subject to widespread regulation by psychoactive drugs. In this study, using an in vivo chronic drug exposure protocol, we describe marked changes (for ∼20% of 384 miRs tested) in the expression profile of rodent miRs after treatment with nicotine, cocaine, or amphetamine. Changes observed were both region- and drug-specific and may therefore reflect the different cellular and molecular mechanisms triggered by each psychostimulant drug. In contrast, expression levels of miRs that were up-regulated in several areas and upon different treatments are therefore likely to be common targets of enhanced neuronal activity and/or plastic changes within these circuits.
To identify which of the miRs targeted by psychostimulants in vivo may regulate neuronal structure, we turned to primary cultures of hippocampal neurons as a well-established model of synapse formation. We first investigated miR expression changes during synaptogenesis. At this stage, neuronal activity results in synchronized Ca2+ transients within the neuronal network (Fig. 2 B; Ogura et al., 1987; Bacci et al., 1999). Previous work has demonstrated that these network Ca2+ signals trigger intracellular cascades and regulate gene expression via Ca2+-dependent transcription factors, such as CREB (Greer and Greenberg, 2008). In fact, it is now apparent that several genes involved in neuronal plasticity, e.g., Bdnf and c-Fos, are regulated in an activity-dependent fashion (Greer and Greenberg, 2008). Emerging data now suggest a role for miRs as fine-tuning regulators of synaptic strength and neuronal plasticity. Furthermore, as miR biogenesis (Lugli et al., 2005) and turnover (Krol et al., 2010) are themselves regulated by synaptic activity, multiple layers of regulation may be present between miRs and neuronal activity. For example many miR loci emerged from a wide screening of the CREB regulon (Vo et al., 2005) or showed activity dependency and/or CREB-responsive elements in their proximal promoter (Impey et al., 2010). In this study, several miRs showed a marked increase between 7 and 14 DIV (Fig. 2 A). In particular, activity-dependent up-regulation was observed for the clusters miR-29a/b and miR-182/183. Further investigation identified a role for miR-29a/b in regulation of synaptic structure. Taken together, confocal imaging and electrophysiological measurements suggested that the effect of miR-29a/b was primarily on postsynaptic spine structure rather than on synaptic transmission by itself. This effect was phenocopied by overexpression of specific siRNA against Arpc3, and, importantly, the effects on spine structure were not observed in neurons overexpressing recombinant Arpc3 mutated in the miR-29a/b seed region. Therefore, our data indicate that Arpc3 and hence actin dynamics are subject to activity-dependent regulation via the miR-29a/b cluster.
The formation and maintenance of spine structure reflect underlying changes in the actin cytoskeleton. Actin turnover in spines is itself a very rapid process, with half-life measured between 30 and 60 s (Star et al., 2002). This requires the concerted effect of numerous actin nucleation proteins, capping proteins, and depolymerizing factors (Hotulainen et al., 2009). Arpc3 belongs to the multisubunit ARP2/3 complex responsible for initiating branching in actin filaments (Goley and Welch, 2006). A pioneering reconstitution study on the ARP2/3 complex described in detail the role of the individual subunits (Gournier et al., 2001). In the absence of Arpc3 (previously named p21 subunit), the complex has no gross abnormalities, is able to cross-link actin filaments into Y-branched arrays, and maintains the regulatory sites activated by exogenous factors. Interestingly though, ARP2/3 depleted of the p21 subunit exhibits reduced nucleation activity (∼10 fold) and decreased branching frequency (Gournier et al., 2001). Therefore, Arpc3 appears to primarily have a regulatory function and represents an ideal target for fine tuning the activity of the complex. Here, we propose that the role of miR-29a/b is to modulate the activity of the ARP2/3 complex by targeting the regulatory subunit Arpc3 (possibly at the synaptodendritic compartment, as shown in Schratt et al. [2006] and Siegel et al. [2009]), thus maintaining flexibility in neuronal networks. In addition, the up-regulation of miR-29a/b in the hippocampus during contextual fear learning suggests a role for these miRs in regulating hippocampal spine remodeling within a more physiological context (Leuner et al., 2003; Knafo et al., 2004; Restivo et al., 2009). Whether the role of these miRs is to facilitate the removal or redirection of existing synaptic connections or to instead provide a homeostatic mechanism to counteract excessive positive synaptogenic stimuli remains to be determined.
The brain adapts to sensory experiences through rapid changes in the number and strength of synaptic connections. It has been recently demonstrated that only a fraction of the synapses formed during development or induced by sensory experiences is stably maintained throughout adult life (Yang et al., 2009), whereas the majority undergoes a process of spine elimination. In this paper, we demonstrate that miR-29a/b target ARPC3 of the Arp2/3 complex, thereby decreasing the rate of actin branching and reducing the efficiency of neurons to drive dendritic spine head enlargement and synaptic consolidation. In light of our findings, it is conceivable that the up-regulation of miRs such as miR-29a/b represents an activity-dependent pathway to counterbalance excessive positive cues driving spine formation and reinforcement of synaptic communication. In support of this hypothesis, miR-29b levels increase steadily in vivo during the early stages of brain development (Hébert et al., 2008) and after context fear conditioning (Fig. 7 F). Therefore, the spatial and temporal regulation of these miRs, combined with the targeting of a key component of the actin remodeling process, would suggest a critical role for miR-29a/b in neuronal structural plasticity.
Materials and methods
Chronic drug treatment in adult mice
Chronic treatments with psychostimulants were performed as described in Ziviani et al. (2011). In brief, adult mice were treated with either saline, 1 mg/kg (−) nicotine hydrogen (+) tartrate, 10 mg/kg cocaine hydrochloride, or 5 mg/kg d-amphetamine-d3 sulfate salt via a single i.p. injection once a day for 5 d. Animals were then sacrificed via decapitation, and brain regions were dissected on ice.
Small RNA extraction and quantification
Total RNA (including the small RNA fraction) was extracted following the manufacturer’s recommendations (miRNeasy kit; QIAGEN). 384-well array microfluidic cards (TaqMan; Applied Biosystems) were used for an initial screen using a real-time PCR system (7900HT Fast; Applied Biosystems). Data were analyzed with StatMiner software (Integromics). The 34 selected miRs were quantified via a custom-made real-time quantitative PCR assay on 96-well plates (Applied Biosystems) following the manufacturer’s recommendations. Samples were retrotranscribed using Megaplex Pool A primers (Applied Biosystems). Individual miRs were quantified via miR assays (TaqMan) following the manufacturer’s recommendations. Real-time PCR data were processed and analyzed using the comparative ΔCT method (Livak and Schmittgen, 2001). The ΔCT values were then expressed relative to the first control sample (defined as 1) and the mean ± SEM calculated from the separate experiments.
Imaging and analysis of dendritic structures
At 14–15 DIV, transfected neurons grown on 28-mm coverslips were mounted on the stage of a confocal microscope (LSM510; Carl Zeiss) and maintained at 37°C in a Krebs/Hepes buffer (KHB; 20 mM Hepes, 130 mM NaCl, 5.4 mM KCl, 1.0 mM MgCl2, 1.8 mM CaCl2, and 10 mM glucose, pH 7.4). Neurons were imaged using a 63× oil immersion lens with an additional 4× optical zoom applied for dendritic regions. Confocal sections of EGFP-MEM–expressing neurons were reconstructed using Volocity software (PerkinElmer). To avoid user bias, imaging and analysis of experiments examining changes in filopodia/spine number were conducted blind. Structures were characterized according to the method of Hotulainen et al. (2009). For experiments requiring the same dendritic region to be imaged on consecutive days, neurons were grown in a 4-well chambered coverglass system (Thermo Fisher Scientific). Spine head volume was calculated from volume-rendered stacks using Volocity software.
Preparation and transfection of primary hippocampal neurons
Primary cultures of hippocampal neurons were prepared from neonatal C57BL6 mice (<1 d old) essentially as previously described (Young et al., 2008). 10–30 µl droplets (containing approximately 50,000–100,000 cells) were seeded onto polylysine-coated coverslips or multiwell plates in Neurobasal A medium supplemented with 2% B27, 1 µg/µl gentamycin, 2 mM glutamax, and 5% FCS. After 24 h, this medium was supplemented with 5 µM arabinofuranosyl cytidine to inhibit glial cell division. After a further 48 h, this was replaced with fresh Neurobasal A supplemented with 2% B27, 1 µg/µl gentamycin, and 2 mM glutamax only.
Except where otherwise indicated, 9-DIV neurons were transfected with 1 µg EGFP-MEM plasmid (Takara Bio Inc.) plus 10 nM miR oligonucleotide, 40 nM siRNA oligonucleotide, or 50 nM miR antagonist oligonucleotide using 2 µl/ml Lipofectamine2000 (Invitrogen) according to the manufacturer’s instructions. For experiments using plasmid expression of miRs, the miR plasmid (GeneCopoeia), which coexpresses a GFP reporter protein, was cotransfected with a plasmid expressing cytosolic tdTomato (provided by M. Schell, Uniformed Services University of the Health Sciences, Bethesda, MD) at a ratio of 2:1 (total DNA was 2 µg). The tdTomato fluorescence was generally brighter than the GFP reporter inherent to the miR plasmid and so was used to identify dendritic structures. Transfections of neurons with miR or siRNA oligonucleotides alone were conducted using 2.5 µl/ml siPORT NeoFX transfection agent (Invitrogen) and the same oligonucleotide concentration described above. For experiments measuring knockdown of protein levels, neurons were transfected at 6 DIV and collected at 9 DIV.
Electrophysiological measurements
Electrophysiological recordings were performed on hippocampal neurons overexpressing miR-29a, -29b, or -Scr in cells cotransfected with EGFP-MEM (identified by GFP fluorescence). Cells were voltage clamped at a holding potential of −60mV. Patch recordings were made using an amplifier (Multiclamp 700B) and pCLAMP 9.2 software (Molecular Devices) sampling at 50 kHz and filtering at 10 kHz. Patch pipettes were pulled from filamented borosilicate glass (with an outer diameter of 1.5 mm and an inner diameter of 0.86 mm; GC150F-7.5; Harvard Apparatus) with a two-stage vertical puller (PC-10; Narishige). Pipettes were used with a final tip resistance of 3–4 MΩ when filled with a solution containing 90 mM K-gluconate, 20 mM KCl, 20 mM Hepes, 0.2 mM EGTA, 1 mM MgCl2, and 0.1 mM CaCl2; pH was adjusted to 7.2 with KOH. Whole-cell access resistances were <20 MΩ and were compensated by 70%. The composition of the bath solutions (normal artificial cerebrospinal fluid) was 125 mM NaCl, 2.5 mM KCl, 26 mM NaHCO3, 10 mM glucose, 1.25 mM NaH2PO4, 2 mM sodium pyruvate, 3 mM myo-inositol, 2 mM CaCl2, 1 mM MgCl2, and 0.5 mM ascorbic acid; pH was 7.4 when continuously bubbled with 95% O2/5% CO2.
Real-time PCR on miR-29 targets
The level of potential miR-29a and -29b targets was assessed via Universal ProbeLibrary assays (TaqMan; Roche) on a LightCycler 480 system (Roche) following the manufacturer’s recommendations. The primers used were the following: ArhGAP28, forward 5′-CCGTCAGGGATGTCAGAGAC-3′ and reverse 5′-GAGTAGCATGCTCACAAGAATCA-3′; ArhGAP29, forward 5′-GGATTCAGAGTCTATAAGTCCAGGAG-3′ and reverse 5′-ATCAGCAGAAGACATGGTTCC-3′; ArhGEF5, forward 5′-TTCATGGACCTCACAAAAACC-3′ and reverse 5′-TCCAGCGAAGCTTTTCACTT-3′; Arpc3, forward 5′-GAAGGAAATGTACACGCTAGGAA-3′ and reverse 5′-GAGGTACGCACGCATCATC-3′; Bace1, forward 5′-AGATGGACTGCAAGGAGACG-3′ and reverse 5′-ATGTTCCAAGGGGTCGTG-3′; Camk2b, forward 5′-GAGGATGAAGATGCCAAAGC-3′ and reverse 5′-GAACTGGAGATTGGCAGGAG-3′; Coro6, forward 5′-TGGAACAGCAATGGTAGCC-3′ and reverse 5′-GTGGGCTGCAAACCTCTC-3′; Daam1, forward 5′-CTGGTTCTTCGCGCTGTC-3′ and reverse 5′-GGCCATGGTTGAATTCTCTC-3′; Dnm3, forward 5′-GATTCCCAGGAAGATGTGGA-3′ and reverse 5′-TGTCCATTTTCATCATTTTCAGTTA-3′; Dok4, forward 5′-CGTCAAGCAAGGCTATGTGA-3′ and reverse 5′-CAGCACCTCCGGTAAATCC-3′; Fez2, forward 5′-CCTCTCGGAGAAAGGGATG-3′ and reverse 5′-GAATGCATGTCCAACTGCTC-3′; Frmd4a, forward 5′-CCTTCAAGCTGGATGAACAAA-3′ and reverse 5′-CTCTCGTTCCAGGCGTTC-3′; Fscn1, forward 5′-GCCAACGAGAGGAACGTG-3′ and reverse 5′-GGTGCGAAAGGCACACTT-3′; Gmip1, forward 5′-CGAGATATCATTGGGGAACG-3′ and reverse 5′-CCAGGTCTTCTCTGCGTACA-3′; Gprin1, forward 5′-GGGTGAGAGAGTCTGGATGC-3′ and reverse 5′-TTAGGGGGCTGGAGTCCTT-3′; Lasp1, forward 5′-TCGTCCTATGGTGGGTACAAG-3′ and reverse 5′-GGCAGCGCTGTAGTCATACAC-3′; Map6d1, forward 5′-AAGCCCTCAAGATCCACAAA-3′ and reverse 5′-TGTGAATTTCCTCACCTCTGG-3′; Ninj2, forward 5′-CAGCAATCCTATTTGATGACTACC-3′ and reverse 5′-GGGAAATGCTAGTTATCCTGACA-3′; Nrsn1, forward 5′-GGGGATTGGAGGGACAGA-3′ and reverse 5′-CATCTTCCCAGCTTCAATGTT-3′; Ophn1, forward 5′-CCATCATCCGTTCGTGCT-3′ and reverse 5′-CCGGGAAGCTGTTTCAAAG-3′; Pcdha1, forward 5′-AAATGATGCTGGCTCTCAAAA-3′ and reverse 5′-CTGGACCAGCCCGTAGAAT-3′; RapGEFL1, forward 5′-GTAGCTGTTGCTTCCTCTGGA-3′ and reverse 5′-ATACCCAGCGTGGTGAAAAC-3′; Rgl3, forward 5′-GCCGTGTACAGCGTCTCC-3′ and reverse 5′-AAGGAAGGTGTCGGTAGCC-3′; Rhobtb, forward 5′-GACACACTTGGTTGCTCTCG-3′ and reverse 5′-ATGCTCACACCACTGACAGC-3′; Spna2, forward 5′-CCTCAGATGAAGTGAGGGAGA-3′ and reverse 5′-CCAGCAGGGCAGTTCTCT-3′; and Stau6, forward 5′-CAAGCCAGCACTCAAATCAG-3′ and reverse 5′-CGTTACTTTCCTTCCGTCTCC-3′.
Luciferase Assay
pGL3-mmArpc3 and pGL3-hsArpc3 were obtained by amplifying the entire 3′-UTR of the mouse and human Arpc3 and then cloning them into pGL3–multiple cloning site (Promega). The primers used were as follows: mmArpc3 nested forward PstI 5′-CTGCAGCTGCAGGAGGAGCCTGGGCAGCAC-3′, mmArpc3 nested reverse EcoRI 5′-GAATTCGAATTCAAGCTTTTTCTCAAAAATGTTTTAATTC-3′, hsArpc3 nested forward NsiI 5′-ATGCATATGCATAGGGAGCCCGGGCAGCCACCG-3′, and hsArpc3 nested reverse EcoRI 5′-GAATTCGAATTCTGGAATGTTAGAAATTTCTTTATTATTAC-3′.
Luciferase experiments were conducted as follows: HEK-293 cells were grown on 96-well clear-bottom white microplates (Corning) at 20–30% confluency. After 18–24 h, miR oligonucleotides or miR antagonists were transfected using 0.5 µl/well siPORT. After a further 8 h, pGL3 constructs were cotransfected with a Renilla plasmid (1:20) via Ca2+/phosphate precipitation. Luciferase chemiluminescence was assessed 24–48 h after transfection using the Dual-Glo Luciferase Assay system (Promega) following the manufacturer’s recommendations. Data were normalized to Renilla luminescence.
Western blotting
Levels of Arpc3, Arp3, and GAPDH proteins were assessed via Western blotting. Samples were collected in 2× loading buffer (0.125 M Tris-HCl, pH 6.8, 4% SDS, 20% glycerol (volume/volume), 0.003% Bromophenol blue, and 10% β-mercaptoethanol), sonicated, and then loaded onto a 13% polyacrylamide gel. Chemiluminescence was detected using ECL Plus (GE Healthcare) and analyzed using ImageJ software (National Institutes of Health). The antibodies used were goat anti-Arpc3 (1:1,000; Abcam), mouse monoclonal anti-Arp3 (1:15,000; Abcam), and mouse monoclonal anti-GAPDH (1:25,000; Santa Cruz Biotechnology, Inc.).
Site-directed mutagenesis
Mouse Arpc3 3′-UTR was mutated using a site-directed mutagenesis kit (Quikchange II; Agilent Technologies) following the manufacturer’s recommendations. The primers used were mArpc3 mutated forward 5′-GGGTGTCGGGTGATTTCGATTTTTGGCAGTTTTAAGCGAACATGTAATATATAATAAATGTCACTGCTTATGTTAGACATT-3′ and reverse 5′-AATGTCTAACATAAGCAGTGACATTTATTATATATTACATGTTCGCTTAAAACTGCCAAAAATCGAAATCACCCGACACCC-3′.
miR-29a sensor
To address the specificity and efficiency of miR-29 oligonucleotides (Invitrogen) and miR antagonists (Exiqon), we designed an miR-29a sensor. Cloning was performed as described in Siegel et al. (2009). The primers used were miR-29a sensor forward 5′-GGCCGCTAACCGATTTCAGATGGTGCTAACTAACCGATTTCAGATGGTGCTAT-3′ and reverse 5′-CTAGATAGCACCATCTGAAATCGGTTAGTTAGCACCATCTGAAATCGGTTAGC-3′. The sensor was cotransfected with EGFP-MEM into primary hippocampal neurons at 4 DIV and imaged after 48–72 h.
Single-cell calcium imaging
Changes in intracellular Ca2+ were measured in hippocampal neurons using fura-4f–acetoxymethyl (Invitrogen). Neuronal cultures grown on 28-mm glass coverslips were loaded with 2 µM fura-4f–acetoxymethyl for 1 h in KHB. Coverslips were then mounted on the stage of an inverted epifluorescence microscope (Axiovert 200; Carl Zeiss) and maintained at 37°C in KHB. Cells were sequentially excited at 340 and 380 nm, and emission fluorescence was collected via a 510-nm band pass filter using a cooled charge-coupled device camera (Hamamatsu Photonics). Measurements of spontaneous network Ca2+ transients in the presence or absence of 50 µM PTx were made by calculating the mean peak minus basal Ca2+ response and are expressed as 340:380 ratio units.
Fear conditioning
The fear conditioning procedure was performed as described in Zanardi et al. (2007). In brief, raining took place in a conditioning chamber (23 × 22 × 24 cm) with gray Plexiglas walls and ceiling. Scrambled shock was delivered by a shock source to a grid floor made of stainless steel bars, which were 2 mm in diameter and spaced 0.5 cm apart. Mice were transferred to the conditioning chamber, and, after an initial acclimatization period of 2 min, were presented with three pairings of the tone with foot shock (0.5 mA for 2 s). The tone was presented for 30 s, and the shock was administered during the last 2 s of the tone. Pairings were separated by 2 min, and mice were removed from the chamber 30 s after the last shock presentation. Mice exposed to the context alone were transferred to the conditioning chamber for 5 min but did not receive the conditioning procedure. Approximately 24 h after conditioning, mice were tested for contextual conditioning. Mice were placed into the conditioning chamber for 5 min, and freezing behavior was scored. Freezing was scored using a time-sampling procedure in which every 10 s a determination was made whether or not a mouse was freezing. Freezing was defined as the absence of all movement except for respiration for a minimum of 1 s.
Statistical analysis
Data were analyzed using an unpaired Student’s t test or a one-way analysis of variance with a Tukey’s post-hoc test (Prism; GraphPad Software). Cumulative frequency histograms were compared using the Kolmogorov–Smirnov test (Axon pCLAMP9; Molecular Devices).
Online supplemental material
Fig. S1. shows initial screening for miRs involved in neuroplasticity using TaqMan low density arrays. Fig. S2 shows that miRs are regulated in a similar fashion by different drugs of abuse and in different brain areas. Fig. S3 shows that pEZX-premiR-29b mimics the effect of oligonucleotides on spine morphology and that miR-29a and -29b do not prevent PTx-induced Ca2+ transients in neuronal networks. Fig. S4 shows the validation of predicted miR-29a and -29b targets in primary cultures of mouse hippocampal neurons and characterization of the dsRed miR-29a sensor plasmid. Fig. S5 shows direct targeting of Arpc3 3′-UTR by miR-29a and -29b. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201103006/DC1.
Acknowledgments
We thank Dalbir K. Dhiraj and Roberta Tufi for helping with Arpc3 Western blots and Kate M. Phillips for helping with miR screening.
G. Lippi, M. Zoli, and S. D’Oro were supported by a Neurocypres grant from the European Commission.
Footnotes
Abbreviations used in this paper:
- DIV
- days in vitro
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- mEPSC
- miniature excitatory postsynaptic current
- miR
- microRNA
- MRE
- miR-responsive element
- NMDA
- N-methyl-d-aspartate
- PTx
- picrotoxin
- TTx
- tetrodotoxin
- UTR
- untranslated region
References
- Bacci A., Verderio C., Pravettoni E., Matteoli M. 1999. Synaptic and intrinsic mechanisms shape synchronous oscillations in hippocampal neurons in culture. Eur. J. Neurosci. 11:389–397 10.1046/j.1460-9568.1999.00440.x [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136:215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekar V., Dreyer J.L. 2009. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol. Cell. Neurosci. 42:350–362 10.1016/j.mcn.2009.08.009 [DOI] [PubMed] [Google Scholar]
- Christensen M., Schratt G.M. 2009. microRNA involvement in developmental and functional aspects of the nervous system and in neurological diseases. Neurosci. Lett. 466:55–62 10.1016/j.neulet.2009.04.043 [DOI] [PubMed] [Google Scholar]
- Dillon C., Goda Y. 2005. The actin cytoskeleton: integrating form and function at the synapse. Annu. Rev. Neurosci. 28:25–55 10.1146/annurev.neuro.28.061604.135757 [DOI] [PubMed] [Google Scholar]
- Dunaevsky A., Tashiro A., Majewska A., Mason C., Yuste R. 1999. Developmental regulation of spine motility in the mammalian central nervous system. Proc. Natl. Acad. Sci. USA. 96:13438–13443 10.1073/pnas.96.23.13438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D., Neilson J.R., Foster K.A., Wang C.F., Seeburg D.P., Batterton M.N., Tada T., Dolan B.M., Sharp P.A., Sheng M. 2010. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 65:373–384 10.1016/j.neuron.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert F., Bonhoeffer T. 1999. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 399:66–70 10.1038/19978 [DOI] [PubMed] [Google Scholar]
- Fineberg S.K., Kosik K.S., Davidson B.L. 2009. MicroRNAs potentiate neural development. Neuron. 64:303–309 10.1016/j.neuron.2009.10.020 [DOI] [PubMed] [Google Scholar]
- Fiore R., Khudayberdiev S., Christensen M., Siegel G., Flavell S.W., Kim T.K., Greenberg M.E., Schratt G. 2009. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 28:697–710 10.1038/emboj.2009.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M., Kaech S., Wagner U., Brinkhaus H., Matus A. 2000. Glutamate receptors regulate actin-based plasticity in dendritic spines. Nat. Neurosci. 3:887–894 10.1038/78791 [DOI] [PubMed] [Google Scholar]
- Fischer A., Sananbenesi F., Schrick C., Spiess J., Radulovic J. 2004. Distinct roles of hippocampal de novo protein synthesis and actin rearrangement in extinction of contextual fear. J. Neurosci. 24:1962–1966 10.1523/JNEUROSCI.5112-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Wang W.Y., Mao Y.W., Gräff J., Guan J.S., Pan L., Mak G., Kim D., Su S.C., Tsai L.H. 2010. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 466:1105–1109 10.1038/nature09271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley E.D., Welch M.D. 2006. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7:713–726 10.1038/nrm2026 [DOI] [PubMed] [Google Scholar]
- Gournier H., Goley E.D., Niederstrasser H., Trinh T., Welch M.D. 2001. Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol. Cell. 8:1041–1052 10.1016/S1097-2765(01)00393-8 [DOI] [PubMed] [Google Scholar]
- Grabrucker A., Vaida B., Bockmann J., Boeckers T.M. 2009. Synaptogenesis of hippocampal neurons in primary cell culture. Cell Tissue Res. 338:333–341 10.1007/s00441-009-0881-z [DOI] [PubMed] [Google Scholar]
- Greer P.L., Greenberg M.E. 2008. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 59:846–860 10.1016/j.neuron.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 466:835–840 10.1038/nature09267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert S.S., Horré K., Nicolaï L., Papadopoulou A.S., Mandemakers W., Silahtaroglu A.N., Kauppinen S., Delacourte A., De Strooper B. 2008. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. USA. 105:6415–6420 10.1073/pnas.0710263105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander J.A., Im H.I., Amelio A.L., Kocerha J., Bali P., Lu Q., Willoughby D., Wahlestedt C., Conkright M.D., Kenny P.J. 2010. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 466:197–202 10.1038/nature09202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P., Llano O., Smirnov S., Tanhuanpää K., Faix J., Rivera C., Lappalainen P. 2009. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J. Cell Biol. 185:323–339 10.1083/jcb.200809046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H.I., Hollander J.A., Bali P., Kenny P.J. 2010. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat. Neurosci. 13:1120–1127 10.1038/nn.2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S., Davare M., Lasiek A., Fortin D., Ando H., Varlamova O., Obrietan K., Soderling T.R., Goodman R.H., Wayman G.A. 2010. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol. Cell. Neurosci. 43:146–156 10.1016/j.mcn.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Han J.J., Siomi M.C. 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10:126–139 10.1038/nrm2632 [DOI] [PubMed] [Google Scholar]
- Knafo S., Ariav G., Barkai E., Libersat F. 2004. Olfactory learning-induced increase in spine density along the apical dendrites of CA1 hippocampal neurons. Hippocampus. 14:819–825 10.1002/hipo.10219 [DOI] [PubMed] [Google Scholar]
- Korkotian E., Segal M. 2001. Regulation of dendritic spine motility in cultured hippocampal neurons. J. Neurosci. 21:6115–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik K.S. 2006. The neuronal microRNA system. Nat. Rev. Neurosci. 7:911–920 10.1038/nrn2037 [DOI] [PubMed] [Google Scholar]
- Krol J., Busskamp V., Markiewicz I., Stadler M.B., Ribi S., Richter J., Duebel J., Bicker S., Fehling H.J., Schübeler D., et al. 2010. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 141:618–631 10.1016/j.cell.2010.03.039 [DOI] [PubMed] [Google Scholar]
- Lamprecht R., LeDoux J. 2004. Structural plasticity and memory. Nat. Rev. Neurosci. 5:45–54 10.1038/nrn1301 [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 75:843–854 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. 2002. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 21:4663–4670 10.1093/emboj/cdf476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuner B., Falduto J., Shors T.J. 2003. Associative memory formation increases the observation of dendritic spines in the hippocampus. J. Neurosci. 23:659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120:15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 433:769–773 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 25:402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lugli G., Larson J., Martone M.E., Jones Y., Smalheiser N.R. 2005. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 94:896–905 10.1111/j.1471-4159.2005.03224.x [DOI] [PubMed] [Google Scholar]
- McClung C.A., Nestler E.J. 2003. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat. Neurosci. 6:1208–1215 10.1038/nn1143 [DOI] [PubMed] [Google Scholar]
- McClung C.A., Nestler E.J. 2008. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology. 33:3–17 10.1038/sj.npp.1301544 [DOI] [PubMed] [Google Scholar]
- Miska E.A., Alvarez-Saavedra E., Townsend M., Yoshii A., Sestan N., Rakic P., Constantine-Paton M., Horvitz H.R. 2004. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 5:R68 10.1186/gb-2004-5-9-r68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura A., Iijima T., Amano T., Kudo Y. 1987. Optical monitoring of excitatory synaptic activity between cultured hippocampal neurons by a multi-site Ca2+ fluorometry. Neurosci. Lett. 78:69–74 10.1016/0304-3940(87)90563-5 [DOI] [PubMed] [Google Scholar]
- Papa M., Segal M. 1996. Morphological plasticity in dendritic spines of cultured hippocampal neurons. Neuroscience. 71:1005–1011 10.1016/0306-4522(95)00490-4 [DOI] [PubMed] [Google Scholar]
- Rajasethupathy P., Fiumara F., Sheridan R., Betel D., Puthanveettil S.V., Russo J.J., Sander C., Tuschl T., Kandel E. 2009. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 63:803–817 10.1016/j.neuron.2009.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restivo L., Vetere G., Bontempi B., Ammassari-Teule M. 2009. The formation of recent and remote memory is associated with time-dependent formation of dendritic spines in the hippocampus and anterior cingulate cortex. J. Neurosci. 29:8206–8214 10.1523/JNEUROSCI.0966-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards D.A., Mateos J.M., Hugel S., de Paola V., Caroni P., Gähwiler B.H., McKinney R.A. 2005. Glutamate induces the rapid formation of spine head protrusions in hippocampal slice cultures. Proc. Natl. Acad. Sci. USA. 102:6166–6171 10.1073/pnas.0501881102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson T.E., Kolb B. 2004. Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology. 47(Suppl 1):33–46 10.1016/j.neuropharm.2004.06.025 [DOI] [PubMed] [Google Scholar]
- Russo S.J., Bolanos C.A., Theobald D.E., DeCarolis N.A., Renthal W., Kumar A., Winstanley C.A., Renthal N.E., Wiley M.D., Self D.W., et al. 2007. IRS2-Akt pathway in midbrain dopamine neurons regulates behavioral and cellular responses to opiates. Nat. Neurosci. 10:93–99 10.1038/nn1812 [DOI] [PubMed] [Google Scholar]
- Russo S.J., Dietz D.M., Dumitriu D., Morrison J.H., Malenka R.C., Nestler E.J. 2010. The addicted synapse: mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci. 33:267–276 10.1016/j.tins.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saneyoshi T., Fortin D.A., Soderling T.R. 2010. Regulation of spine and synapse formation by activity-dependent intracellular signaling pathways. Curr. Opin. Neurobiol. 20:108–115 10.1016/j.conb.2009.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G. 2009. microRNAs at the synapse. Nat. Rev. Neurosci. 10:842–849 10.1038/nrn2763 [DOI] [PubMed] [Google Scholar]
- Schratt G.M., Tuebing F., Nigh E.A., Kane C.G., Sabatini M.E., Kiebler M., Greenberg M.E. 2006. A brain-specific microRNA regulates dendritic spine development. Nature. 439:283–289 10.1038/nature04367 [DOI] [PubMed] [Google Scholar]
- Sempere L.F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V. 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5:R13 10.1186/gb-2004-5-3-r13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G., Obernosterer G., Fiore R., Oehmen M., Bicker S., Christensen M., Khudayberdiev S., Leuschner P.F., Busch C.J.L., Kane C., et al. 2009. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11:705–716 10.1038/ncb1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star E.N., Kwiatkowski D.J., Murthy V.N. 2002. Rapid turnover of actin in dendritic spines and its regulation by activity. Nat. Neurosci. 5:239–246 10.1038/nn811 [DOI] [PubMed] [Google Scholar]
- Tang J., Dani J.A. 2009. Dopamine enables in vivo synaptic plasticity associated with the addictive drug nicotine. Neuron. 63:673–682 10.1016/j.neuron.2009.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda S., Shen H.W., Peters J., Cagle S., Kalivas P.W. 2006. Cocaine increases actin cycling: effects in the reinstatement model of drug seeking. J. Neurosci. 26:1579–1587 10.1523/JNEUROSCI.4132-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N., Klein M.E., Varlamova O., Keller D.M., Yamamoto T., Goodman R.H., Impey S. 2005. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. USA. 102:16426–16431 10.1073/pnas.0508448102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters C.L., Cleck J.N., Kuo Y.-C., Blendy J.A. 2005. μ-Opioid receptor and CREB activation are required for nicotine reward. Neuron. 46:933–943 10.1016/j.neuron.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Wayman G.A., Davare M., Ando H., Fortin D., Varlamova O., Cheng H.Y., Marks D., Obrietan K., Soderling T.R., Goodman R.H., Impey S. 2008. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl. Acad. Sci. USA. 105:9093–9098 10.1073/pnas.0803072105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.E., Griffith E.C., Greenberg M.E. 2002. Regulation of transcription factors by neuronal activity. Nat. Rev. Neurosci. 3:921–931 10.1038/nrn987 [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 75:855–862 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- Winter J., Jung S., Keller S., Gregory R.I., Diederichs S. 2009. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11:228–234 10.1038/ncb0309-228 [DOI] [PubMed] [Google Scholar]
- Yang G., Pan F., Gan W.-B. 2009. Stably maintained dendritic spines are associated with lifelong memories. Nature. 462:920–924 10.1038/nature08577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K.W., Bampton E.T.W., Pinòn L., Bano D., Nicotera P. 2008. Mitochondrial Ca2+ signalling in hippocampal neurons. Cell Calcium. 43:296–306 10.1016/j.ceca.2007.06.007 [DOI] [PubMed] [Google Scholar]
- Zanardi A., Ferrari R., Leo G., Maskos U., Changeux J.P., Zoli M. 2007. Loss of high-affinity nicotinic receptors increases the vulnerability to excitotoxic lesion and decreases the positive effects of an enriched environment. FASEB J. 21:4028–4037 10.1096/fj.07-8260com [DOI] [PubMed] [Google Scholar]
- Ziviani E., Lippi G., Bano D., Munarriz E., Guiducci S., Zoli M., Young K.W., Nicotera P. 2011. Ryanodine receptor-2 upregulation and nicotine-mediated plasticity. EMBO J. 30:194–204 10.1038/emboj.2010.279 [DOI] [PMC free article] [PubMed] [Google Scholar]