Background: Whether RPL11 directly binds to the zinc finger domain of MDM2 still remains elusive.
Results: Mutations of RPL11 or the zinc finger of MDM2 impair the ability of RPL11 to inactivate MDM2 and to activate p53.
Conclusion: RPL11 binds directly the zinc finger of MDM2 via hydrophilic residues.
Significance: Our study unveils the chemical nature for MDM2-RPL11 interactions.
Keywords: Oncogene, p53, Protein-Protein Interactions, Tumor Suppressor Gene, Zinc Finger, H-bonding, HDX-Mass Spectrometry, MDM2, RPL11, Electronic Interactions
Abstract
Ribosomal protein L11 (RPL11) has been shown to activate p53 by binding to MDM2 and negating its p53 suppression activity in response to ribosomal stress. Although a mutation at Cys-305 within the zinc finger domain of MDM2 has been shown to drastically impair MDM2 interaction with RPL11 and thus escapes the inhibition by this ribosomal protein, it still remains elusive whether RPL11 inactivates MDM2 via direct action on this zinc finger domain and what is the chemical nature of this specific interaction. To define the roles of the MDM2 zinc finger in association with RPL11, we conducted hydrogen-deuterium exchange mass spectrometry, computational modeling, circular dichroism, and mutational analyses of the zinc finger domain of MDM2 and human RPL11. Our study reveals that RPL11 forms a stable complex with MDM2 in vitro through direct contact with its zinc finger. This binding is disrupted by single mutations of non-cysteine amino acids within the zinc finger domain of MDM2. Basic residues in RPL11 are crucial for the stable binding and RPL11 suppression of MDM2 activity toward p53. These results provide the first line of evidence for the specific interaction between RPL11 and the zinc finger of MDM2 via hydrophilic residues as well as a molecular foundation for better understanding RPL11 inhibition of MDM2 function.
Introduction
The p53 tumor suppressor protein plays an essential role in protecting all vertebrate animals from undergoing neoplasia and tumorigenesis by inducing cell growth arrest, senescence, DNA repair, autophagy, and apoptosis (1, 2). Also, p53 can play a physiological role in maintaining homeostasis, such as by regulating metabolism (3). Most of these functions are executed via the transcriptional activity of p53 (2), although transcription-independent activity is also described (4). Because of the high toxicity of active p53 to cells, the stability and activity of this nuclear transcriptional factor are tightly monitored via a negative feedback regulation by MDM2 and MDMX (5, 6). MDM2 and MDMX are homologs with highly conserved N-terminal p53-binding domain and C-terminal Ring domain, but the central domains are less conserved between these two proteins (7). They form a complex and function as partners to bind to p53, inhibit its activity, and also mediate its ubiquitination and degradation via E3 ubiquitin ligase activity of MDM2 (8–11). Because of this tight regulation, p53 has a short half-life of ∼30 min and is rarely active in unstressed cells. However, this regulation can be turned off under a stress condition, leading to p53 activation. Distinct stress signals can switch on different cellular mechanisms to activate p53 via post-translational modifications, such as phosphorylation or acetylation of and protein-protein interactions with either p53 itself or MDM2 and MDMX (1, 11, 12).
One of the recently acknowledged stress-signaling pathways is the so-called “ribosomal or nucleolar stress-p53 pathway” (12). Although previously less appreciated, this signaling pathway has been repeatedly verified because RPL11, RPL5, and RPL23 were found to activate p53 by directly binding to MDM2, but not MDMX, and suppressing its activity in response to ribosomal stress (RS)3 caused by actinomycin D and serum starvation (13–17). Also, more RPs, such as RPS7 (18, 19), RPL26 (20–22), RPS3 (23), and other nucleolar proteins (24–26), have been shown to play a similar role in regulating the MDM2-p53 loop. Furthermore, more reagents, such as 5-fluorouracil (27, 28), mycophenolic acid (nucleotide-depleting agent) (29), or glucose depletion (30), and genetic or siRNA-mediated silencing of several genes involving ribosomal biogenesis (31–43) have been shown to cause RS, leading to p53 activation. Remarkably, p53 in RPL11/RPL5-binding defective MDM2 C305F knock-in mice failed to respond to 5-fluorouracil or actinomycin D but still responded to DNA damage signals, demonstrating the physiological role of the RS-p53 pathway (44). It is now clear that the nucleolus is very sensitive to many types of stress, particularly metabolic stress, which activates p53 (45), because ribosomal biogenesis in this dynamic organelle consumes most of the intracellular energy (ATP) (46). Also, the RS-p53 pathway has been recently linked to human genetic disorders characteristic with bone marrow defects, such as the two types of myelodysplastic syndromes (47), 5q syndrome (33), and Diamond Blackfan anemia (48). In these diseases, haploinsufficiency caused by mutations of either RPS14 (49) or other ribosomal protein-encoding genes, such as RPS19 (50) or RPL5 (51), induces ribosomal stress and consequent p53 activation and p53-dependent apoptosis of developing blood cells in bone marrows, thus causing anemia of several blood types. Again, the RPL11-MDM2 binding was shown to account for p53 activation in these myelodysplastic syndromes (47). Therefore, the RS-p53 pathway plays a key role in both physiological and pathological circumstances.
Although much has been learned about the importance of the aforementioned ribosomal proteins in regulating the MDM2-p53 pathway, the molecular insight into the mechanisms underlying their negation of MDM2 activity remains largely obscure. To address this question, we began by focusing on dissecting the interaction between MDM2 and RPL11. Previous studies using a strategy of gross deletion mutation suggested that RPL11, but not RPL5 or RPL23, appears to inhibit MDM2 activity by binding to the zinc finger domain of MDM2 (13, 16, 17, 52). Consistently, a cancer-derived C305S mutation impaired the interaction between MDM2 and RPL11 but not RPL5 or RPL23 (28), suggesting that only RPL11, but not RPL5 or RPL23, may bind to the zinc finger domain. However, another cancer-derived mutant C305F impaired the binding of MDM2 by both RPL11 and RPL5, but not RPL23, in cells (52) and in animals (44). These seemingly conflicting results suggest two possibilities. 1) the zinc finger is a crucial structural component for maintaining the wild type conformation of MDM2. 2) The zinc finger is where RPL11, but not RPL5 or RPL23, physically binds. To address these possibilities, we carried out a set of biochemical, proteomic, computational modeling, mutagenic, and cell-based analyses. Although our results verified that the cancer-related zinc finger cysteine mutations disrupt the binding of MDM2 with RPL11, RPL5, and RPL23, we demonstrate the direct interaction of only RPL11, but not RPL5 or RPL23, with the zinc finger domain of MDM2. Furthermore, we uncover several non-cysteine amino acids within the zinc finger and basic amino acids in MDM2-binding domain of RPL11, which are important for the specific interactions between the two proteins. Mutations of these residues without altering the intact zinc finger motif impair not only their interaction in vitro and in cells but also the ability of RPL11 to inactivate MDM2 and to induce p53 in cells.
EXPERIMENTAL PROCEDURES
Expression Constructs and Mutagenesis
Human MDM2 (amino acids 210–437) cDNAs were amplified by PCR and cloned into the pET-24a vector (Invitrogen). The full-length RPL11 expression plasmids (pPROEX-L11) were provided by Yanping Zhang (University of North Carolina, Chapel Hill). The L11 was cloned into pET-24a from pPOREX-L11 and then transferred to pGEX-4T-1(GE Healthcare) for GST-L11 fusion protein expression. Mutations were induced with Phusion high fidelity DNA polymerase as described in the manufacturer's manual (New England Biolabs) and verified by DNA sequencing.
Preparation of MDM2-RPL11 Complexes
The pET24a plasmid coding for His-tagged human MDM2 (amino acids 210–437) with kanamycin resistance and the pPOREX plasmid coding for His-tagged human RPL11 with ampicillin resistance were co-transformed in BL21 (DE3) cells. Then the two proteins were co-expressed by induction with 0.2 mm isopropyl β-d-1-thiogalactopyranoside at an optical density of 0.6 and further incubation at 16 °C for 20 h. 2L cells were harvested, ground in liquid nitrogen, and lysed in the binding buffer containing 20 mm sodium phosphate, pH 7.5, 20 mm Tris, 300 mm NaCl, 30 mm imidazole, 10% glycerol, 10 mm β-mercaptoethanol, 1 mm PMSF, 1 mm benzamidine, 200 μm pepstatin, and 60 μm leupeptin. Lysates were cleared by centrifugation (30 min, 10,000 × g). Supernatants were incubated with 5 ml of nickel-nitrilotriacetic acid-agarose (Thermo Scientific Pierce) for 4 h at 4 °C. Bound proteins were washed with 5 column volumes of 20 mm Tris, 300 mm NaCl, 30 mm imidazole, 10 mm β-mercaptoethanol and 1 column volume of the same buffer containing 1 m NaCl, and then eluted with the same buffer containing 200 mm imidazole. The eluted proteins were desalted by a HiTrap desalting column (GE Healthcare), then bound to an anion exchange column (Hitrap Q HP, GE Healthcare), and eluted with a gradient buffer containing NaCl from 200 mm to 1 m. Free RPL11 proteins were in the flow-through. MDM2 co-eluted with RPL11 at 300–500 mm NaCl. The final step of the purification over a Superdex 75 10/30 size exclusion column equilibrated with 10 mm Tris, 150 mm NaCl, and 5 mm DTT resulted in 5 mg of pure MDM2-L11 complexes from 5.0 g of cell lysates.
HDX/MS Experiments
5 mg/ml MDM2 and 2 mg/ml MDM2-L11 complex in 40 mm Tris, 150 mm NaCl, pH 8, were preincubated in an ice bath (0 °C) for 30 min. Exchange was initiated by 20-fold dilution of protein samples into D2O buffer (2 mm ammonium acetate at pH 6.8 without pH adjustment for D2O), after 30 s of exchange, the reactions were quenched with equal volumes of ice-cold 1 m citric acid buffer (1 m citric acid, 100% w/v guanidinium hydrochloride, pH 2.2). The quenched samples were allowed to digest with equal molar pepsin in solution, pH 2.2, for 5 min, followed by immediate LC-electrospray ionization MS analysis.
HDX LC-MS System and Peptide Identification
A Shimadzu HPLC, with two LC-10AD pumps, was used to generate a fast gradient with 30 μl/min flow rate, optimized for best sequence coverage. Solvent A was 5% acetonitrile in H2O, 0.1% formic acid, and solvent B consisted of 95% acetonitrile in H2O, 0.1% FA. All components of the setup, including tubing, injector, and column, were submerged in an ice bath at all times to reduce back exchange. For the analysis of proteolytic peptides, 20 μl of chilled digest was injected into a 1.0-mm inner diameter × 50-mm C8 column (Waters Inc.). After desalting for 5 min with 5% solvent B, the peptides were eluted at 30 μl/min with a 5–15% gradient for 0.01 min, 15–30% for 9 min, 30–50% for 1 min, and 50–95% for 1 min. The effluent was infused into a 12-T Varian IonSpec FT-ICR MS (Varian Inc.). For peptide identification, 30-s fractions were collected into a 96-well plate by coupling the HPLC with TriVersa NanoMate (Advion Inc.). Each fraction was run with an internal standard, and the mass spectra were collected by coupling chip-based infusion of the TriVersa NanoMate with the FT-ICR MS. Peptides were identified by a combination of accurate masses and MS/MS. The extent of deuterium incorporation of each peptic peptide was determined by FT-ICR MS from the centroid mass difference between deuterated and nondeuterated samples.
HDX/MS Data Analysis and Presentation
The MS distribution for each peptide was fitted to a Gaussian curve, and the centroid value (xc) was determined using OriginPro8. Changes in deuterium incorporation (ΔHDX) were defined as the difference between the xc values of the complex and apoprotein.
GST Pulldown Assays
GST fusion proteins were purified through glutathione-agarose beads (Thermo Scientific Pierce) according to the manufacturer's instructions. The integrity of purified proteins was confirmed by Coomassie Blue staining and immunoblotting after SDS-PAGE. Glutathione beads conjugated to GST fusion proteins were incubated with His-tagged MDM2 by gently agitating at 4 °C for 2 h in binding buffer (1× PBS, 0.1% Nonidet P-40, and 5% glycerol) and washed four times with wash buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40) and once with the wash buffer without NaCl. The bound protein mixture was resolved by SDS-PAGE and analyzed by immunoblotting and Coomassie staining. The curve fitting and the Kd values of L11 to MDM2 were calculated by the Hill equation using Igor 4.01 (Lake Oswego, OR).
Mammalian Expression Constructs and Plasmid Transfection
N-terminal HA-tagged human MDM2 and FLAG-tagged human L11 expression plasmids have been described previously (53). Mutations were induced with Phusion high fidelity DNA polymerase as described in the manufacturer's manual (New England Biolabs) and verified by DNA sequencing. Plasmids were transiently transfected in H1299 cells via Lipofectin following the manufacturer's protocol (Invitrogen).
Cell Culture, Immunoprecipitation, and Immunoblotting
H1299 cells were seeded, cultured in DMEM supplemented with 10% serum, and transfected with an empty vector or the FLAG-tagged L11 plasmids with HA-tagged MDM2 as indicated and lysed 2 days after transfection. Lysates were centrifuged at 12,000 × g for 10 min. Supernatants were incubated with anti-FLAG M2-agarose (Sigma) for 4 h and washed once with PBS and three times with wash buffer (50 mm Tris-HCl, pH 8.0, 400 mm NaCl, 1% Nonidet P-40). Immunoprecipitates were boiled for 5 min with SDS-sample buffer separated by SDS-PAGE. Proteins were transferred to PVDF membranes by semi-dry blotting. The blots were developed by an enhanced chemiluminescence detection kit (Thermo Scientific), and signals were visualized by Omega 12iC Molecular Image System (UltraLUM). Antibodies used for immunoblotting were mouse monoclonal anti-p53 (DO-1, Santa Cruz Biotechnology) and rabbit polyclonal anti-p21 (M19, Santa Cruz Biotechnology). Anti-FLAG (Sigma) 2A10 monoclonal anti-MDM2 antibodies were described previously (54), and anti-HA (12CA5) has been described (53). The percentage of p53 levels in Fig. 5, C and E, were quantified and expressed as the mean number ± S.D. (n = 3). For each sample, the level (band intensity) of p53 was quantified and normalized against the loading control actin. The percentage of p53 levels for each sample was calculated as the mean value of the average sample value minus negative controls divided by the mean value of positive controls minus the mean value of negative controls, multiplied by 100. Positive controls (100% p53 level) contained p53 only (lane 2), and negative controls (0% p53 level) contained co-expression of p53 and MDM2 protein (lane 3 for WT MDM2 in Fig. 5, C and E, lane 6 for E292A, lane 9 for D294A, and lane 12 for T305A/S306A in Fig. 5C).
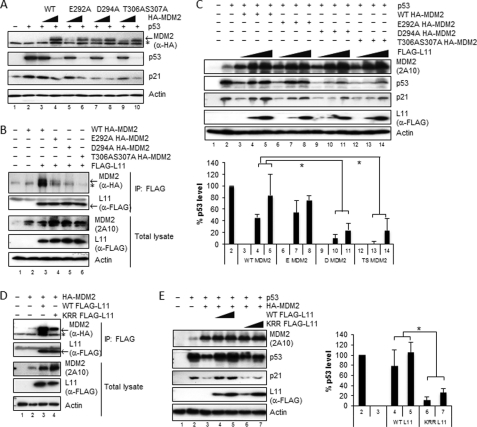
FIGURE 5.
Disruption of the MDM2 and RPL11 binding impairs the ability of RPL11 to inactivate MDM2 and to activate p53. A, MDM2 non-cysteine mutants retain the ability to repress p53 transcriptional activity and degrade p53. Lysates of H1299 cells transfected with p53 and various HA-MDM2 mutants were immunoblotted with anti-HA, anti-p53, and anti-p21 antibodies. The asterisk indicates nonspecific anti-HA antibody-reacting bands. B, MDM2 mutants interfere with the specific binding of MDM2 to RPL11 in cells. FLAG-RPL11 was co-expressed with various HA-tagged MDM2 mutants in H1299 cells. Immunoprecipitations (IP) were carried out using anti-FLAG M2-agarose beads, and precipitates were subjected to immunoblotting with anti-HA or anti-FLAG antibodies. Inputs are shown in the lower panels. C, D294A (D MDM2) and T305A/S306A MDM2 (TS MDM2) mutants are insensitive to RPL11 inhibition compared with wild type MDM2 (WT MDM2). H1299 cells were transfected with p53, HA-MDM2, and FLAG-RPL11 as indicated. After 36 h, cell lysates were generated and immunoblotted with anti-p53, anti-FLAG, and anti-MDM2 antibodies (2A10). D, WT, but not mutant, RPL11 strongly interacts with MDM2 in cells. HA-tagged MDM2 expressed with WT FLAG-RPL11 (WT RPL11) or K52R54R136 FLAG-L11 (KRR-RPL11) in H1299 cells were immunoprecipitated using anti-FLAG M2-agarose beads. Precipitates were immunoblotted with anti-FLAG and anti-HA antibodies. Inputs are shown in the lower panels. E, WT, but not KRR mutant, RPL11 can rescue MDM2-mediated suppression and degradation of p53. Lysates of H1299 cells transfected with p53, HA-MDM2, and FLAG-RPL11 mutant were immunoblotted with anti-FLAG and anti-MDM2 antibodies (2A10). Note: the percentage of the p53 expression level in C and E was quantitated and expressed as the mean number ± S.D. (n = 3) as described under “Experimental Procedure.” p values were calculated using two-tailed Student's t test. * indicates p < 0.01.
Protein-Protein Docking and Structural Modeling
The model of RPL11 complexed with MDM2 was generated using Cluspro 2.0, a fully automated protein-protein docking program (55). The structures that were fed to Cluspro correspond to a solution structure of the zinc finger domain of HDM2 (PDB code 2C6A) (56) and the RPL11 from a crystal structure of the 60 S eukaryotic ribosome proteins (PDB code 3O58, chain K7) (57). The putative protein complexes generated by Cluspro were manually examined using the software PyMOL (DeLano Scientific). Modeling of the zinc finger opening was based on the solution NMR structure of the MDM2 middle domain, and in silico mutations were made with the program Coot (58). Models of both wild type and mutated MDM2 middle domain were used as starting models for simulated annealing at 2000 K with slow cooling using the molecular dynamics simulation model of the program CNS 1.2 (59). Structural representations were generated in PyMOL.
RESULTS
Identification of MDM2(210–437)-RPL11 Complexes by Co-expression and Co-purification
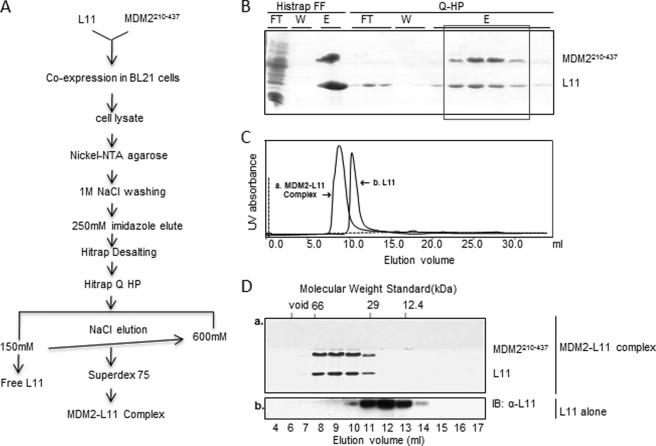
To decipher MDM2 and RPL11 direct interactions, we attempted to co-express MDM2 (amino acids 210–437) and the full-length RPL11 in the same Escherichia coli cells. For stable expression and purification, we co-expressed MDM2 and RPL11 using the pET24a/pPOREX co-expression system as detailed under “Experimental Procedures.” We then co-purified them through a procedure highlighted in Fig. 1A using a nickel-nitrilotriacetic acid affinity column followed by anion exchange and gel filtration chromatography. Free RPL11 proteins were removed by the anion exchange column as the basic RPL11 proteins did not bind to the column. As shown in Fig. 1B, recombinant RPL11 was co-purified with MDM2(210–437) through anion exchange columns. To ensure that they indeed associate with each other in one complex, RPL11-MDM2(210–437)-containing fractions from an anion exchange column were pooled and run on a Superdex 75 gel filtration column, and the elution profile was monitored by UV absorbance and Coomassie staining (Fig. 1, C and D). As shown in Fig. 1C, the RPL11-MDM2(210–437) complex was eluted as a single peak at a molecular mass between 29 and 66 kDa, which is close to the calculated molecular mass of this complex (∼45 kDa), indicating that the complex was formed as a heterodimer. In contrast, the recombinant RPL11 alone was eluted in the fractions between molecular mass 12.4 and 29 kDa as detected by immunoblotting. RPL11 forms a stable and soluble complex with MDM2, as purified RPL11 alone readily precipitated at low concentrations (0.2 mg/ml), but the complex remained stable even at high concentrations (>100 mg/ml).
FIGURE 1.
Recombinant RPL11 protein forms a stable complex with MDM2 (residues 210–437) through co-expression and co-purification from E. coli. A, scheme to purify and identify MDM2-RPL11 complexes. B–D, co-purification of the MDM2-RPL11 complexes was carried out through nickel affinity column followed by anion exchange and size exclusion chromatography. Fractions were visualized on SDS-PAGE (B and D). B, FT indicates flow-through; W indicates washout, and E indicates elution. C and D, size exclusive chromatography of RPL11 alone and MDM2-RPL11 complex. The elution profile was monitored by Coomassie staining or immunoblotting (IB) using anti-L11 antibodies. Panels a and b indicate that these two panels were obtained from two independent experiments for the MDM2-RPL11 complexes (panel a) and RPL11 alone (panel b), respectively.
Mapping of RPL11- and MDM2-binding Domains by HDX-MS
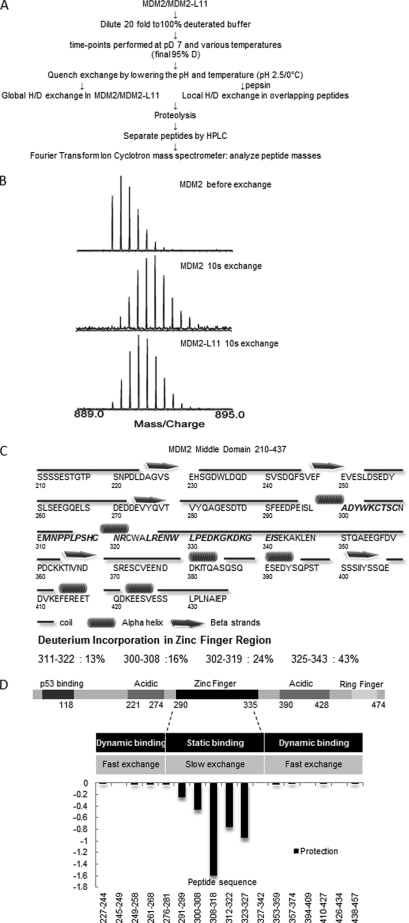
To finely map the RPL11-binding region on MDM2, we analyzed the purified RPL11-MDM2(210–437) complex (Fig. 1) by hydrogen-deuterium exchange mass spectrometry (HDX-MS) as outlined in Fig. 2A. MDM2 peptides generated by peptic digest were identified as described under “Experimental Procedures.” Isobaric peptides with identical masses but different sequences were differentiated by their MS/MS fragmentation. The peptide map after deuterium exchange consisted of 79 MDM2 peptides, corresponding to 75% sequence coverage. As shown in a representative result in Fig. 2B, deuterium incorporation shifted MDM2 peptides to a heavier molecule weight range, whereas the binding of RPL11 to MDM2 pushed them back to a lighter range because the region within the RPL11-MDM2 binding interface is excluded from deuterium exchange.
FIGURE 2.
MDM2 and RPL11 binding interface determined by HDX mass spectrometry. A, plan of the HDX experiment, described under “Experimental Procedures,” is diagrammed in this panel. B, expanded views of the isotopic envelope of one peptide fragment when fully protonated for a 10-s hydrogen/deuterium exchange and a 10-s H/D exchange in the complex. C, predicted secondary structures of MDM2(210–437) with the percent change in deuteration below. D, differences in deuterium incorporation were determined by the mass difference in individual peptides between MDM2(210–437) and the MDM2-bound RPL11 complexes where negative numbers indicate enhanced protection when ligand was bound.
The majority of the peptides within the C4 zinc finger region incorporated relatively less deuterium than other areas consistent with a compact and globular fold described by the NMR data within the range 297–329 (56). As shown in Fig. 2C, deuterium incorporation in the peptide 300–308 was 16%, 302–319 was 24%, and 311–322 was 13%. In contrast, in the same region, the peptide 325–343 displayed 43% deuterium incorporation in good agreement with the NMR data, which indicated an unstructured region in the C-terminal residues following Trp-329 (56). However, the presence of RPL11 altered the deuterium incorporation of MDM2 significantly as shown in Fig. 2D. A number of peptides, for example 291–299, 300–308, 308–318, 312–322, and 323–327, all exhibited reduced deuterium incorporation in the MDM2-L11 complex. These HDX results suggest that the interaction interface between RPL11 and MDM2 consists of residues that span from 291F to 326R, which is in close agreement with the region (284G to 374C) previously suggested by co-immunoprecipitation analyses of cell lysates (13, 52). Among all of these peptides, peptide 291–299 is the only one that does not belong to the C4 zinc finger domain, but it is connected to the N terminus of the C4 zinc finger domain. The MDM2 C4 zinc finger domain contains a defined consensus sequence X4WXCX2–4CX3NX6CX2CX5, where X is any amino acid. In human MDM2, the zinc finger region was defined as the peptide 299LADYWKCTSCNEMNPPLPSHCNRCWALRENWLP331. Hence, our HDX-MS analysis now firmly defines the zinc finger domain containing the 291F to 326R region as the RPL11-binding domain of MDM2.
Structure Modeling of RPL11-MDM2 Interaction
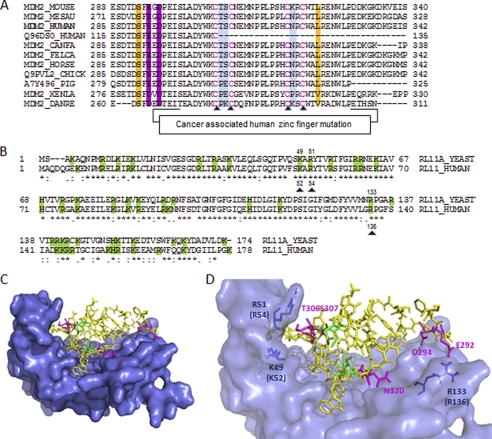
To obtain molecular insights into the MDM2-RPL11 interaction to better understand the mechanism of RPL11 regulation of MDM2 activity, we analyzed the amino acid sequence alignment of RPL11-binding region (amino acids 291–326) of MDM2 from various species (fish to human). This analysis revealed that this evolutionarily conserved region not only harbors a C4 zinc finger motif, which carries mutations in several human cancers (52) and is critical for ribosomal stress response in cells (52) and in animals (44), but it is also enriched with negatively charged residues (Fig. 3A). Sequence analysis of human and yeast RPL11 orthologs indicated that the MDM2-binding regions as grossly mapped previously (53) are highly conserved and enriched with positively charged residues (Fig. 3B). Because the three-dimensional structure for either the zinc finger domain of MDM2 (56) or yeast RPL11 in the content of 60 S ribosome (57) has been determined, they were used to build a protein-protein model using the automated molecular docking program Cluspro 2.0 (55). The putative protein complexes generated by Cluspro were manually examined using the software PyMOL. Information from HDX-MS experiments indicating interacting regions in RPL11 and MDM2 were taken into account in the selection of the models. Because the zinc finger is essential for the RPL11 binding of MDM2 and stress response (52), we predicted that the loss of protein-protein binding upon mutation of one of the four coordinating cysteines that chelate the zinc ion in MDM2 could be due to the disruption of the proper orientation of neighboring residues adjacent to the zinc finger, which might actively participate in the interaction. Therefore, we excluded those models that do not display residues immediately adjacent to the zinc finger but are residing at the interface with RPL11. By this criterion, we selected a structure model for the RPL11-MDM2-zinc finger complex as shown in Fig. 3C. Based on this modeled complex, it appears that the zinc finger domain of MDM2 protrudes into a hydrophilic pocket of RPL11. Interestingly, analysis of the predicted amino acids that reside at the interaction interface between RPL11 and the MDM2 zinc finger domain unveiled possible charge-charge interactions between the two proteins. For example, Glu-292, Asp-294, Thr-306, and Ser-307 within the zinc finger domain of MDM2 might directly contact Arg-133 (human Arg-136), Lys-49 (human Lys-52), and Arg-51 (human Arg-54) of RPL11 via H-bonding and/or salt bridge, respectively (Fig. 3D). This model suggests that electrostatic interactions or H-bonding could be crucial for the formation of a functional complex between MDM2 and RPL11 in regulating the p53-MDM2 feedback loop.
FIGURE 3.
RPL11 interaction with the zinc finger domain of MDM2 via hydrophilic residues. A, multiple sequence alignment of different MDM2 orthologs. Mutated residues for the binding experiments are highlighted in color. B, amino acid sequence comparison between yeast and human of RPL11s. Basic residues are shaded in green. Mutations made in human RPL11 are indicated by black triangles. The sequence alignment suggests that the basic residues in RPL11, which contribute to MDM2 binding, are highly conserved between yeast and human RPL11 orthologs. C, model of the MDM2-RPL11 complex with the zinc finger domain of MDM2 docked on the RPL11 structure. The model was constructed using the yeast RPL11 x-ray structure (PDB code 3O58, chain K7) and the NMR structure of the MDM2 zinc finger domain (PDB code 2C6A). RPL11 is represented as a molecular surface, and the zinc finger domain of MDM2 is shown as sticks. D, magnified view of C, showing the tight molecular interfaces between the zinc finger of MDM2 and the RPL11 basic canyon. The figures were made in PyMOL.
Circular Dichroism Analysis of the Central RP-binding Domain of MDM2 (Amino Acids 210–437)
To provide the evidence for the disruptive conformation by cysteine, but not non-cysteine, mutants in the zinc finger, we analyzed the secondary and tertiary structures of purified wild type and mutant forms of MDM2 by circular dichroism (CD) spectroscopy. As shown in supplemental Fig. S1A, far CD spectra of WT MDM2(210–437) displayed a positive maximum at 195 nm, a large minimum near 208 nm, and a broad minimum near 222 nm, which are characteristic for a structure consisting mostly of unordered secondary structures and a small percentage of α-helical structures. This is consistent with the solution structure of the MDM2 zinc finger domain where it consisted of mostly unordered secondary structures and a one-turn α-helix (56). The far-UV CD spectra of the mutants (D294A, T306A/S307A, and C322R) all superimposed well with that of the WT, indicating that these mutations did not alter the secondary structure content of the zinc finger domain.
Near UV CD spectra of WT MDM2(210–437) displayed broad minima below 270 nm, which are features associated with the presence of Phe (there are three Phe residues in this domain). The spectra also displayed pronounced signals with two maxima at ∼283 and 290 nm, which arise from Trp and/or Tyr (there are four Trp and six Tyr residues in this domain) (supplemental Fig. S1B). We noted that these CD spectra resemble that of colicin E1 channel domain (60), where the maxima at 285 and 292 nm were mostly due to a single Trp residue and that these maxima are more pronounced in polar environment than in apolar environment. Near-UV CD spectra of D294A and T306A/S307A superimposed well with that of the WT. However, the spectra of the C322R mutant showed a pronounced increase of the two characteristic maxima accompanied by slight red shift of the 293 nm peak. The spectral difference between WT and C322R mutant indicates differences in their tertiary structures, and the red shift indicates perturbation of a Trp residue that is situated in an apolar environment. Based on the NMR solution structure of the zinc domain, the most deeply buried Trp residue is Trp-323, which resides in a cleft directly above the zinc ion. Taken together, the data are consistent with the scenario that substitution of Arg for Cys in position 322 resulted in the disruption of the zinc chelation site, thus causing the “zinc cleft” to open (supplemental Fig. S1C).
Acidic and Polar Residues within the Zinc Finger Domain of MDM2 Are Essential for Its Interaction with RPL11
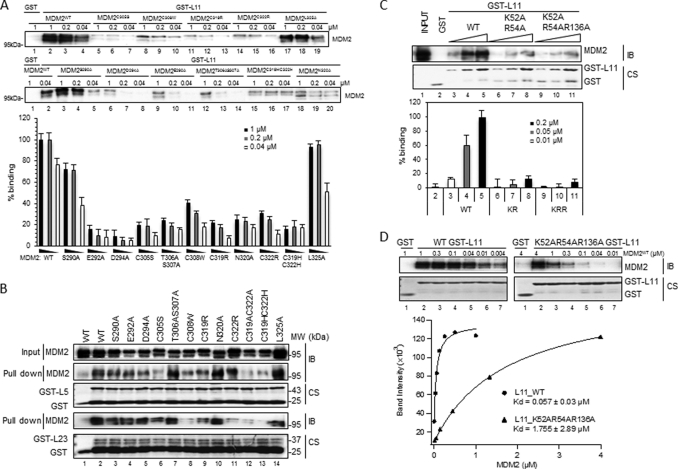
To determine which residues, in addition to cysteines, within the zinc finger domain of MDM2 are also important for the RPL11-MDM2 interaction, we generated individual mutations at different amino acids within the domain in full-length MDM2, including those as predicted in Fig. 3D and each of the four cysteines as controls using His-MDM2-encoding plasmid as a template. His-tagged MDM2 proteins were expressed in and purified from E. coli via nickel beads for GST-RP protein-protein interaction assays. As expected, mutation of each of the four cysteines in the zinc finger domain of MDM2 markedly reduced the interaction of MDM2 with RPL11 (Fig. 4A). Also, substitution of either single Cys with Arg (C322R) or Trp (C308W), or double Cys with Ala and His (C319A/C322A or C319A/C322H) within this domain led to apparent reductions of MDM2-RPL23 and/or MDM2-RPL5 interactions (Fig. 4B). By contrast and interestingly, mutations, including E292A, D294A, T306A/S307A, or N320A, within the zinc finger domain, but not mutations at S290A or L325A, drastically disrupted the interaction of MDM2 with only RPL11 (Fig. 4A) but not RPL5 or RPL23 (Fig. 4B). These results unambiguously argue that the zinc finger domain is where RPL11, but not RPL5 or RPL23, physically contacts when associating with MDM2 and that acidic or polar residues, such as Glu-292, Asp-294, or Thr-306/Ser-307, within this domain are essential for this specific association. These results, consistent with that in supplemental Fig. S1, suggest that mutations of cysteines, particularly double mutations, within this domain can alter the conformation of the central region of MDM2, which could explain why C305S failed to bind to RPL11 only (28), whereas C305F disabled MDM2 to bind to both RPL11 and RPL5 (52).
FIGURE 4.
Mutations of acidic or polar amino acids within the zinc finger domain of MDM2 or basic amino acids of RPL11 impair the interaction between the two proteins. A and B, GST pulldown assays show that RPL11, but not RPL5 and RPL23, directly and strongly binds to MDM2 zinc finger. Fifty nanomoles of GST-tagged ribosomal proteins or GST alone were immobilized on glutathione-agarose beads and then incubated with MDM2 as indicated. Bound MDM2 was detected by immunoblotting with anti-MDM2 (2A10) antibodies. Coomassie stains (CS) of input GST fusion proteins are also shown in B. WT, wild type. Quantification of immunoblots in A with various MDM2 concentrations is shown below. Binding values were justified against background and normalized to the MDM2 protein input with bars indicating the mean of three experiments and error bars representing the S.E. C, same assay as in A was carried out with different concentrations of GST-RPL11 and mutants K52AR54A (KR) or K52AR54AR136A (KRR) as indicated. Quantification of immunoblots is shown below. Values were justified relative to WT GST-RPL11 in the lane 5 (mean ± S.E., n = 3). D, binding affinity of WT GST-RPL11 and K52AR54AR136A to MDM2 as determined by GST pulldown assay and immunoblotting (IB). Each curve represents MDM2 titration with a constant GST RPL11 concentration of 50 nm. The y axis of the chart represents arbitrary units (AU) of the band intensity. The curve fitting and Kd determinations were performed using Igor Pro 4.01A. Kd represents the average of three replicates ± S.E.
Basic Residues of RPL11 Are Crucial for RPL11 to Bind to MDM2
To determine whether the predicted basic residues of RPL11 (Fig. 3D) are crucial for MDM2 binding, we created double (K52A/R54A) or triple mutations (K52A/R54A/R136A) using a GST-RPL11-encoding plasmid as template, because individual single mutations at these residues displayed less effect on the interaction of RPL11 with MDM2 in a GST fusion protein-protein interaction assay (data not shown). In GST-pulldown assays using purified His-MDM2 and GST-RPL11 proteins, we found that the basic residues, Lys-52, Arg-54, or Arg-136, in RPL11 are important for binding to MDM2, as the K52A/R54A mutation markedly inhibited the formation of the RPL11-MDM2 complex in a dose-dependent manner, and an additional mutation at Arg-136 on top of this double mutant further impaired the ability of RPL11 to bind to MDM2 (Fig. 4C). The affinity of this triple mutant of RPL11 was reduced by ∼30-fold compared with its wild type counterpart (Fig. 4D). These results demonstrate that basic residues within the MDM2-binding region of RPL11 are crucial for MDM2 binding and suggest that the chemical nature of the MDM2-RPL11 binding might be electrostatic interactions or H-bonding between hydrophilic residues of their interacting domains.
Hydrophilic Residues of Either MDM2 or RPL11 Are Important for MDM2-RPL11 Binding and p53 Activation in Cells
To consolidate the findings obtained from the above in vitro protein-protein binding assays, we also tested the ability of the aforementioned mutants of either MDM2 or RPL11 to form an RPL11-MDM2 complex and to affect p53 function in cells. We selected the E292A, D294A, and T306A/S307A mutants of MDM2 as well as the triple mutant of RPL11 (K52A/R54A/R136A or KRR-RPL11) for the cell-based analyses because these mutants more dramatically interfered with the formation of MDM2-RPL11 complexes in vitro (Fig. 4).
MDM2 negatively regulates p53 by suppressing the transcriptional activity of p53 through directly binding to the p53 N-terminal transactivation domain (10), and it mediates p53 degradation via ubiquitin-dependent proteolysis as the C terminus of MDM2 possesses an E3 ubiquitin ligase activity (8, 61). To determine whether MDM2 mutants, E292A, D294A, and T305A/S306A, retain the MDM2 functions, we co-expressed p53 and MDM2 mutants in H1299 cells (p53 null). As shown in Fig. 5A, all MDM2 mutants exhibited activities compared with wild type MDM2. Each of the mutants inhibited p53 activity as measured by reduction of endogenous p21 and p53 levels, demonstrating that the mutants do not disrupt the inhibitory activity of MDM2 toward p53. It is noteworthy that MDM2 is capable of suppressing p53 activity without triggering p53 degradation at a certain ratio of p53 to MDM2 (52). D294A partially inhibited p53 transcriptional activity, resulting in decreased p21 levels (Fig. 5A, lane 7), and further markedly reduced p53 protein level at the higher expression level (Fig. 5A, lane 8), suggesting that D294A was comparable with WT, effective in both suppressing p53 activity and mediating p53 degradation. In conclusion, together with CD analyses (supplemental Fig. S1), these non-cysteine mutants of MDM2 do not change the native structure of MDM2 and are still able to repress p53 and mediate p53 degradation.
Consistent with the results of Fig. 4, these three MDM2 mutants failed to bind to RPL11 in cells as analyzed by co-immunoprecipitation assays (Fig. 5B). D294A or T306A/S306A appeared to more markedly impair the ability of MDM2 to form complexes with RPL11 than did E292A (Fig. 5B). In line with these results, ectopically expressed RPL11 was significantly less effective in rescuing the p53 level and activity from the inhibition by D294A or T306A/S306A mutants compared with wild type MDM2 (p < 0.01, Fig. 5C, lanes 9–11 for D294A and lanes 12–14 for T305A/S307A). By contrast, the inhibition of E292A activity by RPL11 was comparable with that for wild type MDM2, as measured by the induction of p53 and p21 levels (Fig. 5C, lanes 6–8), even though this mutant bound to RPL11 less effectively (Fig. 5B). Overall, the results consistently demonstrate that the direct binding of RPL11 to the zinc finger of MDM2 is key for RPL11 to inhibit MDM2 activity toward p53 in cells.
This conclusion was further confirmed by using the triple mutant of RPL11 (K52A/R54A/R136A, KRR-RPL11). This RPL11 mutant failed to bind to MDM2 efficiently in vitro (Fig. 4, C and D) and in cells (Fig. 5D), thus being much less effective in rescuing the reduction of the p53 level by MDM2 in cells (Fig. 5E). Co-transfection of wild type RPL11 (WT RPL11) together with MDM2 resulted in activation and stabilization of p53 in a dose-dependent manner. We detected changes in p21 protein levels parallel to the p53 protein level (Fig. 5E, lanes 4 and 5). The reduction of the p53 level by MDM2 was fully rescued by WT RPL11, and p21 was also fully reactivated. In contrast, KRR-RPL11 was much less effective in rescuing p53 in cells (Fig. 5E, lanes 6 and 7). Taken together, these results demonstrate that the acidic or polar residues within the zinc finger domain of MDM2 and basic residues within the MDM2-binding domain of RPL11 are critical for the MDM2-RPL11 interaction and p53 activation in cells.
DISCUSSION
Although the biological significance of the MDM2-RPL11 binding in regulating the p53-MDM2 feedback loop has been described in cultured cells (52) and using a genetically manipulated mouse model that harbors the C305F mutation of MDM2 (44), it still remains elusive whether RPL11 binds directly to the zinc finger domain of MDM2 for the following reason: 1) distinct outcomes from mutation at Cys-305 residue of MDM2, impairing the binding of MDM2 to both RPL11 and RPL5 upon conversion of C305 to Phe-305; 2) all the assays thus far employed for mapping the RPL11-binding domain of MDM2 were carried out using crude lysates containing MDM2 deletion mutants for co-immunoprecipitation, thus possibly leading to inaccurate results; and 3) random deletion mutations might chop off residues or domains in MDM2 critical for RPL11 binding. Hence, our study as presented here by combining biochemical, proteomic, three-dimensional structure modeling, mutagenesis, and cell-based functional assays unambiguously demonstrates that the zinc finger domain of MDM2 is indeed the domain to which RPL11 directly binds. More interestingly, when examining the direct binding between L11 and the zinc finger domain of MDM2, we identified several non-cysteine acidic or polar residues within the C4 zinc finger domain of MDM2 essential for binding to RPL11 and several basic residues within the MDM2-binding domain of RPL11 crucial for binding to MDM2. These findings suggest that the chemical nature of MDM2-RPL11 interaction must be electrostatic interactions, such as H-bond, between the two proteins. Also mutations of cysteine residues in the C4 zinc finger domain disrupted the binding of MDM2 to RPL5 or RPL23, whereas mutation of any of the non-cysteine residues within the zinc finger domain of MDM2 did not affect these interactions (Fig. 4A). These results indicate the following: 1) RPL5 and RPL23 do not bind to the intact zinc finger domain directly; 2) disruption of the zinc finger domain would make a conformational alteration and affect the integrity of the central acidic region of MDM2. The latter could explain why C305F failed to bind to RPL5 as well (52), and C322R also failed to bind to RPL23 (Fig. 4A). Indeed, CD analysis of this mutant (supplemental Fig. S1) revealed a change in its tertiary, but not secondary structure, suggesting that the intact zinc finger domain of MDM2 is critical for maintaining the wild type three-dimensional structure of its central region. Of note, the local tertiary structure change in the zinc domain did not affect the ability of MDM2 to interact with p53 and to negate its activity (Fig. 5A) (52). Although these findings are quite tempting and important, it remains to be confirmed whether the charged or polar residues identified in MDM2 and RPL11 involve direct contacts with one another in the MDM2-RPL11 complex as predicted in our modeled complex (Fig. 3D). This mystery could and will be unveiled once the crystal structure of the MDM2-RPL11/RPL5 complex is solved in the near future.
Since the discovery of RPL11, RPL5, and RPL23 as direct MDM2 suppressors in response to ribosomal stress, more ribosomal proteins or RP-like proteins have been reported to also bind directly to MDM2 and inactivate its activity toward p53 in cell-based studies (16, 23, 36). Paradoxically, even though most of these MDM2-binding RPs, such as RPL5, RPL23, RPS7, RPL26, RPS3, or RPS27L, bind to the acidic domain of MDM2, cancer-related point mutations have thus far been found only in the RPL11-binding zinc finger domain of MDM2. Consistent with this, homozygous introduction of a cancer-derived single mutation, C305F, of the zinc finger domain into mice led to the impairment of p53 response to ribosomal stress in the animals and also accelerated lymphomagenesis in c-Myc transgenic mice (44). These lines of biological evidence highlight the importance of the zinc finger domain of MDM2 in mediating the physiological response of p53 to ribosomal stress and thus preventing tumorigenesis (44). However, they also raise a key question of whether other MDM2-binding ribosomal proteins, in addition to RPL11, are important for p53 response to this type of stress in vivo or not. In other words, if the zinc finger domain is so important for p53 activation and cancer prevention, why does only RPL11 bind to this domain while so many other RPs interact with central acidic domain of MDM2? Our finding that severely destroying the C4 zinc finger domain of MDM2 via single or double mutations, but not mutations of individual non-cysteine acidic or polar residues of this domain, can impair the binding of MDM2 to RPL5, and RPL23 as well, in addition to RPL11 (Fig. 4A), could offer another explanation. As described above, the mutation of any of the four cysteines could alter a conformational change of the central domain of MDM2 (supplemental Fig. S1), which would disrupt the interaction of MDM2 with either RPL5, such as C305F (52), or RPL5 and RPL23, such as C322R or C319A/C322A, even though RPL5 or RPL23 does not bind to the zinc finger domain directly (Fig. 4A) (15, 16). By the same token, these cysteine mutations might impair the binding of MDM2 to other known MDM2-binding ribosomal proteins as well, although this possibility needs to be verified experimentally. Thus, it is logical and reasonable that several types of human cancers harbor mutations at cysteine residues, instead of acidic or polar residues, within this zinc finger domain, as disruption of the zinc finger domain would easily eliminate the possibility of more ribosomal proteins to interact with MDM2 and to inactivate its activity toward p53 in a cooperative way, such as RPL11 and RPL5 (44, 62). Future studies will be necessary to address how these known and other yet unidentified MDM2-binding ribosomal proteins work together to effectively inactivate MDM2 mechanistically in vitro as well as in response to different types of ribosomal stressors in cells and in animals.
Supplementary Material
Acknowledgments
We greatly appreciate Thomas D. Hurley for help with complex co-expression and co-purification, Mushui Dai for MDM2 middle domain constructs, and Yanping Zhang for the pPOREX-L11 plasmid. We thank Shelya X. Zeng for technical assistance and all of our colleagues for active discussions and help.
This work was supported, in whole or in part, by National Institutes of Health Grants CA127724, CA095441, and CA129828 from NCI (to H. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Fig. S1.
- RS
- ribosomal stress
- RP
- ribosomal protein
- PDB
- Protein Data Bank
- HDX-MS
- hydrogen-deuterium exchange mass spectrometry.
REFERENCES
- 1. Kruse J. P., Gu W. (2009) Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vousden K. H., Prives C. (2009) Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 3. Feng Z., Levine A. J. (2010) Trends Cell Biol. 20, 427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vaseva A. V., Moll U. M. (2009) Biochim. Biophys. Acta 1787, 414–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu X., Bayle J. H., Olson D., Levine A. J. (1993) Genes Dev. 7, 1126–1132 [DOI] [PubMed] [Google Scholar]
- 6. Barak Y., Juven T., Haffner R., Oren M. (1993) EMBO J. 12, 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shvarts A., Steegenga W. T., Riteco N., van Laar T., Dekker P., Bazuine M., van Ham R. C., van der Houven van Oordt W., Hateboer G., van der Eb A. J., Jochemsen A. G. (1996) EMBO J. 15, 5349–5357 [PMC free article] [PubMed] [Google Scholar]
- 8. Haupt Y., Maya R., Kazaz A., Oren M. (1997) Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 9. Honda R., Tanaka H., Yasuda H. (1997) FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 10. Gu J., Kawai H., Nie L., Kitao H., Wiederschain D., Jochemsen A. G., Parant J., Lozano G., Yuan Z. M. (2002) J. Biol. Chem. 277, 19251–19254 [DOI] [PubMed] [Google Scholar]
- 11. Sherr C. J., Bertwistle D., DEN, Besten W., Kuo M. L., Sugimoto M., Tago K., Williams R. T., Zindy F., Roussel M. F. (2005) Cold Spring Harb. Symp Quant Biol. 70, 129–137 [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y., Lu H. (2009) Cancer Cell 16, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M., Vousden K. H. (2003) Cancer Cell 3, 577–587 [DOI] [PubMed] [Google Scholar]
- 14. Bhat K. P., Itahana K., Jin A., Zhang Y. (2004) EMBO J. 23, 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dai M. S., Lu H. (2004) J. Biol. Chem. 279, 44475–44482 [DOI] [PubMed] [Google Scholar]
- 16. Dai M. S., Zeng S. X., Jin Y., Sun X. X., David L., Lu H. (2004) Mol. Cell. Biol. 24, 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y., Wolf G. W., Bhat K., Jin A., Allio T., Burkhart W. A., Xiong Y. (2003) Mol. Cell. Biol. 23, 8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen D., Zhang Z., Li M., Wang W., Li Y., Rayburn E. R., Hill D. L., Wang H., Zhang R. (2007) Oncogene 26, 5029–5037 [DOI] [PubMed] [Google Scholar]
- 19. Zhu Y., Poyurovsky M. V., Li Y., Biderman L., Stahl J., Jacq X., Prives C. (2009) Mol. Cell 35, 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takagi M., Absalon M. J., McLure K. G., Kastan M. B. (2005) Cell 123, 49–63 [DOI] [PubMed] [Google Scholar]
- 21. Ofir-Rosenfeld Y., Boggs K., Michael D., Kastan M. B., Oren M. (2008) Mol. Cell 32, 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y., Wang J., Yuan Y., Zhang W., Guan W., Wu Z., Jin C., Chen H., Zhang L., Yang X., He F. (2010) Nucleic Acids Res. 38, 6544–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yadavilli S., Mayo L. D., Higgins M., Lain S., Hegde V., Deutsch W. A. (2009) DNA Repair 8, 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dai M. S., Sun X. X., Lu H. (2008) Mol. Cell. Biol. 28, 4365–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurki S., Peltonen K., Latonen L., Kiviharju T. M., Ojala P. M., Meek D., Laiho M. (2004) Cancer Cell 5, 465–475 [DOI] [PubMed] [Google Scholar]
- 26. Yu W., Qiu Z., Gao N., Wang L., Cui H., Qian Y., Jiang L., Luo J., Yi Z., Lu H., Li D., Liu M. (2011) Nucleic Acids Res. 39, 2234–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun X. X., Dai M. S., Lu H. (2007) J. Biol. Chem. 282, 8052–8059 [DOI] [PubMed] [Google Scholar]
- 28. Gilkes D. M., Chen L., Chen J. (2006) EMBO J. 25, 5614–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun X. X., Dai M. S., Lu H. (2008) J. Biol. Chem. 283, 12387–12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang F., Hamanaka R. B., Bobrovnikova-Marjon E., Gordan J. D., Dai M. S., Lu H., Simon M. C., Diehl J. A. (2006) J. Biol. Chem. 281, 30036–30045 [DOI] [PubMed] [Google Scholar]
- 31. Pestov D. G., Strezoska Z., Lau L. F. (2001) Mol. Cell. Biol. 21, 4246–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fumagalli S., Di Cara A., Neb-Gulati A., Natt F., Schwemberger S., Hall J., Babcock G. F., Bernardi R., Pandolfi P. P., Thomas G. (2009) Nat. Cell Biol. 11, 501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barlow J. L., Drynan L. F., Hewett D. R., Holmes L. R., Lorenzo-Abalde S., Lane A. L., Jolin H. E., Pannell R., Middleton A. J., Wong S. H., Warren A. J., Wainscoat J. S., Boultwood J., McKenzie A. N. (2010) Nat. Med. 16, 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chakraborty A., Uechi T., Higa S., Torihara H., Kenmochi N. (2009) PLoS One 4, e4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barki M., Crnomarkovi S., Grabusi K., Bogeti I., Pani L., Tamarut S., Cokari M., Jeri I., Vidak S., Volarevi S. (2009) Mol. Cell. Biol. 29, 2489–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Llanos S., Serrano M. (2010) Cell Cycle 9, 4005–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sun X. X., Wang Y. G., Xirodimas D. P., Dai M. S. (2010) J. Biol. Chem. 285, 25812–25821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pellagatti A., Marafioti T., Paterson J. C., Barlow J. L., Drynan L. F., Giagounidis A., Pileri S. A., Cazzola M., McKenzie A. N., Wainscoat J. S., Boultwood J. (2010) Blood 115, 2721–2723 [DOI] [PubMed] [Google Scholar]
- 39. Lindström M. S., Nistér M. (2010) PLoS One 5, e9578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hölzel M., Orban M., Hochstatter J., Rohrmoser M., Harasim T., Malamoussi A., Kremmer E., Längst G., Eick D. (2010) J. Biol. Chem. 285, 6364–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ge J., Rudnick D. A., He J., Crimmins D. L., Ladenson J. H., Bessler M., Mason P. J. (2010) Mol. Cell. Biol. 30, 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sieff C. A., Yang J., Merida-Long L. B., Lodish H. F. (2010) Br. J. Haematol. 148, 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sundqvist A., Liu G., Mirsaliotis A., Xirodimas D. P. (2009) EMBO Rep. 10, 1132–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Macias E., Jin A., Deisenroth C., Bhat K., Mao H., Lindström M. S., Zhang Y. (2010) Cancer Cell 18, 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boulon S., Westman B. J., Hutten S., Boisvert F. M., Lamond A. I. (2010) Mol. Cell 40, 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kressler D., Hurt E., Bassler J. (2010) Biochim. Biophys. Acta 1803, 673–683 [DOI] [PubMed] [Google Scholar]
- 47. Komrokji R. S., List A. F. (2010) Hematol. Oncol. Clin. North Am. 24, 377–388 [DOI] [PubMed] [Google Scholar]
- 48. Ebert B., Lipton J. M. (2011) Semin. Hematol. 48, 73–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ebert B. L., Pretz J., Bosco J., Chang C. Y., Tamayo P., Galili N., Raza A., Root D. E., Attar E., Ellis S. R., Golub T. R. (2008) Nature 451, 335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Devlin E. E., Dacosta L., Mohandas N., Elliott G., Bodine D. M. (2010) Blood 116, 2826–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cmejla R., Cmejlova J., Handrkova H., Petrak J., Petrtylova K., Mihal V., Stary J., Cerna Z., Jabali Y., Pospisilova D. (2009) Hum. Mutat. 30, 321–327 [DOI] [PubMed] [Google Scholar]
- 52. Lindström M. S., Jin A., Deisenroth C., White Wolf G., Zhang Y. (2007) Mol. Cell. Biol. 27, 1056–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dai M. S., Shi D., Jin Y., Sun X. X., Zhang Y., Grossman S. R., Lu H. (2006) J. Biol. Chem. 281, 24304–24313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zeng X., Chen L., Jost C. A., Maya R., Keller D., Wang X., Kaelin W. G., Jr., Oren M., Chen J., Lu H. (1999) Mol. Cell. Biol. 19, 3257–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kozakov D., Hall D. R., Beglov D., Brenke R., Comeau S. R., Shen Y., Li K., Zheng J., Vakili P., Paschalidis ICh., Vajda S. (2010) Proteins 78, 3124–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu G. W., Allen M. D., Andreeva A., Fersht A. R., Bycroft M. (2006) Protein Sci. 15, 384–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ben-Shem A., Jenner L., Yusupova G., Yusupov M. (2010) Science 330, 1203–1209 [DOI] [PubMed] [Google Scholar]
- 58. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 59. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 60. Zakharov S. D., Lindeberg M., Griko Y., Salamon Z., Tollin G., Prendergast F. G., Cramer W. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4282–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 62. Horn H. F., Vousden K. H. (2008) Oncogene 27, 5774–5784 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.