MCPH1, a protein linked to primary microcephaly, directly modulates condensin II to regulate chromosome condensation and shape.
Abstract
Mutations in human MCPH1 (hMCPH1) cause primary microcephaly, which is characterized by a marked reduction of brain size. Interestingly, hMCPH1 mutant patient cells display unique cellular phenotypes, including premature chromosome condensation (PCC), in G2 phase. To test whether hMCPH1 might directly participate in the regulation of chromosome condensation and, if so, how, we developed a cell-free assay using Xenopus laevis egg extracts. Our results demonstrate that an N-terminal domain of hMCPH1 specifically inhibits the action of condensin II by competing for its chromosomal binding sites in vitro. This simple and powerful assay allows us to dissect mutations causing primary microcephaly in vivo and evolutionary substitutions among different species. A complementation assay using patient cells revealed that, whereas the N-terminal domain of hMCPH1 is sufficient to rescue the PCC phenotype, its central domain plays an auxiliary role in shaping metaphase chromosomes by physically interacting with condensin II. Thus, hMCPH1 acts as a composite modulator of condensin II to regulate chromosome condensation and shaping.
Introduction
Autosomal recessive primary microcephaly is a neurodevelopmental disorder characterized by reduced brain size and mental retardation in humans (Thornton and Woods, 2009). At least eight different loci are known to be responsible for this disease, and MCPH1 is one of the seven responsible genes that have been identified so far. Its gene product, MCPH1 (also known as microcephalin or BRIT1), is an 835–amino acid protein that contains three BRCA1 C-terminal (BRCT) domains (Jackson et al., 2002): one (BRCT1) is at the N terminus, whereas the other two (BRCT2 and BRCT3) are tandemly arranged at the C terminus (Fig. 1 A). Although the three BRCT domains and their vicinities are reasonably conserved, the sequences comprising the large central region are highly variable even among vertebrates, implicating that MCPH1 is a rapidly evolving protein (Fig. S1; Ponting and Jackson, 2005).
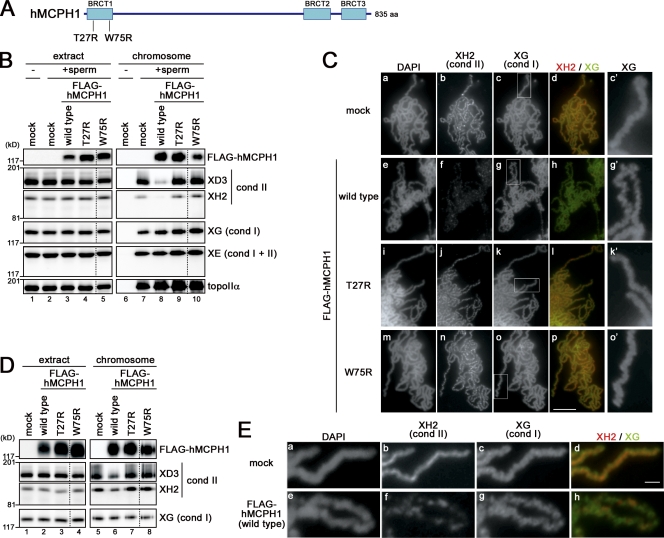
Figure 1.
hMCPH1 inhibits chromosomal binding of condensin II in Xenopus egg extracts. (A) hMCPH1 possesses three BRCT domains (blue boxes). Also shown are two point mutations (T27R and W75R) that cause primary microcephaly in MCPH1 patients. (B) A reticulocyte lysate containing no hMCPH1 (mock) or FLAG-tagged hMCPH1 was mixed with 10 vol metaphase egg extracts and incubated for 30 min. Sperm chromatin was then added and incubated for another 120 min. Chromosome fractions were isolated and analyzed by immunoblotting with the antibodies indicated. Aliquots of the extracts were saved before chromosome isolation and analyzed in parallel. No sperm was added in lanes 1 and 6. (C) Metaphase chromosomes were assembled as described in B, fixed, and stained with DAPI, anti–XCAP-H2 (XH2), and anti–XCAP-G (XG). Close-ups of chromosomal regions indicated by the white rectangles in c, g, k, and o are shown in c′, g′, k′, and o′, respectively. Bar, 5 µm. (D) Sperm chromatin was incubated with Xenopus egg extracts for 120 min to assemble metaphase chromosomes. 0.1 vol reticulocyte lysates containing no hMCPH1 (mock) or FLAG-tagged hMCPH1 was then added and incubated for another 60 min. Chromosome fractions were isolated and analyzed as described in B. (E) Metaphase chromosomes were assembled as described in D and analyzed as in C. Bar, 1 µm. The dotted lines indicate where intervening lanes were removed for presentation purposes. cond, condensin; topoIIα, topoisomerase IIα.
The cellular function of MCPH1 and its relationship to the etiology of microcephaly are not fully understood. In fact, a series of recent studies have been uncovering multiple faces of MCPH1 functions, which are as diverse as DNA damage response (Xu et al., 2004; Wood et al., 2007; Jeffers et al., 2008), cell cycle regulation (Alderton et al., 2006; Tibelius et al., 2009), transcriptional regulation (Yang et al., 2008), and centrosome regulation (Rai et al., 2008). Among them, perhaps one of the best-characterized examples may be γ-H2AX–dependent recruitment of MCPH1 to the sites of DNA double-strand breaks through its BRCT2/3 domains (Wood et al., 2007; Jeffers et al., 2008). It remains to be fully established, however, whether MCPH1 might act upstream or downstream of the checkpoint kinase Chk1 (Alderton et al., 2006; Tibelius et al., 2009).
Another line of recent studies has started to shed light on a distinct, less-appreciated function of MCPH1. Cells from MCPH1 patients were found to display premature chromosome condensation (PCC) in G2 phase of the cell cycle, implicating that MCPH1 might participate in the regulation of chromosome condensation (Trimborn et al., 2004). Central to chromosome condensation in vertebrate cells are two multisubunit complexes, namely, condensin I and II (Swedlow and Hirano, 2003; Hirano, 2005). The two complexes share a pair of structural maintenance of chromosomes (SMC) proteins as their core subunits and possess different sets of non-SMC regulatory subunits (Ono et al., 2003). Depletion of specific subunits of condensin II, but not those of condensin I, from MCPH1 patient cells rescued the PCC phenotype, raising the possibility that PCC might be caused by untimely activation of condensin II in G2 phase (Trimborn et al., 2006). However, direct evidence for inhibition of condensin II by MCPH1 is lacking. Moreover, MCPH1 is known to function as a component of the network that down-regulates Cdk1 in response to DNA damage (Alderton et al., 2006; Tibelius et al., 2009), leaving the alternative and currently prevalent scenario that the apparent condensation defects observed in MCPH1-deficient cells are an indirect consequence of perturbed cell cycle progression.
In the current study, we have developed a cell-free assay using Xenopus laevis egg extracts for studying the action of MCPH1 in vitro. Our results provide a convincing set of evidence that an N-terminal domain of human MCPH1 (hMCPH1) specifically inhibits the action of condensin II under a metaphase-arrested condition, being independent of cell cycle progression. The cell-free assay is demonstrated to be extremely powerful in analyzing microcephaly-causing mutations in humans and dissecting amino acid substitutions among different species. Moreover, a complementation assay using patient cells, combined with a protein–protein interaction assay, reveals that the central domain of hMCPH1, in collaboration with its N-terminal domain, contributes to proper shaping of metaphase chromosomes. Our results suggest that hMCPH1 functions as a composite modulator of condensin II and fine tunes the processes of chromosome condensation and shaping.
Results
hMCPH1 inhibits chromosomal binding of condensin II in Xenopus egg extracts
To dissect the molecular action of hMCPH1, we sought to develop a cell-free assay by using extracts prepared from unfertilized Xenopus eggs. The use of these egg extracts has two big advantages. First, the cell cycle of the unfertilized eggs is naturally arrested at metaphase of meiosis II, at which the activity of cyclin B–Cdk1 is kept high (Morgan, 2007). Second, the dynamics and function of many chromosomal proteins, including condensin I and II, have been characterized extensively in these extracts (e.g., Hirano et al., 1997; Ono et al., 2003). These properties allow us to study the specific role of hMCPH1 in condensin regulation, if any, without being disturbed by other parameters, such as cell cycle progression.
We started with a very simple experiment. Full-length hMCPH1 was produced in a reticulocyte lysate and preincubated with a metaphase extract for 30 min. Sperm chromatin was added into the mixture, and after incubation for another 120 min, chromosome fractions were isolated and analyzed by immunoblotting. Strikingly, we found that exogenously added hMCPH1 effectively inhibited loading of condensin II onto chromosomes. The effect of hMCPH1 was highly specific to condensin II: loading of condensin I and topoisomerase IIα was hardly affected in the same reaction (Fig. 1 B, lanes 7 and 8). We then tested whether two point mutations that had been identified in MCPH1 patients (T27R [Trimborn et al., 2005] and W75R [Gavvovidis et al., 2010]) might affect the activity of hMCPH1 to inhibit condensin II in this cell-free assay. Remarkably, again, both of the mutant proteins barely blocked loading of condensin II (Fig. 1 B, lanes 9 and 10), suggesting that this cell-free assay could indeed recapitulate an aspect of the physiological functions of hMCPH1.
The biochemical data were fully supported by morphological analysis. In this experiment, metaphase chromosomes were assembled in the presence or absence of hMCPH1, fixed, and stained simultaneously with antibodies against condensin I– and condensin II–specific subunits (XCAP-G and -H2, respectively). In a mock-treated extract, both condensin I and II were readily detectable along the entire length of chromatids (Fig. 1 C, a–d) as had been reported previously (Ono et al., 2003). In an hMCPH1-treated extract, however, the signal for condensin II was lost almost completely, whereas the signal for condensin I was unaffected (Fig. 1 C, e–h). We also noticed that the axes of chromosomes assembled in the presence of hMCPH1 were structurally distorted (Fig. 1 C, g′); this zigzag morphology was reminiscent of that observed in chromosomes assembled in an extract depleted of condensin II (Ono et al., 2003). As expected, extracts supplemented with the two mutant forms of hMCPH1 produced chromosomes that were indistinguishable from those assembled in the mock-treated extract (Fig. 1 C, i–p).
We then tested whether hMCPH1 might have an ability to strip condensin II from chromosomes that had already been assembled in the extracts. To this end, metaphase chromosomes were assembled in a control extract, and hMCPH1 was added and incubated for another 60 min. Immunoblotting analysis of chromosome fractions revealed a significant reduction of the level of condensin II but not of condensin I (Fig. 1 D). As judged by morphological analysis, again, hMCPH1-treated chromosomes displayed a zigzag and fragile appearance (Fig. 1 E). Taking these results together, we conclude that exogenously added hMCPH1 blocks the action of condensin II present in the Xenopus egg extracts in a highly specific manner.
An N-terminal domain of hMCPH1 is sufficient to specifically inhibit condensin II in the cell-free assay
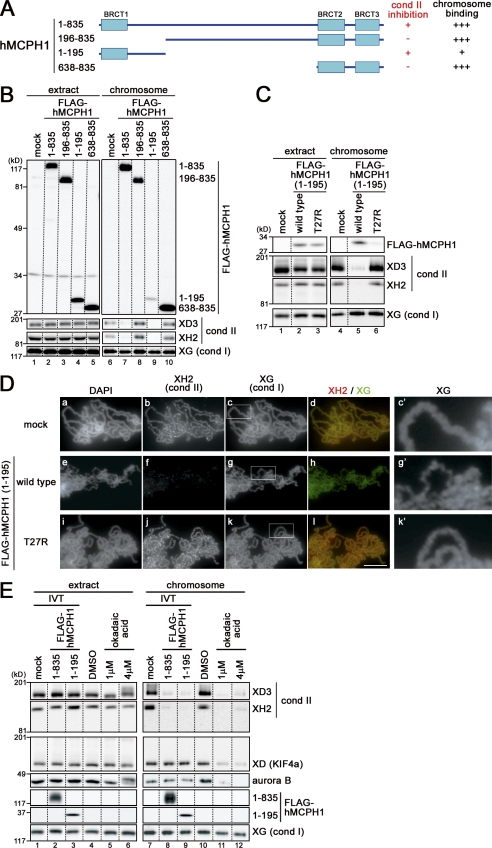
Next, we asked which domains of hMCPH1 might be responsible for the activity to inhibit condensin II observed in the cell-free assay. Selected examples of deletion constructs of hMCPH1 tested are shown in Fig. 2 A. In short, we found that an N-terminal fragment (amino acids 1–195) containing the first BRCT domain (BRCT1) is required and sufficient to inhibit loading of condensin II onto chromosomes (Fig. 2 B, lanes 8 and 9). This N-terminal fragment itself bound to chromosomes, but it does so only weakly when compared with a C-terminal fragment (amino acids 638–835) containing the second and third BRCT domains (Fig. 2 B, lane 10). Introduction of the T27R mutation into the N-terminal domain (amino acids 1–195) not only abolished its condensin II inhibitory activity (Fig. 2, C and D) but also weakened its own chromosome-binding activity (Fig. 2 C, lane 6). None of the shorter N-terminal constructs tested (amino acids 1–95, 1–138, 1–162, and 1–183) inhibited condensin II loading in this assay.
Figure 2.
An N-terminal domain of hMCPH1 is sufficient to specifically inhibit chromosomal loading of condensin II in Xenopus egg extracts. (A) Deletion constructs of hMCPH1 used in this paper. (B) Full-length hMCPH1 and three different deletion constructs were subjected to the cell-free assay. Chromosome fractions and aliquots of the extracts were analyzed by immunoblotting. (C) The same experiment as in B was repeated using the wild-type and mutant (T27R) forms of the N-terminal domain (amino acids 1–195). (D) Metaphase chromosomes were assembled in an extract supplemented with the wild-type and mutant (T27R) forms of the N-terminal domain (amino acids 1–195) as described in B, fixed, and stained with DAPI, anti–XCAP-H2 (XH2), and anti–XCAP-G (XG). Close-ups of chromosomal regions indicated by the white rectangles in c, g, and k are shown in c′, g′, and k′, respectively. Bar, 5 µm. (E) Metaphase egg extracts were mixed with 0.1 vol reticulocyte lysates containing FLAG-hMCPH1 (amino acids 1–835 [full length] or 1–195) or treated with 1 or 4 µM okadaic acid. Sperm chromatin was added and incubated for 120 min to assemble metaphase chromosomes. Chromosome fractions were then isolated and analyzed as described in B. The dotted lines indicate where intervening lanes were removed for presentation purposes. cond, condensin.
A recent study had reported that okadaic acid, a potent inhibitor of PP2A (protein phosphatase 2A), inhibits chromosomal loading of condensin II in Xenopus egg extracts and in human cells (Takemoto et al., 2009). Therefore, we wished to compare the impacts of hMCPH1 and okadaic acid in parallel in our cell-free assay. We found that okadaic acid blocked loading of condensin II, XCAP-D (KIF4a), and aurora B but not of condensin I (Fig. 2 E, lanes 11 and 12) as had been reported by Takemoto et al. (2009). In contrast, hMCPH1 hardly affected loading of XCAP-D and aurora B (Fig. 2 E, lanes 8 and 9), indicating that hMCPH1’s ability to inhibit chromosomal loading of condensin II in Xenopus egg extracts is far more specific than okadaic acid.
The N-terminal domain of hMCPH1 competes for chromosome binding of condensin II in Xenopus egg extracts
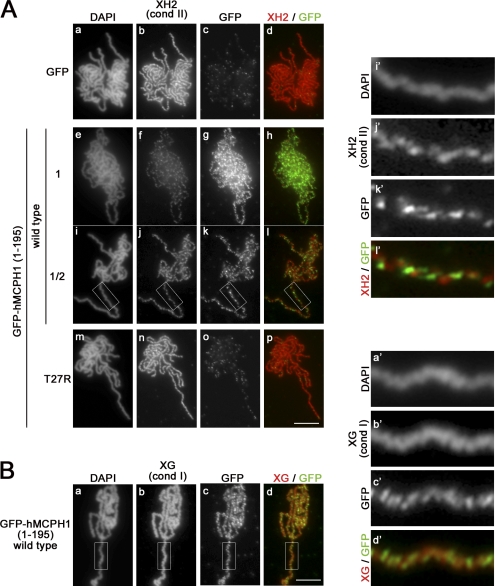
To understand the mechanism by which hMCPH1 inhibits the action of condensin II, we wished to test the possibility that the N-terminal domain of hMCPH1 might compete for binding sites of condensin II on chromosomes. Consistent with this idea, the T27R mutant not only poorly inhibited condensin II but also poorly associated with chromosomes as judged by immunoblotting of chromosomal factions (Fig. 2 C). In experiments shown in Fig. 3, we attempted double immunofluorescent labeling of condensin II and the N-terminal domain of hMCPH1 (a GFP-tagged version) on chromosomes. As expected, GFP alone or the T27R mutant of GFP-hMCPH1 (amino acids 1–195) produced only a background level of its own signals on chromosomes and did not affect loading of condensin II (Fig. 3 A, a–d and m–p). On the other hand, GFP-hMCPH1 (amino acids 1–195) associated with chromosomes and inhibited loading of condensin II in a dose-dependent manner (Fig. 3 A, e–h and i–l). Remarkably, we noticed that condensin II and GFP-hMCPH1 (amino acids 1–195) apparently distributed along chromosomes in a mutually exclusive manner (Fig. 3 A, i′–l′). Under the same condition, we found virtually no sign for displacement of condensin I by GFP-hMCPH1 (amino acids 1–195) exogenously added into the extract (Fig. 3 B). These results suggest that the N-terminal domain of hMCPH1 inhibits loading of condensin II by competing for its binding sites on chromosomes.
Figure 3.
The N-terminal domain of hMCPH1 competes for chromosome binding of condensin II in Xenopus egg extracts. (A) A reticulocyte lysate containing GFP or GFP-hMCPH1 (amino acids 1–195; wild type and T27R) was mixed with 10 vol metaphase egg extracts and incubated for 30 min. Sperm chromatin was then added and incubated for another 120 min. Metaphase chromosomes assembled were fixed and stained with DAPI (a, e, i, and m), anti–XCAP-H2 (XH2; b, f, j, and n), and anti-GFP (GFP; c, g, k, and o). To reduce the dose of GFP-hMCPH1 (amino acids 1–195) to 50% (i–l), the reticulocyte lysate containing the wild-type protein was diluted twofold with a mock lysate before being mixed with the egg extracts. Close-ups of chromosomal regions indicated by the white rectangles in i–l are shown in i′–l′. (B) Metaphase chromosomes assembled in the presence of GFP-hMCPH1 (amino acids 1–195; wild type) as described in A were fixed and stained with DAPI (a), anti–XCAP-G (XG; b), and anti-GFP (GFP; c). Close-ups of chromosomal regions indicated by the white rectangles in a–d are shown in a′–d′. Bars, 5 µm. cond, condensin.
Characterization of Xenopus MCPH1 (xMCPH1) and mouse MCPH1 (mMCPH1) in the cell-free assay
The specific inhibition of condensin II by exogenously added hMCPH1 in the cell-free assay raises the question of whether Xenopus egg extracts might contain endogenous MCPH1. To explore this possibility, we prepared specific antibodies against recombinant fragments of xMCPH1 and found that the extracts indeed contained an appreciable level of endogenous xMCPH1, which was estimated to be comparable with the standard dose of hMCPH1 added exogenously (Fig. S2). We reasoned that, if xMCPH1 also had an activity to inhibit condensin II, depletion of endogenous xMCPH1 from an extract might cause hyperactivation and overloading of condensin II in the cell-free assay. It was found, however, that the presence or absence of endogenous xMCPH1 made little, if any, difference in the morphology of chromosomes or the level of condensin II loaded on them (Fig. 4, A and B).
Figure 4.
Characterization of xMCPH1 and mMCPH1 in the cell-free assay. (A) Sperm chromatin was added to metaphase egg extracts that had been depleted with control IgG (Δmock; lanes 1 and 3) or anti-xMCPH1 (ΔxMCPH1; lanes 2 and 4). After incubation for 120 min, chromosomes were isolated, and their associated polypeptides (lanes 3 and 4) and aliquots of the extracts (lanes 1 and 2) were analyzed by immunoblotting using the antibodies indicated. (B) Metaphase chromosomes were assembled in the mock-depleted (Δmock; a–d) or xMCPH1-depleted (ΔxMCPH1; e–h) extracts, fixed, and double stained with anti–XCAP-H2 (XH2) and anti–XCAP-G (XG). Bulk chromosomal DNA was counterstained with DAPI. (C) A reticulocyte lysate containing no hMCPH1 (−) or FLAG-tagged full-length MCPH1 from human, mouse, and Xenopus was mixed with 10 vol metaphase egg extracts that had been depleted with control IgG (Δmock; rows a–d) or anti-xMCPH1 (ΔxMCPH1; rows e–h) and incubated for 30 min. Sperm chromatin was then added and incubated for another 120 min. The resulting metaphase chromosomes were analyzed as in B. (D) Increasing concentrations of the N-terminal (N-ter.) domains of human, mouse, and Xenopus MCPH1 were mixed with metaphase egg extracts (lanes 1–13) and tested for their ability to inhibit chromosomal loading of condensin (cond) II. The relative concentrations of MCPH1 added were 1:8 (lanes 2, 6, and 10), 1:4 (lanes 3, 7, and 11), 1:2 (lanes 4, 8, and 12), and 1 (lanes 5, 9, and 13); the highest concentration roughly corresponded to the standard one used in other experiments. Sperm chromatin was incubated with these extracts, and chromosome-bound fractions (lanes 14–26) were isolated and analyzed by immunoblotting. Bars, 5 µm.
MCPH1 is a rapidly evolving protein and is implicated in the expansion of brain size during vertebrate evolution (Ponting and Jackson, 2005). It would therefore be of considerable interest if MCPH1 proteins from different vertebrate species were subjected to a comparative study in this particular cell-free assay. To this end, we added the same dose of full-length MCPH1 from human, mouse, and Xenopus into a control extract and an extract depleted of endogenous xMCPH1 (Fig. S2). Intriguingly, we found that, compared with hMCPH1, mMCPH1 or xMCPH1 displayed a much weaker activity to inhibit chromosomal loading of condensin II either in the presence (Fig. 4 C, rows a–d) or absence (Fig. 4 C, rows e–h) of endogenous xMCPH1. Similar results were obtained when the N-terminal domains of hMCPH1, mMCPH1, and xMCPH1 were used. In the experiment shown in Fig. 4 D, the dose of the N-terminal domains of MCPH1 added into metaphase extracts was titrated down starting from the standard dose. The N-terminal domain of hMCPH1 displayed a dose-dependent inhibition of condensin II loading (Fig. 4 D, lanes 15–18), and substantial inhibition was detectable with as low as 25% of the standard dose (Fig. 4 D, lane 16). In contrast, a very low level of inhibition was observed even with the highest dose of the mouse or Xenopus counterpart (Fig. 4 D, lane 22 or lane 26). Although the observation that xMCPH1 has a poor activity to inhibit condensin II in the cell-free assay was somewhat puzzling, it provides a reasonable explanation for why depletion of endogenous xMCPH1 from the extract had little impact on the behavior of condensin II (Fig. 4, A and B).
mMCPH1 can be “humanized” by specific amino acid substitutions
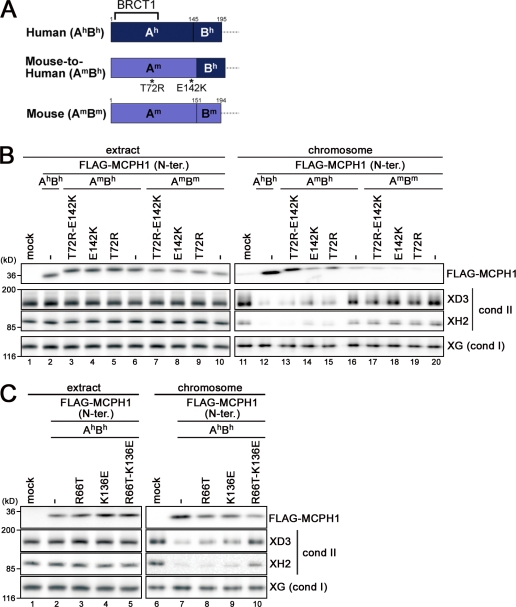
To substantiate the findings described in the previous section, we sought to identify the determinants that would differentiate the activities between hMCPH1 and mMCPH1. Because the N-terminal domains of hMCPH1 and mMCPH1 were relatively divergent in their last quarters, each domain was divided into two subdomains, A (Ah, amino acids 1–145 in hMCPH1; Am, amino acids 1–151 in mMCPH1) and B (Bh, amino acids 146–195 in hMCPH1; Bm, amino acids 152–194 in mMCPH1), and a chimera protein, referred to as AmBh, was created (Fig. 5 A and Fig. S1). We found that AmBh bound to chromosomes slightly better than the authentic mouse sequence (AmBm) but failed to acquire an appreciable level of activity to inhibit condensin II loading (Fig. 5 B, lane 16). Because the simple substitution of the subdomain B was not enough to confer the hMCPH1-like activity on mMCPH1, we took a brute force mutagenesis approach by focusing on nonconserved residues in the subdomain A. A list of substituted residues (singly or in combinations) is shown in Fig. S3 A. In short, we found that mMCPH1 acquired an activity comparable with that of hMCPH1 when two mouse to human substitutions, T72R and E142K, were combined with the subdomain B substitution (Fig. 5 B, compare lanes 12 and 13). A double substitution alone (T72R-E142K) displayed little, if any, effect (Fig. 5 B, lane 17), indicating that successful conversion of the mouse sequence into one with a humanlike activity requires the combination of all three substitutions. Conversely, when two human to mouse substitutions were introduced into the corresponding residues of hMCPH1 (i.e., R66T-K136E), the activity associated with the human protein was greatly reduced (Fig. 5 C). Thus, the cell-free assay successfully allowed us to identify a subset of specific amino acid substitutions that contributes to the functional difference between hMCPH1 and mMCPH1.
Figure 5.
mMCPH1 can be converted into a humanlike, competent form by specific amino acid substitutions. (A) Schematic representation of the constructs used in B. The N-terminal domain of hMCPH1 was divided into two subdomains, Ah (amino acids 1–145) and Bh (amino acids 146–195), and the corresponding domain of mMCPH1 was divided into Am (amino acids 1–151) and Bm (amino acids 152–194), accordingly. A chimera form (AmBh) was created, and single or double point mutations (T72R and E142K) were further introduced in an attempt to confer a humanlike activity on the mouse sequence (mouse to human). (B) Mouse to human conversion. The constructs shown in A were subjected to the cell-free assay. (C) Human to mouse conversion. Two point mutations (R66T and K136E) were introduced singly or doubly into the human sequence, and the resulting constructs were subjected to the cell-free assay. cond, condensin; N-ter., N-terminal; XG, XCAP-G; XH2, XCAP-H2.
Robust interaction with condensin II through its central domain is not required for hMCPH1’s inhibitory activity in the cell-free assay
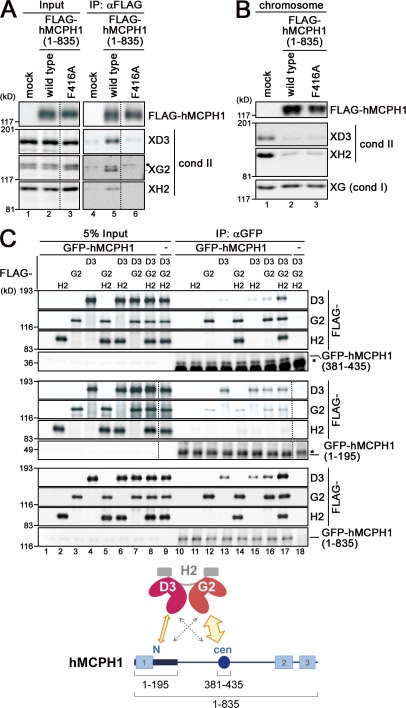
We then tested for physical interactions between hMCPH1 and condensin II. In HeLa cell nuclear extract, endogenous hMCPH1 and human condensin II specifically interacted with each other (Fig. S4, A and B). By extending the previous work by Wood et al. (2008), we found that a central domain of hMCPH1 (amino acids 381–435) was primarily responsible for the condensin II–hMCPH1 interaction (Fig. S4, C and D) and further identified a pair of point mutations (Y412A and F416A) in the central domain that disrupts this interaction (Fig. S4 E). Consistently, full-length hMCPH1 added into Xenopus extracts interacted with endogenous condensin II, and such an interaction was largely abolished when the F416A mutation was introduced into hMCPH1 (Fig. 6 A). The F416A mutant protein, however, was found to retain a full activity to inhibit condensin II loading in the cell-free assay (Fig. 6 B). This observation was surprising at first glance, yet it was consistent with our result that the N-terminal region (amino acids 1–195) of hMCPH1 is sufficient for condensin II inhibition (Fig. 2).
Figure 6.
hMCPH1 physically interacts with condensin II through its N-terminal and central domains. (A) A reticulocyte lysate containing no hMCPH1 (mock) or FLAG-tagged hMCPH1 was mixed with 10 vol metaphase egg extracts and incubated for 60 min. Anti-FLAG beads were then added into the mixtures, and bound fractions and aliquots of the mixtures were analyzed by immunoblotting. Different levels of contrast adjustment were performed between the input and immunoprecipitation (IP) blots. The FLAG data are derived from a different membrane than the condensin (cond) II data. Nonspecific bands that cross reacted with anti–XCAP-G2 (XG2) are indicated by the asterisk. (B) Sperm chromatin was incubated with the mixtures as described in A for 120 min, and chromosome fractions were isolated and analyzed by immunoblotting. (C) FLAG-tagged non-SMC subunits of human condensin II (CAP-D3, -G2, and -H2) were translated individually in reticulocyte lysates and mixed in different combinations as indicated. Three different versions of GFP-tagged hMCPH1 were produced in different reactions, incubated with the aforementioned mixtures, and subjected to immunoprecipitation with anti-GFP. The immunoprecipitates and 5% of input fractions were analyzed by immunoblotting with anti-FLAG and anti-GFP. FLAG data are derived from a different membrane than the GFP data for the top and middle datasets. The asterisks indicate nonspecific bands. The major binding partners of the central (cen; amino acids 381–435) and N-terminal (N; amino acids 1–195) domains of hMCPH1 are CAP-G2 and -D3, respectively (cartoon). The dotted lines indicate where intervening lanes were removed for presentation purposes. XH2, XCAP-H2.
We then sought the possibility that the N-terminal domain of hMCPH1 might possess a minor condensin II–interacting activity independently of its central region. We also wished to identify which subunits of condensin II might be responsible for interactions with hMCPH1. To test this, the three regulatory subunits of human condensin II (CAP-D3, -G2, and -H2) were individually translated in vitro, unmixed or mixed in all possible pairwise combinations, and processed for subunit–subunit interaction assays with full-length or truncated versions of hMCPH1 (Fig. 6 C). In short, we found that the central domain (amino acids 381–435) of hMCPH1 coprecipitated predominantly with CAP-G2, whereas the N-terminal domain (amino acids 1–195) did primarily with the D3 subunit (Fig. S4 F). Together with our previous data showing no direct interaction between CAP-G2 and -D3 subunits (Onn et al., 2007), we conclude that two separate domains of hMCPH1 are capable of interacting with different HEAT subunits of condensin II (Fig. 6 C, cartoon). It should be noted, however, that the interaction between the N-terminal domain of hMCPH1 and CAP-D3 is rather cryptic and not readily detectable in the context of the condensin II holocomplex (Fig. 6 A and Fig. S4 D).
Complementation assay reveals a contribution of the central domain of hMCPH1 to shaping metaphase chromosomes
Finally, we wished to understand to what extent the information obtained from the cell-free assay might be relevant to the in vivo function of hMCPH1. To this end, we set up a complementation assay in which various constructs of hMCPH1 were expressed in MCPH1 patient cells bearing a homozygous truncating mutation by means of a lentivirus vector. In the first set of experiments, GFP-tagged versions of full-length hMCPH1 were introduced into the patient cells, and their expression was confirmed by immunoblotting against total cell lysates (Fig. 7 A). The frequency of prophaselike cells (PLCs) in each cell population was scored (Fig. 7 B; also see Materials and methods). We found that wild-type hMCPH1 rescued the PCC phenotype very efficiently (Fig. 7 B and see Fig. S5 [A and B] for representative images). On the other hand, an hMCPH1 construct with the mild allele T27R failed to fully restore the defective phenotype (Trimborn et al., 2005). Notably, a construct with the F416A mutation displayed a full activity to restore the PCC phenotype, suggesting that the interaction with condensin II mediated through the central domain of hMCPH1 is dispensable for rescuing the PCC phenotype.
Figure 7.
The N-terminal domain of hMCPH1 is sufficient to rescue the PCC phenotype, whereas its central domain is additionally required for shaping metaphase chromosomes in patient cells. (A) MCPH1 patient cells were transduced with GFP-tagged full-length hMCPH1 by means of a lentivirus expression system. Mock-transduced cells and cells transduced with GFP alone were used as negative controls. Lysates were prepared from these cells and analyzed by immunoblotting with the indicated antibodies. (B) The cells described in A were fixed and stained with DAPI. The percentages of prophaselike cells (PLCs) in such populations were scored and plotted (*, P < 0.05; ***, P < 0.001 between each pair; PLCs were scored from three independently prepared coverslips [n = 3]). Representative examples of nuclear morphology in cells expressing GFP alone or GFP-hMCPH1 (wild type) are shown in Fig. S5 (A and B). By definition, the PLCs include not only G2 cells displaying PCC but also early G1 cells displaying delayed chromosome decondensation (Trimborn et al., 2006). In the current study, we follow this definition for the analysis of PLCs. However, when the percentages of the PLCs in patient cells were lowered to the basal level, we describe it as a “PCC rescue” for simplicity. (C) Chromosome spreads were prepared from the cells described in A and stained with DAPI and anti-Smc2. The morphology of chromosomes from 20 spreads was classified into three categories, and each sample was scored accordingly. (a) “Dumpy” is a category of short chromosomes with wavy axes that are commonly observed in the MCPH1 patient cells. (c) “Straight” is a category of long chromosomes with straight axes that are routinely observed in nonpatient cells. (b) “Intermediate” is a category belonging between dumpy and straight. Representative images of each category (stained with anti-Smc2) are shown at the top. Representative data from two independent experiments are shown. Bar, 5 µm. (D) MCPH1 patient cells were transduced with GFP alone or the GFP-tagged N-terminal domain of hMCPH1. Lysates were prepared from these cells and analyzed by immunoblotting as described in A. (E and F) The percentages of PLCs and the morphology of chromosome spreads were scored as described in B and C, respectively. For F, representative data from two independent experiments are shown. (G) A postulated change of condensin (Cond) II activity is shown by the red line. To prevent PCC in G2 phase, the N-terminal (N) domain of MCPH1 is sufficient. To properly shape metaphase chromosomes, both the N-terminal and central (cen) domains are required. Error bars indicate means ± SD.
To study whether hMCPH1 might impact the morphology of metaphase chromosomes, chromosome spreads were prepared and stained with an antibody against Smc2, a core subunit shared by both condensin I and II. In MCPH1 patient cells (either mock treated or transduced with GFP alone), most of metaphase chromosomes displayed an abnormal morphology with very short and thick chromatids. When these so-called “dumpy” chromosomes were stained with anti-Smc2, chromatid axes with a characteristic wavy appearance could be visualized (Fig. 7 C, a and graph, bars 1 and 2). When the patient cells expressing GFP-hMCPH1 were analyzed, the dumpy phenotype was barely observed; ∼80% of chromosomes in the sample displayed a more typical morphology with long, thin, and straight chromatids (Fig. 7 C, c and graph, bar 3). As expected, expression of the T27R mutant protein failed to rescue the dumpy phenotype effectively (Fig. 7 C, bar 4). In contrast, somewhat unexpected was the observation that the F416A protein rescued the phenotype only partially (Fig. 7 C, bar 5).
We then asked whether expression of the N-terminal domain (amino acids 1–195) of hMCPH1 was sufficient to rescue the PCC and/or dumpy phenotypes (Fig. 7 D). The wild-type N-terminal domain efficiently rescued the PCC phenotype, whereas the corresponding domain containing the T27R mutation displayed a partial rescue (Fig. 7 E), an observation similar to that made with the full-length proteins (Fig. 7 B). Two additional constructs (amino acids 196–835 and 638–835) lacking the N-terminal domain failed to rescue the PCC phenotype (Fig. S5, C–E). We noticed, however, that expression of the N-terminal domain alone was not sufficient to fully rescue the dumpy phenotype in patient cells (Fig. 7 F). Collectively, the current results suggest that the N-terminal domain of hMCPH1 is sufficient to rescue the PCC phenotype but that both the N-terminal and central domains are required for properly shaping metaphase chromosomes.
Discussion
In the current paper, we have established a powerful combination of the cell-free and complementation assays to study the functional and physical interactions between MCPH1 and condensin II. In particular, the newly developed cell-free assay provides us with a simple and semiquantitative method for studying the action of MCPH1 in vitro, thereby opening a new avenue in the research field of primary microcephaly.
hMCPH1 specifically inhibits condensin II in Xenopus egg extracts
Previous work showed that depletion of condensin II from MCPH1 patient cells relieved chromosome condensation defects, leading to the proposal that MCPH1 might act as a negative regulator of condensin II (Trimborn et al., 2006). It has remained unclear, however, whether MCPH1 directly inhibits condensin II or indirectly down-regulates its activity by perturbing its upstream cell cycle regulators (e.g., Alderton et al., 2006; Tibelius et al., 2009). The cell-free assay established in the current study utilizes a Xenopus egg extract whose cell cycle state is stably maintained at metaphase (Morgan, 2007). Our current results therefore provide the most compelling evidence available, so far, that hMCPH1 has the ability to inhibit the action of condensin II independently of the parameter of cell cycle progression. We also show that the N-terminal domain (amino acids 1–195) of hMCPH1 is required and sufficient for this inhibitory activity. Importantly, the mutations identified in MCPH1 patients (T27R and W75R) compromise the ability of hMCPH1 to inhibit condensin II in the cell-free assay, arguing that loss of this inhibitory activity (hence, misregulation of condensin II) might directly be relevant to the etiology of MCPH1 microcephaly.
Then, mechanistically, how might the N-terminal domain of hMCPH1 inhibit the action of condensin II? Although many, if not all, BRCT domains are known to function as phosphopeptide-binding motifs (Glover et al., 2004), hMCPH1’s BRCT1 does not possess key residues involved in such molecular recognition. It is therefore unlikely that hMCPH1’s action on condensin II involves phosphopeptide binding. Instead, our results show that the N-terminal domain of hMCPH1 associates with chromosomes and effectively competes for binding sites of condensin II in the cell-free assay. In fact, analyses of the T27R mutation causing primary microcephaly in humans (Figs. 2 and 3) and evolutionary substitutions among different organisms (Figs. 4 and 5) further confirm and establish that the chromosome-binding activity of this domain is tightly coupled to its condensin II inhibitory activity. The potential mechanism by which condensin II is targeted to chromosomes remains to be determined (Hirano, 2005). We anticipate that hMCPH1 will be an excellent tool for addressing this important problem in the future. Moreover, it should be added that the weak interaction observed between the N-terminal domain of hMCPH1 and the CAP-D3 subunit (Fig. 6) might also contribute, at least in part, to specific inhibition of condensin II.
hMCPH1 regulates chromosome condensation and shaping in human cells
The morphology of metaphase chromosomes is determined by a delicate balance of actions between condensin I and condensin II (Ono et al., 2003; Shintomi and Hirano, 2011). It is reasonable to speculate that the loss of function of hMCPH1 would disturb such a balance, thereby leading to the formation of dumpy chromosomes. Our current data suggest that the central domain of hMCPH1 contributes to proper shaping of metaphase chromosomes by using its ability to associate with condensin II (Fig. 7 G). Although it remains to be determined how the central domain might collaborate with the N-terminal domain to execute this function, one possibility would be that combined actions of the two domains help increase the mobility of condensin II and thereby promote its proper distribution along chromosomes. Thus, rather than being a simple inhibitor, MCPH1 is likely to act as a composite modulator of condensin II that fine tunes its function and distribution, presumably throughout the cell cycle. In fact, condensin II’s action is not limited to mitosis and contributes to a wide range of interphase chromosome functions (Gosling et al., 2007; Hartl et al., 2008). Condensin II therefore must have a basal level of activity to fulfill these nonmitotic functions within the interphase nucleus. We infer that MCPH1 keeps this basal activity of condensin II below a certain threshold during G2 phase, thereby preventing PCC from occurring before mitotic entry (Fig. 7 G). This idea would explain why a low level of MCPH1 activity (e.g., T27R) is able to partially suppress PCC during G2 phase in human cells but barely inhibits a fully activated form of condensin II in metaphase egg extracts. Similarly, although mMCPH1 possesses a low level of activity as judged by the cell-free assay (see the following section), it seems sufficient to suppress PCC in mouse cells (Wood et al., 2008; Trimborn et al., 2010).
Evolutionary implications
MCPH1 is a rapidly evolving protein (Ponting and Jackson, 2005). The current study is the first to compare the activities of MCPH1 from different vertebrates in a single functional assay in vitro. Unexpectedly, we find that mMCPH1 or xMCPH1 displays a substantially lower level of activities compared with hMCPH1. Assessing MCPH1’s ability to inhibit condensin II in the cell-free assay is technically straightforward and highly reproducible, having allowed us to quickly identify a specific combination of amino acid substitutions that confer the functional difference between hMCPH1 and mMCPH1. Notably, one of the key residues identified (R66 in humans) is absolutely invariable among primates, whereas another key residue (K136 in humans) and subdomain B are more variable even among primates (Fig. S3 B). Apart from these residues, it was noticed that only two residues (M96 and S101) are unique to humans in the N-terminal domain of MCPH1. To test the possibility that these two human-specific residues might confer the high inhibitory activity on hMCPH1, we substituted them with the nonhuman-type residues. It was found, however, that the resulting mutant protein (M96T-S101P) retained the same level of inhibitory activity as wild-type hMCPH1 in the cell-free assay (Fig. S3 C).
To what extent might the functional difference between hMCPH1 and mMCPH1 observed in the cell-free assay be physiologically significant? We are fully aware that one needs to be very cautious about discussing functional evolution of MCPH1 on the basis of limited information currently available and that any hypotheses must vigorously be validated in vivo. Nevertheless, it seems reasonable to argue that the cell-free assay described here will be instrumental in addressing the evolutionary aspect of this class of proteins in future studies.
Beyond chromosome condensation: might condensin II function in brain development?
The etiology of MCPH1 primary microcephaly is not fully understood (Thornton and Woods, 2009). Remarkably, we find that the mutations identified in MCPH1 patients (T27R and W75R) compromise the ability of hMCPH1 to inhibit condensin II in our cell-free assay. These results suggest that loss of this inhibitory activity might indeed be relevant to the etiology of MCPH1 microcephaly. Then, how might misregulation of condensin II potentially be linked to microcephaly in humans? We consider two possibilities, both of which are admittedly highly speculative at present. First, it is possible that MCPH1 mutations cause up-regulation of condensin II, which in turn perturbs the program of gene expression supporting normal development of the brain. Consistent with this notion, accumulating lines of evidence suggest that condensin II may play a direct role in gene regulation (Hartl et al., 2008) and/or cell differentiation (Gosling et al., 2007). Second, condensin II might function, along with MCPH1, as a component of the signaling network that responds to DNA damage (Alderton et al., 2006; Tibelius et al., 2009). In this sense, it is interesting to note that condensin II has been hypothesized to be a potential target of the surveillance system (the so-called antephase checkpoint) that helps reverse an early phase of chromosome condensation and prevents mitotic entry in response to a range of cellular stresses (Hirano, 2005; Chin and Yeong, 2010). Perturbation of such putative checkpoint functions could specifically affect the unique mode of cell divisions during neurogenesis (Thornton and Woods, 2009). In any case, to fully understand the physiological functions of MCPH1, we need to learn more about its molecular functions. The current study paves the way toward this goal by demonstrating that MCPH1 functions as a specific modulator of condensin II, thereby establishing a potential link between chromosome condensation and brain development in vertebrates.
Materials and methods
Antibodies
Rabbit antisera were raised against a C-terminal recombinant fragment (amino acids 378–835) of hMCPH1. We also prepared rabbit polyclonal antisera against two different recombinant fragments (amino acids 118–328 and 458–791) of Xenopus mcph1. All recombinant proteins were expressed from the pRSETA vector (Invitrogen) in Escherichia coli as hexahistidine (His6) fusions. Soluble proteins were purified on a Ni-Sepharose 6 Fast Flow (GE Healthcare), whereas insoluble proteins were purified by electroelution after SDS-PAGE. For affinity purification of rabbit antibodies (Hirano et al., 1997), a rabbit antiserum was passed over a column packing Affi-Gel (Bio-Rad Laboratories) coupled to recombinant protein three times, and the column was washed initially with TBS (20 mM Tris-HCl, pH 7.5, and 0.15 M NaCl) and then with TBS containing 0.5 M NaCl and 0.2% Triton X-100 and again with TBS. The IgG bound to the column was then eluted with 0.2 M glycine-HCl, pH 2.0, containing 0.15 M NaCl. Peak fractions were pooled, dialyzed against TBS, and then used as purified antibodies. Other antibodies used in this study were described previously (Hirano et al., 1997; Losada et al., 2002; Ono et al., 2003). The following antibodies were obtained from commercial sources: anti–α-tubulin (clone DM1A; Sigma-Aldrich), anti-GFP (rabbit polyclonal, ab6556; Abcam), anti-GFP (mouse monoclonal, ab38689; Abcam), and anti-FLAG M2 (F3165; Sigma-Aldrich).
Plasmid construction and mutagenesis
For expression of GFP-tagged hMCPH1 (or its truncated forms) in 293T cells, the pEGFP-C2 vector (Takara Bio Inc.) was used. For in vitro transcription/translation reactions, the pTnT vector (Promega), a pTnT derivative with a 3×FLAG sequence (Shintomi and Hirano, 2009), and a pTnT derivative with a GFP-encoding sequence were used. For lentiviral expression, a vector (GFP.IN.pCL1THPC) was used that expresses GFP and the neomycin-resistant gene under the control of the spleen focus-forming virus U3 promoter. Point mutations were introduced with a site-directed mutagenesis kit (QuikChange XL; Agilent Technologies).
The cell-free assay using Xenopus egg extracts
Metaphase egg extracts and demembranated sperm chromatin were prepared as described previously (Hirano et al., 1997). In brief, low-speed supernatants were prepared by crushing the unfertilized eggs of Xenopus in XBE2 buffer (10 mM K-Hepes, pH 7.7, 0.1 M KCl, 2 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA, and 50 mM sucrose) and further fractionated by centrifugation at 50,000 rpm at 4°C for 120 min in a rotor (TLS55; Beckman Coulter) in an ultracentrifuge (Optima TLX; Beckman Coulter). The soluble fractions were collected and centrifuged again at 50,000 rpm for 30 min. The resulting supernatants (i.e., high-speed supernatants) were used as metaphase egg extracts. Sperm chromatin was prepared in SMH buffer (20 mM K-Hepes, pH 7.5, 2 mM MgCl2, and 250 mM sucrose) containing 0.4% BSA and 30% glycerol. For biochemical analysis, metaphase extracts were diluted twofold with XBE8 (XBE2 containing 8 mM rather than 2 mM MgCl2), and sperm chromatin was added at a final concentration of 4,000 nuclei/µl (Ono et al., 2003). After incubating at 22°C for 120 min, the mixtures were overlaid on a 30% sucrose cushion made in XBE2 and spun at 13,000 rpm for 15 min at 4°C in a rotor (TMS-21; TOMY) in a microfuge (MX-305; TOMY). After removing soluble fractions, chromosome fractions recovered in the pellets were analyzed by immunoblotting. For morphological analysis, metaphase extracts were mixed with sperm chromatin at a final concentration of 1,000 nuclei/µl. After incubating at 22°C for 120 min, the assembly mixtures were fixed with 10 vol of 2% formaldehyde in XBE2 containing 0.1% Triton X-100 for 10 min at room temperature and centrifuged at 7,000 rpm in a swinging-bucket rotor (JS13.1; Beckman Coulter) in a centrifuge (Avanti HP-25; Beckman Coulter) for 15 min onto coverslips through a 30% glycerol cushion made in XBE2. Chromosomes spun on the coverslips were processed for immunofluorescence as described previously (Ono et al., 2003). In brief, the coverslips were blocked with 1% BSA in TBS containing 0.1% Triton X-100 and then incubated with the primary antibodies. Alexa Fluor 568– and Alexa Fluor 488–labeled IgG (Invitrogen) were used as secondary antibodies. The samples were counterstained with DAPI, mounted on slides with Vectashield (Vector Laboratories), and observed under a microscope (BX51; Olympus) with an UPlanSApo 100×/1.40 NA oil objective lens (Olympus). All grayscale images were obtained by a cooled charge-coupled device camera (DP30; Olympus) with DP Controller software (Olympus), pseudocolored, and merged using Photoshop (Adobe). To produce recombinant proteins in reticulocyte lysates, the TNT Quick Coupled Transcription/Translation System (Promega) was used. A typical reaction mixture (50 µl) containing 1 µg plasmid DNA was supplemented with 20 µM methionine and incubated at 30°C for 90 min.
Immunoprecipitation
For in vitro subunit–subunit interaction assays, reticulocyte lysates containing recombinant proteins were diluted 10-fold with a dilution buffer (20 mM K-Hepes, pH 7.7, 100 mM KCl, 2.5 mM MgCl2, 0.1% Tween 20, and 0.5 mg/ml BSA), rotated at 4°C for 60 min, and spun at 13,000 rpm in a microfuge at 4°C for 10 min. The supernatants were incubated with 1 µg affinity-purified antibody on ice for 60 min. Immunoprecipitates were recovered on 10 µl rProtein A Sepharose beads (GE Healthcare), washed three times with the dilution buffer and once with the dilution buffer without Tween 20 and BSA, and analyzed by immunoblotting. For the experiment shown in Fig. 6 A, egg extracts were supplemented with 0.1 vol reticulocyte lysates and incubated at 22°C for 60 min. The mixtures were incubated with 5 µl FLAG M2 beads (Sigma-Aldrich) at 4°C for 60 min with occasional mixing. The beads were washed three times with KMH (20 mM K-Hepes, pH 7.7, 100 mM KCl, and 2 mM MgCl2) containing 0.1% Triton X-100, twice with KMH, and subjected to SDS-PAGE.
Transduction of MCPH1 patient cells with recombinant lentiviruses
SV40-transformed fibroblasts were derived from a patient with a homozygous MCPH1 truncating mutation (p.Thr143AsnfsX5; Trimborn et al., 2006). The lentiviral system used in this study was designed to express GFP-tagged hMCPH1 under the control of the spleen focus-forming virus U3 promoter. All recombinant lentiviruses were produced by transient transfection of 293FT cells as follows. Subconfluent 293FT cells were cotransfected with GFP.IN.pCL1THPC containing an hMCPH1 insert, pCAG-HIVgp, and pCMV-VSV-G-RSV-Rev (Miyoshi et al., 1998; the latter two plasmids were provided by H. Miyoshi, RIKEN BioResource Center, Tsukuba, Ibaraki, Japan) by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After overnight incubation, the culture medium was replaced with a fresh one, and the cell cultures were continued for another 36 h. Culture supernatants containing recombinant lentiviruses were then collected and used to transduce MCPH1 patient cells. 2 d after the transduction, cell populations resistant to G418 (at a final concentration of 250 µg/ml) were selected for 5 d before being subjected to the analyses of PLCs and chromosome spreads. For immunoblotting analysis of total lysates, 2.5 × 105 cells were resuspended in 50 µl SDS sample buffer and heat denatured for 5 min, and a 5-µl aliquot was subjected to SDS-PAGE.
Analysis of PLCs
Cells were grown on coverslips coated with poly-l-lysine, fixed with 2% paraformaldehyde in PBS, pH 7.4, for 15 min, permeabilized in 0.5% Triton X-100 in PBS for 5 min at room temperature, and then stained with DAPI. The coverslips were mounted on slides with Vectashield. The cell images were captured with the high-throughput imaging device (CELAVIEW-RS100; Olympus) with LUCPlanFLN 20×/0.45 NA objective lens (Olympus). To refine the analysis of PLCs, cell populations displaying nuclear GFP signals above a certain threshold were selected from G418-resistant cells by using the CELAVIEW software (Olympus). According to this strategy, ∼20% of total cells were judged as GFP positive in most cases. Approximately 1,000 cells were chosen randomly from this GFP-positive population, and mitotic cells, except those in prophase, were removed manually. PLCs were then judged by DAPI stain, and its percentage was scored. The statistical significance in mean values among multiple sample groups was examined with Tukey’s multiple comparison test after one-way analysis of variance test. Values shown in Figs. 7 (B and E) and S5 E represent the means ± SD.
Preparation of chromosome spreads
Control and transduced cells were treated with colcemid at a final concentration of 0.02 µg/ml for 3 h before mitotic cells were collected by tapping culture dishes. The collected cells were then treated with 75 mM KCl at 37°C for 30 min, and centrifuged onto poly-l-lysine–coated coverslips at 1,000 rpm for 10 min in a cytocentrifuge (Cytospin 2; Thermo Fisher Scientific). The chromosome spreads were fixed and permeabilized as described in the previous section, blocked with 3% BSA in PBS, and incubated for 2 h with an anti-Smc2 antibody at a final concentration of 2 µg/ml. Alexa Fluor 568–labeled goat anti–rabbit IgG (Invitrogen) was used as a secondary antibody at a 1:500 dilution. The samples were counterstained with DAPI and mounted on slides with Vectashield. The images of ∼20 chromosome spreads of each sample were obtained with the microscope (BX51) as described in The cell-free assay using Xenopus egg extracts section, and the phenotypes were categorized as shown in Fig. 7 (C and F).
Extract preparation and immunoprecipitation of human cells
For siRNA-mediated depletion of hMCPH1, HeLa cells were transfected twice with 100 nM siRNA duplexes (Thermo Fisher Scientific) by means of Oligofectamine (Invitrogen) at 24 and 48 h after seeding. Control cells were transfected with a transfection mixture containing no siRNAs. The sequences of the sense strand of the siRNA duplexes were as follows: sihMCPH1 #1, 5′-AGGAAGUUGGAAGGAUCCAdTdT-3′; and sihMCPH1 #2, 5′-CUCUCUGUGUGAAGCACCUdTdT-3′ (in which dT stands for deoxythymidine). For immunoblotting analysis of total lysates from HeLa cells, 2 × 105 cells were suspended in 50 µl of SDS sample buffer and heat denatured for 5 min. A 5-µl aliquot was resolved by SDS-PAGE and analyzed by immunoblotting. HeLa nuclear extracts were prepared in buffer B (20 mM K-Hepes, pH 8.0, 100 mM KCl, 2 mM MgCl2, 0.2 mM EDTA, 20% glycerol, 1 mM β-mercaptoethanol, and 0.5 mM PMSF) as described previously (Losada et al., 2000). In brief, asynchronously growing HeLa cells were hypotonically treated, homogenized, and centrifuged. The pellet containing nuclei was extracted with buffer B containing 300 mM KCl, homogenized, sonicated, and centrifuged at 16,500 rpm at 4°C for 30 min in a rotor (SS34; Sorval). The supernatant was dialyzed against buffer B and clarified by another centrifugation at 16,500 rpm for 20 min, and then, the resulting supernatant was used as a HeLa nuclear extract. For immunoprecipitation, 20 µl of the extracts was incubated with 1 µg affinity-purified antibody at 4°C for 60 min with occasional mixing. Immunoprecipitates were recovered on 10 µl rProtein A Sepharose beads, washed three times with buffer B supplemented 0.1% NP-40 and twice with buffer B, and analyzed by immunoblotting. For immunoprecipitation from total cell extracts, 293T cells transiently expressing GFP-hMCPH1 were resuspended in a lysis buffer (20 mM K-Hepes, pH 7.7, 100 mM KCl, 0.1% NP-40, 2 mM MgCl2, 10% glycerol, 40 mM β-glycerophosphate, 0.5 mM PMSF, and 10 µg/ml each of leupeptin, chymostatin, and pepstatin A) at a concentration of 107 cells/ml, incubated at 4°C for 30 min, sonicated briefly, and centrifuged at 13,000 rpm in a microfuge for 10 min. 1 µg affinity-purified antibody was added to 200 µl extracts and incubated on ice for 60 min. Immunoprecipitates were recovered on 10 µl rProtein A Sepharose beads, washed several times with wash buffer (20 mM K-Hepes, pH 7.7, 100 mM KCl, 2 mM MgCl2, and 10% glycerol), and analyzed by immunoblotting.
Immunodepletion of Xenopus egg extracts
10 µg of the affinity-purified antibodies against xMCPH1 was mixed with 25 µl rProtein A Sepharose beads and incubated for 60 min at 4°C. The antibody-coupled beads were washed and equilibrated with XBE2. 50 µl of the egg extracts was incubated with 25 µl antibody beads on ice for 60 min with occasional mixing. After incubation, the supernatants were recovered by brief spin and used as depleted extracts. For mock depletion, the same amount of control rabbit IgG (Sigma-Aldrich) was used.
Online supplemental material
Fig. S1 shows primary structures of vertebrate MCPH1. Fig. S2 shows that the amounts of exogenously added MCPH1 proteins are comparable with that of endogenous xMCPH1. Fig. S3 shows our attempts to identify amino acid residues that might confer a high level of condensin II inhibitory activity on hMCPH1. Fig. S4 shows additional data for physical interactions between hMCPH1 and condensin II. Fig. S5 shows that the N-terminal domain of hMCPH1 is necessary and sufficient to rescue the PCC phenotype in patient cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201106141/DC1.
Acknowledgments
We appreciate H. Miyoshi for providing us with the vectors for lentivirus production. We also thank the Support Unit for Bio-material Analysis, RIKEN, Brain Science Institute Research Resources Center, for help with DNA sequencing analysis and distributing FANTOM3 clones. We are grateful to A. Matsuura for technical assistance, J. Lee for advice, and other members of the Hirano laboratory for critically reading the manuscript.
This work was supported by the RIKEN President’s Discretionary Fund, Grant-in-Aid for Specially Promoted Research (to T. Hirano), Grant-in-Aid for Scientific Research C (to T. Ono), and Grant-in-Aid for Young Scientists B (to D. Yamashita). D. Yamashita and K. Shintomi are RIKEN Special Postdoctoral Researchers.
Footnotes
Abbreviations used in this paper:
- BRCT
- BRCA1 C terminal
- hMCPH1
- human MCPH1
- mMCPH1
- mouse MCPH1
- PCC
- premature chromosome condensation
- PLC
- prophaselike cell
- SMC
- structural maintenance of chromosomes
- xMCPH1
- Xenopus MCPH1
References
- Alderton G.K., Galbiati L., Griffith E., Surinya K.H., Neitzel H., Jackson A.P., Jeggo P.A., O’Driscoll M. 2006. Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat. Cell Biol. 8:725–733 10.1038/ncb1431 [DOI] [PubMed] [Google Scholar]
- Chin C.F., Yeong F.M. 2010. Safeguarding entry into mitosis: the antephase checkpoint. Mol. Cell. Biol. 30:22–32 10.1128/MCB.00687-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavvovidis I., Pöhlmann C., Marchal J.A., Stumm M., Yamashita D., Hirano T., Schindler D., Neitzel H., Trimborn M. 2010. MCPH1 patient cells exhibit delayed release from DNA damage-induced G2/M checkpoint arrest. Cell Cycle. 9:4893–4899 10.4161/cc.9.24.14157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J.N., Williams R.S., Lee M.S. 2004. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem. Sci. 29:579–585 10.1016/j.tibs.2004.09.010 [DOI] [PubMed] [Google Scholar]
- Gosling K.M., Makaroff L.E., Theodoratos A., Kim Y.H., Whittle B., Rui L., Wu H., Hong N.A., Kennedy G.C., Fritz J.A., et al. 2007. A mutation in a chromosome condensin II subunit, kleisin beta, specifically disrupts T cell development. Proc. Natl. Acad. Sci. USA. 104:12445–12450 10.1073/pnas.0704870104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl T.A., Smith H.F., Bosco G. 2008. Chromosome alignment and transvection are antagonized by condensin II. Science. 322:1384–1387 10.1126/science.1164216 [DOI] [PubMed] [Google Scholar]
- Hirano T. 2005. Condensins: organizing and segregating the genome. Curr. Biol. 15:R265–R275 10.1016/j.cub.2005.03.037 [DOI] [PubMed] [Google Scholar]
- Hirano T., Kobayashi R., Hirano M. 1997. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 89:511–521 10.1016/S0092-8674(00)80233-0 [DOI] [PubMed] [Google Scholar]
- Jackson A.P., Eastwood H., Bell S.M., Adu J., Toomes C., Carr I.M., Roberts E., Hampshire D.J., Crow Y.J., Mighell A.J., et al. 2002. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am. J. Hum. Genet. 71:136–142 10.1086/341283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers L.J., Coull B.J., Stack S.J., Morrison C.G. 2008. Distinct BRCT domains in Mcph1/Brit1 mediate ionizing radiation-induced focus formation and centrosomal localization. Oncogene. 27:139–144 10.1038/sj.onc.1210595 [DOI] [PubMed] [Google Scholar]
- Losada A., Yokochi T., Kobayashi R., Hirano T. 2000. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J. Cell Biol. 150:405–416 10.1083/jcb.150.3.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Hirano M., Hirano T. 2002. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev. 16:3004–3016 10.1101/gad.249202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H., Blömer U., Takahashi M., Gage F.H., Verma I.M. 1998. Development of a self-inactivating lentivirus vector. J. Virol. 72:8150–8157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D.O. 2007. The Cell Cycle: Principles of Control. New Science Press, London: 297 pp [Google Scholar]
- Onn I., Aono N., Hirano M., Hirano T. 2007. Reconstitution and subunit geometry of human condensin complexes. EMBO J. 26:1024–1034 10.1038/sj.emboj.7601562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Losada A., Hirano M., Myers M.P., Neuwald A.F., Hirano T. 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 115:109–121 10.1016/S0092-8674(03)00724-4 [DOI] [PubMed] [Google Scholar]
- Ponting C., Jackson A.P. 2005. Evolution of primary microcephaly genes and the enlargement of primate brains. Curr. Opin. Genet. Dev. 15:241–248 10.1016/j.gde.2005.04.009 [DOI] [PubMed] [Google Scholar]
- Rai R., Phadnis A., Haralkar S., Badwe R.A., Dai H., Li K., Lin S.Y. 2008. Differential regulation of centrosome integrity by DNA damage response proteins. Cell Cycle. 7:2225–2233 10.4161/cc.7.14.6303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K., Hirano T. 2009. Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl-Pds5 and Sgo1. Genes Dev. 23:2224–2236 10.1101/gad.1844309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K., Hirano T. 2011. The relative ratio of condensin I to II determines chromosome shapes. Genes Dev. 25:1464–1469 10.1101/gad.2060311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow J.R., Hirano T. 2003. The making of the mitotic chromosome: modern insights into classical questions. Mol. Cell. 11:557–569 10.1016/S1097-2765(03)00103-5 [DOI] [PubMed] [Google Scholar]
- Takemoto A., Maeshima K., Ikehara T., Yamaguchi K., Murayama A., Imamura S., Imamoto N., Yokoyama S., Hirano T., Watanabe Y., et al. 2009. The chromosomal association of condensin II is regulated by a noncatalytic function of PP2A. Nat. Struct. Mol. Biol. 16:1302–1308 10.1038/nsmb.1708 [DOI] [PubMed] [Google Scholar]
- Thornton G.K., Woods C.G. 2009. Primary microcephaly: do all roads lead to Rome? Trends Genet. 25:501–510 10.1016/j.tig.2009.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibelius A., Marhold J., Zentgraf H., Heilig C.E., Neitzel H., Ducommun B., Rauch A., Ho A.D., Bartek J., Krämer A. 2009. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J. Cell Biol. 185:1149–1157 10.1083/jcb.200810159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimborn M., Bell S.M., Felix C., Rashid Y., Jafri H., Griffiths P.D., Neumann L.M., Krebs A., Reis A., Sperling K., et al. 2004. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am. J. Hum. Genet. 75:261–266 10.1086/422855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimborn M., Richter R., Sternberg N., Gavvovidis I., Schindler D., Jackson A.P., Prott E.C., Sperling K., Gillessen-Kaesbach G., Neitzel H. 2005. The first missense alteration in the MCPH1 gene causes autosomal recessive microcephaly with an extremely mild cellular and clinical phenotype. Hum. Mutat. 26:496 10.1002/humu.9382 [DOI] [PubMed] [Google Scholar]
- Trimborn M., Schindler D., Neitzel H., Hirano T. 2006. Misregulated chromosome condensation in MCPH1 primary microcephaly is mediated by condensin II. Cell Cycle. 5:322–326 10.4161/cc.5.3.2412 [DOI] [PubMed] [Google Scholar]
- Trimborn M., Ghani M., Walther D.J., Dopatka M., Dutrannoy V., Busche A., Meyer F., Nowak S., Nowak J., Zabel C., et al. 2010. Establishment of a mouse model with misregulated chromosome condensation due to defective Mcph1 function. PLoS ONE. 5:e9242 10.1371/journal.pone.0009242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.L., Singh N., Mer G., Chen J. 2007. MCPH1 functions in an H2AX-dependent but MDC1-independent pathway in response to DNA damage. J. Biol. Chem. 282:35416–35423 10.1074/jbc.M705245200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.L., Liang Y., Li K., Chen J. 2008. Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J. Biol. Chem. 283:29586–29592 10.1074/jbc.M804080200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Lee J., Stern D.F. 2004. Microcephalin is a DNA damage response protein involved in regulation of CHK1 and BRCA1. J. Biol. Chem. 279:34091–34094 10.1074/jbc.C400139200 [DOI] [PubMed] [Google Scholar]
- Yang S.Z., Lin F.T., Lin W.C. 2008. MCPH1/BRIT1 cooperates with E2F1 in the activation of checkpoint, DNA repair and apoptosis. EMBO Rep. 9:907–915 10.1038/embor.2008.128 [DOI] [PMC free article] [PubMed] [Google Scholar]