Background: SSeCKS/GRAVIN/AKAP12 is a major protein kinase C substrate and binding protein.
Results: SSeCKS binding of PKC via two homologous domains is required for attenuation of PKC activity and transduction of phorbol ester-induced cytoskeletal remodeling and survival signals.
Conclusion: SSeCKS controls PKC signaling via direct scaffolding interactions.
Significance: PKC signaling is regulated by the spatiotemporal scaffolding activity of SSeCKS.
Keywords: Apoptosis, Cytoskeleton, Phorbol Esters, Protein Kinase C (PKC), Threonine/Serine Protein Kinase, SSeCKS/GRAVIN/AKAP12, Prostate Cancer
Abstract
The product of the SSeCKS/GRAVIN/AKAP12 gene (“SSeCKS”) is a major protein kinase (PK) C substrate that exhibits tumor- and metastasis-suppressing activity likely through its ability to scaffold multiple signaling mediators such as PKC, PKA, cyclins, calmodulin, and Src. Although SSeCKS and PKCα bind phosphatidylserine, we demonstrate that phosphatidylserine-independent binding of PKC by SSeCKS is facilitated by two homologous SSeCKS motifs, EG(I/V)(T/S)XWXSFK(K/R)(M/L)VTP(K/R)K(K/R)X(K/R)XXXEXXXE(E/D) (amino acids 592–620 and 741–769). SSeCKS binding to PKCα decreased kinase activity and was dependent on the two PKC-binding motifs. SSeCKS scaffolding of PKC was increased in confluent cell cultures, correlating with significantly increased SSeCKS protein levels and decreased PKCα activity, suggesting a role for SSeCKS in suppressing PKC activation during contact inhibition. SSeCKS-null mouse embryo fibroblasts displayed increased relative basal and phorbol ester (phorbol 12-myristate 13-acetate)-induced PKC activity but were defective in phorbol 12-myristate 13-acetate-induced actin cytoskeletal reorganization and cell shape change; these responses could be rescued by the forced expression of full-length SSeCKS but not by an SSeCKS variant deleted of its PKC-binding domains. Finally, the PKC binding sites in SSeCKS were required to restore cell rounding and/or decreased apoptosis in phorbol ester-treated LNCaP, LNCaP-C4-2, and MAT-LyLu prostate cancer cells. Thus, PKC-mediated remodeling of the actin cytoskeleton is likely regulated by the ability of SSeCKS to control PKC signaling and activity through a direct scaffolding function.
Introduction
The protein kinase C (PKC) family consists of so-called conventional (cPKC3; α, β1, β2, and γ), novel (nPKC; δ, ϵ, η, and θ), and atypical (ι and λ) PKC isoforms with inherent serine/threonine kinase activity. The rapid activation of one or more PKC isoforms induces major changes in the dynamics of the actin cytoskeletal organization, resulting in changes in cell morphology that affect cell motility, outgrowth of cellular processes such as neurites, proliferation, and cell survival. For example, phorbol esters such as phorbol 12-myristate 13-acetate (PMA) or diacylglycerol can enter cells and bind directly to and activate cPKC and nPKC isoforms, resulting in rapid and massive remodeling of the actin cytoskeleton (1).
In some tissues and cell types, the chronic treatment with PMA is linked to the induction of cancer initially through the activation of proliferative pathways such as Ras-Raf-MEK-ERK but also through the control of actin cytoskeletal dynamics via activation of non-PKCs such as the chimaerins and Ras guanyl-releasing protein (2) and through the down-regulation of specific PKC isoforms such as PKCδ (3). Altered PKC expression levels and activities have been reported in various types of malignancies including breast, prostate, thyroid, pituitary, and lung cancers and various leukemias (4), typically with increased activity levels of PKCα, -δ, and -ϵ associated with increased proliferation, cell motility, or apoptosis (1). However, roles for specific PKC isoforms are governed by tissue context. For example, PKCα can be tumor-promoting in some tissue such as the skin but tumor-suppressing in colon cancer (5, 6).
PKC-induced mitogenesis and cytoskeletal reorganization is controlled by substrates and binding partners of PKC. Indeed, “substrates that interact with protein kinase C” (STICKs) fulfill both roles, interacting with PKC via domains rich in phosphatidylserine (PS), calmodulin, and F-actin binding sites (7). In contrast, “receptors for activated C kinases” (RACKs) bind only activated forms of PKCs and thus are thought to promote phosphorylation of substrates. A third regulatory mechanism involves the direct interaction of certain PKC isoforms with signaling structures. For example, PKCα is enriched in caveolae, whereas PKCβ and -ϵ bind to F-actin. A small group of scaffolding proteins including SSeCKS and AKAP79 are capable of binding PKC, PKA, calmodulin, PS, and phosphoinositol bisphosphates and thus may play a central role in regulating several cross-talking pathways (8).
The list of known PKC substrates includes signal transduction proteins thought to work at plasma membrane sites (e.g. MARCKS and SSeCKS), cytoskeletal proteins (GAP43, adducin, fascin, ERM proteins (ezrin, radixin, moesin), AFAP-110, myosin light chain, and vinculin), and nuclear structural proteins (e.g. lamin B). Many of these substrates tether microtubules to plasma membrane sites or facilitate actin fiber assembly. For example, the ability of PKC to phosphorylate myosin light chain directly affects the ability of the myosin light chain kinase to phosphorylate myosin light chain (9), resulting in altered cell motility and cell shape change (9–13). However, little is known regarding the specific mechanism by which PKC substrates mediate cell shape change or the resulting biologies associated with cancer progression such as invasiveness, proliferation, and survival. Prostate cancer (CaP) progression has been linked to the activation and/or overexpression of specific PKC isoforms such as α, β, ϵ, η, and ζ (14, 15). However, the transgenic expression of PKCϵ, but not PKCα or -δ, was sufficient to induce CaP (16). Further confounding the issue, PKCα promotes proliferation of androgen receptor-negative PC-3 CaP cells, whereas it along with PKCδ promotes apoptosis (17) in the androgen-responsive cell line LNCaP (18–22). Our laboratory identified SSeCKS (for Src-suppressed C kinase substrate) as a substrate of PKC whose expression is severely down-regulated in src- and ras-transformed cells (23, 24). Subsequently, Jaken and co-workers (25, 26) identified STICK72, which encodes a “>200-kDa” PKC substrate with PS binding activity, as identical to SSeCKS. SSeCKS has many similarities to the well studied PKC substrate MARCKS, although they have quite different sequences and molecular weights. These include transcriptional suppression by v-src, PS-induced binding to PKC, N-terminal myristoylation, a predicted rodlike shape, heat stability, severely retarded mobility on SDS-PAGE, Ca2+-dependent calmodulin binding, and PMA-induced translocation to perinuclear sites (24, 27). We recently showed that SSeCKS can associate with PKCα, -δ, and -ϵ isozymes when overexpressed (28).
Here, we map PS-independent PKC-binding domains on SSeCKS and show that this binding inhibits PKC kinase activity. Furthermore, in the absence of SSeCKS, PMA-induced PKC activation failed to induce cell shape change and cytoskeletal reorganization. The finding that full-length SSeCKS, but not an SSeCKS mutant deleted of its PKC-binding domains, can rescue PMA-induced cell shape change in MEF and prostate cancer cells and can prevent PMA-induced apoptosis in LNCaP cells strongly suggests that SSeCKS mediates PKC-induced cytoskeletal signaling through a direct physical interaction with PKC.
EXPERIMENTAL PROCEDURES
Cell Culture
WT- and SSeCKS-null (KO)-MEF (28) and θNX packaging cells (29) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum, 2 mm l-glutamate, 100 μg/ml penicillin-streptomycin, 1% sodium pyruvate, 10 mm HEPES, pH 7.0, and 1% non-essential amino acids. NIH3T3 cells were grown in DMEM supplemented with 2 mm glutamine and 10% calf serum. S2-6 cells are NIH3T3 cells that encode a tetracycline-regulated transactivator (Tet-Off); S24 cells are S2-6 cells stably transfected with a tetracycline-regulated SSeCKS expression vector (30). MLL/2-6 and MLL/V-2 cells are MAT-LyLu rat prostate epithelial cells expressing a tetracycline-regulated αSSeCKS cDNA (2–6) or vector (V-2) (31). Cells were maintained in DMEM containing 10% heat-inactivated calf serum, 0.7 μg/ml tetracycline, 1 μg/ml hygromycin, and 1 μg/ml puromycin. LNCaP (ATCC CRL-1740) and LNCaP-C4-2 (gift of L. Chung, Cedars-Sinai Medical Center) were grown in DMEM containing 10% fetal calf serum (FCS) and 5 nm R1881 (LNCaP only).
Plasmids and Constructs
All GST-SSeCKS truncation mutants were made by PCR amplification using the forward and reverse primers listed in supplemental Table 1 and then confirmed by sequencing. Forward and revere primers encode 5′-GCCGAATTC sequence extensions containing an EcoRI site (underlined) to facilitate cloning into the pGEX-5Xa, -b, or -c vectors (depending on the reading frame). PKC isozyme-GFP constructs were obtained from the Soh laboratory (Inha University, South Korea). SSeCKS constructs containing the Δ553–900 deletion in either pBlueScript II-KS/SSeCKS (32) or pcDNA3.1/αSSeCKS-GFP (33) were produced by a long range inverse PCR technique we described previously (33) using “553” and “900” primers containing an SpeI site (underlined): 553 primer, 5′-ATGCACTAGTGCAGAGCT CTCTCCCTTCT G-3′; 900 primer, 5′-ATGCACTAGTTGAACACACAGCGGGAGA A-3′. The His-TAT-SSeCKS fusion construct was produced by PCR amplification of pBlueScript/SSeCKS with forward primer 5′-CGGGATCCTACGCACGAGCAGCAGCACGACAggcacgagcaggcgcaggcagttccacc-3′ (BamHI site underlined, TAT sequence in bold, and SSeCKS sequence in lowercase) and reverse primer 5′-GGAATTCCTTTGGTCCGTTGGTGTC-3′ (EcoRI site underlined) and subcloned into BamHI/EcoRI-cut pET28a, which encodes an N-terminal His tag. The forward primer encodes the 11-aa YARAAARQARA TAT protein transduction domain (34). pTAT-P1, encoding a His-TAT-DHRS12-P1 fusion protein, was kindly provided by Lionel Coignet (Roswell Park Cancer Institute).
GST Fusion and TAT-SSeCKS Protein Preparation
GST-SSeCKS fusion proteins were expressed in BL21-pLysS bacteria by induction with 1 mm isopropyl β-d-thiogalactopyranoside at 30 °C for 1 h and purified as described previously (32) using 250 mm NaCl, 50 mm HEPES, pH 7.9, 5 mm EDTA, 0.1% Nonidet P-40, 0.5 mm DTT, 1 mm PMSF, and 20 mm reduced glutathione for protein elution from glutathione-Sepharose columns (GE Healthcare). TAT-SSeCKS was expressed in BL21-pLysS bacteria by induction with isopropyl β-d-thiogalactopyranoside at 30 °C for 3 h followed by resuspension of bacterial pellets in lysis buffer (50 mm NaH2PO4, 300 mm NaCl, and 10 mm imidazole), addition of lysozyme (0.2 mg/ml final concentration), and incubation for 30 min on ice followed by sonication. The lysates were boiled for 3 min and centrifuged at 14,000 × g for 20 min at 4 °C; His-TAT-SSeCKS protein was retained in the supernatant based on the predicted lack of SSeCKS secondary structure (32).
Immunoblotting (IB)
Radioimmune precipitation assay lysates containing 20–60 μg of cell protein/gel lane were immunoblotted with various antibodies (Abs) as described previously (35). Primary rabbit or mouse polyclonal or monoclonal Abs (1:1000 dilution) were specific for GST, PKCα, PKCβI, PKCβII, PKCγ, PKCδ, PKCϵ, PKCη, pan-PKC (sc-80; recognizes α, β, and γ) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), His6 tag, GAPDH (Chemicon/Millipore, Billerica, MA), or SSeCKS (32). Secondary Abs (1:5000 dilution) included horseradish peroxidase- (HRP; Chemicon) or Alexa Fluor 680 (Invitrogen/Molecular Probes, Carlsbad, CA)-conjugated anti-rabbit or anti-mouse IgG. The HRP-labeled Abs were identified by incubating the blots with Lumi-Light chemiluminescence substrate (Roche Applied Science) and then imaging and quantifying using GeneTools software on a Chemi-Genius2 bioimaging system (Syngene, Frederick, MD), whereas the Alexa Fluor signals were identified and quantified using an Odyssey Infrared BioImager (Li-Cor Biosciences, Lincoln, NE).
Immunoprecipitation (IP)
Cells were harvested, washed twice with ice-cold PBS, incubated for 30 min on ice in 0.2–0.5 ml of Nonidet P-40 lysis buffer (140 mm NaCl, 20 mm NaPO4, pH 7.8, 5 mm EDTA, 1% Nonidet P-40, 10 μg/ml aprotinin, 10 g/ml leupeptin, 10 μg/ml pepstatin, 10 mm sodium fluoride, 1 mm DTT, and 1 mm PMSF) to allow cells to swell, and then lysed by forcing the cell suspension five times through a hypodermic needle (25 gauge). The lysates were cleared by centrifugation (14,000 rpm, 4 °C, 10 min) whereupon the supernatant (500 μg of protein/sample) was precleared with a 10-min incubation at 4 °C with 50 μl of Protein A/G-agarose (Thermo/Pierce, Rockford, IL) and then incubated 2 h to overnight with 25 μl of Protein A/G-agarose-Ab beads (Protein A/G preincubated for 1 h with 5–10 μg of IP Ab and then washed twice with 1 ml of lysis buffer) plus lysis buffer to bring the final volume to 500 μl. The pellets were washed thrice with lysis buffer and boiled for 3 min in 20 μl of 2× Laemmli buffer, and associated proteins were separated by SDS-PAGE. Transfer of the protein to PVDF and IB analysis were as above.
Pulldown Assay
GST fusion proteins (1 μg each) were diluted in 200 μl of ice-cold Binding Buffer (20 mm HEPES, pH 7.9, 1 mm MgCl2, 40 mm KCl, 0.1 mm EDTA, 0.1% Nonidet P-40, 1 mm DTT, and 1 mm PMSF) and then incubated with 25 μl of washed glutathione-SepharoseTM 4B beads (GE Healthcare) with gentle agitation at 4 °C for 1 h. The beads were washed thrice with radioimmune precipitation assay buffer and once with Binding Buffer (1 ml/wash). After resuspending in 245 μl of Binding Buffer, either 5 μl of in vitro transcribed/translated (TnT, Promega, Madison, WI) protein or 500 μg of cell lysate were added, and the mixture was agitated gently at 4 °C for 2 h, washed four times (1 ml each) with Binding Buffer, boiled in Laemmli buffer, and then analyzed by SDS-PAGE.
Lipid/Protein Overlay Assays
Nitrocellulose membranes containing PS (Sigma-Aldrich) were produced as follows. PS was dissolved in chloroform and then diluted with an equal volume of 6% Triton X-100 in water followed by sonication thrice (5 s each). 10 μl of various PS concentrations were spotted and dried at room temperature (RT) for 1 h followed by storage at 4 °C in the dark. PS/phosphatidylethanolamine/phosphatidylglycerol/phosphatidic acid membrane strips (Echelon Biosciences Inc., Salt Lake City, UT) were blocked with 3% BSA in PBS for 1 h at RT and then incubated for 1 h with the probe protein (1 μg/ml) in blocking buffer. After three washes (5 min/wash) with PBS and 0.1% Triton X-100 (PBS/T), the membranes were processed with primary and secondary antibodies in standard IB (above).
PKC/SSeCKS Overlay Assay
Purified GST or GST-SSeCKS fusion proteins (1 μg/slot in 250-μl volumes) were loaded onto PVDF membranes using a Slot-Blot apparatus (Bio-Rad) whereupon the membranes were dried at room temperature and stored at 4 °C. Membranes were blocked with 3% BSA in PBS for 0.5 h at room temperature and then washed three times with PBS/T. Membranes were incubated with purified PKC (1 μg/ml) in 3% BSA in PBS/T for 1 h at RT, washed, and processed for PKC-specific IB.
In Vitro Transcription and Translation
One microgram of plasmid DNA was added to T7-TnT reaction assays (Promega) containing 25 μl of rabbit reticulocyte lysate, 2 μl of TnT reaction buffer, 1 μl of amino acid mixture minus methionine (1 mm), 2 μl of [35S]methionine (10 μCi/μl), 1 μl of RNase inhibitor (40 units/μl), 1 μl of T7 RNA polymerase, and double distilled H2O to make 50 μl total and incubated at 30 °C for 90 min.
PKC Kinase Activity Assay
Total cellular PKC kinase activity was measured using a solid phase enzyme-linked immunoabsorbent assay kit (Assay Designs/Stressgen, Ann Arbor, MI) according to the manufacturer's specifications. Absorbance of the chromogenic substrate product was measured at 450 nm, and the relative PKC kinase activity was calculated as follows: (average sample absorbance − average blank absorbance)/volume (μl) × dilution factor. PKCα kinase activity either from IPs or using baculovirus-expressed enzyme (Biomol) was assayed in 60-μl reactions containing PKCα in 20 μl of kinase buffer (50 mm HEPES, pH 7.5, 10 mm MgCl2, 1 mm dithiothreitol, 2.5 mm EGTA, 1 mm sodium fluoride, 0.1 mm Na3VO4, and 10 mm β-glycerophosphate) plus 40 μl of kinase buffer containing 5 μg of myelin basic protein (MBP) or histone H1 substrate and 5 μCi of [γ-32P]ATP. The reactions were incubated at 30 °C for 30 min and terminated by adding SDS sample buffer followed by boiling for 3 min. The reaction products were analyzed by SDS-PAGE and phosphorimaging (Typhoon ImageQuant, GE Healthcare).
PMA-induced Cell Shape Change
Phorbol 12-myristate 13-acetate (Avanti Polar Lipids Inc., Alabaster, AL) was dissolved in dimethyl sulfoxide as a 5 mm stock solution and stored in aliquots at −20 °C. Cultures of MEF were plated 18 h before PMA treatment, which was 100 nm for various lengths of time.
Immunofluorescence Analysis
Cells plated on glass coverslips (22 mm2) were fixed at −20 °C for 20 min with 60% acetone and 3.7% paraformaldehyde in PBS (for F-actin staining) or in 4% paraformaldehyde and 0.5% Triton X-100 at room temperature for 10 min followed by two PBS washes. The slides were blocked with 3% nonfat dry milk in PBS for 30 min at room temperature. F-actin was stained with FITC-phalloidin (Sigma), and nuclei were stained with DAPI (Invitrogen; 1:500). Fluorescent images were captured using a Nikon TE2000-E inverted microscope equipped with a Roper CoolSnap HQ charge-coupled device camera.
Apoptosis/Cell Death Assay
Apoptosis and cell death were measured, respectively, in LNCaP cells labeled with phycoerythrin-Annexin V and 7-aminoactinomycin D using a V-PE Apoptosis Detection kit (BD Biosciences/Pharmingen) according to the manufacturer's specifications. The stained cells were detected in the Roswell Park Cancer Institute Flow and Image Cytometry Core Lab (Paul Wallace, Director) using a BD Biosciences FACSCalibur and FCS Express 3.0 software (De Novo Software, Los Angeles, CA).
RESULTS
PS-dependent and -independent SSeCKS/PKC Binding
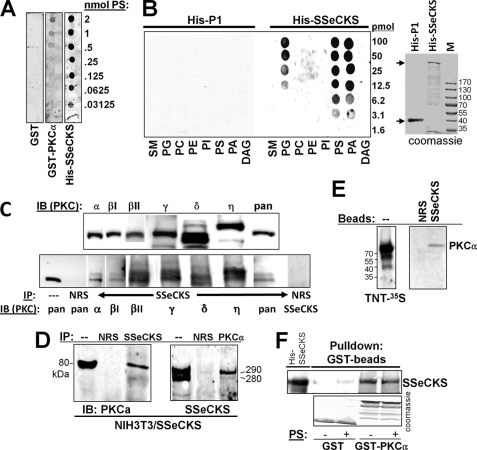
The original finding that SSeCKS binds PKC comes from overlay assays using high, non-physiologic levels (10–100 μg/ml) of PS (25). We also showed (32) that the PS-independent co-IP of SSeCKS and PKC could be enhanced by the addition of high levels of PS (123 μm). In contrast, Piontek and Brandt (36) showed that the ability of GRAVIN (the human SSeCKS/AKAP12 orthologue) to co-immunoprecipitate PKCα or -βII was not enhanced by PS. Thus, we investigated whether SSeCKS encodes distinct PS-dependent and -independent binding activities and if so whether these were encoded by different SSeCKS protein domains. Nitrocellulose strips containing increasing concentrations of PS were incubated with either equal amounts of GST, GST-PKCα, or His-SSeCKS, and after stringent washing, the blots were immunoblotted for either GST or SSeCKS. SSeCKS and PKCα bound levels of PS (Fig. 1A) that are physiologically relevant given that 16–20% of the phospholipids of a cell are PS and that cell membranes typically consist of 6–10% PS (37). SSeCKS also bound phosphatidic acid and phosphatidylglycerol with similar affinities but failed to bind phosphatidylethanolamine, phosphatidylcholine, phosphatidylinositol, sphingomyelin, or diacylglycerol (Fig. 1B). As a control, the His-DHRS12-P1 protein encoding a mitochondrial dehydrogenase (a gift from Lionel Coignet, Roswell Park Cancer Institute) failed to bind to any of the lipids, indicating that the His-SSeCKS binding was not due to the shared His-TAT N-terminal tag. Taken together, these data suggest that SSeCKS binding to PS may have physiologic significance.
FIGURE 1.
SSeCKS association with PKC isoforms and with PS. A, SSeCKS binds PS. Shown is the overlay assay of GST, GST-PKCα, or His-TAT-SSeCKS on membranes containing dilutions of PS probed with anti-GST or anti-SSeCKS Ab. B, SSeCKS binds multiple phospholipids. Left panel, overlay assay of control His-TAT-DHRS12-P1 or His-TAT-SSeCKS protein on membranes containing dilutions of sphingomyelin (SM), phosphatidylglycerol (PG), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), PS, phosphatidic acid (PA), or diacylglycerol (DAG) probed with anti-His6 mAb (right panel, Coomassie-stained gel showing the 37-kDa His-TAT-DHRS12-P1 and the 300-kDa His-TAT-SSeCKS proteins; M, marker proteins). C, SSeCKS associates with cPKC and nPKC isoforms. Upper panel, IB of SSeCKS-overexpressing NIH3T3 lysates using PKC isoform-specific mAbs; lower panel, IP of SSeCKS-overexpressing NIH3T3 lysates with anti-SSeCKS or NRS followed by IB with isoform-specific PKC Abs, pan-PKC (recognizes α, β, and γ), or SSeCKS Ab. D, reciprocal co-IP between SSeCKS and PKCα. Lysates from S24 cells (NIH3T3[Tet-Off/SSeCKS] (30)) grown in the absence of tetracycline were immunoprecipitated with NRS, anti-SSeCKS, or anti-PKCα Ab, and then the IPs were probed by IB using the reciprocal Ab. E, left panel, autoradiography of 35S-labeled full-length PKCα produced in TnT reactions; right panel, pulldown of labeled PKCα proteins using beads containing NRS or SSeCKS (after IP from S24 cell lysates). F, beads with GST or GST-PKCα (Coomassie-stained in the bottom panel) were used to precipitate His-SSeCKS protein in the presence or absence of PS (20 μg/ml) followed by SSeCKS IB (top panel).
In an attempt to determine which PKC isoform classes could associate with SSeCKS in a PS-independent manner, IPs of SSeCKS from lysates of SSeCKS-overexpressing NIH3T3 cells (30) were immunoblotted with PKC isoform-specific Ab or with a pan-PKC serum that recognizes PKCα, -β, and -γ isoforms. These cell lysates are not PS-free given that roughly two-thirds of cellular PS is not membrane-associated (37). SSeCKS readily associated with several cPKC (α, βI, βII, and γ) and nPKC (δ and η) isoforms without the need for added PS (Fig. 1C), although no association with the atypical PKC isoform λ was detected (not shown). As a negative control, IP with normal rabbit serum (NRS) resulted in no reactivity to a pan-PKC serum. Thus, SSeCKS binding of PKC is not isoform-specific. The SSeCKS/PKC association could be demonstrated by reciprocal co-IPs from SSeCKS-overexpressing NIH3T3 lysates (Fig. 1D) by pulldown of 35S-labeled PKCα using SSeCKS beads (versus NRS beads) (Fig. 1E) or by pulldown of overexpressed SSeCKS using GST-PKCα versus GST beads (Fig. 1F). Note that in the latter example, the addition of PS failed to enhance the association of SSeCKS and PKCα.
The PS-independent binding sites on SSeCKS were identified by performing pulldown assays with bait beads containing fusions of GST to non-overlapping SSeCKS fragments of roughly 200 aa (Fig. 2A, bottom panel) incubated with cell lysates from HEK293T cells forced to overexpress PKCα. After probing these pulldowns with anti-PKCα, two SSeCKS regions exhibited association: aa 553–720 and 721–900 (Fig. 2A, top panel). The inability of other GST-SSeCKS fusions and of GST alone to pull down PKCα indicates that the binding by the 553–720 and 721–900 regions was specified by their SSeCKS sequences. To further determine whether these regions are required for SSeCKS-PKC binding, an in-frame aa 553–900 SSeCKS deletion mutant was produced (“Δ553–900”). We then assessed the binding to GST or GST-PKCα beads by 35S-labeled full-length and Δ553–900 SSeCKS products synthesized in coupled in vitro transcription/translation systems. Whereas the full-length SSeCKS product could be pulled down by the GST-PKCα beads, loss of the aa 553–900 domain ablated this binding (Fig. 2B).
FIGURE 2.
SSeCKS/PKC binding: domain mapping and effects of culture confluence. A, domains of SSeCKS that associate with PKCα. Lysates of HEK293T cells transiently transfected with a PKCα-encoding expression vector were incubated with beads containing GST alone or GST fusions of various SSeCKS domains (lower panel, Coomassie-stained GST proteins). The beads were then subjected to IB analysis and probed with anti-PKCα Ab (upper panel). B, SSeCKS aa 553–900 are required or PKCα binding. Left panel, 35S-labeled FL SSeCKS and SSeCKS[Δaa553–900] were produced in TnT reactions; right panel, IP of 35S-labeled proteins with beads containing GST or GST-PKCα. C, effect of PKC activity and PS (100 μg/ml) on SSeCKS domain binding. Left panel, slot blots containing GST or GST-SSeCKS fusion proteins blotted with anti-GST Ab; right panel, overlay assays incubated with various PKC preparations and probed by IB with PKC Abs. D, upper panel, homologous sequences within the aa 553–720 and 721–900 domains (*, identical; #, homologous). Ser-613, putative PKC phosphorylation site (underlined); bold, residues represented in the peptides in the lower panel. Lower panel, peptide competitors used in E (homologous residues of the 599 and 748 peptides in bold). E, aa 596–605 and 745–753 required for SSeCKS to bind PKCα. GST- or GST-SSeCKS beads were used to bind cell lysates of HEK293T cells transiently overexpressing PKCα followed by IB for PKCα. Δ596–605 and Δ745–753 are internal deletions to GST-SSeCKS[553–720] and SSeCKS[721–900], respectively. Right side, addition of molar excess (100×) of peptides 599, 748, and 835 described in D. The relative levels of bound PKC are shown in the lower box; *, S.E. from two independent experiments. F, Coomassie-stained SDS-PAGE of GST and GST-SSeCKS fusion proteins used in E. *, purified GST proteins.
Because the pulldown assays in Fig. 2, A and B, contain endogenous levels of PS from cell lysates, we sought to determine whether the two SSeCKS domains, aa 553–720 and 721–900, could bind PKC as purified proteins. Thus, produced slot blots containing equal amounts (1 μg/slot) of GST or GST-SSeCKS fusion protein were overlaid with various PKC preparations (or GST as a negative control) followed by IB with various PKC Abs. A commercial enzymatically active PKCα enzyme preparation (Biomol) bound to both domains (Fig. 2C), whereas bacterially expressed GST-PKCα bound only to 721–900 (Fig. 2B). Importantly, the GST-PKCα is not enzymatically active (supplemental Fig. S1) likely because of the absence of phosphorylation in the activation loop (1). PKCα labeled with 35S in TnT reactions only bound the 721–900 SSeCKS domain (Fig. 2C). In our hands, this form of PKC had no discernable PKC activity (data not shown), and thus, there is a correlation between the lack of enzymatic activity and failure to bind the aa 553–720 SSeCKS domain. Interestingly, preincubation of the bacterial GST-PKCα with 100 μg/ml PS, which marginally increased its enzymatic activity level (supplemental Fig. S1), induced binding to both 553–720 and 721–900 (Fig. 2C). It is noteworthy that enzymatically active PKCϵ only bound to the aa 721–900 domain (first panel on the right), and we speculate that because this isoform has a much lower affinity for PS than PKCα (38) it does not undergo a PS-mediated conformation change that would allow it to recognize the aa 553–720 domain. Nonetheless, in contrast to previous results by Piontek and Brandt (36) using pulldowns, this result plus the data in Fig. 1C and our recently published data (28) show that under these saturated binding conditions PKCϵ can bind to at least one SSeCKS domain. Taken together, our results strongly suggest that the 721–900 domain on SSeCKS binds PKC irrespective of enzyme activity, whereas the 553–720 domain binds a second PKC conformer associated with enzyme activity and possibly PS binding in the case of PKCα.
Although neither of the PKC binding SSeCKS regions shares homology with the PS-independent PKC-binding domain on RACK (39), they share a 28-aa sequence, EG(I/V)(T/S)XWXSFK(K/R)(M/L)VTP(K/R)K(K/R)X(K/R)XXXEXXXE(E/D), having 43% identity and 71% homology (Fig. 2D, upper panel). To show that the two homologous regions facilitate the PKCα binding activity of the 553–720 and 721–900 domains, GST fusions were produced lacking these regions (Δ596–605 and Δ745–753). Beads loaded with GST or these SSeCKS domains were used to pull down overexpressed PKCα from 293T cell lysates as in Fig. 2A. Loss of either homologous region ablated the PKCα binding activity of the respective 553–720 and 721–900 domains (Fig. 2E). Moreover, PKCα binding by each domain could be competed by incubating with molar excess levels of peptides covering either homologous sequence (“599” or “748”; Fig. 2D, lower panel) but not by a non-homologous SSeCKS peptide (“835”) (Fig. 2E). These data are consistent with SSeCKS encoding two independent yet homologous PKCα binding sites.
Effect of Culture Confluence on SSeCKS/PKC Association
Data from our laboratory and others suggest that the scaffolding activity of SSeCKS for PKC is antagonized by the phosphorylation of SSeCKS by PKC and other mitogen-activated kinases (32). Although the abundance of SSeCKS protein rises precipitously in confluence (40), the relative phosphorylation of SSeCKS decreases (41). Moreover, PMA or serum can induce serine phosphorylation of SSeCKS in cycling cells but not in confluent cultures (41). The increasing abundance of SSeCKS protein in confluent versus subconfluent WT-MEF correlated with decreased levels of total cellular PKC activity (Fig. 3A). Although PKCα proteins levels were unaffected by confluence in WT-MEF or in KO-MEF isolated from an SSeCKS-null mouse model (42), KO-MEF had roughly half the PKCα levels of WT-MEF (Fig. 3, A (upper panel) and B). Importantly, however, subconfluent KO-MEF exhibited roughly 3-fold more total PKC activity (based on an ELISA using a universal PKC substrate; Fig. 3A) or PKCα activity (based on the in vitro kinase activity of immunoprecipitated PKCα against MBP substrate; Fig. 3B) than WT-MEF, and this activity increased in confluent KO-MEF (Fig. 3A). More PKCα co-immunoprecipitated with SSeCKS from lysates of confluent cells (Fig. 3C, inset panel), and that population had roughly 40% less relative kinase activity than the co-immunoprecipitated PKCα from subconfluent lysates (Fig. 3C). PMA was capable of activating PKC to similar extents in subconfluent cultures of KO- and WT-MEF (Fig. 3D), ruling out the possibility of selective sensitivity in the KO-MEF. (Note that cultures of KO-MEF become contacted-inhibited (not shown), and thus, the lack of SSeCKS or the lack of PKC attenuation is not sufficient to override contact inhibition.) Taken together, these data suggest that the relatively underphosphorylated SSeCKS found in confluent cells has increased scaffolding activity for PKC and that SSeCKS binding in confluent cells inhibits PKC activity.
FIGURE 3.
SSeCKS inhibits PKC activity through its scaffolding domains. A, confluence-induced suppression of PKC activity correlates with increased SSeCKS levels. Upper panel, IB of lysates of WT- or KO-MEF for SSeCKS, PKCα, or GAPDH; lower panel, relative PKC specific activity (total activity normalized for PKCα protein levels) in subconfluent versus confluent conditions. Error bars, S.E. from triplicate experiments. B, increased relative PKCα activity in KO-MEF based on a 32P in vitro kinase assay using immunoprecipitated PKCα and MBP substrate. Relative protein or phosphorylation levels (numbers below the lower panels) reflect a typical result of three independent experiments. C, SSeCKS/PKCα association increases with culture confluence and correlates with decreased kinase activity. Lysates from subconfluent (subconfl.) and confluent cultures of NIH3T3 cells were directly blotted for PKCα (inset panel, “input”) or immunoprecipitated with anti-SSeCKS Ab and then probed by IB with anti-PKCα (lower inset panel) or subjected to PKC kinase assays (graph) as in A. Error bars, S.E. from triplicate experiments. D, PKC can be activated by PMA in the absence of SSeCKS. Lysates of serum-starved subconfluent WT- or KO-MEF treated with PMA (50 nm) or DMSO vehicle (“Ctrl”) for 1 or 5 min (′) were assayed for PKC activity. Error bars, S.E. from triplicate experiments. E, the PKCα-inhibitory domain on SSeCKS maps to aa 553–900. HA IPs of HEK293T lysates co-transfected with HA-PKCα, pEGFP, and either FL, Δ553–900, or Δ2–553 SSeCKS expression vectors were added to a 32P in vitro kinase assay as in B. Error bars, S.E. from triplicate experiments. F, expression of FL or Δ553–900 SSeCKS products in WT- (“+/+”) or KO (“−/−”)-MEF probed with anti-SSeCKS Ab. G, FL but not Δ553–900 SSeCKS suppresses PKC activity in cells. Lysates of WT- or KO-MEF transfected with pCMV vector (“V”) or expression vectors encoding FL or Δ553–900 SSeCKS were assayed for total PKC specific activity as in A. Error bars, S.E. from triplicate experiments. H, the homologous motifs aa 596–605 and 745–753, respectively, are required for SSeCKS aa 553–720 and 721–900 domains to inhibit PKCα activity (act.). Molar excesses (25×) of GST or GST-SSeCKS proteins (shown in Fig. 2F) were added to 32P in vitro kinase (IVK) assays with commercial (Biomol) PKCα and MBP substrate (lower panel, total versus 32P levels of MBP). Error bars, S.E. from triplicate experiments. *, p < 0.01. I, phosphorylation levels of the GST or GST-SSeCKS competitors used in H.
SSeCKS Inhibits PKC Activity
To investigate whether SSeCKS could directly inhibit PKC activity, 293T cells were co-transfected with HA-PKCα plus expression vectors for pEGFP or SSeCKS constructs, and then PKCα activity was monitored in HA IPs using the in vitro kinase/MBP assay described in Fig. 3B. Whereas full-length (FL) SSeCKS inhibited PKCα activity 2.5-fold, deletion of the two major PKC-binding regions (aa 553–900) abrogated this effect (Fig. 3E). In contrast, deletion of an upstream region (aa 2–553) shown not to encode PKC binding activity (Fig. 2, A and C) did not decrease PKC inhibition.
To determine whether SSeCKS scaffolding activity for PKC was responsible for the confluence-mediated PKC attenuation, KO-MEF were co-transfected with expression vectors encoding GFP fusions of either full-length SSeCKS or SSeCKS deleted of its PKC-binding domains (Δ553–900) (Fig. 3F). After isolation of the transfectants by FACS for GFP, aliquots of the cells were cultured under subconfluent conditions for an additional 24 h, and then cell lysates were assayed for total PKC activity. Full-length but not Δ553–900 SSeCKS could rescue PKC attenuation (Fig. 3G), indicating that SSeCKS likely attenuates PKC during contact inhibition through a direct scaffolding activity.
We sought to determine whether the PKC kinase-inhibitory activity of SSeCKS mapped to the two binding motifs, aa 596–605 and 745–753. Thus, the in vitro kinase activity of purified, enzymatically active PKCα against MBP substrate was assessed in the presence of a molar excess (25×) of either GST or GST-SSeCKS proteins. Whereas the individual PKC-binding domains in SSeCKS, aa 553–720 or 721–900, could inhibit PKCα activity, deletion of the aa 596–605 and 745–753 motifs in these proteins, respectively, abrogated this inhibition (Fig. 3H). As a control, the addition of a C-terminal SSeCKS domain encoding the PKA-Regulatory Subunit II-binding (AKAP) motif (43) failed to inhibit the kinase activity. Given that SSeCKS is also a major PKC substrate (26, 32), the GST and GST-SSeCKS fusion proteins from the PKCα kinase assay described in Fig. 3H were phosphorimaged. Only the aa 533–720 domain of SSeCKS was robustly phosphorylated (Fig. 3I). Importantly, although it was in molar excess, the relative phosphorylation of this SSeCKS domain was roughly 12% of that of the MBP substrate (data not shown). The fact that deletion of aa 596–605 severely decreased PKCα-mediated phosphorylation of the aa 553–720 domain along with the inability of the aa 721–900 domain to be phosphorylated (Fig. 3I) strongly suggests that PKCα phosphorylates Ser-613 (Ser-627 in rat and Ser-614 in human) and not a motif shared with the aa 741–769 PKC-binding domain (Fig. 2D). Indeed, phosphorylation of SSeCKS Ser-613 has been documented in at least 25 proteomics studies on PhosphoSitePlus. Taken together, these data suggest that SSeCKS inhibits PKC activity via direct scaffolding to its two PKC-binding domains, aa 596–605 and 745–753, and not via substrate competition.
SSeCKS/PKC Association Required to Transduce PMA-induced Cytoskeletal Reorganization
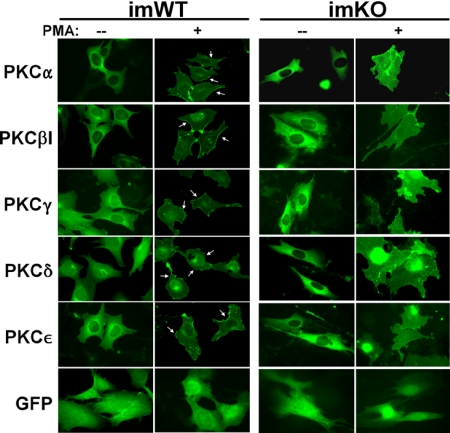
It is well established that PKC activation by short term PMA treatment induces major remodeling of the actin cytoskeleton in many cell types, starting with the collapse of actin fiber networks to the perinucleus and a loss of peripheral focal adhesions, resulting in increased cell rounding (1). Given that SSeCKS is an in vitro and in vivo PKC substrate (26, 32, 44) and that short term PMA treatment induces a co-translocation of SSeCKS and actin fibers to the perinucleus (32, 36, 45), we addressed whether KO-MEF were defective in PMA-induced PKC translocation, cytoskeletal remodeling, and cell rounding. Although KO-MEF exhibit increased PMA-induced PKC activation (Fig. 3D), immortalized KO (imKO)-MEF (28) showed little plasma membrane translocation of GFP-PKC isozymes or cell rounding after short term PMA treatment, whereas PKCα, -βI, -γ, -δ, or -ϵ moved to the cell periphery in PMA-treated immortalized WT-MEF concomitant with increased cell rounding (Fig. 4, arrows). As a control, pEGFP-transfected immortalized WT (imWT)-MEF showed a PMA-induced cell shape change but no membrane enrichment of GFP. These data strongly suggest that SSeCKS is required to scaffold activated PKC isozymes to sites of activity such as those involving substrate phosphorylation regulating actin cytoskeleton remodeling.
FIGURE 4.
Defective plasma membrane translocation of PKC isozymes in SSeCKS-null cells. imWT-MEF or imKO cells transfected with GFP fusion constructs of PKC isozymes (versus GFP alone) were treated with 100 nm PMA for 30 min and then imaged. Membrane translocation of PKC is shown by arrows.
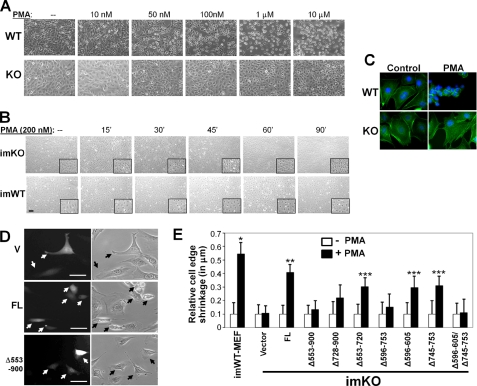
PMA induced cell rounding in a dose-dependent manner in confluent cultures of WT-MEF treated for 1 h with 100 nm, showing cell rounding in >80% of cells (counted in three independent fields of at least 200 cells in two independent experiments; Fig. 5A, top panel). Cell rounding was defined as the loss of flat cytoplasmic surfaces extending from the perinuclear area and the concomitant increase in phase-contrast refractility. In contrast, KO-MEF treated with 10 nm–1 μm PMA showed minimal cell shape change with only slight cell rounding at 10 μm PMA (Fig. 5A, bottom panel). Treatment of subconfluent cultures for even shorter periods (15–30 min) with 100 nm PMA yielded similar results, namely cell rounding in WT-MEF but little effect in KO-MEF (data not shown), consistent with our previous finding that culture confluence suppresses the ability of PMA to activate PKC (41). Longer treatment with 200 nm PMA induced cell rounding and the loss of F-actin stress fibers after 45 min in imWT- but not in imKO-MEF (Fig. 5B). Moreover, PMA failed to cause disassembly of F-actin stress fibers in the KO-MEF (Fig. 5C).
FIGURE 5.
PKC-binding domains in SSeCKS required to facilitate PMA-induced cell shape change. A, PMA fails to induce cell shape change in KO-MEF. Confluent cultures of WT- or KO-MEF were treated with various PMA concentrations for 1 h. B, treatment of imWT-MEF or imKO-MEF with 200 nm PMA for various times (′, minutes; insets, 4× magnifications. C, cell rounding and loss of actin stress fibers in WT- but not KO-MEF treated with 100 nm PMA for 0.5 h. Stress fibers, FITC-phalloidin; nuclei, DAPI. D, paired photomicrographs of GFP fluorescence (left panels) and phase-contrast (right panels) microscopy of KO-MEF transiently transfected with pBABE vector, FL SSeCKS, or Δ553–900 expression vectors. Size bars, 10 μm. Black/white arrows, transfected cells. E, PMA-induced cell shape change (relative shrinkage of cell edge) in imKO-MEF transfected with FL or SSeCKS deletion mutants compared with control imWT-MEF. Error bars, S.E. from triplicate experiments. *, p = 7.6 × 10−6; **, p = 0.0002; ***, p < 0.05.
To address whether the PKC scaffolding activity of SSeCKS is required for PMA-induced cell shape change, KO-MEF transiently co-transfected with expression vectors for SSeCKS plus EGFP were treated with 100 nm PMA for 30 min. Rounded versus flat GFP-expressing cells were scored by simultaneous phase-contrast and fluorescence microscopy (Fig. 5D, supplemental Fig. S2, and supplemental Fig. S4 for real time video microscopy) with the untransfected cells acting as internal controls. The expression of FL SSeCKS significantly restored the ability of PMA to induce cell rounding (Fig. 5, D and E). In contrast, expression of the Δ553–900 SSeCKS mutant failed to restore PMA-induced cell rounding; mutants lacking either PKC-binding domain, aa 596–605 or 745–753, partially restored cell rounding, whereas a mutant lacking both was incapable of restoration (Fig. 5E and supplemental Fig. S3). This suggests that the two SSeCKS PKC-binding domains work independently in cells. Taken together, these findings are consistent with a role for SSeCKS in mediating PKC-induced cytoskeletal reorganization through a direct association with PKC.
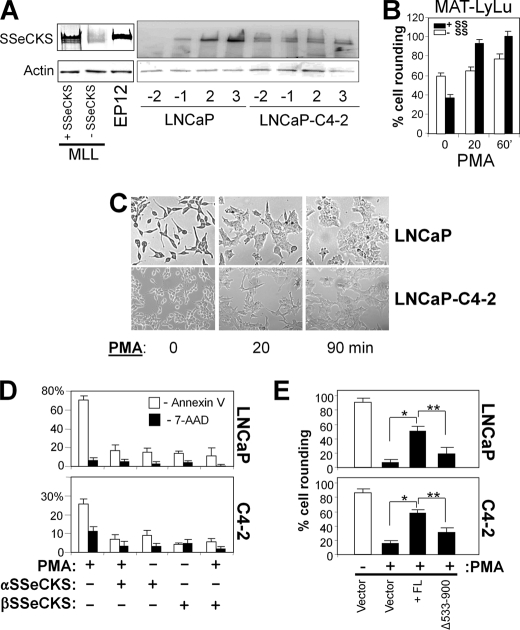
We investigated whether the deficiency in SSeCKS levels in rat and human CaP cell lines that we reported previously (31, 46) might play a direct role in facilitating PMA-induced cytoskeletal reorganization. The relative SSeCKS protein level in MAT-LyLu (MLL) cells, an androgen-responsive rat CaP cell line, was roughly 8-fold lower than in the untransformed, SV40 T antigen immortalized rat prostate epithelial cell line EP12 (47) (Fig. 6A, left panel). The forced re-expression of FL SSeCKS induced cell flattening prior to PMA treatment (Fig. 6B) consistent with our previously reported role for SSeCKS in re-establishing an actin-based cytoskeletal architecture and cell-cell adherence in MLL cells (31). Addition of PMA to MLL/vector cells (Fig. 6B, “− SS”) induced only marginal cell shape change between 20 and 60 min of treatment, whereas even short term (20 min) treatment of the SSeCKS-re-expressing cells (Fig. 6B, “+ SS”) caused a significant increase in cell rounding. These findings suggest that SSeCKS may facilitate the ability of PMA to induce cytoskeletal reorganization in MLL cells.
FIGURE 6.
SSeCKS facilitates PMA-induced CaP cell shape change. A, SSeCKS levels in CaP cells. IB for SSeCKS or β-actin from lysates of MLL cells +/− SSeCKS (2–6 cells grown, respectively, in regular or tetracycline-containing medium) versus EP12 cells (immortalized, untransformed rat prostate epithelial). Right panel, LNCaP or LNCaP-C4-2 cells were grown under pre- (−2 and −1) and confluent (2 and 3) conditions with the numbers representing days before or after confluence. B, SSeCKS increases PMA-induced cell rounding in MLL cells. Subconfluent cultures of MLL/2-6 cells grown in the absence or presence of tetracycline (+ and − SSeCKS (SS), respectively) treated with PMA (100 nm). ′, minutes. Error bars, S.E. from triplicate experiments. C, D, and E, SSeCKS prevents PMA-induced apoptosis and rescues PMA-induced cell rounding in LNCaP or LNCaP-C4-2 cells. Phase-contrast microscopy of subconfluent LNCaP cultures shows PMA-induced cell flattening (C). Cells were transfected transiently with GFP fusions of either the α or β SSeCKS isoform (24, 55), treated for 24 h with 100 nm PMA or DMSO vehicle, and then analyzed by FACS after staining for phycoerythrin-Annexin V (white columns) or 7-aminoactinomycin D (7-AAD) (black columns) and then gating for double staining with GFP-positive cells (D). Error bars, S.E. from triplicate experiments. Cells transfected with empty vector (pEGFP), FL SSeCKS-GFP or SSeCKS[Δ533–900]-GFP were analyzed by fluorescence microscopy for cell rounding (E). Error bars, S.E. from triplicate experiments. *, p < 0.002; **, p < 0.01.
To strengthen this notion, we reproduced this analysis in human LNCaP cells, which exhibit severe down-regulation of SSeCKS (25-fold) compared with the untransformed, SV40 T antigen immortalized human epithelial cell line P69SV40T as we reported previously (48). Even with this down-regulation, SSeCKS is likely to exert some regulatory functions in this cell type because SSeCKS protein levels were still induced by culture confluence (Fig. 5A, right panel). In contrast, the metastatic, androgen-independent derivative, LNCaP-C4-2 (49), showed no confluence-induced SSeCKS. PMA treatment of LNCaP or LNCaP-C4-2 cells induced severe cell flattening (Fig. 6, C and E) and apoptosis (Fig. 6D) in agreement with previous publications (21, 22, 50–52). However, the re-expression of FL SSeCKS α or β isoform rescued these cells from PMA-induced apoptosis (Fig. 6D), and FL αSSeCKS, but not the SSeCKS[Δ553–900] mutant, could facilitate PMA-induced cell rounding (Fig. 6E). Taken with the data for MLL cells and KO-MEF, these findings indicate that SSeCKS is sufficient to facilitate PMA-induced cell shape change likely through a direct interaction with PKC.
DISCUSSION
The phenomenon of PMA-induced actin cytoskeletal remodeling has been well described in the literature for some time, and there is clear evidence that the reorganization of the cytoskeletal architecture, mediated in major part through the activation of PKC isoforms, controls oncogenic processes such as proliferation, cell survival, and invasiveness (1). Here we show that a major PKC substrate, SSeCKS/GRAVIN/AKAP12, is required in MEF and CaP cells to mediate PMA-induced cell shape change and in the case of CaP cells to suppress PMA-induced apoptosis. Moreover, we show that a region in SSeCKS containing at least two PKC-binding domains is required to rescue the PMA effect on the actin cytoskeleton. These data strongly suggest that PKC signals to the cytoskeleton are mediated through a direct scaffolding interaction between SSeCKS and PKC. Consistent with this notion, SSeCKS-null MEF were defective in PMA-induced cytoskeletal reorganization and cell shape change. This deficiency was unaffected by cell confluence, and moreover, these cells were not defective in PKC kinase activation by PMA, although they were defective in the ability to translocate PKC isozymes to the plasma membrane.
Previous attempts to demonstrate PKC-SSeCKS binding involved overlay protocols requiring high, non-physiologic levels of PS, specifically 100–400 μg/ml (25, 44). Indeed, we showed previously that the addition of high levels of PS enhanced the formation of PKC-SSeCKS complexes in pulldown experiments from endogenous protein lysates (32). Here, we confirm that SSeCKS binds PS independently and at levels likely to be physiologically relevant. The affinity of SSeCKS for PS is likely facilitated by the potential PS-binding motif, 506FSSSGLKKLSGKKQR520, which is homologous (bold residues) to the known PS-binding motif in PKC, FXFXLKXXXXKXXR (53). Interestingly, SSeCKS also binds phosphatidic acid and phosphatidylglycerol albeit at affinities 2–4-fold lower than for PS. It should be noted that the direct binding affinity of SSeCKS for PS is at least 2 orders of magnitude lower than what is required to facilitate the overlay experiments described above. This suggests that the PKC/SSeCKS association in the overlay assays may be indirect, i.e. facilitated by the independent association of PKC and SSeCKS to PS in structures promoted by high concentrations of PS (e.g. micelles). Indeed, our current results indicate that the addition of PS did not enhance the overall binding level between bacterially produced GST-PKCα and His-SSeCKS, although it did induce binding site preference (below). This is the first description of a PS-independent protein-protein interaction between PKC and SSeCKS. A role for PS in facilitating PKC/SSeCKS association in cells cannot be ruled out, however, because we show reciprocal co-IP between SSeCKS and various cPKC or nPKC family members from cell lysates, which likely contain physiologic levels of free PS (non-membranous). This agrees with previous findings from Piontek and Brandt (36) in which endogenous human SSeCKS (GRAVIN) and PKCα, -βI, and -βII could be co-immunoprecipitated from lysates of NT2-N1 neurons without the addition of PS.
Our data identify two independent PKC-binding domains on SSeCKS, aa 596–605 and 745–753, which share the common motif EG(I/V)(T/S)XWXSFK(K/R)(M/L)VTP(K/R)K(K/R)X(K/R)XXXEXXXE(E/D). The aa 745–753-containing domain bound all three preparations of PKCα we tested irrespective of kinase activity. This domain also bound a commercial enzymatically active preparation of PKCϵ, a finding that contrasts with a previous report (36), although it should be noted that our conditions are at saturating levels. In contrast, enzymatically inactive preparations of PKCα failed to bind to the aa 596–605-containing domain, whereas the enzymatically active commercial PKCα preparation bound this domain. Moreover, the preincubation of bacterially expressed GST-PKCα with high PS levels, a treatment that marginally activates PKC, induced binding to the aa 596–605-containing domain. Thus, the 596–605 domain might have higher affinity for enzymatically active or PS-stimulated conformations of PKCα.
SSeCKS association with PKCα increases in confluent cultures, a condition in which SSeCKS protein accumulation is induced significantly (40) and in which SSeCKS phosphorylation by PKC (following PMA addition) is minimized (54). Our data suggest that SSeCKS binding under these cellular conditions inhibits PKCα activity. This is based on our finding that the ability of the aa 553–720 and 721–900 domains to inhibit PKCα activity in vitro was dependent on retention of their PKC-binding domains (i.e. deletion of either aa 596–605 or 745–753 abrogated the inhibitory activities). The finding that total cellular PKC activity in SSeCKS-null MEF was more than double that found in WT-MEF strengthens the notion that a function of SSeCKS in cells is to attenuate PKC activation through its scaffolding activity.
Our model suggests that SSeCKS regulates PKC activity in a dynamic, spatiotemporal process: during contact inhibition when SSeCKS-PKC binding is saturated, SSeCKS inhibits PKC activity, whereas in mitogenically activatable cells, SSeCKS binding to PKC facilitates its translocation to sites where it can phosphorylate substrates involved in cytoskeletal remodeling and cell survival. Continued mitogenic stimulation results in the phosphorylation of SSeCKS by PKC and other kinases, antagonizing its binding to PKC and acting as a negative feedback to disengage PKC from mediating signals to the actin cytoskeleton. This might explain why in CaP lacking SSeCKS PMA fails to induce the cytoskeletal remodeling that regulates normal cell cycle progression, resulting in a reactive program of apoptosis. In this context, SSeCKS down-regulation promotes CaP malignancy by facilitating PKC-induced invasiveness and proliferation at the cost of increased apoptosis.
Acknowledgments
We thank Jennifer Black for critical reading of the manuscript. We thank Jeff Streb, Joe Miano, Jae-Won Soh, and Lionel Coignet for sharing plasmid reagents.
This work was supported, in whole or in part, by National Institutes of Health Grants CA94108 and CA116430 (to I. H. G.) and Comprehensive Cancer Center Grant P30-CA016056 from the NCI.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1–S4.
- cPKC
- conventional PKC
- nPKC
- novel PKC
- PMA
- phorbol 12-myristate 13-acetate
- STICK
- substrate that interacts with protein kinase C
- PS
- phosphatidylserine
- RACK
- receptor for activated C kinases
- MARCKS
- myristoylated alanine-rich protein kinase C substrate
- CaP
- prostate cancer
- SSeCKS
- Src-suppressed C kinase substrate
- MEF
- mouse embryo fibroblasts
- MLL
- MAT-LyLu
- aa
- amino acid
- IB
- immunoblotting
- Ab
- antibody
- IP
- immunoprecipitation
- MBP
- myelin basic protein
- NRS
- normal rabbit serum
- AKAP
- A-Kinase Anchoring Protein
- FL
- full-length
- imKO
- immortalized KO
- imWT
- immortalized WT
- EGFP
- enhanced GFP.
REFERENCES
- 1. Larsson C. (2006) Cell. Signal. 18, 276–284 [DOI] [PubMed] [Google Scholar]
- 2. Kazanietz M. G. (2000) Mol. Carcinog. 28, 5–11 [DOI] [PubMed] [Google Scholar]
- 3. Hornia A., Lu Z., Sukezane T., Zhong M., Joseph T., Frankel P., Foster D. A. (1999) Mol. Cell. Biol. 19, 7672–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hofmann J. (2004) Curr. Cancer Drug Targets 4, 125–146 [DOI] [PubMed] [Google Scholar]
- 5. Leitges M. (2007) Biochem. Soc. Trans. 35, 1018–1020 [DOI] [PubMed] [Google Scholar]
- 6. Oster H., Leitges M. (2006) Cancer Res. 66, 6955–6963 [DOI] [PubMed] [Google Scholar]
- 7. Jaken S., Parker P. J. (2000) BioEssays 22, 245–254 [DOI] [PubMed] [Google Scholar]
- 8. Diviani D., Scott J. D. (2001) J. Cell Sci. 114, 1431–1437 [DOI] [PubMed] [Google Scholar]
- 9. Turbedsky K., Pollard T. D., Bresnick A. R. (1997) Biochemistry 36, 2063–2067 [DOI] [PubMed] [Google Scholar]
- 10. Bogatcheva N. V., Verin A. D., Wang P., Birukova A. A., Birukov K. G., Mirzopoyazova T., Adyshev D. M., Chiang E. T., Crow M. T., Garcia J. G. (2003) Am. J. Physiol. Lung Cell. Mol. Physiol. 285, L415–L426 [DOI] [PubMed] [Google Scholar]
- 11. Weber L. P., Seto M., Sasaki Y., Swärd K., Walsh M. P. (2000) Biochem. J. 352, 573–582 [PMC free article] [PubMed] [Google Scholar]
- 12. Venema R. C., Raynor R. L., Noland T. A., Jr., Kuo J. F. (1993) Biochem. J. 294, 401–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Singer H. A. (1990) J. Pharmacol. Exp. Ther. 252, 1068–1074 [PubMed] [Google Scholar]
- 14. Koren R., Ben Meir D., Langzam L., Dekel Y., Konichezky M., Baniel J., Livne P. M., Gal R., Sampson S. R. (2004) Oncol. Rep. 11, 321–326 [PubMed] [Google Scholar]
- 15. Cornford P., Evans J., Dodson A., Parsons K., Woolfenden A., Neoptolemos J., Foster C. S. (1999) Am. J. Pathol. 154, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Benavides F., Blando J., Perez C. J., Garg R., Conti C. J., DiGiovanni J., Kazanietz M. G. (2011) Cell Cycle 10, 268–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stewart J. R., O'Brian C. A. (2005) Mol. Cancer Ther. 4, 726–732 [DOI] [PubMed] [Google Scholar]
- 18. Gonzalez-Guerrico A. M., Meshki J., Xiao L., Benavides F., Conti C. J., Kazanietz M. G. (2005) J. Biochem. Mol. Biol. 38, 639–645 [DOI] [PubMed] [Google Scholar]
- 19. Gonzalez-Guerrico A. M., Kazanietz M. G. (2005) J. Biol. Chem. 280, 38982–38991 [DOI] [PubMed] [Google Scholar]
- 20. Fujii T., García-Bermejo M. L., Bernabó J. L., Caamaño J., Ohba M., Kuroki T., Li L., Yuspa S. H., Kazanietz M. G. (2000) J. Biol. Chem. 275, 7574–7582 [DOI] [PubMed] [Google Scholar]
- 21. Garcia-Bermejo M. L., Leskow F. C., Fujii T., Wang Q., Blumberg P. M., Ohba M., Kuroki T., Han K. C., Lee J., Marquez V. E., Kazanietz M. G. (2002) J. Biol. Chem. 277, 645–655 [DOI] [PubMed] [Google Scholar]
- 22. Tanaka Y., Gavrielides M. V., Mitsuuchi Y., Fujii T., Kazanietz M. G. (2003) J. Biol. Chem. 278, 33753–33762 [DOI] [PubMed] [Google Scholar]
- 23. Gelman I. H. (2002) Front. Biosci. 7, d1782–1797 [DOI] [PubMed] [Google Scholar]
- 24. Gelman I. H. (2010) Genes Cancer 1, 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hyatt S. L., Liao L., Aderem A., Nairn A. C., Jaken S. (1994) Cell Growth Differ. 5, 495–502 [PubMed] [Google Scholar]
- 26. Chapline C., Cottom J., Tobin H., Hulmes J., Crabb J., Jaken S. (1998) J. Biol. Chem. 273, 19482–19489 [DOI] [PubMed] [Google Scholar]
- 27. Lin X., Gelman I. H. (2002) Biochem. Biophys. Res. Commun. 290, 1368–1375 [DOI] [PubMed] [Google Scholar]
- 28. Akakura S., Nochajski P., Gao L., Sotomayor P., Matsui S., Gelman I. H. (2010) Cell Cycle 9, 4656–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pear W. S., Nolan G. P., Scott M. L., Baltimore D. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gelman I. H., Lee K., Tombler E., Gordon R., Lin X. (1998) Cell Motil. Cytoskeleton 41, 1–17 [DOI] [PubMed] [Google Scholar]
- 31. Xia W., Unger P., Miller L., Nelson J., Gelman I. H. (2001) Cancer Res. 61, 5644–5651 [PubMed] [Google Scholar]
- 32. Lin X., Tombler E., Nelson P. J., Ross M., Gelman I. H. (1996) J. Biol. Chem. 271, 28430–28438 [DOI] [PubMed] [Google Scholar]
- 33. Bu Y., Gelman I. H. (2007) J. Biol. Chem. 282, 26725–26739 [DOI] [PubMed] [Google Scholar]
- 34. Ho A., Schwarze S. R., Mermelstein S. J., Waksman G., Dowdy S. F. (2001) Cancer Res. 61, 474–477 [PubMed] [Google Scholar]
- 35. Moissoglu K., Gelman I. H. (2003) J. Biol. Chem. 278, 47946–47959 [DOI] [PubMed] [Google Scholar]
- 36. Piontek J., Brandt R. (2003) J. Biol. Chem. 278, 38970–38979 [DOI] [PubMed] [Google Scholar]
- 37. Selkirk J. K., Elwood J. C., Morris H. P. (1971) Cancer Res. 31, 27–31 [PubMed] [Google Scholar]
- 38. Medkova M., Cho W. (1998) Biochemistry 37, 4892–4900 [DOI] [PubMed] [Google Scholar]
- 39. Stebbins E. G., Mochly-Rosen D. (2001) J. Biol. Chem. 276, 29644–29650 [DOI] [PubMed] [Google Scholar]
- 40. Lin X., Nelson P., Gelman I. H. (2000) Mol. Cell. Biol. 20, 7259–7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nelson P. J., Gelman I. H. (1997) Mol. Cell. Biochem. 175, 233–241 [DOI] [PubMed] [Google Scholar]
- 42. Akakura S., Huang C., Nelson P. J., Foster B., Gelman I. H. (2008) Cancer Res. 68, 5096–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nauert J. B., Klauck T. M., Langeberg L. K., Scott J. D. (1997) Curr. Biol. 7, 52–62 [DOI] [PubMed] [Google Scholar]
- 44. Chapline C., Mousseau B., Ramsay K., Duddy S., Li Y., Kiley S. C., Jaken S. (1996) J. Biol. Chem. 271, 6417–6422 [DOI] [PubMed] [Google Scholar]
- 45. Yan X., Walkiewicz M., Carlson J., Leiphon L., Grove B. (2009) Exp. Cell Res. 315, 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Su B., Zheng Q., Vaughan M. M., Bu Y., Gelman I. H. (2006) Cancer Res. 66, 5599–5607 [DOI] [PubMed] [Google Scholar]
- 47. Yamazaki K., Pienta K. J. (1995) In Vivo 9, 427–432 [PubMed] [Google Scholar]
- 48. Bae V. L., Jackson-Cook C. K., Brothman A. R., Maygarden S. J., Ware J. L. (1994) Int. J. Cancer 58, 721–729 [DOI] [PubMed] [Google Scholar]
- 49. Wu H. C., Hsieh J. T., Gleave M. E., Brown N. M., Pathak S., Chung L. W. (1994) Int. J. Cancer 57, 406–412 [DOI] [PubMed] [Google Scholar]
- 50. Young C. Y., Murtha P. E., Zhang J. (1994) Oncol. Res. 6, 203–210 [PubMed] [Google Scholar]
- 51. Garzotto M., White-Jones M., Jiang Y., Ehleiter D., Liao W. C., Haimovitz-Friedman A., Fuks Z., Kolesnick R. (1998) Cancer Res. 58, 2260–2264 [PubMed] [Google Scholar]
- 52. Henttu P., Vihko P. (1998) Biochem. Biophys. Res. Commun. 244, 167–171 [DOI] [PubMed] [Google Scholar]
- 53. Igarashi K., Kaneda M., Yamaji A., Saido T. C., Kikkawa U., Ono Y., Inoue K., Umeda M. (1995) J. Biol. Chem. 270, 29075–29078 [DOI] [PubMed] [Google Scholar]
- 54. Nelson P. J., Moissoglu K., Vargas J., Jr., Klotman P. E., Gelman I. H. (1999) J. Cell Sci. 112, 361–370 [DOI] [PubMed] [Google Scholar]
- 55. Streb J. W., Kitchen C. M., Gelman I. H., Miano J. M. (2004) J. Biol. Chem. 279, 56014–56023 [DOI] [PubMed] [Google Scholar]