Abstract
Ionotropic glutamate receptors (iGluRs), including the NMDA receptor subtype, are ligand-gated ion channels critical to fast signaling in the CNS. NMDA receptors are obligate heterotetramers composed of two GluN1 and typically two GluN2 subunits. However, the arrangement of GluN subunits in functional receptors—whether like subunits are adjacent to (N1/N1/N2/N2) or diagonal to (N1/N2/N1/N2) one another—remains unclear. Recently, a crystal structure of a homomeric AMPA receptor revealed that the four identical subunits adopt two distinct and subunit-specific conformations termed A/C and B/D with subunits of like conformations (e.g., A/C) diagonal to one another. In the structure, the two conformers were notable at the level of the linkers (S1–M1, M3–S2, and S2–M4) that join the ligand-binding domain to the transmembrane ion channel with the M3–S2 linker positioned more proximal to the central axis of the channel pore in the A/C conformation and S2–M4 more proximal in the B/D conformation. Using immunoblots and functional assays, we show that introduced cysteines in the M3/M3–S2 linker of GluN1, but not GluN2, show dimer formation and oxidation-induced changes in current amplitudes predictive of the A/C conformation. Conversely, introduced cysteines in the S2–M4 linker of GluN2, but not GluN1, showed similar functional effects, suggesting that the GluN2 subunit adopts the B/D conformation. Thus, we show that NMDA receptors, like AMPA receptors, possess distinct subunit-specific conformations with GluN1 approximating the A/C and GluN2 the B/D conformation. GluN subunits are therefore positioned in a N1/N2/N1/N2 arrangement in functional NMDA receptors.
Introduction
Glutamate-activated NMDA receptors are integral in the transduction and modulation of synaptic activity underlying neurodevelopment (Mattson, 2008) and higher-order cognitive functions (Lau and Zukin, 2007; Citri and Malenka, 2008). Perturbations in glutamatergic transmission exacerbate numerous brain diseases, including psychiatric, neurodegenerative, and excitotoxic disorders (Kalia et al., 2008). NMDA receptors are obligate heterotetramers typically composed of the ubiquitous GluN1 subunit and either GluN2(A–D) and/or GluN3(A and B) subunits (Cull-Candy and Leszkiewicz, 2004; Paoletti and Neyton, 2007). The resulting diversity in receptor composition contributes to differences in NMDA receptor biogenesis, trafficking, posttranslational modifications, cellular distribution, and biophysical properties (Cull-Candy and Leszkiewicz, 2004; Paoletti and Neyton, 2007; Piña-Crespo et al., 2010; Traynelis et al., 2010).
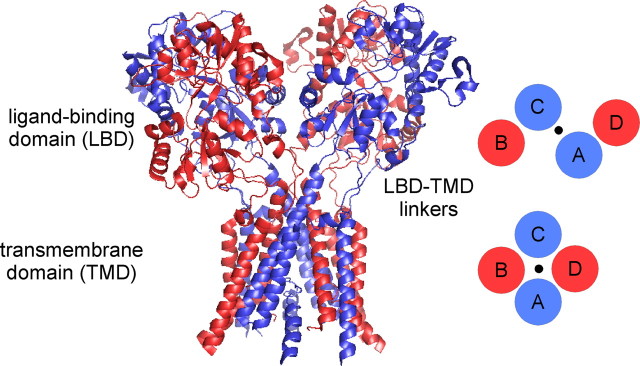
At most synapses, functional NMDA receptors are formed by two GluN1 and two GluN2 subunits arranged as a dimer of dimers. Although the dimer pair is almost certainly GluN1/GluN2 (Furukawa et al., 2005; Gielen et al., 2008; Lee and Gouaux, 2011), the arrangement of subunits in functional tetrameric complexes—whether identical subunits are positioned adjacent to (N1/N1/N2/N2) (Schorge and Colquhoun, 2003) or diagonal to (N1/N2/N1/N2) (Sobolevsky et al., 2009) one another—remains uncertain. Surprisingly, in a crystal structure of a homomeric AMPA receptor, four identical GluA2 subunits adopted two distinct conformations, termed A/C and B/D, with like conformers diagonal to one another (Fig. 1) (Sobolevsky et al., 2009). These two distinct conformations were notable at the level of the linkers (S1–M1, M3–S2, and S2–M4) that couple the ligand-binding domain (LBD) (S1, S2) to the ion channel formed by the transmembrane domain (TMD) (M1–M4). These linkers accommodate the symmetry mismatch between the LBD (two-fold symmetry) and the TMD (four-fold symmetry) (Fig. 1) with those linkers in diagonal subunits adopting a common spatial relationship.
Figure 1.
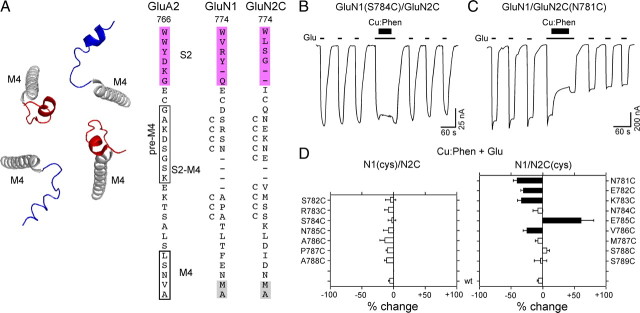
Individual subunits within tetrameric ionotropic glutamate receptors adopt two distinct subunit conformations. Structure of a homomeric glutamate receptor (GluA2cryst, PDB accession code 3KG2) (Sobolevsky et al., 2009) depicting the organization of individual subunits at the level of the LBD and the TMD. The LBD is organized as a dimer of dimers comprised of individual dimer pairs A/D and B/C, whereas the ion channel within the TMD is organized as a tetramer displaying four-fold symmetry (schematics on the right). Four identical subunits showed two distinct subunit conformations termed A/C (blue) and B/D (red) with like conformers diagonal to one another. The two distinct subunit conformations are notable at the level of the LBD–TMD linkers that mediate the transition from the two-fold symmetry of the LBD to the four-fold symmetry of the TMD.
For obligate heteromeric NMDARs, if like subunits (e.g., GluN1) are diagonal to one another (N1/N2/N1/N2), then they must adopt either an A/C or a B/D conformation (Sobolevsky et al., 2009). In contrast, if like subunits are adjacent to one another (N1/N1/N2/N2), then the adjacent subunits (e.g., GluN1 and GluN1) must adopt different conformations, one A/C and the other B/D. Sobolevsky et al. (2009) proposed that like subunits in NMDA receptors are diagonal to one another with GluN1 adopting the A/C and GluN2 the B/D conformation. However, these experiments are limited in scope. Only a small number of introduced cysteines were tested for cross-linking (as an index of proximity). Further, the identified receptors, assayed solely by immunoblots, do not necessarily represent functional receptors. During biogenesis, many positions can transiently exist in close proximity to one another (e.g., Clarke and Fersht, 1993), thus forming disulfide bonds independent of their positioning in a mature, functional receptor. Finally, the results provided no positive evidence for the predicted B/D conformation of the GluN2 subunit.
Taking advantage of substituted cysteines in the LBD–TMD linkers, we find strong subunit-specific patterns of cross-linking in functional NMDA receptor subunits. The patterns of cross-linking are consistent with GluN1 and GluN2 approximating the A/C and B/D conformations, respectively, therefore positioning like subunits diagonal to one another in functional tetrameric complexes.
Materials and Methods
Materials
The GluN1 glycine-site antagonist, 5,7-dichlorokynurenic acid (DCKA), was purchased from Tocris Bioscience. All other reagents including the GluN2 glutamate-site antagonist, dl-2-amino-phosphonopentanoic acid (APV), CuSO4, phenanthroline, and dithiothreitol (DTT), were purchased from Sigma Chemicals.
Mutagenesis and expression
Cysteine substitutions in the rat GluN1a (accession #P35439), GluN2A (Q00959), and GluN2C (Q00961) subunits were generated as described previously (see Talukder et al., 2010) (and references therein). All numbering is for the mature protein using signal peptides of lengths of 18 (GluN1), 19 (GluN2A), and 19 (GluN2C) aa. The GluN2A background we used in oocytes had an endogenous cysteine replaced with alanine (C399A) (Choi et al., 2000). Although this mutation has been used previously to prevent MTS reagents from reacting with the GluN2A subunit (e.g., Talukder et al., 2010), it does not alter the effects of reducing or oxidizing agents on NMDA receptors. Wild-type and mutant GluN1 and GluN2A or GluN2C mRNA (0.01–0.1 μg/μl with 50–75 nl injected per oocyte) were coexpressed in female Xenopus laevis oocytes (Sobolevsky et al., 2002). Wild-type and mutant GluN1 and GluN2A DNA (4 μg) were transfected into HEK 293 cells using Fugene (Roche Molecular Biochemicals).
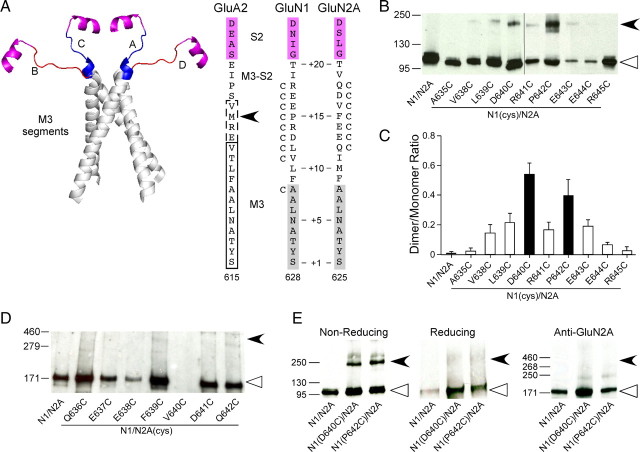
The α-helical extent of the M3 segment at its C-terminal end depends on the specific conformer with M3 in the A/C conformer extended by about one turn of an α-helix (Fig. 2A, right, dashed box) compared to the B/D conformer (Fig. 2A, right, solid box). Correspondingly, the M3–S2 linker (from the C-terminal end of M3 to Helix E in the S2 segment) also varies in length between conformers. Our experiments in this region encompass positions in the C-terminal portion of the M3 segment as well as the M3–S2 linker. Thus, to encompass all positions tested in the present study, we refer to them as the M3/M3–S2 linker.
Figure 2.
Receptors with cysteines substituted in the GluN1 but not in the GluN2A M3/M3–S2 linker yield dimers. A, Backbone structure (left) of the M3 transmembrane segment (gray)—the major pore-forming domain—and the M3/M3–S2 linkers (A/C, blue; B/D, red) that connect the M3 transmembrane segment to helix E of the LBD (magenta) (GluA2cryst, PDB accession code 3KG2). Sequence alignment (right) of residues in and around the M3/M3–S2 linker in AMPA GluA2 and NMDA GluN1 and GluN2A. Proximal S2 (helix E) is shown in magenta. For GluA2, the boxed regions indicate the α-helical extent of the A/C (dashed lines) or B/D (solid lines) conformation of M3 (Sobolevsky et al., 2009). For NMDA receptor subunits, M3, as defined by hydrophobicity (Biology Workbench), is highlighted in gray. Substitution of M629 with cysteine in GluA2 (arrowhead) yielded dimers in immunoblots (Sobolevsky et al., 2009). Positions substituted with cysteine are indicated by a “C” adjacent to the native residue. Numbering is for the mature protein (see Materials and Methods). We also used a relative numbering system, referencing the initial serine (S) in the highly conserved SYTANLAAF motif as +1 (Jones et al., 2002). B, D, Immunoblots of membrane-purified proteins isolated from Xenopus oocytes injected with wild-type or cysteine-substituted GluN1 (B) or GluN2A (D) subunits. Protein expression was assayed using antibodies against the N-terminal domain of GluN1 (B) or GluN2A (D). Open triangles and arrowheads indicate approximate location of monomer (GluN1, 114 kDa; GluN2A, 173 kDa) and dimer (GluN1, 228 kDa; GluN2A, 346 kDa) bands, respectively. The GluN2A gel was overexposed to illustrate the lack of subunit-specific dimers. Note receptors containing GluN2A(V640C) (D) showed functional currents and positive monomer signal when probed with the anti-GluN1, but we could not detect a monomer band using the anti-GluN2A antibody. C, Quantification of band intensity (see Materials and Methods) for wild-type and GluN1 cysteine-substituted mutants (n > 4 for each). Filled bars indicate values statistically different from GluN1/GluN2A (p < 0.05). E, Immunoblots of membrane-purified proteins isolated from HEK 293 cells probed for anti-GluN1 under nonreducing (left) or reducing (100 mm DTT, middle) conditions or for anti-GluN2A (right) demonstrating subunit-specific dimer formation. Open triangles and arrowheads indicate approximate location of monomer (GluN1, 114 kDa; GluN2A, 173 kDa) and dimer (GluN1, 228 kDa; GluN2A, 346 kDa) bands, respectively.
Whole-cell current recordings and data analysis
Whole-cell currents of Xenopus oocytes were recorded at room temperature (20°C) using two-microelectrode voltage clamp (DAGAN TEV-200A, DAGAN) with Cell Works software (npi electronic) (Sobolevsky et al., 2002). When recording GluN1/GluN2C, the external solution consisted of the following (in mm): 115 NaCl, 2.5 KCl, 0.18 CaCl2, and 5 HEPES (pH 7.2, NaOH). When recording GluN1/GluN2A, the same solution was used except BaCl2 was substituted for CaCl2 (to prevent Ca2+-dependent desensitization) and 100 μm EDTA (to minimize Zn2+-mediated modification) was added to the external solution. All reagents, including glutamate (200 μm), glycine (20 μm), APV (100 μm), DCKA (10 μm), copper(II):phenanthroline (Cu:Phen) (2:50 μm), and DTT (either 1 or 4 mm), were applied with the bath solution.
Experimental protocols
NMDAR wild-type and cysteine-substituted mutant channels were probed from the extracellular side with oxidizing (Cu:Phen) and reducing (DTT) reagents. Stock solutions of phenanthroline (0.5 m) and CuSO4 (0.1 m) were diluted to the experimental concentrations in the external solution immediately before the experiment.
Steady-state reactions were quantified at a holding potential of −60 mV (see Fig. 3B,C,E). Baseline glutamate-activated current amplitudes (Ipre) were established by three to five 15 s applications of glutamate and glycine. All agonist or any other reagent applications were separated by 30–120 s washes in glutamate-free solution. Cu:Phen (or DTT) was applied for 60 s either in the presence of agonists or in their absence (but in the presence of the competitive antagonists APV and DCKA). After exposure to redox reagent, current amplitudes (Ipost) were determined again using three to five agonist applications. The change in glutamate-activated current amplitude, expressed as a percentage (%change), was calculated as follows: %change = 100 × (Ipost − Ipre)/Ipre. In certain instances, we corrected for observed current amplitude rundown by fitting a single exponential function to a minimum of three pre-redox reagent glutamate-activated current amplitudes.
Figure 3.
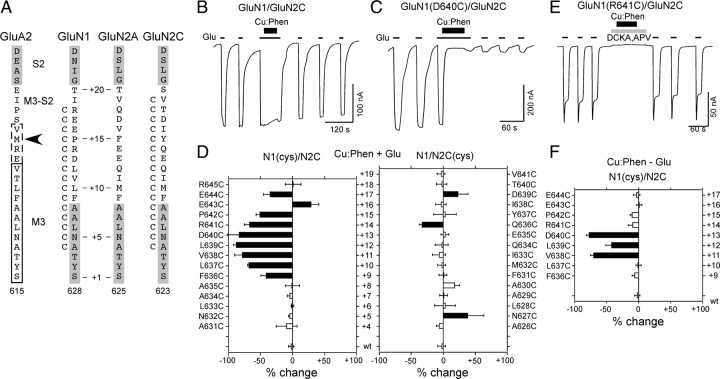
Receptors with substituted cysteines in the M3/M3–S2 linker of GluN1 but not GluN2C show widespread current inhibition following oxidation. A, Same as Figure 2A (right) but including GluN2C. Proximal portion of Helix E (S2) is indicated in gray. B, C, Example current recordings from oocytes showing the effect of Cu:Phen (2:50 μm, thick lines, 60 s) in the presence of agonists (open and closed states) on GluN1/GluN2C (B) or GluN1(D640C)/GluN2C (C). Currents were elicited by glycine (20 μm) and glutamate (200 μm) (thin lines labeled with “Glu”) at a holding potential of −60 mV. D, Mean percentage change (±2 SEM) (n ≥ 3 for each) of glutamate-activated current amplitudes measured before (Ipre) and after (Ipost) exposure of wild-type and cysteine-substituted GluN1 (left) or GluN2C (right) to Cu:Phen in the presence of agonists [%change = 100 × (Ipost − Ipre)/Ipre]. Left and right pointing bars indicate inhibition and potentiation, respectively. Filled bars indicate values significantly different from wild-type GluN1/GluN2C (p < 0.05). The small current potentiation seen for GluN2C(N627C) and GluN2C(A630C) is transient and reflects the effect of Cu:Phen in the presence of agonists. E, Example recording of Cu:Phen applied (60 s) in the absence of agonists (closed state), but in the presence of the competitive antagonists DCKA (10 μm) and APV (100 μm) (gray box). F, Mean percentage change (± 2 SEM) (n > 3) of glutamate-activated current amplitudes, measured in the absence of agonists, for GluN1 cysteine-substituted positions that showed significant effects in the presence of agonists. Results are shown as in D.
Protein chemistry
Fractionation of membrane proteins.
Ten to twelve healthy oocytes were injected with 1–2 ng of mRNA. Two to three days after transfection (HEK 293 cells)/injection (oocytes) cells were washed in PBS, fixed in PBS containing 0.58 mmN-ethylmaleimide (NEM, Pierce), homogenized in lysis buffer (20 mm Tris, 0.58 mm NEM), and centrifuged at 3000 RPM (Eppendorf Centrifuge 5417R) for 3 min at 4°C. The supernatant was recovered and centrifuged at 40,000 RPM (Beckman TLA 120.2 rotor) for 10 min at 4°C. The pellet was rinsed in PBS and recentrifuged at 40,000 RPM. The resulting pellet was resuspended and first sonicated in solubilization buffer [20 mm Tris, 50 mm NaCl, 1/1000 protease inhibitor cocktail (Sigma), 0.58 mm NEM] without detergent and then incubated with detergent (0.03% Na-deoxycholate, 1% Triton X-100) for 1 h at 4°C. Solubilized proteins were centrifuged at 40,000 RPM for 20 min at 4°C and membrane proteins contained in the supernatant were separated by SDS-PAGE under nonreducing or reducing (100 mm DTT) conditions.
Oocytes treated with Cu:Phen were harvested in the same manner as NEM-treated oocytes except for two differences. First, oocytes were washed in PBS and then fixed in PBS containing only Cu:Phen (1:50 μm) and the competitive antagonists APV (100 μm) and DCKA (10 μm). Additionally, Cu:Phen/APV/DCKA and NEM were present in the lysis and solubilization solutions.
Immunoblotting.
Proteins were transferred from the gel to 0.45 mm nitrocellulose membranes by semidry transfer (Bio-Rad) using Bjerrum–Schaffer–Nielsen buffer. Blots were blocked and incubated with primary antibody, either anti-GluN1 (1:500, Millipore MAB363) or anti-GluN2A (1:500, Millipore AB1555P), overnight. Blots were washed before incubation with HRP-conjugated goat anti-mouse (sc-2302) or HRP-conjugated goat anti-rabbit (sc-2030) and developed using WB luminol reagent (sc-2048, all reagents Santa Cruz Biotechnologies) before exposure to chemiluminescence Biomax Film (Kodak).
Quantification of immunoblots.
Immunoblot films were scanned using an EPSON flat scanner (Epson Perfection V700 Photo) in an 8 bit gray scale mode, 1200 dpi, reflective and quantified using NIH ImageJ (1.38×) on a Windows XP platform. To quantify band densities, we defined a box encompassing the size of the largest monomer band. For each lane in the gel, this box was placed in the presumed location of the monomer and dimer bands as well as in a region in the leading edge (no protein sample) to define background. The measured density of the background was subtracted out from the density for the monomer and dimer bands, and a ratio of the background-subtracted dimer and monomer densities (dimer/monomer ratio) was expressed in arbitrary units.
To minimize differences in saturation levels in monomer/dimer band densities, we tried to make the monomer band densities approximately equal between groups (i.e., NEM- and Cu:Phen-treated samples). To accomplish this, we typically ran an initial gel where every sample was loaded at 10 μl, and would subsequently run a second gel where loading volumes were varied to generate comparable monomer band densities (typically 5 μl for NEM-treated samples and 15 μl for Cu:Phen-treated samples). Even with this correction, the monomer bands tended to have higher density for the NEM-treated condition (average density was ∼550,000 arbitrary units for NEM-treated compared to ∼450,000 for Cu:Phen-treated). A higher monomer band density tended to favor the dimer/monomer ratio, further validating the statistical outcomes in Figure 4F (where dimer/monomer ratios were higher for certain constructs in Cu:Phen-treated condition).
Figure 4.
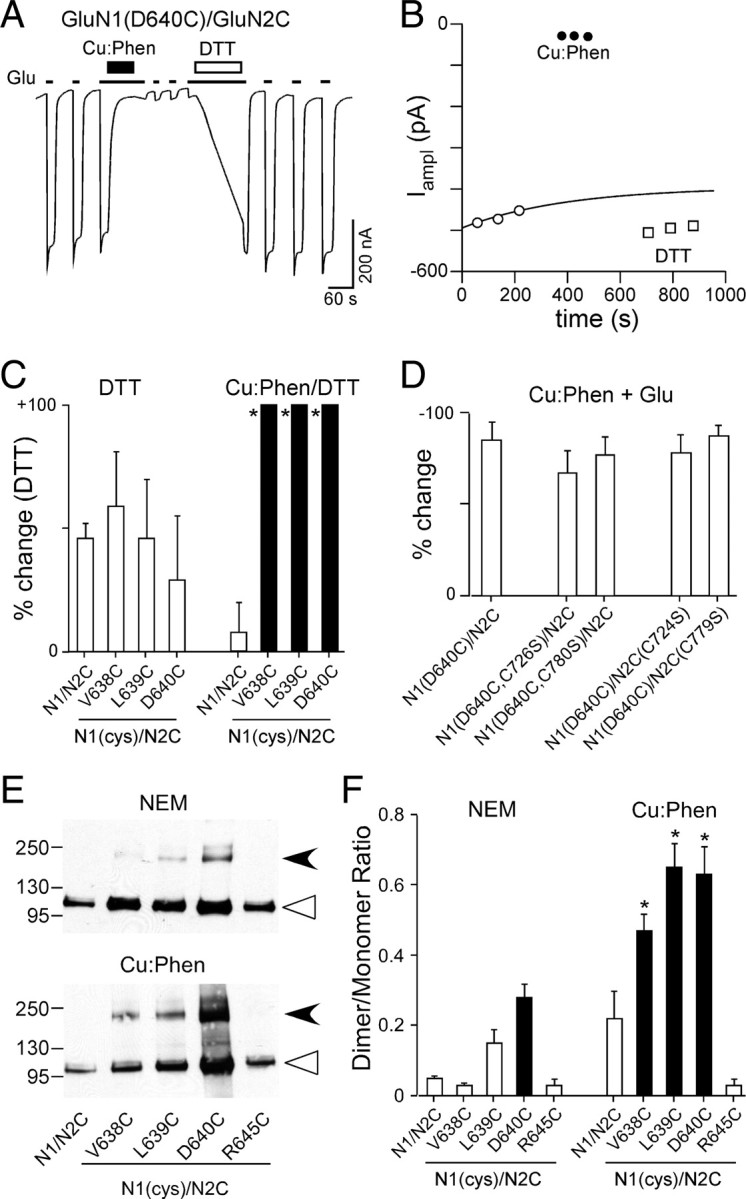
Current inhibition reflects cross-linking of substituted cysteines. A, Effect of DTT (1 mm), applied in the presence of agonists, on Cu:Phen-induced inhibited currents in GluN1(D640C)/GluN2C. B, Glu-activated current amplitudes for GluN1(D640C)/GluN2C recorded before (open circles) and after Cu:Phen (filled circles) or DTT (open squares) (amplitudes are from trace shown in A). C, Mean percentage change (±2 SEM) (n > 3) of glutamate-activated current amplitudes for wild-type and GluN1 cysteine-substituted positions that showed significant effects of Cu:Phen in the absence of agonists (Fig. 3F). %change is measured following DTT (Ipost) either relative to currents before any treatment (DTT) (Table 2) or to currents following Cu:Phen (Cu:Phen/DTT) (as in Fig. 4A,B). Filled bars indicate values significantly different from GluN1/GluN2C within treatment groups (p < 0.05). Asterisks indicate values statistically different between treatment groups for equivalent positions (p < 0.05). The mean %change for DTT in the Cu:Phen/DTT condition was 540 ± 100%, n = 5 (V638C), 150 ± 15%, n = 4 (L639C), and 520 ± 170%, n = 8 (D640C). D, Mean percentage change (± 2 SEM) (n > 4) of glutamate-activated current amplitudes for GluN1(D640C)/GluN2C or the same constructs with endogenous cysteines neutralized to serine (S) measured before or after Cu:Phen treatment in the presence of agonists. None of the values were significantly different from that for GluN1(D640C)/GluN2C, indicating that the effect of Cu:Phen arises from introduced cysteines. E, Immunoblots of membrane-purified proteins isolated from Xenopus oocytes injected with wild-type or cysteine-substituted GluN1 subunits coexpressed with GluN2C treated without (NEM, top) or with (Cu:Phen, bottom) Cu:Phen. Protein expression was assayed using antibodies against the N-terminal domain of GluN1. Open triangles and arrowheads indicate approximate location of monomer (114 kDa) and dimer (228 kDa) bands, respectively. F, Quantification of band intensity (see Materials and Methods) for wild-type and GluN1 cysteine-substituted receptors (n > 4 for each) either without (left) or with (right) Cu:Phen treatment. Filled bars indicate values statistically different from GluN1/GluN2C within treatment groups (p < 0.05). Asterisks indicate values statistically different between treatment groups for equivalent positions (p < 0.05).
Data analysis
Data analysis was done using Igor Pro (Wavemetrics) and Microsoft Excel. For analysis and illustration, leak currents were subtracted from total currents. An ANOVA or Student's t test was used to define statistical differences. The Tukey or Dunnett's test was used for multiple comparisons. Significance was assumed at p < 0.05.
Results
NMDA receptors with introduced cysteines in the M3/M3–S2 linker of GluN1 but not GluN2A yield dimers
In the closed state for AMPA receptors, the C-terminal end of the M3 segment and the M3–S2 linker, referred to as the M3/M3–S2 linker (see Materials and Methods), are proximal in the A/C conformer (Fig. 2A, left) with the closest approach in the C-terminal end occurring at M629 (distance between α-carbons 12 Å) (Fig. 2A, right, arrowhead). The M3/M3–S2 linkers of the B/D conformer (Fig. 2A, left) are more distal (M629 separated by 30.2 Å) (Table 1) (Sobolevsky et al., 2009). Introduced cysteines at the homologous position in the NMDA receptor GluN1 subunit (P642, position +15) (Fig. 2A), but not in the GluN2A subunit (F639), yielded dimers on immunoblots, suggesting that the M3/M3–S2 linkers are proximal in GluN1 (Sobolevsky et al., 2009). However, distal to the highly conserved SYTANLAAF motif (positions +1 to +9) in the M3 transmembrane segment, there is divergence in the identity and nature of the residues in the M3/M3–S2 linkers between AMPA and NMDA receptor subunits, as well as among NMDA receptor subunits (Fig. 2A, right). Hence, the detailed arrangement of the linkers may differ between subunits. We therefore introduced cysteines over a wider range of positions in NMDA receptor subunits (Fig. 2A, right) and characterized these constructs by immunoblots (Fig. 2B,D,E).
Table 1.
Distance between α-carbons for homologous positions in the M3 segment and M3–S2 linker in the A/C and B/D subunits in GluA2cryst
| GluA2 | Distance between αCs (Å) |
GluN1 | GluN2A | GluN2C | |
|---|---|---|---|---|---|
| A/C | B/D | ||||
| P632 | 26.6 | 49.6 | R | Q | T |
| S631 | 19.7 | 42.7 | E | D | D |
| V630 | 18.2 | 35.8 | E | V | I |
| M629 | 12.0 | 30.2 | P | F | Y |
| R628 | 16.9 | 22.7 | R | E | Q |
| E627 | 19.1 | 19.6 | D | E | E |
| V626 | 12.7 | 12.2 | L | Q | Q |
| T625 | 11.3 | 10.2 | V | I | I |
| L624 | 14.9 | 15.9 | L | M | M |
| F623 | 17.0 | 17.2 | F | F | F |
| A622 | 10.4 | 10.8 | A | A | A |
Shown are residues in GluA2cryst (Sobolevsky et al., 2009) (PDB ID 3KG2) and homologous positions in GluN1, GluN2A, or GluN2C. Underlined residues for GluA2cryst are part of the M3 helix for the A/C and B/D subunits. Bold residues are the additional helical component of the A/C subunit. Average distances for positions shown are 16.3 Å (A/C) and 24.3 Å (B/D).
For GluN1 (Fig. 2B) and consistent with previous results (Sobolevsky et al., 2009), a dimer band (filled arrowhead) was prominent for P642C. In addition, D640C also showed a prominent dimer, whereas dimers of decreased and inconsistent intensity were observed for V638C, L639C, R641C, E643C, and E644C (Fig. 2B,C). No dimers were detected for wild-type (GluN1/GluN2A), A635C, or R645C (Fig. 2B,C). Observed dimers for D640 and P642 were not dependent on expression system, were absent under reducing conditions, and were specific to GluN1 with no dimers detected when probed with anti-GluR2A (Fig. 2E). In contrast, no dimer bands were identified for the tested M3/M3–S2 GluN2A positions (Fig. 2D).
These results are consistent with the M3/M3–S2 linkers of GluN1 being positioned proximal to one another as in the A/C conformer. Also notable is that positions presumably located even more proximal to one another (e.g., A635 in GluN1 corresponding to A622 in A/C or B/D conformer of GluA2, ∼10.6 Å) (Table 1) do not yield dimers, indicating that other factors (e.g., membrane, local environment, and backbone rigidity) also affect cross-linking. Still, the present results do not address functional receptors.
Functional assays of proximity
To test the general arrangement of the linkers in functional NMDA receptor subunits, we introduced individual cysteines in the M3/M3–S2 linker of either GluN1 or GluN2C (Fig. 3A) and tested the effects of DTT, a reducing agent, or Cu:Phen, an oxidizing agent, on current amplitudes. For these experiments, we preferentially used GluN2C since it shows limited desensitization compared to GluN2A (Krupp et al., 1996).
For wild-type GluN1/GluN2C, DTT applied in the presence of agonists produced an ∼46% potentiation of current amplitudes (46 ± 3%, n = 7; mean %change ± SEM, number of recordings) (Table 2). For all cysteine-substituted M3/M3–S2 positions in GluN1 and GluN2C, DTT applied in the presence of agonists did not produce a significant effect on glutamate-activated currents compared to wild type (Table 2), including GluN1(D640C) and GluN1(P642C).
Table 2.
Effect of DTT on current amplitudes in wild-type and cysteine-substituted GluN1 and GluN2C subunits
| N1(cys)/N2C |
Relative position | N1/N2C(cys) |
||||
|---|---|---|---|---|---|---|
| Construct | %change | n | Construct | %change | n | |
| N1/N2C | 46 ± 3 | 7 | ||||
| N1(R645C)/N2C | 61 ± 6 | 8 | +18 | N1/N2C(T640C) | 60 ± 9 | 5 |
| N1(E644C)/N2C | 68 ± 3 | 8 | +17 | N1/N2C(D639C) | 36 ± 4 | 7 |
| N1(E643C)/N2C | 67 ± 8 | 5 | +16 | N1/N2C(I638C) | 41 ± 9 | 10 |
| N1(P642C)/N2C | 35 ± 1 | 5 | +15 | N1/N2C(Y637C) | 41 ± 12 | 5 |
| N1(R641C)/N2C | 31 ± 8 | 8 | +14 | N1/N2C(Q636C) | 47 ± 17 | 6 |
| N1(D640C)/N2C | 29 ± 4 | 10 | +13 | N1/N2C(E635C) | 42 ± 8 | 5 |
| N1(L639C)/N2C | 46 ± 4 | 9 | +12 | N1/N2C(Q634C) | 47 ± 10 | 4 |
| N1(V638C)/N2C | 59 ± 12 | 11 | +11 | N1/N2C(I633C) | 36 ± 11 | 5 |
| N1(L637C)/N2C | 46 ± 3 | 7 | +10 | N1/N2C(M632C) | 41 ± 11 | 5 |
| N1(F636C)/N2C | 57 ± 7 | 6 | +9 | N1/N2C(F631C) | 38 ± 12 | 5 |
| N1(A635C)/N2C | 42 ± 10 | 6 | +8 | N1/N2C(A630C) | 17 ± 5 | 7 |
Values shown are mean ± SEM. %change was obtained in the presence of agonist as in Figure 3B (with DTT replacing Cu:Phen) and with DTT applied at 1 or 4 mm. None of the values are significantly different from that seen in wild type (ANOVA, p < 0.05). Relative position references the initial serine (S) in the highly conserved SYTANLAAF motif as +1 (Fig. 3A).
An oxidizing agent produces current inhibition of receptors containing substituted cysteines in the M3/M3–S2 linker in GluN1 but not GluN2
To test the effects of the oxidizing agent Cu:Phen on functional receptors (Fig. 3B,C), we compared glycine- and glutamate-activated (referred to as glutamate-activated) current amplitudes before (Ipre) and after (Ipost) exposure to extracellular Cu:Phen (2:50 μm, thick lines) applied in the presence of agonists, where the channel exists both in the open and closed states. For wild-type GluN1/GluN2C (Fig. 3B), Cu:Phen had no significant effect on glutamate-activated current amplitudes (−2 ± 2%, n = 9). In contrast, for GluN1(D640C)/GluN2C (Fig. 3C), Cu:Phen induced a strong and persistent inhibition of current amplitudes (−83 ± 9%, n = 5).
Figure 3D summarizes the effect of Cu:Phen applied in the presence of agonists (Cu:Phen + Glu) on glutamate-activated current amplitudes. For GluN1, numerous (nine) M3/M3–S2 positions (F636 to E644) showed significant effects, either current inhibition (leftward pointing bars) or potentiation (rightward pointing). On the other hand, for homologous positions in GluN2C, only a limited number of positions (three) showed significant effects. Of these, positions N627 and D639 showed significant current potentiation that rapidly reversed over time (data not shown) and may reflect a transient effect of Cu:Phen on glutamate-activated current amplitudes.
The AMPA receptor crystal structure, where the M3/M3–S2 linker is proximal in the A/C conformation, was generated in the presence of a competitive antagonist (Sobolevsky et al., 2009). We therefore tested the reactivity of Cu:Phen in the absence of agonists, but in the presence of the GluN1 and GluN2 competitive antagonists (DCKA and APV, respectively) (Fig. 3E) (closed state), focusing on those positions that showed a significant effect in the presence of agonists (Fig. 3D). As summarized in Figure 3F, a subset of GluN1 positions (V638, L639, and D640) showed significant reactivity to Cu:Phen in the closed state. In contrast, GluN2C(Q636C), the single GluN2C position showing persistent effects (and inhibition) in the presence of agonists, did not show a significant effect in the closed state (−10 ± 3%, n = 6, data not shown), compared to the observed effect in wild-type (−5 ± 2%, n = 8).
Cu:Phen-induced current inhibition reflects cross-linking of introduced cysteines
Persistent changes in current amplitudes induced by Cu:Phen in GluN1 are presumably due to cross-linking of introduced cysteines. To test this assumption, we performed a number of control experiments. Initially, we characterized the effect of the reducing agent DTT on Cu:Phen-induced inhibited currents. If the observed current inhibition following treatment with Cu:Phen is the result of cross-linking of introduced cysteines in the M3/M3–S2 linker of GluN1, then DTT should potentiate current amplitudes more strongly than in wild-type receptors. As illustrated for GluN1(D640C)/GluN2C (Fig. 4A,B), an initial application of Cu:Phen in the presence of agonists strongly reduced current amplitudes (Fig. 4B, solid circles), as shown previously (Fig. 3D). Subsequent application of DTT dramatically potentiated these Cu:Phen-inhibited current amplitudes (Fig. 4A,B, open squares) (516 ± 170%, n = 8).
Figure 4C summarizes the effect of DTT on wild-type and the three GluN1 cysteine-substituted positions (V638, L639, and D640) that showed significant effects following Cu:Phen in the absence of agonists (Fig. 3F). For wild-type receptors, DTT applied in the presence of agonists without any Cu:Phen pretreatment (labeled “DTT” in Fig. 4C) potentiated current amplitudes by ∼46% (46 ± 3%, n = 7), an effect also seen for all three GluN1 cysteine-substituted positions (Table 2; open bars, Fig. 4C, DTT). For wild-type receptors, application of DTT following Cu:Phen treatment [as for GluN1(D640C)/GluN2C in Fig. 4A] potentiated current amplitudes by ∼8% (7.7 ± 6%, n = 4) (Fig. 4C, Cu:Phen/DTT). For the three GluN1 cysteine-substituted positions, DTT significantly potentiated the Cu:Phen-inhibited current amplitudes compared to wild-type recorded under the same conditions (solid bars, Fig. 4C, Cu:Phen/DTT) and compared to the same position measured with DTT alone (asterisks). These results strongly support the idea that current inhibition reflects cross-linking of substituted cysteines.
We also tested whether the introduced cysteine at D640 interacted with endogenous cysteines in GluN1 or GluN2C. In NMDA receptor subunits, four endogenous cysteines are located in the linkers and/or proximal face of the ligand-binding domain (Furukawa et al., 2005): GluN1(C726), GluN1(C780), GluN2C(C724), and GluN2C(C779). Replacing these cysteines with serine (S) did not significantly alter current inhibition for D640C consistent with Cu:Phen inducing cross-linking between introduced cysteines (Fig. 4D).
As an additional test that Cu:Phen treatment induced disulfide cross-linking among GluN1 subunits, we analyzed immunoblots of proteins purified in the absence (NEM) or presence (Cu:Phen) of Cu:Phen for V638, L639, and D640, as well as a control position that did not show a significant effect of Cu:Phen, R645 (Fig. 4E,F). As summarized in Figure 4F, when treated with NEM, position D640C showed a significant dimer/monomer ratio (solid bar) compared to wild-type, as was found for GluN1/GluN2A (Fig. 2C). Further, when treated with Cu:Phen, introduced cysteines at positions V638, L639, and D640 showed significant increases in the dimer/monomer ratio compared to both wild-type treated with Cu:Phen (solid bars) and to the same positions treated with NEM (asterisks).
In summary, these results indicate that current inhibition induced by Cu:Phen is due to cross-linking of introduced cysteines in the M3/M3–S2 linker of the GluN1 subunits. Since this cross-linking occurs only for the GluN1 and not the GluN2C subunit (Fig. 3D,F), the present results strongly support a more proximal positioning of the M3/M3–S2 linker in GluN1 than in the GluN2C subunit. This proximal positioning of the M3–S2 in GluN1 is consistent with it adopting the A/C conformation.
Cross-linking of introduced cysteines in the GluN1 M3/M3–S2 linker is not dependent on the GluN2 subunit
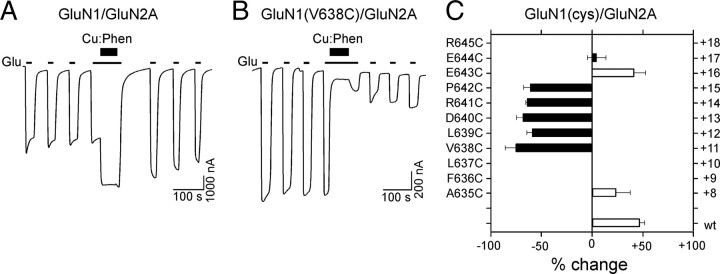
To verify that the observed cross-linking of the M3/M3–S2 linker in GluN1 was independent of the nature of the GluN2 subunit, we coexpressed GluN1 cysteine substituted receptors with GluN2A and tested for the effect of Cu:Phen (Fig. 5). Wild-type GluN1/GluN2A (Fig. 5A,C), in contrast to wild-type GluN1/GluN2C (Fig. 3B,D), showed a significant potentiation (46 ± 5%, n = 7) following Cu:Phen treatment in the presence of agonists. Nevertheless, although the degree of potentiation or inhibition may differ, all tested cysteine-substituted GluN1 positions when coexpressed with GluN2A showed a functional profile (Fig. 5C) similar to that of GluN1(cys)/GluN2C (Fig. 3D). These findings support the conclusion that the observed effects of Cu:Phen on introduced cysteines in the M3/M3–S2 linker of GluN1 are a result of cross-linking among the GluN1 subunits and that this behavior is not dependent on the specific GluN2 subtype.
Figure 5.
Cu:Phen has comparable effects on receptors with substituted cysteines in the GluN1 M3/M3–S2 linker when coexpressed with GluN2A. A, B, Example current recordings from Xenopus oocytes injected with GluN1/GluN2A (A) or GluN1(V638C)/GluN2A (B) mRNA. Cu:Phen was applied in the presence of agonists (e.g., Fig. 3B,C). C, Mean percentage change (±2 SEM) (n > 4) in glutamate-activated current amplitudes measured before (Ipre) and after (Ipost) exposure of wild-type and cysteine-substituted GluN1 coexpressed with GluN2A in the presence of agonists. Left and right pointing bars indicate inhibition and potentiation, respectively. Filled bars indicate values significantly different from wild-type GluN1/GluN2A (p < 0.05). Note that with respect to wild-type GluN1/GluN2C (Fig. 3D), treatment with Cu:Phen produces a persistent potentiation in GluN1/GluN2A; however, the effect of Cu:Phen on the positions tested (except for E643) in both GluN2C (Fig. 3D) and GluN2A was identical.
Cross-linking of substituted cysteines in GluN2C but not GluN1 S2–M4 linker
The proximal positioning of the M3/M3–S2 linker in GluN1 relative to GluN2C is consistent with GluN1 subunits adopting the A/C conformation. However, the M3/M3–S2 results do not provide positive evidence for the positioning of the GluN2 subunits. In the AMPA receptor crystal structure, the S2–M4 linkers are considerably more proximal in the B/D conformation (19.3 Å) (from K776 to K783) than in the A/C conformation (54.6 Å) (Fig. 6A, left, Table 3). Note however, that the specific arrangement of S2–M4 linkers in NMDA receptor subunits is different from that of AMPA receptor subunits due to gaps in the NMDA receptor sequence (Fig. 6A, right). If the GluN2 subunits approximate the B/D conformation, we would then anticipate that introduced cysteines in the S2–M4 linker of GluN2, but not GluN1, might cross-link and affect current amplitudes. We therefore tested a range of substituted cysteines in the S2–M4 linkers of GluN1 and GluN2C (Fig. 6A).
Figure 6.
Receptors with substituted cysteines in the S2–M4 linker of GluN2C but not GluN1 show current inhibition following oxidation. A, Backbone structure (left) of the M4 transmembrane segment (gray) and the S2–M4 linkers of the A/C (blue) and B/D (red) subunits (GluA2cryst, PDB accession code 3KG2). Sequence alignment (right) of residues in and around the S2–M4 linker in AMPA GluA2 and NMDA GluN1 and GluN2C with introduced cysteines indicated by a “C” adjacent to the native residue. The pre-M4 helix in GluA2 is boxed (Sobolevsky et al., 2009). Dashes indicate gaps in aligned sequence. Other features are displayed as in Figure 2A with M4, as defined by hydrophobicity (Biology Workbench), highlighted in gray and helix K of the S2 domain highlighted in magenta. B, C, Example recordings of Cu:Phen applied in the presence of agonists for S2–M4 cysteine substitutions either GluN1(S784C)/GluN2C (B) or GluN1/GluN2C(N781C) (C). D, Mean percentage change (±2 SEM) (n > 3) of glutamate-activated current amplitudes measured before (Ipre) and after (Ipost) exposure of wild-type and cysteine-substituted GluN1 (left) or GluN2C (right) subunits to Cu:Phen in the continuous presence of agonists. Results are shown as in Figure 3D.
Table 3.
Distance between α-carbons for homologous positions in the S2–M4 linker in the A/C and B/D subunits in GluA2cryst
| GluA2 | Distance between αCs (Å) |
GluN1 | GluN2A | GluN2C | |
|---|---|---|---|---|---|
| A/C | B/D | ||||
| K776 | 61.1 | 18.8 | D | H | Q |
| D777 | 59.5 | 18.5 | S | N | N |
| S778 | 61.8 | 23.5 | R | E | E |
| G779 | 56.6 | 22.3 | S | K | K |
| S780 | 51.4 | 13.4 | N | N | N |
| K781 | 52.9 | 18.8 | A | E | E |
| E782 | 47.2 | 18.4 | P | V | V |
| K783 | 45.9 | 21.2 | A | M | M |
| T784 | 40.7 | 24.4 | T | S | S |
| S785 | 34 | 25.2 | L | S | S |
| A786 | 31 | 29.6 | T | Q | K |
| L787 | 29.9 | 28.9 | F | L | L |
| S788 | 35.6 | 34.6 | E | D | D |
Shown are residues in GluA2cryst and homologous positions in GluN1, GluN2A, or GluN2C. Positions highlighted in bold are part of the pre-M4 helix. Based on sequence alignment, there are gaps in the vicinity of the pre-M4 helix in NMDA receptor subunits (see Fig. 6A for additional details). Average distances for positions shown are 46.7 Å (A/C) and 22.9 Å (B/D).
Consistent with the predicted outcome, substituted cysteines in the S2–M4 linker of GluN2C, but not GluN1, showed significant, albeit small, effects of Cu:Phen applied in the presence of agonists (Fig. 6B–D). A subset of these GluN2C positions also showed current inhibition when Cu:Phen was applied in the absence of agonists (Fig. 7A,B), replicating the proximal positioning of the S2–M4 linker as observed in the AMPA receptor crystal structure (Sobolevsky et al., 2009).
Figure 7.
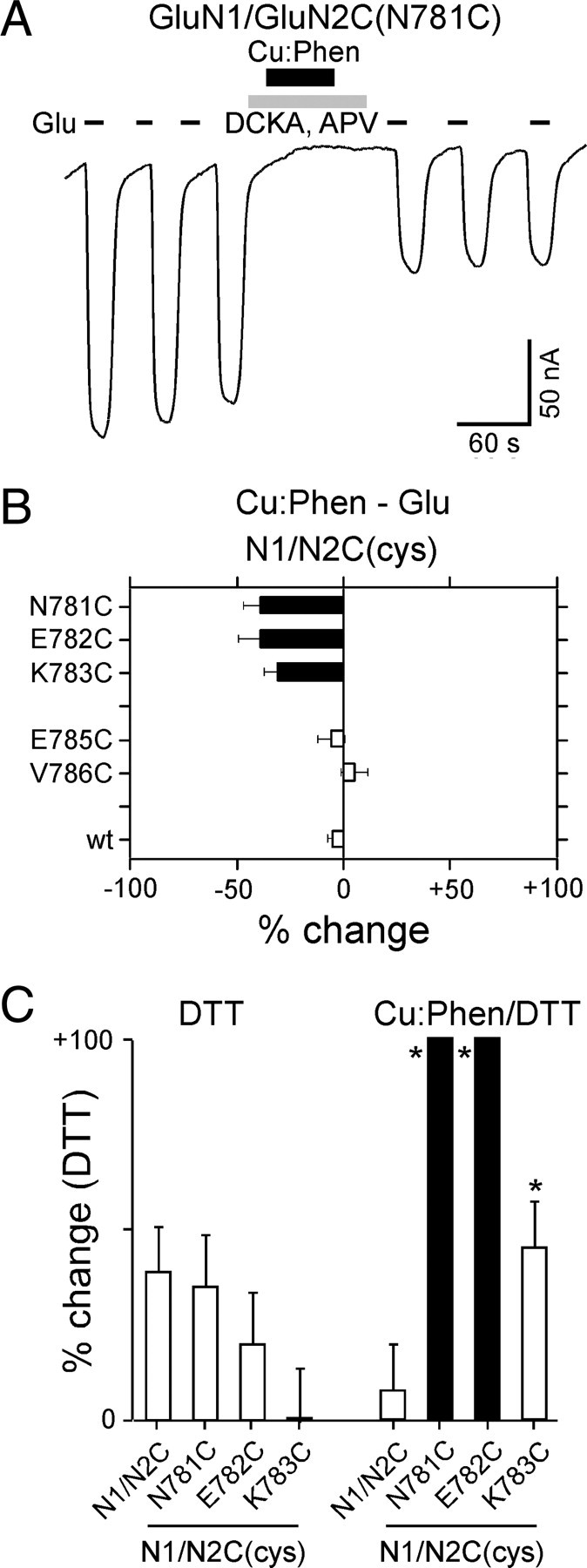
A subset of positions in the GluN2C S2–M4 linker are reactive in the absence of agonists. A, Example recording of Cu:Phen applied in the absence of agonists on GluN1/GluN2C(N781C). B, Mean percentage change (±2 SEM) of glutamate-activated current amplitudes, measured in the absence of agonists, for GluN2C S2–M4 cysteine-substituted positions that showed significant effects in the presence of agonists (Fig. 6D). C, Mean percentage change (±2 SEM) (n > 3) of glutamate-activated current amplitudes for wild-type and GluN2 cysteine-substituted positions that showed significant effects of Cu:Phen in the absence of agonists (Fig. 7B). %change is measured following DTT (Ipost) relative to currents before any treatment (DTT) (raw data not shown) or to currents following Cu:Phen (Cu:Phen/DTT). Filled bars indicate values significantly different from GluN1/GluN2C within treatment groups (p < 0.05). Asterisks indicate values statistically different between treatment groups for equivalent positions (p < 0.05). The mean %change for DTT in the Cu:Phen/DTT condition was 100 ± 30%, n = 6 (N781C), 110 ± 25%, n = 9 (E782C), and 45 ± 12%, n = 5 (K783C).
To verify that the current inhibition was due to disulfide cross-linking, we tested the effect of the reducing agent DTT on those S2–M4 GluN2C positions that were reactive in the absence of agonists (N781, E782, K783) (Fig. 7B) either without initial Cu:Phen treatment (Fig. 7C, DTT) or following Cu:Phen treatment (Fig. 7C, Cu:Phen/DTT). Compared to the potentiation in wild-type, application of DTT without Cu:Phen treatment did not elicit any significant changes in the current amplitudes for the cysteine-substituted receptors (Fig. 7C, DTT), though potentiation was largely absent in K783. In contrast, following Cu:Phen-induced current inhibition, DTT significantly potentiated current amplitudes for both N781C and E782C compared to wild-type when recorded under the same conditions (solid bars, Fig. 7C, Cu:Phen/DTT) and for each N781C, E782C, and K783C compared to the same position measured with DTT alone (asterisks). Interestingly, K783C did not show a significant effect when compared to wild-type (open bar, Fig. 7C, Cu:Phen/DTT), but did show a significant effect when compared to DTT alone (asterisk), suggesting that the mutation itself might alter the redox state of the receptor. Nevertheless, the fact that the DTT potentiation of Cu:Phen-inhibited currents is significantly greater than DTT alone for K783C as well as N781C and E782C supports the idea that current inhibition is due to cross-linking of substituted cysteines.
The reduced magnitude of current inhibition for GluN2C S2–M4 positions reactive in the absence of agonists (Fig. 7B) relative to those for GluN1 M3/M3–S2 (Fig. 3F) may reflect a generally more remote relative positioning: average distance for GluN2C S2–M4 positions (assuming B/D conformation) is 22.9 Å (Table 3), whereas it is 16.3 Å for GluN1 M3/M3–S2 positions (assuming A/C conformation) (Table 1).
In summary, these experiments suggest that the S2–M4 linker in GluN2C is more proximal than that of GluN1. In the context of the AMPAR structure, these results are consistent with the GluN2 subunit approximating the B/D conformation. In combination with the results for the M3/M3–S2 linker, which suggest that GluN1 approximates the A/C conformation, like subunits in the functional tetrameric complex are diagonal to one another in a N1/N2/N1/N2 arrangement.
Discussion
Defining the subunit arrangement in functional ionotropic glutamate receptors (iGluR) is an important advance in understanding mechanisms of iGluR biogenesis, trafficking, and gating. Activation of iGluRs occurs when agonist-induced conformational changes in the LBD are propagated to the TMD resulting in ion channel opening (Sun et al., 2002; Erreger et al., 2004). Three polypeptide linkers (S1–M1, M3–S2, and S2–M4) that join the LBD to the ion channel mediate these agonist-induced effects on ion channel opening/closure. The recent crystal structure of a homomeric AMPA receptor (GluA2) showed that identical subunits adopted two distinct conformations, termed A/C and B/D, which were notable at the level of the LBD–TMD linkers because they took on subunit specific conformations (Sobolevsky et al., 2009). Using immunoblots and whole-cell electrophysiology, we show that functional obligate heteromeric NMDA receptors take on distinct, subunit-specific conformations with GluN1 approximately the A/C conformer and GluN2 approximating the B/D conformer with subunits therefore arranged as N1/N2/N1/N2 (Rambhadran et al., 2010).
Conformers of NMDA receptor subunits
Given the overall sequence homology among the iGluR subtypes and subunits (Traynelis et al., 2010), it is likely that iGluRs (e.g., NMDA and AMPA) share a common LBD–TMD linker structure. Compared to AMPA receptors, the linkers in NMDA receptors vary in their primary sequence and/or length (e.g., S2–M4) (Figs. 3A, 6A), but these differences probably result in small local structural differences rather than changing the overall general arrangement of the linkers. Nevertheless, because of these small differences and the absence of an full-length NMDA receptor structure, NMDA receptor subunits can only be considered to approximate rather than adopt the A/C and B/D conformations.
In this study, introduced cysteines in the M3/M3–S2 linker of GluN1, but not GluN2, showed both dimer formation (Figs. 2B,D, 4E) and oxidation-induced alteration of current amplitudes (Fig. 3D) consistent with a more proximal positioning of the M3/M3–S2 linker in the GluN1 subunit. Spontaneous disulfide bond formation only occurs when sulfhydryl moieties are within 2 Å of one another (Careaga and Falke, 1992; Clarke and Fersht, 1993). Hence, assuming proper orientation for a rigid structure, the α-carbons of introduced cysteines must be positioned within 6–8 Å of one another to cross-link. If either the backbone is flexible or one uses a strong oxidation agent (e.g., Cu:Phen), then cross-linking can occur among cysteines over a greater range of separation (∼20 Å) (Careaga and Falke, 1992; Clarke and Fersht, 1993). On average in the crystal structure, the distance between M3/M3–S2 positions (A622 to P632) of the A/C and B/D conformations is 16.3 and 24.3 Å, respectively (Table 1). Based on these considerations, there are positions in the GluN2 M3/M3–S2 linker whose analogous position in the crystal structure is quite proximal in the B/D conformer (e.g., GluN2A(Q636)) (Table 1) that do not show dimer formation (Fig. 2D) nor significant oxidized-induced current inhibition (Fig. 3D). Additionally, the observed oxidation-induced inhibition in the S2–M4 linker of the GluN2 but not GluN1 subunit (Fig. 6D) is consistent with a more proximal positioning of the GluN2 S2–M4 linker. It is important to note that the local S2–M4 structure of NMDA receptor subunits is almost certainly different from that in AMPA receptor subunits due to gaps in the sequence alignments (Fig. 6A, right). Still, we do not believe these differences greatly alter the relative spatial positioning of the conformers—especially given the dramatic difference in typical distances in the A/C (46.7 Å) compared to the B/D (22.9 Å) conformations (Table 3). Further, we concede that Cu:Phen likely induces rarely visited conformations; however, this point is not critical here, since we are only interested in proximity rather than gating states.
An alternative interpretation of the present results is that the GluN1 and GluN2 subunits are positioned adjacent to each other in a N1/N1/N2/N2 arrangement. Although we cannot completely rule out this alternative, it is highly unlikely if one accepts that the general arrangement of NMDA receptor subunits including at the LBD–TMD linkers is comparable to that of AMPA receptors. Specifically, if like subunits (e.g., GluN1 and GluN1) are adjacent to one another, then one subunit must adopt the A/C conformation and the other the B/D conformation. Thus, a N1/N1/N2/N2 arrangement would result in GluN1/GluN1 and GluN2/GluN2 dimers at the level of the linkers having identical conformational symmetry. With regards to our experimental protocol, if like subunits adopted a N1/N1/N2/N2 arrangement, then we would expect cross-linking for GluN1 and GluN2 to show identical patterns functionally—that is, introduced cysteines in the M3/M3–S2 linkers of both GluN1 and GluN2 should show cross-linking due to their identical conformations. However, we find that the pattern of cross-linking is highly subunit-specific showing cross-linking either in M3/M3–S2 (GluN1) (Figs. 2, 3) or S2–M4 (GluN2) (Fig. 6). As such, our results suggest that like subunits, sharing a common conformer, are positioned diagonal to one another, which we interpret as A/C (GluN1) and B/D (GluN2) leading to a N1/N2/N1/N2 arrangement.
The present study does not address the subunit-specific conformations of external iGluR elements, specifically the amino-terminal domain (ATD). Indeed, the ATD in NMDA receptor subunits may take on a twisted closed-cleft conformation (Karakas et al., 2009, 2011; Stroebel et al., 2011) that is distinct to the conformation of the same domain in AMPA (Clayton et al., 2009; Sobolevsky et al., 2009) and kainate (Kumar et al., 2009) receptors, possibly resulting in differences in the pattern of domain swapping. However, additional work is required to determine the arrangement of subunit domains at the level of the ATD, as well as the functional relationship between the ATDs and the A/C and B/D conformations at the level of the LBD–TMD linkers (Gielen et al., 2009; Yuan et al., 2009).
Subunit-specific effects on gating and biogenesis
Because cross-linking occurred only for specific subunits, it appears that the conformation approximated by NMDA receptor subunits in the tetrameric complex is invariant, either A/C (GluN1) or B/D (GluN2). Nevertheless, the functional significance of the different conformers and how they might arise and be constrained during biosynthesis is unknown. In terms of functional properties, subunit-specific differences in the LBD–TMD linker structure may underlie subunit-specific contributions to ion permeation (Watanabe et al., 2002) and channel gating (Banke and Traynelis, 2003; Sobolevsky et al., 2007; Blanke and VanDongen, 2008a,b; Kussius and Popescu, 2009). Recent work from our laboratory suggests that despite subunit-specific differences in the structure of the LBD–TMD linkers, NMDA receptors undergo concerted gating that appears tightly coupled at the level of these linkers (Talukder and Wollmuth, 2011). Still, how these different LBD–TMD linker arrangements couple the LBD to pore opening remain unknown.
NMDA receptors can form di-heteromeric (e.g., GluN1/GluN2 and/or GluN1/GluN3) or tri-heteromeric (e.g., GluN1/GluN2/GluN3) receptors that differ widely in their biophysical and pharmacological properties (Traynelis et al., 2010). The initial step mediating the formation of these iGluR heteromers is dimer formation. However, much controversy exists regarding the nature of this initial dimer—whether it is a homodimer (e.g., GluN1) (Papadakis et al., 2004; Farina et al., 2011) or a heterodimer (e.g., GluN1/GluN2) (Schüler et al., 2008)—and the order in which the dimers assemble to form a functional tetrameric receptor. The amino-terminal domain in iGluRs is critical for initial dimer formation (Paoletti and Neyton, 2007; Shanks et al., 2010; Farina et al., 2011). Interestingly, heteromeric AMPA receptors (e.g., GluA1/GluA2), like NMDA receptors, are arranged in an alternating subunit-specific manner (A1/A2/A1/A2) (Mansour et al., 2001). Therefore, the presence of subunit-specific conformations in heteromeric assemblies may be a necessary component not only of dimer formation, but also for tetramerization and/or for stabilization of the tetrameric assembly. Thus, the existence of two distinct conformations may serve a role in ensuring the proper biosynthesis and trafficking of iGluRs to the membrane, but the role of the specific conformations in these processes remain unknown.
Footnotes
This work was supported by an NIH R01 grant from NIMH (MH066892) (L.P.W.), a minority supplement from NIMH (C.L.S.), an NIH National Research Service Award from NINDS (NS073382) (C.L.S.) and Howard Hughes Medical Institute (Grant 52005887) and Undergraduate Research and Creative Activities fellowships (P.B.). We thank Drs. Myles Akabas, Hiro Furakawa, Mark Bowen, and Iehab Talukder for helpful discussions and/or comments on the manuscript. We thank Janet Allopena for technical assistance.
References
- Banke TG, Traynelis SF. Activation of NR1/NR2B NMDA receptors. Nat Neurosci. 2003;6:144–152. doi: 10.1038/nn1000. [DOI] [PubMed] [Google Scholar]
- Blanke ML, VanDongen AM. Constitutive activation of the N-methyl-D-aspartate receptor via cleft-spanning disulfide bonds. J Biol Chem. 2008a;283:21519–21529. doi: 10.1074/jbc.M709190200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanke ML, VanDongen AM. The NR1 M3 domain mediates allosteric coupling in the N-methyl-D-aspartate receptor. Mol Pharmacol. 2008b;74:454–465. doi: 10.1124/mol.107.044115. [DOI] [PubMed] [Google Scholar]
- Careaga CL, Falke JJ. Structure and dynamics of Escherichia coli chemosensory receptors. Engineered sulfhydryl studies. Biophys J. 1992;62:209–219. doi: 10.1016/S0006-3495(92)81806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YB, Tenneti L, Le DA, Ortiz J, Bai G, Chen HS, Lipton SA. Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nat Neurosci. 2000;3:15–21. doi: 10.1038/71090. [DOI] [PubMed] [Google Scholar]
- Citri A, Malenka RC. Synaptic plasticity: multiple forms, functions, and mechanisms. Neuropsychopharmacology. 2008;33:18–41. doi: 10.1038/sj.npp.1301559. [DOI] [PubMed] [Google Scholar]
- Clarke J, Fersht AR. Engineered disulfide bonds as probes of the folding pathway of barnase: increasing the stability of proteins against the rate of denaturation. Biochemistry. 1993;32:4322–4329. doi: 10.1021/bi00067a022. [DOI] [PubMed] [Google Scholar]
- Clayton A, Siebold C, Gilbert RJ, Sutton GC, Harlos K, McIlhinney RA, Jones EY, Aricescu AR. Crystal structure of the GluR2 amino-terminal domain provides insights into the architecture and assembly of ionotropic glutamate receptors. J Mol Biol. 2009;392:1125–1132. doi: 10.1016/j.jmb.2009.07.082. [DOI] [PubMed] [Google Scholar]
- Cull-Candy SG, Leszkiewicz DN. Role of distinct NMDA receptor subtypes at central synapses. Sci STKE. 2004;2004:re16. doi: 10.1126/stke.2552004re16. [DOI] [PubMed] [Google Scholar]
- Erreger K, Chen PE, Wyllie DJ, Traynelis SF. Glutamate receptor gating. Crit Rev Neurobiol. 2004;16:187–224. doi: 10.1615/critrevneurobiol.v16.i3.10. [DOI] [PubMed] [Google Scholar]
- Farina AN, Blain KY, Maruo T, Kwiatkowski W, Choe S, Nakagawa T. Separation of domain contacts is required for heterotetrameric assembly of functional NMDA receptors. J Neurosci. 2011;31:3565–3579. doi: 10.1523/JNEUROSCI.6041-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KS, VanDongen HM, VanDongen AM. The NMDA receptor M3 segment is a conserved transduction element coupling ligand binding to channel opening. J Neurosci. 2002;22:2044–2053. doi: 10.1523/JNEUROSCI.22-06-02044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia LV, Kalia SK, Salter MW. NMDA receptors in clinical neurology: excitatory times ahead. Lancet Neurol. 2008;7:742–755. doi: 10.1016/S1474-4422(08)70165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009;28:3910–3920. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 2011 doi: 10.1038/nature10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp JJ, Vissel B, Heinemann SF, Westbrook GL. Calcium-dependent inactivation of recombinant N-methyl-D-aspartate receptors is NR2 subunit specific. Mol Pharmacol. 1996;50:1680–1688. [PubMed] [Google Scholar]
- Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol. 2009;16:631–638. doi: 10.1038/nsmb.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussius CL, Popescu GK. Kinetic basis of partial agonism at NMDA receptors. Nat Neurosci. 2009;12:1114–1120. doi: 10.1038/nn.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci. 2007;8:413–426. doi: 10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
- Lee CH, Gouaux E. Amino terminal domains of the NMDA receptor are organized as local heterodimers. PLoS One. 2011;6:e19180. doi: 10.1371/journal.pone.0019180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour M, Nagarajan N, Nehring RB, Clements JD, Rosenmund C. Heteromeric AMPA receptors assemble with a preferred subunit stoichiometry and spatial arrangement. Neuron. 2001;32:841–853. doi: 10.1016/s0896-6273(01)00520-7. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Glutamate and neurotrophic factors in neuronal plasticity and disease. Ann N Y Acad Sci. 2008;1144:97–112. doi: 10.1196/annals.1418.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Neyton J. NMDA receptor subunits: function and pharmacology. Curr Opin Pharmacol. 2007;7:39–47. doi: 10.1016/j.coph.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Papadakis M, Hawkins LM, Stephenson FA. Appropriate NR1-NR1 disulfide-linked homodimer formation is requisite for efficient expression of functional, cell surface N-methyl-D-aspartate NR1/NR2 receptors. J Biol Chem. 2004;279:14703–14712. doi: 10.1074/jbc.M313446200. [DOI] [PubMed] [Google Scholar]
- Piña-Crespo JC, Talantova M, Micu I, States B, Chen HS, Tu S, Nakanishi N, Tong G, Zhang D, Heinemann SF, Zamponi GW, Stys PK, Lipton SA. Excitatory glycine responses of CNS myelin mediated by NR1/NR3 “NMDA” receptor subunits. J Neurosci. 2010;30:11501–11505. doi: 10.1523/JNEUROSCI.1593-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambhadran A, Gonzalez J, Jayaraman V. Subunit arrangement in N-methyl-D-aspartate (NMDA) receptors. J Biol Chem. 2010;285:15296–15301. doi: 10.1074/jbc.M109.085035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorge S, Colquhoun D. Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J Neurosci. 2003;23:1151–1158. doi: 10.1523/JNEUROSCI.23-04-01151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüler T, Mesic I, Madry C, Bartholomäus I, Laube B. Formation of NR1/NR2 and NR1/NR3 heterodimers constitutes the initial step in N-methyl-D-aspartate receptor assembly. J Biol Chem. 2008;283:37–46. doi: 10.1074/jbc.M703539200. [DOI] [PubMed] [Google Scholar]
- Shanks NF, Maruo T, Farina AN, Ellisman MH, Nakagawa T. Contribution of the global subunit structure and stargazin on the maturation of AMPA receptors. J Neurosci. 2010;30:2728–2740. doi: 10.1523/JNEUROSCI.5146-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Beck C, Wollmuth LP. Molecular rearrangements of the extracellular vestibule in NMDAR channels during gating. Neuron. 2002;33:75–85. doi: 10.1016/s0896-6273(01)00560-8. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Prodromou ML, Yelshansky MV, Wollmuth LP. Subunit-specific contribution of pore-forming domains to NMDA receptor channel structure and gating. J Gen Physiol. 2007;129:509–525. doi: 10.1085/jgp.200609718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D, Carvalho S, Paoletti P. Functional evidence for a twisted conformation of the NMDA receptor GluN2A subunit N-terminal domain. Neuropharmacology. 2011;60:151–158. doi: 10.1016/j.neuropharm.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Talukder I, Wollmuth LP. Local constraints in either the GluN1 or GluN2 subunit equally impair NMDA receptor pore opening. J Gen Physiol. 2011;138:179–194. doi: 10.1085/jgp.201110623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder I, Borker P, Wollmuth LP. Specific sites within the ligand-binding domain and ion channel linkers modulate NMDA receptor gating. J Neurosci. 2010;30:11792–11804. doi: 10.1523/JNEUROSCI.5382-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe J, Beck C, Kuner T, Premkumar LS, Wollmuth LP. DRPEER: a motif in the extracellular vestibule conferring high Ca2+ flux rates in NMDA receptor channels. J Neurosci. 2002;22:10209–10216. doi: 10.1523/JNEUROSCI.22-23-10209.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci. 2009;29:12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]