Background: The biosynthetic pathway for the unique antibiotic roseoflavin from Streptomyces davawensis is unknown.
Results: A novel N,N-8-amino-8-demethyl-d-riboflavin dimethyltransferase (RosA) was purified from S. davawensis.
Conclusion: RosA catalyzes the two terminal steps of roseoflavin biosynthesis.
Significance: RosA is the first enzyme of roseoflavin biosynthesis to be identified.
Keywords: Antibiotics, Bacteria, Biosynthesis, Enzyme Catalysis, Flavin, Flavin Analogs
Abstract
Streptomyces davawensis synthesizes the antibiotic roseoflavin (RoF) (8-dimethylamino-8-demethyl-d-riboflavin). It was postulated that RoF is synthesized from riboflavin via 8-amino- (AF) and 8-methylamino-8-demethyl-d-riboflavin (MAF). In a cell-free extract of S. davawensis, an S-adenosyl methionine-dependent conversion of AF into MAF and RoF was observed. The corresponding N,N-8-amino-8-demethyl-d-riboflavin dimethyltransferase activity was enriched by column chromatography. The final most active fraction still contained at least five different proteins that were analyzed by enzymatic digestion and concomitant de novo sequencing by MS/MS. One of the sequences matched a hypothetical peptide fragment derived from an as yet uncharacterized open reading frame (sda77220) located in the middle of a (putative) gene cluster within the S. davawensis genome. Expression of ORF sda77220 in Escherichia coli revealed that the corresponding gene product had N,N-8-amino-8-demethyl-d-riboflavin dimethyltransferase activity. Inactivation of ORF sda77220 led to a S. davawensis strain that synthesized AF but not MAF or RoF. Accordingly, as the first identified gene of RoF biosynthesis, ORF sda77220 was named rosA. RosA (347 amino acids; 38 kDa) was purified from a recombinant E. coli strain (as a His6-tagged protein) and was biochemically characterized (apparent Km for AF = 57.7 ± 9.2 μm; apparent KD for AF = 10.0 μm; kcat = 0.37 ± 0.02 s−1). RosA is a unique enzyme and may be useful for a variety of applications.
Introduction
The Gram-positive bacterium Streptomyces davawensis was first isolated from a Philippine soil sample in a screening program for antibiotic-producing organisms. S. davawensis synthesizes the red riboflavin (vitamin B2) analog roseoflavin (RoF)2 (Scheme 1; compound 12) (1, 2). RoF is the only known natural riboflavin analog with antibiotic activity. RoF is toxic to Gram-positive but also to Gram-negative bacteria provided that the compound is able to enter the cell (3, 4). The molecular mechanism of action of RoF and its biosynthesis as well as the resistance mechanism of the producer strain are currently not understood (5). The synthesis of presumably inactive flavin cofactors, i.e. roseoflavin 5′-monophosphate and roseoflavin adenine dinucleotide, may at least in part explain its antibiotic properties (6). In addition, roseoflavin-5′-monophosphate may block FMN-riboswitches, rendering cells riboflavin auxotrophic (7–9). The genome of S. davawensis has been sequenced in our laboratory to near completion (99%), and publication of these data is planned for early 2012.
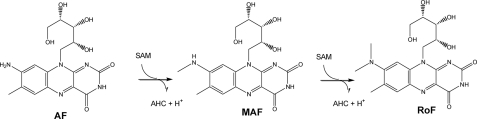
SCHEME 1.
The enzymatic conversion of AF and MAF into RoF. The SAM-dependent reaction releases S-adenosyl-l-homocysteine (AHC).
It was postulated that RoF is synthesized from GTP and ribulose 5-phosphate through riboflavin, 8-amino-8-demethyl-d-riboflavin (AF), and 8-methylamino-8-demethyl-d-riboflavin (MAF) (10, 11). The major lines of evidence were 1) incorporation of 14C of [2- and U-14C]guanine and [2-14C]riboflavin into RoF, 2) no incorporation of 14C of [8-14C]guanine into RoF, and 3) formation of [2-14C]roseoflavin upon the addition of [2-14C]AF or [2-14C]MAF (11). Accordingly, S. davawensis is able to convert a vitamin into an antibiotic vitamin analog. This strategy for the synthesis of an antimicrobial compound seems to be very economical as the precursors for antibiotic synthesis are readily available. Furthermore, many microorganisms have efficient vitamin transporters that catalyze the uptake of vitamins but also vitamin analogs. Thus, the delivery of the antivitamin to the target molecules in the cytoplasm is ensured. Last, antibiotic vitamin analogs have multiple cellular targets. In the case of RoF, incorporating the potentially inactive cofactors roseoflavin 5′-monophosphate or roseoflavin adenine dinucleotide may in principle inactivate all flavoenzymes within a cell (e.g. in bacteria about 1–3% of all enzymes are flavoenzymes (12)). Accordingly, for NAD(P)H:flavin oxidoreductase (EC 1.5.1.30) from Beneckea harveyi (now Vibrio harveyi) (13) roseoflavin-5′-monophosphate was found to be an inactive cofactor (14). For FAD-dependent d-amino acid oxidase (EC 1.4.3.3) from Sus scrofa, roseoflavin adenine dinucleotide was found to be an inactive cofactor (6). Other flavoenzymes are currently under investigation. As an important step toward the analysis of the antibiotic roseoflavin, the first enzyme (an N,N-8-amino-8-demethyl-d-riboflavin dimethyltransferase) active within the RoF biosynthetic pathway was partially purified using a novel assay. The corresponding gene was identified, biochemically characterized, and named rosA. The gene was found to be located in a gene cluster, which possibly contains other genes involved in RoF biosynthesis. Inactivation of rosA produced a MAF/RoF-deficient strain.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The plasmids and strains used in this work are shown in Table 1. S. davawensis (Streptomyces strain 768) was aerobically grown at 37 °C and pH 7.2 in a nutrient broth (YS) containing yeast extract (2 g/liter) and soluble potato starch (10 g/liter). Escherichia coli was cultivated on LB. For growth of precultures or small scale cultures (up to 200 ml), baffled Erlenmeyer flasks were used for good aeration and cell dispersion. The cultures were agitated at 220 rpm in an orbital shaker. For growth on solid media agar-agar (16 g/liter) was added. To produce spore suspensions, a maltose soybean growth medium was prepared containing maltose (20 g/liter), soybean meal (20 g/liter), and agar-agar (20 g/liter). At a larger scale (for recombinant protein production) E. coli was grown in a 15-liter bioreactor (Bioengineering, Wald, Switzerland) at 1000 rpm, 37 °C, and an aeration of 1.0 vvm in a total volume of 10 liters. S. davawensis was also grown in a 15-liter bioreactor. A total of 5 × 108 S. davawensis spores were used to inoculate 10 liter of YS medium. Growth conditions were kept constant at 700 rpm, 37 °C, and an aeration of 0.3 vvm in a total volume of 10 liter until stationary phase, where production of RoF occurred.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| S. davawensis | Wild-type strain | Japan Society for Culture Collections, Streptomyces strain no. 768 |
| S. albus | Wild-type strain | DSMZ strain no. 41208 |
| S. lividans 66 TK24 | str-6, SLP2−, SLP3− | John Innes Centre, Norwich, UK |
| E. coli Rosetta 2 (DE3) | Expression strain | Merck KGaA, Darmstadt, Germany |
| E. coli ET12567(pUB307) | dam-13::Tn9, dcm-6, hsdM, conjugative plasmid conferring Kanr (pUB307) | Ref. 19 |
| pET24a(+) | Expression vector | Merck KGaA, Darmstadt, Germany |
| pFJ01 | pET24a(+)rosA | This work |
| pFJ02 | pET24a(+)rosA-His6 | This work |
| pOJ436 | acc(3)IV(Aprr), (cos)3λ, IncPα oriT, colE1 origin, intφC31 | Ref. 20 |
| pFJ03 | pOJ436 carrying a 38,277-bp insert containing the putative ros gene cluster | This work |
| pSET152 | acc(3)IV(Aprr), oriT(RK2), pUC18 origin, intφC31 | Ref. 20 |
| pFJ04 | pSET152 carrying a 10,321-bp insert with the putative ros gene cluster derived from pFJ03 | This work |
| pUC18 | Cloning vector | Ref. 16 |
| pIJ773 | Source of the apramycin resistance gene aac(3)IV and oriT | 18 |
| pFJ05 | pUC18 cloning vector containing the apramycin resistance gene aac(3)IV and oriT from pIJ773 | This work |
| pFJ06 | pFJ05 containing a 509-bp fragment internal to rosA; used for inactivation of rosA | This work |
Isolation of Total DNA and Other Molecular Biology/Cloning Techniques
For the isolation of total DNA from S. davawensis, a modified protocol of the Kirby Mix Procedure was used (15). Other molecular biology techniques were carried out according to standard protocols (16).
N,N-8-Amino-8-demethyl-d-riboflavin Dimethyltransferase Assay and HPLC Analysis of Flavins
N,N-8-Amino-8-demethyl-d-riboflavin dimethyltransferase (RosA) activity was measured in a final volume of 1 ml of 50 mm Tris-HCl (pH 8.0) containing 200 μm AF and 2 mm S-adenosyl methionine (SAM). The mixture was preincubated at 37 °C for 5 min, and the reaction was started by the addition of SAM. After appropriate time intervals, an aliquot was removed and applied directly to an HPLC column (REPROSIL-PUR C18-AQ, 5-μm particle size, 250 × 4 mm; Dr. A. Maisch HPLC-GmbH, Ammerbuch-Entringen, Germany). The following solvent system was used at a flow rate of 0.8 ml/min: 40% (v/v) methanol, 100 mm formic acid, 100 mm ammonium formate (pH 3.7). AF, MAF, and RoF were detected photometrically at 485, 495, and 509 nm, respectively. HPLC/MS analysis was performed using 35% (v/v) methanol, 20 mm formic acid, 20 mm ammonium formate (REPROSIL-PUR C18-AQ, 5-μm particle size, 250 × 2 mm; Dr. A. Maisch HPLC-GmbH) with the Agilent 1260 Infinity system and a 6130 Quadrupole ESI-MS (Agilent Technologies, Santa Clara, CA). RosA activity is expressed as μmol of RoF formed/min from AF and SAM. Assays were carried out by varying the concentration of the substrate AF (ranging from 16 to 326 μm) to determine the Km and kcat of the first methylation step under saturating conditions of SAM (2 mm). To determine the Km and kcat for SAM, AF was kept constant at saturating conditions (200 μm), and the concentration of SAM was varied (ranging from 25 to 500 μm). The initial reaction rates were plotted against the substrate concentrations, and the kinetic constants Km and Vmax were evaluated from the best fit to the Michaelis-Menten equation using nonlinear regression in the SigmaPlot software. The turnover numbers, kcat, were calculated with the subunit molecular mass of 38 kDa for RosA. The pH/temperature dependence was determined using the rate of change of absorption at 505 nm in a spectrophotometer (Ultrospec 3100 pro, GE Healthcare).
Purification of Native N,N-8-Amino-8-demethyl-d-riboflavin Dimethyltransferase from S. davawensis
To wet-packed stationary phase cells of S. davawensis suspended in 50 mm Tris-HCl (pH 8.0), EDTA, free Complete Protease Inhibitor Mixture (Roche Applied Science) was added according to the recommendations of the manufacturer. Cells were then disintegrated in a French press system at 2000 bar and 10 °C. Cell debris was removed by centrifugation at 10,000 × g for 30 min and at 100,000 × g for 1 h. Solid (powdered) ammonium sulfate was added to the cell-free extract to achieve 30% saturation. After centrifugation at 10,000 × g, the supernatant was brought to 60% ammonium sulfate saturation and centrifuged as described above. The protein pellet was dissolved in loading buffer (1 m ammonium sulfate, 50 mm Tris-HCl, 5 mm DTT, 1 mm EDTA (pH 8.0)) for hydrophobic interaction chromatography with phenyl-Sepharose™ High Performance (GE Healthcare). All the following chromatographic steps were performed using the ÄKTApurifier™ system (GE Healthcare). After equilibration of the column and application of the sample, dimethyltransferase eluted within a step gradient of 50% elution buffer (50 mm Tris-HCl, 5 mm DTT, 1 mm EDTA, 10% glycerol (pH 8.0)). Active fractions were pooled, and the buffer was changed to anion exchange chromatography loading buffer (50 mm Tris-HCl, 2 mm DTT, 1 mm EDTA, pH 8.0, 10% glycerol) with Hiprep Desalting columns (GE Healthcare). The sample was loaded on a Mono Q 5/50 GL column (GE Healthcare) previously equilibrated with AEX (GE Healthcare) loading buffer and eluted by a linear gradient of 0–0.125 m ammonium sulfate in 50 mm Tris-HCl, 2 mm DTT, 1 mm EDTA, pH 8.0, and 10% glycerol. Active fractions were pooled and concentrated with Vivaspin concentrators (Sartorius Stedim Biotec, Goettingen, Germany) with a molecular weight cutoff of 10,000 at 4,000 × g. The last purification step was size exclusion chromatography on a HiLoad 16/60 Superdex 200 Prep Grade column (GE Healthcare) with 50 mm Tris-HCl, 2 mm DTT, 1 mm EDTA (pH 8.0) and 10% glycerol. Finally active fractions were concentrated with Vivaspin concentrators as described above and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie Brilliant Blue R-250. The purest fraction was used for subsequent protein identification by MS/MS.
De Novo Peptide Sequencing by MS/MS
The Protein band was excised from SDS-PAGE and destained using ammonium bicarbonate and acetonitrile. In-gel digestion was performed overnight at 37 °C using trypsin after saturation of the gel pieces. Peptides were extracted from the gel, and peptide sequences were determined using Ultraflex Maldi TOF/TOF (Bruker, Ettlingen, Germany). Peptide sequences were compared with protein sequence data deduced from the genomic sequence of S. davawensis.
Heterologous Overexpression of S. davawensis rosA in E. coli
To construct the expression vectors pET24a(+)rosA (pFJ01) and pET24a(+)rosA-His6 (pFJ02) introducing an additional C-terminal His6 tag, NdeI/XhoI-treated pET24a(+) (Merck KGaA, Darmstadt, Germany) and the modifying oligonucleotides rosAfw (5′-ACT TAC ATA TGC GGC CGG AAC CGA CC-3′) and rosArev (5′-ATA ACT CGA GTC AGC CGG CCG TGC C-3′) or rosArevHis (5′-ATA ACT CGA GGC CGG CCG TGC CG-3′) were used. E. coli Rosetta 2 (DE3) (Merck KGaA) was transformed with pET24a(+)rosA (pFJ01) and pET24a(+)rosA-His6 (pFJ02). For rosA and rosA-His6 expression, the resulting strains were grown at 37 °C on LB medium. Expression was induced by adding 0.1 mm isopropylthiogalactopyranoside after the culture had reached an absorbance of 0.6 (A600). After 5 h of further aerobic incubation, cells were harvested by centrifugation.
Purification of Recombinant His-tagged RosA from E. coli
Chromatographic steps were performed using the ÄKTApurifier™ system (GE Healthcare). Frozen cell paste of E. coli Rosetta 2 (DE3) (6 g) overproducing RosA was resuspended in 30 ml of HisTrap (GE-Healthcare) binding buffer (50 mm Na2HPO4 (pH 7.4), 500 mm NaCl, 20 mm imidazole). Cells were passed twice through a French press at 1500 bar. All subsequent centrifugation steps were performed at 10,000 × g and 4 °C. Centrifugation for 45 min removed cell debris and unbroken cells. The clear lysate was filtered (0.2 μm) and applied to a 5-ml HisTrap column after equilibration with loading buffer. When the UV signal returned to base line, elution of His6-tagged protein was induced with 50% elution buffer (50 mm Na2HPO4 (pH 7.4), 500 mm NaCl, 500 mm imidazole). Aliquots of the fractions were analyzed by SDS-PAGE and staining with Coomassie Brilliant Blue R-250. The apparently homogeneous fractions were tested directly for N,N-8-amino-8-demethyl-d-riboflavin dimethyltransferase activity and stored at −20 °C. The enzyme was stable for at least five months under these conditions.
Ligand Binding Titrations
Titrations were performed in 100 mm Tris-HCl, pH 7.4, by systematically varying the RosA concentration. The spectra were recorded between 300 and 650 nm with a Uvikon 933 spectrophotometer (BioTek Kontron Instruments AG). The dissociation constant (KD) for AF was estimated by determining the concentration of free and bound AF by means of membrane centrifugation in Microcon filter devices (Millipore). Flavins were analyzed by HPLC.
Isolation of Total RNA from S. davawensis and RT-PCR Analysis
S. davawensis was grown in YS to mid-exponential growth phase for 15 h (no RoF was present in the culture medium) and to the late stationary phase for 24 h (until RoF was present in the culture medium). Total RNA was isolated using a modified method of Chomczynski and Sacchi (17). The mycelium was harvested by centrifugation at 4 °C, immediately frozen in liquid nitrogen, and broken by shearing in a mortar. About 400 mg of the lysate was suspended in 4 ml of buffer A (4 m guanidinium thiocyanate, 25 mm sodium citrate (pH 7), 0.5% N-lauroyl sarcosine, 100 mm β-mercaptoethanol). Subsequently, 400 μl of 2 m sodium acetate (pH 4), 4 ml of phenol (pH 4), and 800 μl of chloroform/isoamyl alcohol (49:1; v/v) were added, and the mixture was incubated on ice for 10 min. After centrifugation (30 min, 10,000 × g, 4 °C) the aqueous phase was mixed with an equal volume of isopropyl alcohol and incubated at −20 °C for 1 h. After centrifugation (20 min, 10,000 × g, 4 °C) the precipitate was washed with 70% ethanol, air-dried, and resuspended in 100 μl of RNase free water. Dissolved RNA was purified with the RNeasy RNA isolation kit (Qiagen, Hilden, Germany), and on-column DNase I digest was performed twice according to the protocol of the manufacturer to eliminate chromosomal DNA contamination. All aqueous solutions were prepared using diethylpyrocarbonate-treated water. RNA was used as the template for RT-PCR to probe for the presence of rosA (and gltA, see below) mRNA. Oligonucleotides rosA401bpfw (5′-TCG TTC GAC TTC GGG CGC TTC A-3′) and rosA401bprv (5′-ACC AGC ACC AGC ATG TCG AGG T-3′) were used for detection of the rosA transcript, and the oligonucleotides gltA408bpfw (5′-ACA CCG CCG CCT ATA AAT CCG C-3′) and gltA408bprv (5′-ACG GGT GGC CGA TGG ACT TCT T-3′) were used to probe for the transcript of the S. davawensis citrate synthase gene gltA (control).
Gene Disruption of rosA
A 1284-bp DNA fragment containing an apramycin resistance cassette and Streptomyces oriT was generated from pIJ773 (18) by XbaI treatment. The latter fragment was blunted by Klenow treatment and ligated to PscI-digested (and subsequently blunted) pUC18 (16) to generate pFJ05. A 509-bp fragment internal to rosA was amplified by PCR using the modifying oligonucleotides rosKnOu01fw (5′-TAT CAA GCT TGA TGG GCG ACC TGT TG-3′) and rosKnOu01rv (5′-TAT GGA TCC CCC AGT CCT CCA GCA C-3′) and was ligated to HindIII/BamHI-digested pFJ05 to give pFJ06. The resulting plasmid was conjugated to S. davawensis using the non-methylating E. coli strain ET12567 and the helper plasmid pUB307 (19). Exconjugants were selected with 50 μg/ml apramycin and 20 μg/ml nalidixic acid on MS-agar. Spores of the rosA disruption mutant and the wild-type strain were used to inoculate 25 ml of TSB-medium. After 48 h of growth, 10 ml of the precultures were used to inoculate a total volume of 100 ml of YS medium (roseoflavin production medium). The cultures were grown for 72 h to the stationary phase, and the production of flavins was monitored using HPLC/MS. The site-specific integration of the plasmid was verified by PCR using oligonucleotides SEQSDknou1fw (5′-GAC CGG GGT GCT GGT GGA CG-3′), SEQSDknou1rv (5′-CGA TGC CGC TCG CCA GTC GA-3′), SEQSDknou2fw (5′-GGG AAT AAG GGC GAC ACG GAA ATG-3′), and SEQSDknou2rv (5′-ATC GCC GCG TCG AAC TCG TCG-3′), and genomic DNA was derived from S. davawensis wild type or the rosA disruption strain. The corresponding PCR products covered the plasmid sequence and rosA at both junctions.
Construction of a Genome Library
Total DNA (108 μg) of S. davawensis was partially digested with MboI to fragments with approximate sizes of 40 kb. The 40-kb fragments were dephosphorylated with FastAP (Fermentas, St. Leon-Rot, Germany) and extracted once using phenol:chloroform:isoamyl alcohol (25:24:1) followed by chloroform extraction and ethanol precipitation. The shuttle cosmid vector pOJ436 (20) was digested with KspAI(HpaI), dephosphorylated, and subsequently digested with BamHI. The resulting two fragments were extracted once using phenol:chloroform:isoamyl alcohol (25:24:1) followed by chloroform extraction and ethanol precipitation and ligated to the prepared genomic fragments. The ligation reaction was used for in vitro packaging with Max-Plax™ Lambda Packaging Extracts. E. coli EPI100™-T1R cells (Epicenter Biotechnologies, Hess, Oldendorf, Germany) were used for subsequent transfection. After growth at 37 °C for 16 h, colony lifts were performed according to a standard protocol (16).
Screening the Genomic Library with a Homologous rosA Probe
An internal fragment of the gene rosA of S. davawensis was amplified by PCR using the oligonucleotides rosAfw (5′-ACT TAC ATA TGC GGC CGG AAC CGA CC-3′) and rosArev (5′-ATA ACT CGA GTC AGC CGG CCG TGC C-3′). The corresponding PCR product was purified by preparative gel electrophoresis and used as a template for random hexanucleotide primed-labeling with digoxigenin-dUTP and Klenow enzyme (High Prime DNA Labeling and Detection Kit II; Roche Diagnostics). Hybridization was performed according to the instructions of the manufacturer.
Cloning and Heterologous Expression of the Putative S. davawensis Roseoflavin Gene Cluster in Streptomyces albus and Streptomyces lividans
Cosmid pFJ03, identified by colony hybridization with a rosA probe, carries a 38-kbp insert including the putative roseoflavin biosynthetic (ros) gene cluster. The isolated cosmid was digested with EcoRV and AvrII, and the resulting 10,321-bp fragment containing the putative ros gene cluster and its probable promoter structure was excised from a preparative agarose gel. Integrative shuttle plasmid pSET152 (20) was digested with EcoRV, treated with shrimp alkaline phosphatase (Fermentas), subsequently digested with XbaI, and ligated to the 10,321-bp fragment. The resulting ligation reaction was used to transform E. coli NEB10β cells (New England Biolabs, Frankfurt, Germany). The corresponding plasmid pFJ04 could be isolated from a transformant strain. The plasmid pFJ04 was used to transform S. lividans (TK24) by PEG-assisted protoplast transformation (15). The plasmid pFJ04 was also used to transform E. coli ET12567(pUB307) (19). The latter strain subsequently was conjugated to S. albus according to standard protocols (15) to ensure plasmid transfer. Transformants and exconjugants were grown to stationary phase and analyzed with respect to RoF synthesis using HPLC. Both, culture supernatants and cell-free extracts were analyzed.
Protein Concentration Determination
Protein concentration was estimated by the method of Bradford (21).
Nucleotide Sequence Accession Numbers
The sequence reported here has been deposited in the GenBankTM data base under the accession number FR750395.
RESULTS
N,N-8-Amino-8-demethyl-d-riboflavin Dimethyltransferase Activity Is Present in a Cell-free Extract from S. davawensis in the RoF Production Phase
Under laboratory conditions using a starch and yeast extract containing culture broth, RoF synthesis in S. davawensis was observed in the stationary phase of growth. It was postulated by Juri et al. (10) that two subsequent methylation reactions convert AF to RoF via MAF (Scheme 1). Indeed, by using a cell-free extract from RoF producing S. davawensis cells, we could measure the SAM-dependent synthesis of both MAF and RoF from AF employing a novel enzyme assay containing SAM and AF. The specific activity for the formation of RoF was 27 microunits mg−1 of total protein. Under these reaction conditions, both products, MAF and RoF, could be detected and isolated by HPLC with subsequent identification by mass spectrometry (Fig. 1). Using a cell-free extract from exponentially growing S. davawensis cells lacking RoF production, formation of MAF and RoF was not observed.
FIGURE 1.
The in vitro formation of MAF and RoF from AF. An assay mixture containing a cell-free extract from RoF-producing S. davawensis cells and 200 μm AF and 2.0 mm SAM was incubated at 37 °C for 20 h. An aliquot was removed, and the compounds were separated on an HPLC column. Peak intensity is given in arbitrary absorbance units (AU). The chromatogram (top) shows the control reaction not containing SAM. The major peak is the substrate AF. The bottom chromatogram shows the formation of roseoflavin and the intermediate of the methylation reaction, MAF. The insets (top, control reaction containing AF; bottom, 20 h sample containing AF, MAF, and RoF) show the assay mixtures, which were analyzed by HPLC.
Purification of N,N-8-Amino-8-demethyl-d-riboflavin Dimethyltransferase (AF Dimethyltransferase) from S. davawensis and Identification of the Corresponding Gene
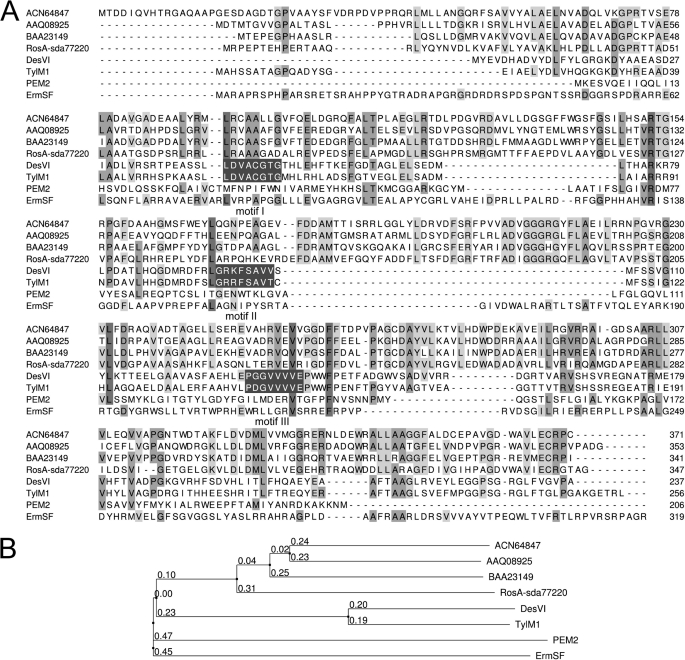
An AF dimethyltransferase activity was enriched from a cell-free S. davawensis extract (stationary phase culture) by a three-step chromatography procedure. The final active fractions still contained at least six different proteins (as judged by Coomassie Brilliant Blue R-250 staining) (Fig. 2A). From one of the major proteins present in one of the final active fractions with an estimated molecular mass of about 40 kDa, a peptide sequence of GVLEDWADADAVR was determined by mass spectrometry. This sequence matched a predicted peptide fragment of a hypothetical protein deduced from a yet uncharacterized open reading frame (sda77220; GenBankTM FR750395) located in the middle of a (hypothetical) ros gene cluster within the S. davawensis genome (Fig. 3). The deduced primary structure of sda77220 was compared with structures of other proteins in the public databases (BLASTp) and was found to be weakly similar to other dimethyltransferases (Fig. 4). In particular, the putative dimethyltransferase did show weak sequence similarity to TylM1 and DesVI, two well characterized N,N-dimethyltransferases from Streptomyces fradiae and Streptomyces venezuelae (23), respectively. The latter enzymes have an aminohexose sugar substrate and are active in the biosynthesis of mycaminose and desosamine, found in several macrolide antibiotics. TylM1 and DesVI each possess all three of the consensus sequence motifs (I-III) (Fig. 4) typical for methyltransferases that use SAM as co-substrate (24). Motif I of TylM1 and DesVI is thought to be a variant of the canonical methyltransferase motif GXGXG, which is found in glycine N-methyltransferases and forms part of the SAM binding pocket (25). Motifs I-III are not present in the deduced primary structure of sda77220. Still, this comparison on a sequential level suggested that ORF sda77220 may encode for a methyltransferase activity necessary to convert AF into RoF. To confirm this, ORF sda77220 was expressed in E. coli using pET-24a(+) (pFJ01). Because the G+C content (70.64%) of our target gene is very high and contains rare codons that may hamper efficient expression, an E. coli strain providing the necessary rare tRNAs was employed. A cell-free extract of a pFJ01-containing strain was tested for AF dimethyltransferase activity and compared with controls harboring the empty expression vector pET-24a(+). A cell-free extract of the strain containing pFJ01 had a specific activity for the formation of RoF of 2.0 milliunits mg−1 of total protein. The control strain did not produce AF dimethyltransferase activity. We concluded from these experiments that ORF sda77220 very likely codes for an enzyme, which is active in the final two steps of the RoF biosynthetic pathway. Thus, ORF sda77220 tentatively was named rosA being the first gene of the RoF pathway to be identified.
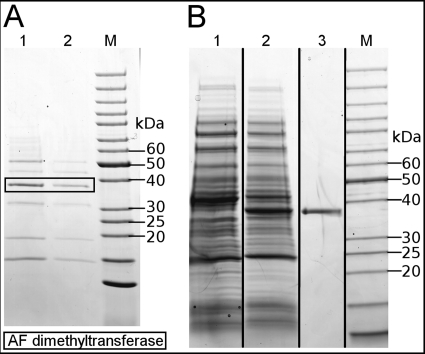
FIGURE 2.
A, enrichment of RosA from a cell-free extract from S. davawensis is shown. SDS-PAGE/Coomassie Brilliant Blue R-250 staining of protein samples collected from the final steps during RosA purification is shown. M, molecular mass marker; lane 1, HiLoad 16/60 Superdex 200 prep grade SEC chromatography, active fraction 1; lane 2, HiLoad 16/60 Superdex 200 prep grade column, active fraction 2. B, overproduction and purification (immobilized metal affinity chromatography) of recombinant S. davawensis RosA (His6-tagged) is shown. SDS-PAGE/Coomassie Brilliant Blue R-250 staining of cell-free extracts of isopropylthiogalactopyranoside-induced E coli strains harboring pET24a(+)rosA-His6 (pFJ02) (lane 2) is shown. Lane 1, control with E. coli Rosetta 2 (DE3) containing the empty plasmid pet24a(+); lane 3, purified His6-tagged RosA. M, molecular weight marker.
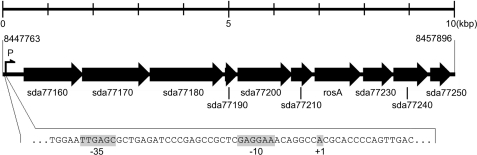
FIGURE 3.
The putative ros gene cluster of S. davawensis containing sda77160, sda77170, sda77180, sda77190, sda77200, sda77210, rosA (sda77220), sda77230, sda77240, and sda77250. The bottom part shows the putative promoter sequence of the gene cluster (−35 and −10 sequences) and the possible transcription start (+1). Possible functions of the putative genes are shown in Table 2.
FIGURE 4.
A, shown is multiple sequence alignment of representative primary structures of N,N- and O-methyltransferases. ACN64847, putative O-methyltransferase, Streptomyces diastatochromogenes; AAQ08925, putative O-methyltransferase, Streptomyces griseus; BAA23149, putative O-methyltransferase, Actinomadura hibisca; RosA-sda77220, N,N-dimethyltransferase from S. davawensis involved in the biosynthesis of roseoflavin; DesVI, N,N-dimethyltransferase from S. venezuelae involved in the biosynthesis of desosamine (23); TylM1, N,N-dimethyltransferase from S. fradiae involved in the biosynthesis of mycaminose (23); PEM2, methyltransferase of the phosphatidylethanolamine methylation pathway in Saccharomyces cerevisiae (28); ErmSF, N,N-dimethyltransferase from S. fradiae introducing two methyl groups into a single base within 23 S rRNA conferring resistance to macrolide and lincosamide antibiotics (29). The first and the last residue numbers of the sequences in the alignment are indicated, and the total length of the protein is shown at the end. TylM1 and DesVI each possess all three of the consensus sequence motifs I-III (white letters, dark gray background) typical for methyltransferases that use SAM as co-substrate (see “Results”). Conserved residues are highlighted in gray. B, shown are evolutionary distances of representative N,N- and O-methyltransferases.
Overproduction, Purification, and Characterization of Recombinant S. davawensis RosA from E. coli
Both the native form (using pFJ01) and a C-terminal His6-tagged version of RosA (using pFJ02) were produced in E. coli. Cell-free extracts of the corresponding recombinant E. coli strains (both produced similar amounts of RosA as judged by SDS-PAGE) were compared with respect to their specific RosA activities (2.0 milliunits mg−1 of total protein using pFJ01; 1.9 milliunits mg−1 of total protein using pFJ02). Thus, no significant difference in specific activity was detected for the two enzyme versions, indicating that the C-terminal His6 tag did not negatively affect the activity of RosA. His6-tagged RosA was purified by immobilized metal affinity chromatography to apparent homogeneity using a standard protocol (Fig. 2B). His6-tagged RosA was employed for the subsequent biochemical characterization of this novel enzyme. The identity of recombinant RosA was confirmed by N-terminal peptide sequencing and de novo peptide sequencing using MS/MS. The apparent subunit molecular mass of about 40 kDa, as revealed by SDS-PAGE (Fig. 2B, lane 3), correlated well with the predicted value of 37,935 Da based on the translated peptide sequence. Gel filtration with molecular weight standards suggested that His6-tagged RosA exists as a monomer. The apparent pH optimum of RosA was 8.8, and the apparent temperature optimum of RosA was 52 °C. N,N-8-Amino-8-demethyl-d-riboflavin dimethyltransferase from S. davawensis is a colorless enzyme with no additional absorbance between 400 nm and the protein peak at 280 nm.
Kinetic Characterization of His6-tagged RosA
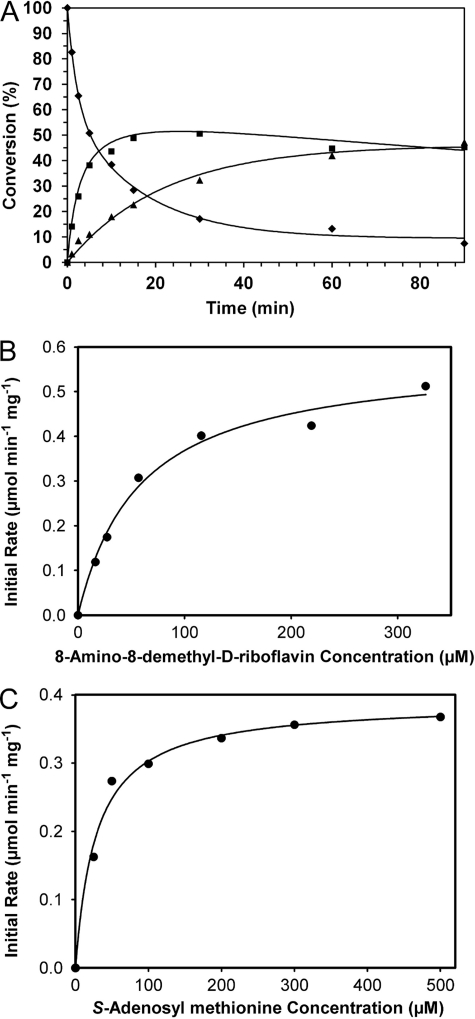
The purified His6-tagged RosA protein of S. davawensis was kinetically characterized. First of all, the time course of RosA-catalyzed mono- and dimethylation reactions of AF were studied to determine the rate constants of each step. The progress of the reaction was monitored over a period of 90 min by taking aliquots from the incubation at appropriate time intervals followed by HPLC analysis. The percent conversion of substrate to monomethylated intermediate and dimethylated product was estimated on the basis of the integration of the corresponding HPLC peaks. The incubation was conducted under saturating concentrations of SAM. Monotonic consumption of the substrate AF and the concomitant formation of the dimethylation product RoF were observed (Fig. 5A). The concentration of the monomethylated intermediate increased to 50% of overall conversion when the reaction was initiated, and the reaction rate of the first methylation step decreased slightly, whereas the reaction rate of the second methylation step increased slowly. When the data were fit to the rate law of irreversible unimolecular consecutive reactions, rate constants of 0.07 min−1 and 0.02 min−1 for the respective mono- and dimethylation steps of the RosA reaction were deduced.
FIGURE 5.
Time course of RosA-catalyzed N-methyl transfer reactions (A) and determination of steady-state kinetic parameters (B and C) for the first methylation of A) catalyzed by RosA. A, ♦, AF; ■, MAF; ▴, roseoflavin. B, rates of individual reactions were obtained by incubating varying amounts of AF (16–326 μm) with 2.3 μm RosA and 2.0 mm SAM. The initial rates were plotted against the AF concentrations. To determine the kinetic constants Km and Vmax, the data were fit to the Michaelis-Menten equation using SigmaPlot. C, as above, rates of individual reactions were obtained by incubating varying amounts of SAM (25–500 μm) with 2.3 μm RosA and 200 μm AF. The initial rates were plotted against the SAM concentrations. To determine the kinetic constants Km and Vmax, the data were fit to the Michaelis-Menten equation using SigmaPlot.
The kinetic parameters for the monomethylation step catalyzed by RosA were determined by following the rate of substrate (AF) consumption. The values of Km and Vmax were calculated from the best fit to the Michaelis-Menten equation using nonlinear regression in the SigmaPlot software. In these experiments the concentration of one substrate (AF or SAM) was kept fixed, whereas that of the other varied. When the concentration of SAM was maintained at 2.0 mm (saturation), a Km of 57.7 ± 9.2 μm and a kcat of 22.0 ± 1.1 min−1 (Fig. 5B) (Vmax of 0.58 ± 0.03 μmol min−1 mg protein−1) were determined for AF. Similarly, when the concentration of AF was maintained at 200 μm (saturation), the Km for SAM was determined to be 28.6 ± 4.2 μm, and the kcat (assuming a RosA monomer of 38 kDa) was estimated to 14.8 ± 0.5 min−1 (Fig. 5C). MAF was not available to our laboratory, and therefore, the kinetic parameters for the second half of methylation by following the rate of formation of the dimethylated product RoF could not be performed. Riboflavin and 8-hydroxy-8-demethyl-d-riboflavin (in this compound the 8-amino group of AF is replaced by a hydroxyl group) were available and tested as substrates. Using the latter substrates, no activity was found.
Ligand Binding Titrations
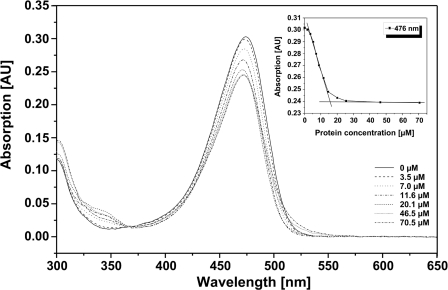
His6-tagged RosA was purified from a recombinant E. coli strain and used for the subsequent titration experiment. A constant amount of the substrate AF was incubated with varying amounts of RosA (Fig. 6). A dissociation constant (KD) for AF of 10 μm was estimated by determining the concentration of free and bound AF by means of membrane centrifugation in Microcon filter devices (Millipore).
FIGURE 6.
Titration of RosA with AF. Different amounts of RosA (0–70.5 μm) were incubated with AF (200 μm), and the absorbance spectra were recorded between 300 and 650 nm. The arrow indicates the absorbance maximum (476 nm). The inset shows the change in absorbance at 476 nm due to the formation of the RosA-AF complex as a function of the concentration of protein. AU, absorbance units.
Transcriptional Analysis of rosA in S. davawensis
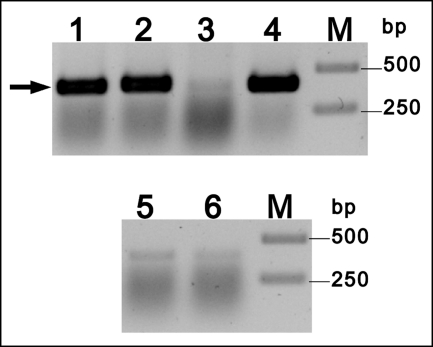
Total RNA was prepared from two different S. davawensis cultures in the standard yeast extract/starch medium. One culture was grown to the stationary phase and produced RoF. The other culture was harvested in the exponential growth phase where no RoF was present in the cultures. Total RNA from both cultures was analyzed by reverse transcription PCR using specific rosA primers. A signal was detected in total RNA derived from RoF-producing cells only (Fig. 7, lane 1). This finding suggests that rosA is an active gene and that it is associated with RoF biosynthesis. Sequence analysis upstream of ORF sda77160 revealed a single sequence similar to the described Streptomyces (vegetative) consensus promotor TTGACR (−35) and TASRRT (−10) (Fig. 3) (26).
FIGURE 7.
Change in expression of the gene rosA in the time course of growth of a S. davawensis culture. The level of rosA-transcript was assessed by RT-PCR. Lane 1, the template RNA was isolated during the RoF production phase, and rosA specific primers were used. A band corresponding to a fragment of the rosA gene was detected (arrow). Lane 2, the template RNA was isolated during the RoF production phase, and gltA-specific primers were used (positive control, gltA is a constitutively expressed gene coding for citrate synthase). Lane 3, the template RNA was isolated during exponential growth phase (no RoF synthesis) and rosA-specific primers were used. Lane 4, the template RNA was isolated during exponential growth phase, and gltA-specific primer were used. Lanes 5 and 6 show the RT-PCR control reactions not containing reverse transcriptase and rosA specific primers. The template RNA was isolated during exponential growth phase and roseoflavin production phase, respectively. Lanes M, 1-kb markers.
Inactivation of ORF sda77220 Leads to a S. davawensis Strain That Synthesizes AF but Not MAF or RoF
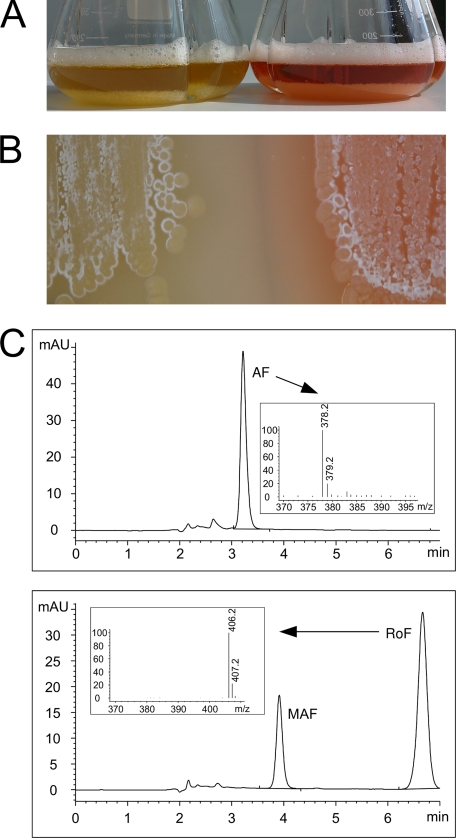
The gene rosA (ORF sda77220) was inactivated by insertion of a cassette conferring resistance to the antibiotic apramycin. The corresponding rosA-deficient strain was grown to the stationary phase in standard YS-broth. The culture supernatant and a cell-free extract were analyzed with respect to flavin synthesis by HPLC/MS. Neither MAF nor RoF was detected (Fig. 8). Instead, accumulation of AF was observed (Fig. 8) showing that inactivation of rosA did not affect synthesis of the remaining enzymes of the pathway. Site-specific integration of the cassette was checked by PCR. BLASTn analysis did not reveal any other likely integration site in the S. davawensis genome other than rosA.
FIGURE 8.
A, wild-type S. davawensis (flask to the right) and a rosA-deficient S. davawensis strain (flask to the left) were grown to the stationary phase (72 h) in a starch/yeast extract containing broth. The wild-type strain (right) produced red RoF. The rosA-deficient strain (left) apparently did not produce a red compound. Instead, the culture appeared yellow (due to the synthesis of AF). B, both strains (wild-type S. davawensis, right; rosA-deficient S. davawensis, left) were grown on a solid growth medium and a similar phenotype (see A) was observed. C, culture supernatants of both strains (see A) were analyzed by HPLC/MS with respect to their flavin content. For the rosA-deficient S. davawensis strain (upper panel), neither roseoflavin (red) nor MAF was detected. Instead, the latter strain was found to synthesize the yellow roseoflavin precursor AF (apparent molecular mass 378.2 (+1)). The wild-type strain (lower panel) synthesized RoF (apparent molecular mass 406.2 (+1)) and MAF (apparent molecular mass 392.2 (+1); MS data are not shown in the inset). mAU, milliabsorbance units.
The rosA Gene Cluster from S. davawensis Does Not Support RoF Production in S. albus and S. lividans
To study the function of the putative RoF gene cluster in S. davawensis expression, experiments employing the closely related species S. albus and S. lividans, which are both unable to produce RoF, were carried out. To obtain a subgenomic fragment containing S. davawensis ORFs sda77160, sda77170, sda77180, sda77190, sda77200, sda77210, rosA, sda77230, sda77240, and sda77250, a cosmid library was prepared using the integrative Streptomyces vector pOJ436. About 4000 colonies of the library were screened by colony lifts using a digoxigenin-labeled rosA probe. One of the lysed colonies produced a strong hybridization signal. The cosmid from the corresponding strain (pFJ03) was isolated and characterized using restriction endonucleases. The restriction pattern of the subgenomic DNA insert perfectly matched the expected restriction pattern, suggesting that an intact subgenomic fragment has been cloned. DNA sequencing revealed that the cosmid pFJ03 contained an insert of 38,277 bp. A DNA fragment containing the putative ros gene cluster (sda77160, sda77170, sda77180, sda77190, sda77200, sda77210, rosA, sda77230, sda77240, and sda77250) and the natural putative promotor (see Fig. 3) was excised from plasmid FJ03 using restriction endonucleases and ligated to integrative Streptomyces plasmid pSET152 carrying no additional promoter sequence in front of the multiple cloning site (FJ04) (27). Expression experiments produced negative results, i.e. the corresponding recombinant S. albus and S. lividans strains (transformed with FJ04) were unable to synthesize RoF. In the latter strains, RosA activity (0.5 microunits mg protein−1) was detected showing that expression of the heterologous gene cluster indeed occurred. These results suggest that the gene cluster sda77160, sda77170, sda77180, sda77190, sda77200, sda77210, rosA, sda77230, sda77240, and sda77250 does not contain all genes necessary for RoF biosynthesis. The functions of the remaining ORFs are unclear; analysis of their primary structures revealed best hits, which are shown in Table 2.
TABLE 2.
Hypothetical gene products within the ros gene cluster were analyzed using BLASTp (22)
aa, amino acids.
| ORF | Amino acids (no.) | Similarity |
|---|---|---|
| sda77160 | 435 | 35% identity (aa 134–379) to “radical SAM domain-containing protein from Methanosaeta thermophila PT” (GenBankTM accession no. YP_842640) |
| sda77170 | 508 | 26% identity (aa 115–441) to “radical SAM domain protein from Desulfonatronospira thiodismutans ASO3–1” (GenBankTM accession no. EFI34877) |
| sda77180 | 553 | 36% identity (aa 204–560) to “AMP-dependent synthetase and ligase from Rhodoferax ferrireducens T118” (GenBankTM accession no. YP_523400) |
| sda77190 | 91 | 29% identity (aa 21–72) to “acyl carrier protein from Acholeplasma laidlawii PG-8A” (GenBankTM accession no. ABX80867) |
| sda77200 | 406 | 37% identity (aa 157–406) to β-ketoacyl-acyl carrier protein synthase II from Thermodesulfovibrio yellowstonii DSM 11347” (GenBankTM accession no. ACI20203) |
| sda77210 | 167 | 34% identity (aa 41–119) to “thiolesterase superfamily protein Rhodopseudomonas palustris DX-1” (GenBankTM accession no. EFC27112) |
| sda77220 | 347 | RosA |
| sda77230 | 222 | 30% identity (aa 62–202) to “fructose-2,6-bisphosphatase from Pelotomaculum thermopropionicum SI” (accession no. BAF59496) |
| sda77240 | 261 | 53% identity (aa 135–253) to “phytanoyl-CoA dioxygenase from Streptomyces bingchenggensis BCW-1” (GenBankTM accession no. ADI09555) |
| sda77250 | 157 | 76% identity (aa 115–151) to “hypothetical protein from Streptomyces scabiei” (GenBankTM accession no. YP_003494084) |
DISCUSSION
It was suggested earlier by Juri et al. (10) that AF was an intermediate in the biosynthesis of the riboflavin analog RoF in S. davawensis. The results reported here now show that this indeed is the case. We were able to partially purify a novel N,N-8-amino-8-demethyl-d-riboflavin dimethyltransferase from S. davawensis, which converts AF in two steps to RoF. Both methylation reactions depend on the methyl group donor SAM. The corresponding gene identified in the genome of S. davawensis was named rosA and was found to be located in a cluster comprising a total of 10 genes. The inactivation of rosA led to a MAF/RoF-deficient strain accumulating AF, unequivocally showing that rosA is responsible for the terminal two steps in RoF biosynthesis. The primary structure of RosA is similar (up to 35% at the amino acid level) to several SAM-dependent N-methyl- and O-methyl-transferases (Fig. 4). RosA apparently is a new member of a small family of enzymes that is capable of catalyzing a N,N-dimethylation reaction. RosA shares low similarities to several characterized N,N-dimethyltransferases, such as PEM2 (28), ErmSF (29), TylM1, and DesVI (23).
RosA activity and rosA transcripts were only detectable in the RoF production phase. AF apparently is a good substrate for the enzyme (Km = 57.7 ± 9.2 μm; KD = 10.0 μm) and thus most likely also is the natural substrate in S. davawensis. It was reported earlier (23) that reactions catalyzed by dimethyltransferases are generally slow, with kcat values in the range of 1 s−1. The kcat = 0.37 ± 0.02 s−1 of RosA is within this range.
The functions of the other putative genes (Table 2) of the ros gene cluster (Fig. 3) are unclear. No obvious candidate genes responsible for reactions of the roseoflavin biosynthetic pathway are present in the cluster.
Heterologous expression of the putative roseoflavin gene cluster in S. albus and S. lividans did not lead to RoF production, although RosA activity was observed (0.5 microunits mg protein−1), which indicates that heterologous gene expression was successful. However, the latter activity is relatively weak as compared with the RosA activity in S. davawensis (27 microunits mg protein−1) and shows that the expression level of the cluster in the heterologous hosts was low. Possibly, this low expression level was not enough to support RoF synthesis. Our conclusion, however, is that not all the genes required for RoF biosynthesis are located within the ros gene cluster. In many cases the genes involved in the synthesis of secondary metabolites were found to be clustered. Known exceptions to this are, e.g. the genes involved in the synthesis of moenomycin (31), ansamitocin (32), and pristinamycin (33).
S. davawensis (in contrast to S. lividans and S. albus) is RoF-resistant. S. lividans and S. albus, both, were transformed with the putative ros gene cluster (using the plasmids described above); however, the resulting recombinant strains were not RoF-resistant. This indicates that the gene(s) for RoF resistance is not present within this first ros gene cluster.
In summary, we have reported the identification of a novel biosynthetic gene cluster and the identification and characterization of the first enzyme of the RoF biosynthetic pathway in S. davawensis. Our discoveries also have practical implications for research in the field of streptomycetes. For example, the probably unique enzyme RosA could be used to determine promoter activity in bacteria of the genus Streptomyces, as other reporter systems, e.g. lacZ-based assays, did not perform well in these bacteria. Because both RoF and especially AF have antibacterial (9, 30) and also antimalarial activity, the identification of the corresponding pathway is also important with respect to the construction of a AF/RoF-overproducing strain and the possibility to generate chemically altered compounds with improved properties. Furthermore, it is now possible to synthesize 13C or 14C-labeled MAF and RoF. The latter antibiotic flavin derivatives may be important in identifying flavoenzymes inhibited by these compounds.
This work was supported by the Anneliese Konanz-Stiftung and the German Federal Ministry of Education and Research.
This work is dedicated to Wolfgang Buckel on the occasion of his 70th birthday.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) FR750395.
- RoF
- roseoflavin
- AF
- 8-amino-8-demethyl-d-riboflavin
- MAF
- 8-methylamino-8-demethyl-d-riboflavin; SAM, S-adenosyl methionine.
REFERENCES
- 1. Otani S., Takatsu M., Nakano M., Kasai S., Miura R. (1974) J. Antibiot. (Tokyo) 27, 88–89 [PubMed] [Google Scholar]
- 2. Otani S., Kasai S., Matsui K. (1980) Methods Enzymol. 66, 235–241 [DOI] [PubMed] [Google Scholar]
- 3. Grill S., Yamaguchi H., Wagner H., Zwahlen L., Kusch U., Mack M. (2007) Arch. Microbiol. 188, 377–387 [DOI] [PubMed] [Google Scholar]
- 4. Vogl C., Grill S., Schilling O., Stülke J., Mack M., Stolz J. (2007) J. Bacteriol. 189, 7367–7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abbas C. A., Sibirny A. A. (2011) Microbiol. Mol. Biol. Rev. 75, 321–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grill S., Busenbender S., Pfeiffer M., Köhler U., Mack M. (2008) J. Bacteriol. 190, 1546–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee E. R., Blount K. F., Breaker R. R. (2009) RNA Biol. 6, 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ott E., Stolz J., Lehmann M., Mack M. (2009) RNA Biol. 6, 276–280 [DOI] [PubMed] [Google Scholar]
- 9. Mansjo M., Johansson J. (2011) RNA Biol. 8, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Juri N., Kubo Y., Kasai S., Otani S., Kusunose M., Matsui K. (1987) J. Biochem. (Tokyo) 101, 705–711 [DOI] [PubMed] [Google Scholar]
- 11. Matsui K., Juri N., Kubo Y., Kasai S. (1979) J. Biochem. (Tokyo) 86, 167–175 [PubMed] [Google Scholar]
- 12. Macheroux P., Kappes B., Ealick S. E. (2011) FEBS J. 278, 2625–2634 [DOI] [PubMed] [Google Scholar]
- 13. Thompson C. C., Vicente A. C., Souza R. C., Vasconcelos A. T., Vesth T., Alves N., Jr., Ussery D. W., Iida T., Thompson F. L. (2009) BMC Evol. Biol. 9, 258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walsh C., Fisher J., Spencer R., Graham D. W., Ashton W. T., Brown J. E., Brown R. D., Rogers E. F. (1978) Biochemistry 17, 1942–1951 [DOI] [PubMed] [Google Scholar]
- 15. Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000) Practical Streptomyces Genetics, pp. 168–169, The John Innes Foundation, Norwich, UK [Google Scholar]
- 16. Sambrook J., Fritsch E., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 17. Chomczynski P., Sacchi N. (1987) Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 18. Gust B., Challis G. L., Fowler K., Kieser T., Chater K. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flett F., Mersinias V., Smith C. P. (1997) FEMS Microbiol. Lett. 155, 223–229 [DOI] [PubMed] [Google Scholar]
- 20. Bierman M., Logan R., O'Brien K., Seno E. T., Rao R. N., Schoner B. E. (1992) Gene 116, 43–49 [DOI] [PubMed] [Google Scholar]
- 21. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 22. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen H., Yamase H., Murakami K., Chang C. W., Zhao L., Zhao Z., Liu H. W. (2002) Biochemistry 41, 9165–9183 [DOI] [PubMed] [Google Scholar]
- 24. Kagan R. M., Clarke S. (1994) Arch. Biochem. Biophys. 310, 417–427 [DOI] [PubMed] [Google Scholar]
- 25. Cheng X., Kumar S., Posfai J., Pflugrath J. W., Roberts R. J. (1993) Cell 74, 299–307 [DOI] [PubMed] [Google Scholar]
- 26. Strohl W. R. (1992) Nucleic Acids Res. 20, 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blaesing F., Mühlenweg A., Vierling S., Ziegelin G., Pelzer S., Lanka E. (2005) J. Biotechnol. 120, 146–161 [DOI] [PubMed] [Google Scholar]
- 28. Kodaki T., Yamashita S. (1989) Eur. J. Biochem. 185, 243–251 [DOI] [PubMed] [Google Scholar]
- 29. Zalacain M., Cundliffe E. (1989) J. Bacteriol. 171, 4254–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Otani S., Matsui K., Kasai S. (1997) Osaka City Med. J. 43, 107–137 [PubMed] [Google Scholar]
- 31. Ostash B., Doud E. H., Lin C., Ostash I., Perlstein D. L., Fuse S., Wolpert M., Kahne D., Walker S. (2009) Biochemistry 48, 8830–8841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu T. W., Bai L., Clade D., Hoffmann D., Toelzer S., Trinh K. Q., Xu J., Moss S. J., Leistner E., Floss H. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7968–7973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mast Y. J., Wohlleben W., Schinko E. (2011) J. Biotechnol. 155, 63–67 [DOI] [PubMed] [Google Scholar]