Abstract
Circadian rhythms are generated in central and peripheral tissues by an intracellular oscillating timing mechanism known as the circadian clock. Several lines of evidence show a strong and bidirectional interplay between metabolism and circadian rhythms. Receptor interacting protein 140 (RIP140) is a coregulator for nuclear receptors and other transcription factors that represses catabolic pathways in metabolic tissues. Although RIP140 functions as a corepressor for most nuclear receptors, mounting evidence points to RIP140 as a dual coregulator that can repress or activate different sets of genes. Here, we demonstrate that RIP140 mRNA and protein levels are under circadian regulation and identify RIP140 as a modulator of clock gene expression, suggesting that RIP140 can participate in a feedback mechanism affecting the circadian clock. We show that the absence of RIP140 disturbs the basal levels of BMAL1 and other clock genes, reducing the amplitude of their oscillations. In addition, we demonstrate that RIP140 is recruited to retinoid-related orphan receptor (ROR) binding sites on the BMAL1 promoter, directly interacts with RORα, and increases transcription from the BMAL1 promoter in a RORα-dependent manner. These results indicate that RIP140 is not only involved in metabolic control but also acts as a coactivator for RORα, influencing clock gene expression.
Keywords: nuclear receptor, coactivator, coregulator, RIP140, Nrip1, BMAL1, RORA, Rev-erbα
Many physiological processes exhibit diurnal variations that persist even in the absence of environmental timing cues. These rhythmic changes in physiology that operate to anticipate the needs of the organism and show a periodicity of about 24 hours are called circadian rhythms. Circadian rhythms are present in central and peripheral tissues and generated by an intracellular oscillating timing mechanism known as the circadian oscillator or molecular clock (Duguay and Cermakian, 2009; Kohsaka and Bass, 2007; Yang et al., 2007).
The mammalian molecular clock relies on a transcriptional-translational feedback loop in which BMAL1 and CLOCK bind as heterodimers to regulatory elements on target genes, stimulating their transcription (Darlington et al., 1998; Ripperger and Schibler, 2006). Among those genes are the repressor proteins PERIOD (PER) and CRYPTOCHROME (CRY) families. Once PER and CRY have reached a critical concentration and are appropriately modified, they inhibit the activity of BMAL1-CLOCK, thereby repressing their own transcription and that of the other BMAL1-CLOCK target genes. Following additional modifications, PER and CRY are degraded, and the cycle begins again (Dardente and Cermakian, 2007; Dibner et al., 2010; Dunlap, 1999; Griffin Jr. et al., 1999; Lee et al., 2001). A second interconnecting feedback loop involving orphan nuclear receptors of the Rev-erb and ROR families regulates the levels of expression of BMAL1, CLOCK, and CRY1. In this secondary loop, ROR members activate both BMAL1 and Rev-erbα expression, while in turn, Rev-erbα represses transcription of both itself and BMAL1, closing the loop (Akashi and Takumi, 2005; Duguay and Cermakian, 2009; Liu et al., 2008; Sato et al., 2004). These nuclear receptors are not essential for circadian rhythms but are thought to improve the robustness of the clock, affecting its period length and phase-shifting properties (Akashi and Takumi, 2005; Liu et al., 2008). In addition, these nuclear receptors are thought to be links between circadian rhythms and metabolism (Duguay and Cermakian, 2009; Schmutz et al., 2010).
Several lines of evidence show a strong interplay between metabolism and circadian rhythms (Duguay and Cermakian, 2009; Eckel-Mahan and Sassone-Corsi, 2009; Froy, 2010). Transcriptome profiling studies revealed that several key genes involved in metabolism are rhythmically expressed (Panda et al., 2002; Yang et al., 2006). Mice devoid of a functional circadian oscillator develop metabolic syndrome, and mutations or ablations of clock genes are often associated with metabolic problems (Eckel-Mahan and Sassone-Corsi, 2009; Green et al., 2008; Turek et al., 2005). Conversely, the dominance of feeding cycles as a zeitgeber (time cue) for peripheral clocks implies that metabolic signals impact the clock machinery itself (Arble et al., 2009; Damiola et al., 2000; Eckel-Mahan and Sassone-Corsi, 2009).
Glucose and lipid metabolism in the liver and adipose tissue exhibit circadian variations due, at least in part, to circadian expression of key enzymes in these pathways (Lamia et al., 2008; Panda et al., 2002; So et al., 2009; Zvonic et al., 2006). The expression of these genes is regulated by several pathways including nuclear receptor signaling (Christian et al., 2006; Laitinen et al., 2005; Lau et al., 2008; Sonoda et al., 2008), and interestingly, a number of nuclear receptors are themselves expressed in a circadian manner. Thus, in addition to Rev-erbα, the expression of peroxisome proliferator–activated receptors (PPARs), estrogen-related receptors (ERRs), and thyroid hormone receptors (TRs) is subject to circadian control in liver, muscle, and adipose tissues (Yang et al., 2006). Furthermore, the ability of nuclear receptors to stimulate the expression of metabolic genes depends on the recruitment of coactivators whose activity and/or expression not only varies in response to environmental stimuli but that are also expressed in a circadian manner (Asher et al., 2008; Liu et al., 2007; Nakahata et al., 2008; Panda et al., 2002). For example, PGC1α and PGC1β, which are coactivators for ERRs and PPARs, are responsible for the activation of catabolic gene expression in metabolic tissues (Handschin and Spiegelman, 2006). The expression of these 2 cofactors is increased following exercise and stress or in cold temperatures, but it is also subject to circadian control (Handschin and Spiegelman, 2006; Liu et al., 2007).
RIP140 is a corepressor for many catabolic genes and antagonizes the positive effects of PGC1 coactivators (Christian et al., 2006; Hallberg et al., 2008; White et al., 2008). Thus, catabolism is increased in mice devoid of RIP140, and they are lean and protected against diet-induced obesity and insulin resistance (Leonardsson et al., 2004). On the other hand, RIP140 functions as a coactivator for a number of genes involved in triglyceride synthesis (Herzog et al., 2007), growth factors of the EGF family (Nautiyal et al., 2010), and inflammatory cytokines (Zschiedrich et al., 2008). Transcriptome profiling has shown that RIP140 mRNA is subject to circadian oscillations in the liver (Hughes et al., 2009; Panda et al., 2002), and we have previously noted alterations in the daily patterns of rest/activity in RIP140-null mice kept in normal 12-hour light/12-hour dark conditions (Hudson-Davies et al., 2008). In view of these observations, we have examined the relationship between this metabolic cofactor and the molecular clock. We have found that RIP140 is not only subject to circadian gene expression but also functions as a positive regulator of the molecular clock by potentiating the activity of retinoid-related orphan receptor α (RORA).
Materials and Methods
Animal Samples
Three-month-old wild-type and RIP140 knockout male mice of the C57BL/6J background were kept in a 12-hour light/12-hour dark cycle with regular chow provided ad libitum. Animals were sacrificed in groups of 3 of each genotype per day on 2 consecutive days (to avoid excessive time difference between sample 1 and 6 of the same genotype), and tissue samples were preserved in RNALate (Invitrogen, Carlsbad, CA) for RNA extraction.
Quantitative Real-Time RT-PCR
Total RNA was purified from tissues and cells using TriReagent (Sigma-Aldrich, St. Louis, MO). For reverse transcription (RT), RNA (1 µg) was reverse transcribed to cDNA using SuperScript II Reverse Transcriptase (Invitrogen) and random hexamers (Invitrogen). Quantitative PCR (QPCR) was performed on diluted cDNA samples with SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) in an ABI Step One Plus QPCR system (Applied Biosystems, Carlsbad, CA) using the following program: 95 °C for 10 minutes, 40 cycles at 95 °C for 15 seconds, 60 °C for 30 seconds, and 72 °C for 30 seconds. Primers used are listed in Supplementary Table S1 and were designed using Primer3 (http://frodo.wi.mit.edu/primer3). RT-QPCR results were analyzed using the 2−ΔΔCt method as described by Livak and Schmittgen (2001). The geometric mean of 3 housekeeping genes—β-2 microglobulin (B2m), ribosomal protein L13A (RPL13a), and TATA box binding protein (Tbp, for mice) or ribosomal protein L19 (RPL19, for humans)—was used as a calibrator after confirming that the genes were not affected by the treatment (Livak and Schmittgen, 2001).
Cell Culture
All cells were cultured under humidified 10% CO2 at 37 °C in high glucose Dulbecco’s modified Eagle’s medium (cat. no. 41966, Invitrogen) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 µg/mL streptomycin, 0.25 µg/mL amphotericin, and Glutamax (Invitrogen).
Synchronization by Serum Pulse
Cells were cultured at confluence for 2 days in 10% FBS-DMEM. Synchronization was achieved by incubation in DMEM supplemented with 50% horse serum (PAA) for 2 hours. After this incubation, cells were washed with PBS and placed in serum-free medium (time zero). Protein and mRNA samples were collected every 3 hours for 51 hours.
Mouse embryonic fibroblasts (MEFs) were isolated from 12.5-day-old embryos produced by crossing heterozygous RIP140 knockout mice. In brief, embryos were carefully detached from the amniotic sac, and each embryo was placed in individual 3.5-cm dishes and decapitated. Bodies were finely minced with scissors in 1X Trypsin/EDTA (Invitrogen), and the fragments were pipetted up and down using a 5-mL plastic Pasteur pipette. Trypsin was then blocked by adding 10% FBS-DMEM. Cellular debris was allowed to settle for 2 minutes, and the medium containing the cell suspension was transferred to a 25-cm flask. Embryos were genotyped during expansion (passage 2) and used for experiments between passages 3 and 12. Each MEF line corresponds to an individual embryo.
Immunoblotting and Antibodies
Cell lysates were harvested by the addition of SDS lysis buffer (2% SDS, 30 mM NaCl, 10 mM HEPES, pH 7.4, 20 mM NaF, 1 mM NaPPi, 1 mM PMSF, and 1X Complete Protease Inhibitor Cocktail [Roche, Basel, Switzerland]). Equal amounts of protein from lysates were resolved by SDS-PAGE, immunoblotted, and detected with an ECL or ECLplus kit (GE Healthcare, Little Chalfont, UK). For antibodies, anti-RIP140 monoclonal antibody (6D7) generated against residues 301 to 478 of human RIP140 was described previously (Herzog et al., 2007). The following antibodies were used: anti-RORA (sc-6062, Santa Cruz Biotechnology, Santa Cruz, CA), anti–β-actin (ab8226, Abcam, Cambridge, UK), anti-HA (11867423, Roche), anti-V5 (R960-25, Invitrogen), and normal mouse or goat immunoglobulin G (IgG) (sc-2025 or sc-2028, Santa Cruz Biotechnology).
Transient Transfection
pCIEF-V5RIP140 expression vector was described previously (Nautiyal et al., 2010). The BMAL1 and 2XmtROREs-BMAL1 promoter reporters and pcDNA3-RORA1 and pcDNA3-Rev-erbα expression constructs were described (Akashi and Takumi, 2005) and kindly provided by Professor T. T. Takumi. pSPORT6-RORC was purchased from I.M.A.G.E. Consortium (Manassas, VA). RNA duplexes to knock down RIP140, RORA, Rev-erbα, and noncoding control siRNA duplexes were purchased from NBS Biologicals (Huntingdon, UK), and sequences are shown in Supplementary Table S2.
Cells were plated in 24-well plates, 5 × 104 cells per well, in phenol red–free Optimem (Invitrogen) supplemented with 2% FBS 1 day prior to transfection. Cells were transfected using FuGene HD (Roche) or Lipofectamine 2000 (in RNAi experiments) (Invitrogen) according to the instructions of the manufacturer. In all cases, less than 100 ng of DNA was used. Empty pCIEF and/or pcDNA3 vectors were used to adjust the same amount of DNA in all treatments. There were 50 ng of BMAL1 or 2XmtROREs-BMAL1 promoter reporter and 5 ng of pcDNA4-eGFP used in all the experiments. The amount of RORA and RORC expression vectors varied between 0.1 and 1 ng, Rev-erbα expression vector between 10 and 30 ng, and the amount of RIP140 expression vector between 3 and 27 ng. Cells were harvested for luciferase assay approximately 36 hours after transfection. Firefly luciferase was measured with Steadylite HTS (PerkinElmer, Waltham, MA) according to the instructions of the manufacturer. In all cases, cells were cotransfected with pcDNA4-eGFP, and 480/535 fluorescence was used as an internal control to correct for differences in transfection efficiencies.
Real-Time Luciferase Assay
RIP140–/– cells were cotransfected with pBMAL1-dLUC (Gamsby et al., 2009) and pBABE- puro (Addgene, Cambridge, MA) using FuGene 6 (Roche) following the manufacturer’s instructions. Clones of stably transfected cells were then generated by adding puromycin (1.5 µg/mL) for approximately 1 month, and individual colonies were selected and assayed for luciferase activity in a Turner TD-20e luminometer using 1 mM luciferin in PBS (Gold Biotechnology, St. Louis, MO). Luciferase-positive clones were then assayed for clock function in a LumiCycle (Actimetrics, Wilmette, IL) as previously described (Gamsby et al., 2009). The period length (τ) was calculated using the sine fit (damped) function from the LumiCycle analysis software package (Actimetrics), and the phase (φ) was calculated using the second peak after release from serum shock as a reference.
Immunoprecipitation
Proteins were crosslinked using 2 mM DSP (Pierce, Waltham, MA) according to the manufacturer’s instructions for 30 minutes. Cells were lysed in SDS lysis buffer (2% SDS, 30 mM NaCl, 10 mM HEPES, pH 7.4, 20 mM NaF, 1 mM NaPPi, 1 mM PMSF, and 1X Complete Protease Inhibitor Cocktail [Roche]). Immunoprecipitation was carried out using Dynabeads Protein A Immunoprecipitation Kit (Invitrogen) following the manufacturer’s instructions. After immunoprecipitation, samples were decrosslinked by heating at 95 °C in 10% β-mercaptoethanol for 5 minutes.
Chromatin Immunoprecipitation (ChIP) Assay
Cells were crosslinked (1% formaldehyde for 15 minutes) and nuclei isolated by scraping the cells in swelling buffer (25 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, and 0.1% Nonidet, supplemented with protease inhibitor cocktail [Roche] and 1 mM PMSF) followed by centrifugation for 5 minutes at 14,000g. Nuclei were sonicated in lysis buffer (1% SDS, 1% Triton-X100, 0.5% deoxycholate, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1) for 30 minutes. Chromatin (500 µg) was immunoprecipitated using Dynabeads Protein A Immunoprecipitation Kit (Invitrogen) following the manufacturer’s instructions with minor changes. Washes were performed as follows: 1X low salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM TRIS-HCl, pH 8.1, 150 mM NaCl); 2X high salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM TRIS-HCl, pH 8.1, 500 mM NaCl); 1X LiCl buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM TRIS-HCl, pH 8.1); and 2X TE buffer (10 mM TRIS-HCl, pH 8, 1 mM EDTA). Samples were eluted in 100 L of EB buffer (1% SDS and 1 mL 0.1 M NaHCO3). For rechip experiments, samples were eluted in 50 mL of 10 mM DTT at 37 °C for 30 minutes and then diluted in 2 mL of low salt buffer and immunoprecipitated as indicated above. After reversal of crosslinking, DNA fragments were purified from the samples with a QIAquick PCR purification kit (Qiagen, Venlo, the Netherlands) and used as templates in QPCRs. Primers used are listed in Supplementary Table S3 and were designed using Primer3 (http://frodo.wi.mit.edu/primer3).
Statistical Analysis
Results were analyzed with Prism 4 (GraphPad Software, La Jolla, CA) using the Student t test or ANOVA followed by the Student Newman-Keuls multiple comparison test according to experimental design. p values lower than 0.05 were considered evidence for statistical significance.
Results
RIP140 Protein and mRNA Levels Exhibit Circadian Oscillations
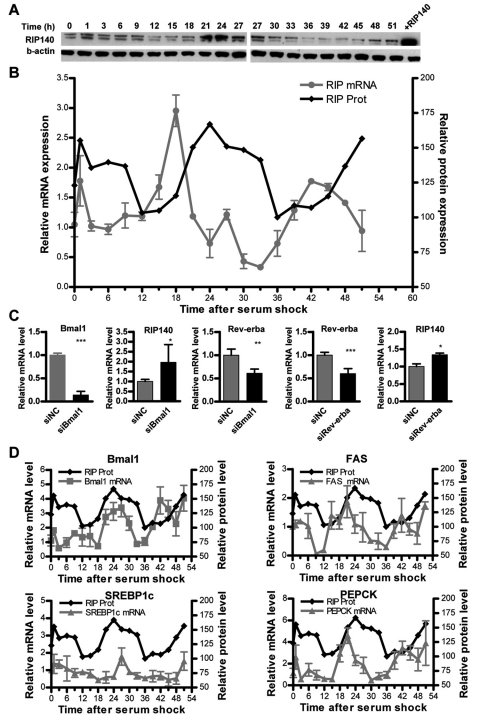
Inspection of a circadian gene expression database from mouse liver indicated that RIP140 was subject to circadian oscillation (Hughes et al., 2009; Panda et al., 2002). To investigate whether this periodicity was cell autonomous, we analyzed RIP140 expression in synchronized HepG2 human liver cells. Cells were synchronized by a serum pulse, and samples of mRNA and protein were collected every 3 hours for 51 hours. We found that RIP140 mRNA and protein were expressed in a circadian fashion, indicating that this is an intrinsic property of liver cells (Fig. 1A and 1B). It is interesting to note that there was a phase difference between RIP140 mRNA and protein that is typical for many clock genes (Dunlap, 1999; Liu et al., 1999). Initially, we investigated whether BMAL1 directly controls RIP140 expression by depleting it from U2OS cells using RNA interference (RNAi). As expected, the depletion of BMAL1 led to a decrease in mRNA of its well-known target Rev-erbα, but interestingly, it resulted in an increase in RIP140 expression, indicating that RIP140 expression is not activated by BMAL1 but may be subject to repression by Rev-erbα (Fig. 1C). While canonical E-boxes were not evident within the RIP140 locus, ROR binding elements (RORE) were noted in intronic regions of the gene (Heim et al., 2009). Consistent with these observations, the depletion of Rev-erbα by RNAi increased the levels of RIP140 mRNA (Fig. 1C). Thus, Rev-erbα seems to repress the expression of RIP140 and may mediate the effects of BMAL1 on RIP140 expression, but whether this repressive effect is direct cannot be ascertained from these results alone.
Figure 1.
Circadian expression of RIP140. (A) Western blots for RIP140 and β-actin, representative of 2 independent experiments. (B) Levels of RIP140 mRNA, measured by real-time PCR, and protein (measured by densitometry of A). Points represent mean ± SEM (n = 4). Figure representative of 3 independent experiments. Cells were synchronized by 50% serum pulse as described in Materials and Methods. (C) mRNA levels for BMAL1, RIP140, and Rev-erbα after depletion of BMAL1 using a specific siRNA for BMAL1 and levels of Rev-erbα and RIP140 mRNA after depletion of Rev-erbα using a specific siRNA for Rev-erbα. U2OS cells were transfected with siRNA, and mRNA was collected 48 hours after transfection. Bars represent mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 versus control. Student t test (n = 6). Figure representative of 3 independent experiments. (D) Levels of BMAL1, fatty acid synthase (FAS), sterol regulatory element-binding protein 1c (SREBP1c), and phospho enol pyruvate carboxykinase (PEPCK) mRNA, superimposed on RIP140 protein levels. Points represent mean ± SEM of 4 biological replicates.
A number of metabolic genes, such as FAS, SREBP1c, PEPCK, G6Pase, PDK4, and others, that display circadian oscillations in the liver are known targets for RIP140 (Christian et al., 2006; Herzog et al., 2007; Panda et al., 2002; White et al., 2008). The pattern of expression of these genes overlapped with the changes in RIP140 protein, indicating that RIP140 and its associated metabolic pathways are controlled in a coordinated fashion. There was also a particularly marked overlap between RIP140 protein and BMAL1 expression (Fig. 1D).
RIP140 Regulates the Basal Expression of Clock Genes In Vivo and In Vitro
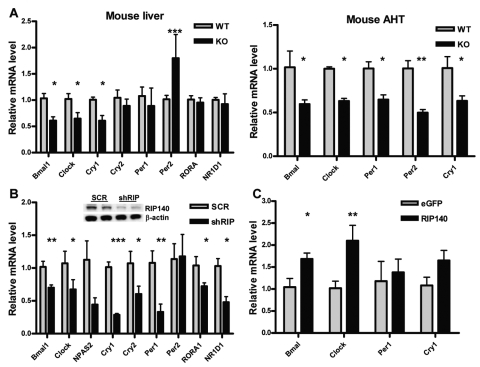
Given the oscillations in RIP140 expression, we examined the possibility that it might participate in a feedback mechanism to signal back to the molecular clock. We investigated the impact of RIP140 on the expression of BMAL1 and other clock genes, determining their mRNA levels in the liver and anterior hypothalamus (AHT) of wild-type (WT) and RIP140 knockout (KO) mice. Samples were collected at ZT2, and mRNA levels for different clock genes were measured by RT-QPCR. At this time point, there was a consistent reduction in the RNA levels for all clock genes studied in the AHT and of BMAL1, CLOCK, and CRY1 in the liver of KO animals (Fig. 2A). The only exception was PER2, which was increased in the liver (Fig. 2A). Similarly, the depletion of RIP140 from HepG2 cells by constitutively expressing RIP140 shRNA also led to a reduction in clock gene expression as compared with a scrambled shRNA control cell line (Fig. 2B). Conversely, exogenous expression of RIP140 led to an increase in the levels of BMAL1 and CLOCK (Fig. 2C). Thus, we conclude that RIP140 functions as a positive regulator of clock gene expression.
Figure 2.
RIP140 absence reduces the basal expression of clock genes. (A) mRNA level of different clock genes in the liver and anterior hypothalamus (AHT) of wild-type (WT) and RIP140 knockout (KO) mice. Samples were collected at ZT2 (2 hours after lights are turned on), and mRNA was extracted and quantified by RT-QPCR. Bars represent mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 versus control. Student t test (n = 6). (B) mRNA levels of clock genes in HepG2 human liver cell lines depleted of RIP140 quantified by RT-QPCR. RIP140 knockdown cells (shRIP) were generated by stably transfecting HepG2 cells with a vector expressing RIP140-specific shRNA. HepG2 cells constitutively expressing a nonspecific scrambled shRNA (scr) were used as a control. (Inset) Western blot showing RIP140 expression in the different cell lines. Bars represent mean ± SEM (n = 6). *p < 0.05, **p < 0.01, and ***p < 0.001 versus control. Student t test. (C) mRNA levels of clock genes in HuH7 human liver cells transiently transfected with a RIP140 expressing vector or equal amounts of an eGFP expressing vector. Bars represent mean ± SEM (n = 3). *p < 0.05 and **p < 0.01 versus control. Student t test.
The Absence of RIP140 Influences the Level of Expression of Clock Genes in Synchronized MEFs
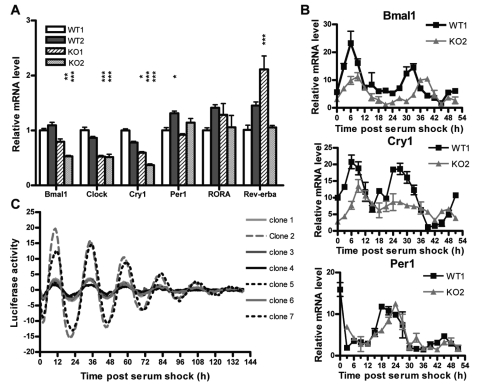
To investigate the role of RIP140 in circadian gene expression, we compared the expression of clock genes in mouse embryonic fibroblasts (MEFs) from WT and KO littermates. As in the mouse liver, AHT, and RIP140-depleted HepG2s, unsynchronized MEFs devoid of RIP140 express less mRNA for several clock genes including BMAL1, CLOCK, and CRY1 (Fig. 3A). After serum pulse synchronization, the circadian expression of BMAL1, CRY1, and PER1 was maintained in the absence of RIP140, but the amplitude of the oscillations was reduced for BMAL1 and CRY1. Interestingly, a slight phase difference in BMAL1 and CRY1 expression between WT and KO MEFs was also observed in the RT-QPCR data (Fig. 3B). However, this observation was not consistent with the phase of expression seen for PER1. Because phase and period estimates based on sampling of RNAs followed by RT-QPCR are limited by both the density of time points and the duration of experiments, we generated stably transfected lines of RIP140 KO MEFs in which the BMAL1 promoter (pBMAL1-dLuc) was used to drive rhythmic expression of firefly luciferase. In 7 independently derived clones, experiments monitored during 6 days provided consistent phase and period determinations of BMAL1 reporter activity in the RIP140 KO MEFs (Fig. 3C). The phase of peak BMAL1 promoter activity on day 2 was 34.32 ± 0.62 hours (2 standard deviations) after serum shock and is in agreement with prior estimates of phase from the NIH 3T3 cell line (33.99 ± 0.22) (Gamsby et al., 2009). This phase falls within the region spanning the peaks for BMAL1 mRNA in RIP140 KO and WT littermate MEFs (Fig. 3B), suggesting that the apparent phase differences mainly reflect biological variability. Thus, while RIP140 is not required to maintain circadian rhythmicity, it contributes to the overall expression and oscillation of BMAL1 and CRY1.
Figure 3.
Clock gene expression and circadian rhythms in RIP140 knockout (KO) mouse embryonic fibroblasts (MEFs). The MEFs were generated as described in Materials and Methods by crossing RIP140 KO heterozygous animals. (A) Level of expression of clock genes in unsynchronized MEFs generated from 2 wild-type (WT1/2) and 2 knockout (KO1/2) embryos measured by real-time PCR. Bars represent mean ± SEM (n = 6). *p < 0.05, **p < 0.01, and ***p < 0.001 versus WT1. One-way ANOVA followed by Student Newman-Keuls post hoc multiple comparison test. Figure representative of 3 independent experiments. (B) Daily oscillations in the mRNA levels of BMAL1, CRY1, and PER1 in synchronized WT and KO MEFs measured by real-time PCR. Cells were synchronized by 50% serum pulse as described in Materials and Methods. (C) RIP140 KO MEFs were stably transfected with the BMAL1 promoter driving luciferase. Seven individual clones (clone 1-7) were assayed for rhythmicity. After synchronization by serum pulse, luciferase activity was measured in a real-time luminometer for 6 days. τ and φ were calculated as described in Materials and Methods (N = 7).
RIP140 Affects BMAL1 Expression by Coactivation of RORA
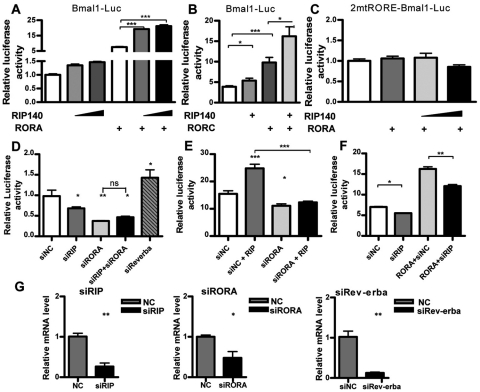
RIP140 has been reported to interact with most, if not all, nuclear receptors (White et al., 2008) and a number of other transcription factors, including p65, CREB, and c-jun (Nautiyal et al., 2010; Zschiedrich et al., 2008). Retinoid-related orphan receptor α (RORA) is a nuclear receptor well known to stimulate transcription from the BMAL1 promoter (Akashi and Takumi, 2005). Therefore, we investigated the possibility that RIP140 might function as a regulator of its activity. First, we examined whether RIP140 was able to modulate the ability of RORA to stimulate transcription from a BMAL1 reporter in HuH7 human liver cells (Fig. 4A). RIP140 expression alone led to a small increase in BMAL1 promoter activity, potentially by means of endogenous RORs, and significantly increased the activation of the promoter by exogenously expressed RORA. RIP140 also increased the activity of RORC on the BMAL1 promoter, suggesting that this coregulator could function as a general coactivator for this group of nuclear receptors (Fig. 4B). Importantly, there was no increase in reporter activity when the ROR binding sites were mutated to abolish recruitment of the receptor to the promoter (Fig. 4C). Depletion of RIP140 using RNAi (Fig. 4G) reduced the basal activity of the BMAL1 promoter, as did the depletion of RORA (Fig. 4D). However, depletion of RIP140 in the absence of RORA failed to further reduce BMAL1 reporter activity, indicating that the presence of RORA is required for RIP140 function on this promoter (Fig. 4D). Moreover, the depletion of RORA using RNAi completely prevents any positive effect of RIP140 (Fig. 4E). Depletion of RIP140 partially reduced the activation by RORA (Fig. 4F), suggesting that, while RIP140 serves as a positive activator, other cofactors also contribute to BMAL1 promoter activity.
Figure 4.
RIP140 activates the BMAL1 promoter. HuH7 cells were cotransfected with a reporter vector containing an intact (A and B, BMAL1-Luc) or a ROREs mutant (C, 2xmtROREBMAL1-Luc) BMAL1 promoter with RORA or RORC and increasing concentrations of RIP140 expression vectors. (D) HuH7 cells were cotransfected with a BMAL1 reporter and siRNA duplexes targeting RIP140, RORA, Rev-erbα, or a noncoding (siNC) oligo as control. (E) Cotransfection of a BMAL1 reporter with RIP140 and siRNA oligos targeting RORA. (F) Cotransfection of a BMAL1 reporter with RORA and siRNA oligos targeting RIP140. (G) Knockdown of RIP140, RORA, and Rev-erbα was confirmed by RT-QPCR. Figures representative of at least 3 independent experiments. Bars represent mean ± SEM (n = 6). *p < 0.05, **p < 0.01, and ***p < 0.001 versus control. Two-way ANOVA followed by Student Newman-Keuls multiple comparison test.
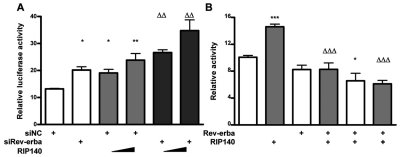
The orphan nuclear receptor Rev-erbα reduces the expression of BMAL1 and other genes by displacing RORA and recruiting corepressors such as NCoR (Yin and Lazar, 2005). To investigate whether RIP140 could increase BMAL1 expression by suppressing Rev-erbα repressive activity, the ability of Rev-erbα knockdown and overexpression to modulate a BMAL1 reporter was investigated. Depletion of Rev-erbα by RNAi not only increased basal activity but also RIP140-stimulated BMAL1 promoter activity (Fig. 5A). Overexpression of Rev-erbα, on the other hand, reduced the basal activity of the reporter and prevented the positive effect of RIP140 (Fig. 5B). These results indicate that RIP140 is capable of acting as an activator independently of Rev-erbα.
Figure 5.
Rev-erbα antagonizes the effect of RIP140. (A) HuH7 cells were cotransfected with a BMAL1 reporter, siRNA duplexes targeting Rev-erbα (siRev-erbα), or a noncoding oligo (siNC) as control and increasing amounts of a RIP140 expressing vector. (B) Cotransfection of a BMAL1 reporter with an expression vector for RIP140 and increasing concentrations of a Rev-erbα expression vector. Figures representative of 3 independent experiments. Bars represent mean ± SEM (n = 6). *p < 0.05, **p < 0.01, and ***p < 0.001 versus control. ΔΔp < 0.01 and ΔΔΔp < 0.001 versus siRev-erbα (A) or RIP140 (B). Two-way ANOVA followed by Student Newman-Keuls multiple comparison test.
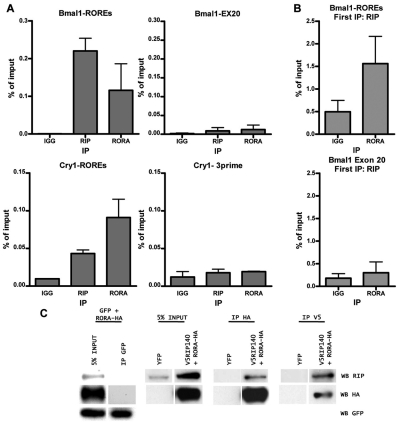
Next, we examined the endogenous BMAL1 promoter in HepG2 cells by performing ChIP experiments. We found that RIP140 was present in the vicinity of ROREs on the BMAL1 promoter, as well as in other RORE-containing genes (Ueda et al., 2005), but not in different regions of the same genes that were examined as negative controls (Fig. 6A). By sequentially immunoprecipitating for RIP140-bound chromatin and then for RORA-bound chromatin, we were able to demonstrate co-occupancy of RIP140 and RORA in the vicinity of the BMAL1 ROREs (Fig. 6B). Finally, we investigated the interaction between RIP140 and RORA by expressing epitope-tagged proteins and performing coimmunoprecipitation/Western blotting experiments using antibodies to the epitope tags. Thus, we expressed RIP140-V5 and RORA-HA and demonstrated that the 2 proteins were able to interact either by immunoprecipitating with V5 antibodies followed by Western blotting with HA antibodies or vice versa but not with an anti-GFP antibody used as a control (Fig. 6C). We also demonstrated that the endogenous proteins were able to interact, albeit weakly, but this might reflect the relative avidities of the anti-RIP140 and anti-RORA antibodies to their specific epitopes. Thus, we conclude that RIP140 stimulates transcription of BMAL1 and other clock genes by functioning as a coactivator for RORA.
Figure 6.
RIP140 is recruited to ROR binding elements and interacts with RORA. (A) Chromatin immunoprecipitation assay for RIP140 and RORA performed with nuclear extracts of unsynchronized HepG2 cells. Purified DNA from precipitated chromatin was amplified by real-time QPCR using primers encompassing the ROR binding elements on the BMAL1 or CRY1 genes. Distal regions of these genes lacking ROR binding elements were used as control. Bars represent mean ± SEM of 3 independent experiments. (B) Sequential chromatin immunoprecipitation assay (rechip). DNA immunoprecipitated with an anti-RIP140 antibody was then immunoprecipitated with an anti-RORA antibody. DNA purified after the second precipitation was amplified by real-time QPCR using primers encompassing the ROR binding elements on the BMAL1 gene. A distal region of this gene lacking ROR binding elements was used as control. Bars represent mean ± SEM (n = 3). (C) HEK293 cells were transiently transfected with V5-tagged RIP140 and HA-tagged RORA expression vectors. Cells cotransfected with YFP expression vector and empty vector or GFP- and HA-tagged RORA expression vectors were used as controls. Total cell lysates were immunoprecipitated using anti-V5, anti-HA, or anti-GFP antibodies, and Western blots were performed using specific RIP140, HA, or GFP antibodies.
Discussion
RIP140 plays an important role as a coregulator for nuclear receptors in controlling energy expenditure in adipose tissue and muscle (Leonardsson et al., 2004; Morganstein et al., 2008; Seth et al., 2007). In the liver, it regulates lipid and glucose homeostasis, acting either as a corepressor or as a coactivator, depending on the target gene (Herzog et al., 2007). Mounting evidence shows that the maintenance of circadian rhythms is essential to sustain a normal metabolic status (Eckel-Mahan and Sassone-Corsi, 2009; Froy, 2010; Johnston et al., 2009; Kohsaka and Bass, 2007) and that an intact circadian clock in the liver is required to maintain systemic glucose homeostasis (Lamia et al., 2008). Here, we show that the metabolic regulator RIP140, while it is not essential for the maintenance of circadian rhythmicity in cells, modulates clock gene expression by potentiating the activity of ROR nuclear receptors.
RIP140 expression is circadian in the liver and synchronized liver cells. Interestingly, in synchronized liver cells, RIP140 protein oscillated in phase with BMAL1 mRNA, pointing to a possible transactivation of this gene by RIP140. We were unable to identify conserved E-boxes within the RIP140 gene. Depletion of BMAL1 led to an increase of RIP140, rather than a decrease, suggesting that it is a higher order clock-controlled gene instead of a direct target for BMAL1/CLOCK. The RIP140 locus extends over 100 Kbp with regulatory regions both intragenic and in the upstream promoter (Augereau et al., 2006; Heim et al., 2009; Nichol et al., 2006). RIP140 contains binding regions for RORs/Rev-erbs and other circadianly expressed transcription factors (Heim et al., 2009). Depletion of Rev-erbα in U2OS cells led to an increase in RIP140, suggesting that this nuclear receptor is a potential repressor of RIP140 expression. As RIP140 seems to be repressed by Rev-erbα, a well-known BMAL1 target gene, we propose that RIP140 could be a second-order clock-controlled gene.
Under normal 12:12 light/dark cycles, RIP140 KO mice show significantly more activity recorded during the light phase, when the mice would normally be resting, as compared with WT controls (Hudson-Davies et al., 2008). These data suggested the possibility of alterations in the circadian system. Here, we report many and varied effects of RIP140 on the expression of clock genes: BMAL1, CLOCK, and CRY1 were all found to be downregulated in the liver and AHT of RIP140 KO mice, while the effects on PER1 and PER2 varied depending on the tissue. As these measurements were carried out at a single time point (ZT2) instead of during a complete 24-hour period, we cannot conclude that this effect will be seen, or in fact will not change, at other times of the day. However, consistent with these data, the absence of RIP140 gave rise to a constant downregulation of BMAL1 and CRY1 in synchronized MEFs, and the knockdown of RIP140 in human liver cells led to the downregulation of several core clock genes, indicating that RIP140 can impact elements within the circadian clock.
Despite the reduced expression of several clock genes in synchronized RIP140 KO cells, these lines displayed relatively normal circadian periods and phases of oscillations in BMAL1 gene reporters, indicating that RIP140 is not necessary for the oscillatory function of the molecular clock. Nevertheless, it has been suggested that smaller amplitudes of expression of clock genes, such as those seen here, can lead to decreased stability of the clock, culminating in either increased sensitivity to time cues or loss of rhythmicity after introduction of constant conditions (Eckel-Mahan and Sassone-Corsi, 2009; Liu et al., 2008; Liu et al., 2007; Yoo et al., 2004). Thus, it is an interesting possibility to be tested in the future whether loss of RIP140 may render animals more sensitive to external factors, such as nutrition, that can affect the circadian system.
There are several metabolic similarities between the RIP140 KO mice and the staggerer mice (sg/sg) that express a dominant negative form of RORA. Both strains are lean and protected against diet-induced obesity and insulin resistance (Lau et al., 2008; Leonardsson et al., 2004). In addition, RIP140 and RORA share several target genes in the liver (Jetten, 2009; Journiac et al., 2009). Here, we show that in liver cells, RIP140 interacts with RORA and potentiates RORs activity on the BMAL1 promoter, providing a mechanism to explain the lower levels of clock genes in RIP140 KO animals and cells. It is important to note that not only BMAL1 but also CLOCK and CRY1 are consistently downregulated in the absence of RIP140 in tissues and cells and that all these genes are also known targets for RORA (Liu et al., 2008). Rev-erbα, the negative component in the subsidiary feedback loop of the molecular clock, competes with RORA for the ROR binding sites and represses the expression of BMAL1 by recruiting corepressors (Yin and Lazar, 2005). Therefore, we tested the alternative possibility that RIP140 may potentiate BMAL1 expression by blocking the ability of Rev-erbα to function as a repressor. The positive effect of RIP140 was not affected by a reduction on Rev-erbα levels, indicating that the presence of this nuclear receptor is not necessary for RIP140 action. On the other hand, overexpression of Rev-erbα, displacing RORA from its binding sites, blocked the effect of RIP140.
It has been reported that other nuclear receptor coregulators, among them PGC1α, can coactivate RORA, affecting clock gene expression in the liver and other organs (Liu et al., 2007). In other tissues, particularly in adipose tissue and muscle, RIP140 is known to suppress the catabolic effects of PGC1α (Christian et al., 2006; Hallberg et al., 2008; White et al., 2008). Given these observations, it may seem surprising that both RIP140 and PGC1α are capable of serving as coactivators for the molecular clock in the liver. However, both proteins can be induced in the liver by similar signals (Berriel et al., 2008; Puigserver, 2005), suggesting that a certain degree of cooperation between them may exist. Moreover, it has been shown that RIP140 and PGC1α can physically interact and that they can be simultaneously recruited to endogenous promoters (Hallberg et al., 2008). On the other hand, RIP140 and PGC1α oscillate with different phases (Panda et al., 2002; http://expression.gnf.org/circadian), which might lead to temporal compartmentalization and therefore absence of competition/interaction. The precise relation between these 2 cofactors remains to be elucidated. Finally, although RIP140 seems to regulate BMAL1 and other clock genes via direct interaction with RORA, several other indirect mechanisms such as changes in the energy balance/oxidative status of the RIP140-depleted cells cannot be ruled out as possible causes of changes in clock function.
In summary, we showed that RIP140 is a clock-controlled gene and protein whose circadian regulation is cell autonomous and that its absence reduces the basal levels of expression of clock genes that are targets for RORs. This suggests that RIP140 can participate in a feedback loop that connects metabolism to circadian rhythms and that the absence of RIP140 might affect the robustness of the system and its sensitivity to external stimuli.
Supplementary Material
Acknowledgments
We thank Dr. Jaya Nautiyal for her kind help with the RIP140 KO mice, Mary P. Hever for providing excellent technical assistance, and Kelly C. Heim and Roger White for helpful discussion. This work was supported by Wellcome Trust program grant 079200/Z/06/Z, BBSRC grant BB/C504327/1, and National Institutes of Health grant RO1 GM083336.
Footnotes
Supplementary material for this article is available on the Journal of Biological Rhythms website: http://jbr.sagepub.com/supplemental.
References
- Akashi M, Takumi T. (2005) The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol 12:441-448 [DOI] [PubMed] [Google Scholar]
- Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. (2009) Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 17:2100-2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134:317-328 [DOI] [PubMed] [Google Scholar]
- Augereau P, Badia E, Fuentes M, Rabenoelina F, Corniou M, Derocq D, Balaguer P, Cavailles V. (2006) Transcriptional regulation of the human NRIP1/RIP140 gene by estrogen is modulated by dioxin signalling. Mol Pharmacol 69:1338-1346 [DOI] [PubMed] [Google Scholar]
- Berriel DM, Krones-Herzig A, Metzger D, Ziegler A, Vegiopoulos A, Klingenspor M, Muller-Decker K, Herzig S. (2008) Nuclear receptor cofactor receptor interacting protein 140 controls hepatic triglyceride metabolism during wasting in mice. Hepatology 48:782-791 [DOI] [PubMed] [Google Scholar]
- Christian M, White R, Parker MG. (2006) Metabolic regulation by the nuclear receptor corepressor RIP140. Trends Endocrinol Metab 17:243-250 [DOI] [PubMed] [Google Scholar]
- Damiola F, Le MN, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. (2000) Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14:2950-2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardente H, Cermakian N. (2007) Molecular circadian rhythms in central and peripheral clocks in mammals. Chronobiol Int 24:195-213 [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. (1998) Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280:1599-1603 [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. (2010) The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72:517-549 [DOI] [PubMed] [Google Scholar]
- Duguay D, Cermakian N. (2009) The crosstalk between physiology and circadian clock proteins. Chronobiol Int 26:1479-1513 [DOI] [PubMed] [Google Scholar]
- Dunlap JC. (1999) Molecular bases for circadian clocks. Cell 96:271-290 [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan K, Sassone-Corsi P. (2009) Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol 16:462-467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froy O. (2010) Metabolism and circadian rhythms: implications for obesity. Endocr Rev 31:1-24 [DOI] [PubMed] [Google Scholar]
- Gamsby JJ, Loros JJ, Dunlap JC. (2009) A phylogenetically conserved DNA damage response resets the circadian clock. J Biol Rhythms 24:193-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. (2008) The meter of metabolism. Cell 134:728-742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EA, Jr., Staknis D, Weitz CJ. (1999) Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286:768-771 [DOI] [PubMed] [Google Scholar]
- Hallberg M, Morganstein DL, Kiskinis E, Shah K, Kralli A, Dilworth SM, White R, Parker MG, Christian M. (2008) A functional interaction between RIP140 and PGC-1alpha regulates the expression of the lipid droplet protein CIDEA. Mol Cell Biol 28:6785-6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Spiegelman BM. (2006) Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev 27:728-735 [DOI] [PubMed] [Google Scholar]
- Heim KC, Gamsby JJ, Hever MP, Freemantle SJ, Loros JJ, Dunlap JC, Spinella MJ. (2009) Retinoic acid mediates long-paced oscillations in retinoid receptor activity: evidence for a potential role for RIP140. PLoS One 4:e7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog B, Hallberg M, Seth A, Woods A, White R, Parker MG. (2007) The nuclear receptor cofactor, receptor-interacting protein 140, is required for the regulation of hepatic lipid and glucose metabolism by liver X receptor. Mol Endocrinol 21:2687-2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson-Davies, Pocock V, White R, Parker M, Milligan SR. (2008) Disturbances in core body temperature in RIP140-null mice. J Therm Biol 34:100-108 [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. (2009) Harmonics of circadian gene transcription in mammals. PLoS Genet 5:e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten AM. (2009) Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal 7:1-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JD, Frost G, Otway DT. (2009) Adipose tissue, adipocytes and the circadian timing system. Obes Rev 10 Suppl 2:52-60 [DOI] [PubMed] [Google Scholar]
- Journiac N, Jolly S, Jarvis C, Gautheron V, Rogard M, Trembleau A, Blondeau JP, Mariani J, Vernet-der GB. (2009) The nuclear receptor ROR{alpha} exerts a bi-directional regulation of IL-6 in resting and reactive astrocytes. Proc Natl Acad Sci U S A 106:21365-21370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka A, Bass J. (2007) A sense of time: how molecular clocks organize metabolism. Trends Endocrinol Metab 18:4-11 [DOI] [PubMed] [Google Scholar]
- Laitinen S, Fontaine C, Fruchart JC, Staels B. (2005) The role of the orphan nuclear receptor Rev-Erb alpha in adipocyte differentiation and function. Biochimie 87:21-25 [DOI] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. (2008) Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A 105:15172-15177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. (2008) The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem 283:18411-18421 [DOI] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. (2001) Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107:855-867 [DOI] [PubMed] [Google Scholar]
- Leonardsson G, Steel JH, Christian M, Pocock V, Milligan S, Bell J, So PW, Medina-Gomez G, Vidal-Puig A, White R, Parker MG. (2004) Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc Natl Acad Sci U S A 101:8437-8442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. (2008) Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet 4:e1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Li S, Liu T, Borjigin J, Lin JD. (2007) Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature 447:477-481 [DOI] [PubMed] [Google Scholar]
- Liu Y, Heintzen C, Loros J, Dunlap JC. (1999) Regulation of clock genes. Cell Mol Life Sci 55:1195-1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402-408 [DOI] [PubMed] [Google Scholar]
- Morganstein DL, Christian M, Turner JJ, Parker MG, White R. (2008) Conditionally immortalized white preadipocytes: a novel adipocyte model. J Lipid Res 49:679-685 [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. (2008) The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134:329-340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal J, Steel JH, Rosell MM, Nikolopoulou E, Lee K, Demayo FJ, White R, Richards JS, Parker MG. (2010) The nuclear receptor cofactor receptor-interacting protein 140 is a positive regulator of amphiregulin expression and cumulus cell-oocyte complex expansion in the mouse ovary. Endocrinology 151:2923-2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol D, Christian M, Steel JH, White R, Parker MG. (2006) RIP140 expression is stimulated by estrogen-related receptor alpha during adipogenesis. J Biol Chem 281:32140-32147 [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109:307-320 [DOI] [PubMed] [Google Scholar]
- Puigserver P. (2005) Tissue-specific regulation of metabolic pathways through the transcriptional coactivator PGC1-alpha. Int J Obes (Lond) 29:S5-S9 [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. (2006) Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet 38:369-374 [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. (2004) A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43:527-537 [DOI] [PubMed] [Google Scholar]
- Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. (2010) The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev 24:345-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A, Steel JH, Nichol D, Pocock V, Kumaran MK, Fritah A, Mobberley M, Ryder TA, Rowlerson A, Scott J, et al. (2007) The transcriptional corepressor RIP140 regulates oxidative metabolism in skeletal muscle. Cell Metab 6:236-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AY, Bernal TU, Pillsbury ML, Yamamoto KR, Feldman BJ. (2009) Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc Natl Acad Sci U S A 106:17582-17587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Pei L, Evans RM. (2008) Nuclear receptors: decoding metabolic disease. FEBS Lett 582:2-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. (2005) Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308: 1043-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. (2005) System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet 37:187-192 [DOI] [PubMed] [Google Scholar]
- White R, Morganstein D, Christian M, Seth A, Herzog B, Parker MG. (2008) Role of RIP140 in metabolic tissues: connections to disease. FEBS Lett 582:39-45 [DOI] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. (2006) Nuclear receptor expression links the circadian clock to metabolism. Cell 126:801-810 [DOI] [PubMed] [Google Scholar]
- Yang X, Lamia KA, Evans RM. (2007) Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb Symp Quant Biol 72:387-394 [DOI] [PubMed] [Google Scholar]
- Yin L, Lazar MA. (2005) The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol 19:1452-1459 [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. (2004) PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 101:5339-5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschiedrich I, Hardeland U, Krones-Herzig A, Berriel DM, Vegiopoulos A, Muggenburg J, Sombroek D, Hofmann TG, Zawatzky R, Yu X, et al. (2008) Coactivator function of RIP140 for NFkappaB/RelA-dependent cytokine gene expression. Blood 112:264-276 [DOI] [PubMed] [Google Scholar]
- Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, Wu X, Goh BC, Mynatt RL, Gimble JM. (2006) Characterization of peripheral circadian clocks in adipose tissues. Diabetes 55:962-970 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.