Background: Hyaluronan (HA) modulates key cancer cell functions through interaction with its CD44 and RHAMM receptors.
Results: Low molecular weight HA (LMWHA) significantly increased (p ≤ 0.01) the adhesion capacity of HT1080 cells in a RHAMM-dependent manner.
Conclusion: RHAMM/HA interaction regulates fibrosarcoma cell adhesion via the activation of FAK and ERK1/2 signaling pathways.
Significance: Identification of a novel HA-signaling pathway.
Keywords: Cancer Biology, ERK, Glycosaminoglycan, Hyaluronate, Receptor Regulation, Hyaluronan, RHAMM, Cell Adhesion
Abstract
Hyaluronan (HA) modulates key cancer cell functions through interaction with its CD44 and receptor for hyaluronic acid-mediated motility (RHAMM) receptors. HA was recently found to regulate the migration of fibrosarcoma cells in a manner specifically dependent on its size. Here, we investigated the effect of HA/RHAMM signaling on the ability of HT1080 fibrosarcoma cells to adhere onto fibronectin. Low molecular weight HA (LMWHA) significantly increased (p ≤ 0.01) the adhesion capacity of HT1080 cells, which high molecular weight HA inhibited. The ability of HT1080 RHAMM-deficient cells, but not of CD44-deficient ones, to adhere was significantly decreased (p ≤ 0.001) as compared with control cells. Importantly, the effect of LMWHA on HT1080 cell adhesion was completely attenuated in RHAMM-deficient cells. In contrast, adhesion of RHAMM-deficient cells was not sensitive to high molecular weight HA treatment, which identifies RHAMM as a specific conduit of the LMWHA effect. Western blot and real time-PCR analyses indicated that LMWHA significantly increased RHAMM transcript (p ≤ 0.05) and protein isoform levels (53%, 95 kDa; 37%, 73 kDa) in fibrosarcoma cells. Moreover, Western blot analyses showed that LMWHA in a RHAMM-dependent manner enhanced basal and adhesion-dependent ERK1/2 and focal adhesion kinase (FAK) phosphorylation in HT1080 cells. Utilization of a specific ERK1/2 inhibitor completely inhibited (p ≤ 0.001) LMWHA-dependent adhesion, suggesting that ERK1/2 is a downstream effector of LMWHA/RHAMM signaling. Likewise, the utilization of the specific ERK1 inhibitor resulted in a strong down-regulation of FAK activation in HT1080 cells, which identifies ERK1/2 as a FAK upstream activator. In conclusion, our results suggest that RHAMM/HA interaction regulates fibrosarcoma cell adhesion via the activation of FAK and ERK1/2 signaling pathways.
Introduction
The extracellular matrices (ECMs),3 which mainly consist of collagens, glycoproteins, proteoglycans, and hyaluronan, are complicated structures that surround and support cells within tissues. The ECM acts as a physical scaffold to which tumor cells attach and migrate and thus is crucial for the regulation of cell motility, proliferation, invasion, and metastasis. Hyaluronan (HA), a glycosaminoglycan, is a ubiquitous component of the extracellular matrix that provides tissue homeostasis and is known to have a fundamental role in maintaining the ECM architecture.
High levels of HA reported in tumor cells and peri-tumor stroma are suggested to be strong independent prognostic indicators of poor outcome in breast, ovarian, gastric, and colorectal cancers (1, 2). In tumor cell microenvironment systems, HA is able to transmit signals originating from the ECM into the cell, and changes in its metabolism have been linked to the promotion of cell motility, adhesion, migration, and metastasis (3–5). Considerable evidence indicates that HA mediates these biological processes mostly via specific interactions with its receptors, a family of proteins termed hyaladherins (4, 6–10). CD44 is the best characterized HA receptor and is indicated to be the principal mediator of HA signaling (11, 12). RHAMM receptor (receptor for hyaluronic acid-mediated motility) is unique among the hyaladherins due to its variable distribution on the cell surface, within the cytoplasm, in the nucleus, or secreted to the ECM (13–16). Different RHAMM isoforms have been detected due to alternative splicing (10). Transcript variants are suggested to be expressed in a specific cell type manner (10). RHAMM protein has been reported to lack a typical transmembrane domain and is glycosylphosphatidylinositol-anchored to the cell membrane (17–19). Cell surface RHAMM interacts with CD44 and modulates cell motility, wound healing, and signal transduction. More importantly, cell surface RHAMM can have invasive functions similar to CD44 and can even substitute for CD44 functions (20). Intracellular RHAMM binds to actin filaments, podosomes, the centrosome, microtubules, and the mitotic spindle (4, 21, 22), thereby affecting crucial cellular processes in tumorogenesis (22–24).
Various studies demonstrated an overexpression of RHAMM during tumor development and a prognostic significance of its expression in breast, colon, brain, prostate, endometrial and pancreatic cancers, as well as in leukemia, aggressive fibromatosis, multiple myeloma and melanoma (23, 25–33). Previous publications have shown that the interaction of HA with RHAMM can induce a number of cellular signaling molecules, including focal adhesion kinase (FAK), extracellular signal-regulated kinase (ERK1/2), protein kinase C (PKC), pp60-c-Src, NFκB, Ras, and phosphatidylinositol kinase (PI3K) (26, 34–40).
Fibrosarcoma is a rare malignant tumor originating from fibroblasts. Different cell lines of fibroblastic origin have been shown to have an abundant ECM with a high content and turnover of HA and proteoglycans (41). Cell migration is a highly coordinated process crucially dependent on cell adhesion to the ECM (42). Both cell functions are necessary for efficient tumor cell invasion and metastasis. Recently, we demonstrated that the treatment of fibrosarcoma (HT1080) cells with various molecular weight HA preparations resulted in regulation of their migration capacity in a manner strictly dependent on HA size (43). In this study, we examined the effects of HA on fibrosarcoma cell adhesion and focused on the mechanism of its action. Our results demonstrate that HA regulates fibrosarcoma cell adhesion through interaction with its RHAMM receptor and consecutive activation of FAK and ERK1/2 signaling pathways.
EXPERIMENTAL PROCEDURES
Materials
Low molecular weight hyaluronan (LMWHA; 15–40 kDa; GLR001) was obtained from R&D Diagnostics and produced by microbial fermentation of Streptococcus pyogenes, and its molecular weight was achieved by acid hydrolysis of higher molecular weight native sodium hyaluronate (measured by multiangle laser light scattering). The commercial name of the high molecular weight hyaluronan (HMWHA; 2.5 × 106 Da) preparation used is Visthesia 1.5% (0.8 ml of sodium hyaluronate 15 mg/ml and lidocaine hydrochloride 10 mg/ml, Zeiss). All secondary antibodies (anti-goat, anti-rabbit, and anti-mouse) as well as the primary antibodies polyclonal goat anti-actin (I19, sc-1616), anti-RHAMM (E-19, sc-16170), anti-FAK (A-17, sc-557), and anti-CD44 (HCAM, sc-9960) were purchased from Santa Cruz Biotechnology. p-FAK (MAB1144) was purchased from Millipore, and anti-ERK1/2 (9102) and p-ERK1/2 (9101) were purchased from Cell Signaling Technology. Hyaluronidase (Streptomyces hyalurolyticus), chondroitinase ABC (Proteus vulgaris), chondroitinase ACII Arthro (Arthrobacter aurescens), and Δ-disaccharides of HA and CS were purchased from Seikagaku Corp., Japan. Oligosaccharides of HA were the kind gift of Dr. P. Heldin, Ludwig Institute for Cancer Research, Uppsala, Sweden. Cell culture reagents were obtained from Biosera, East Sussex, UK, unless stated otherwise.
Cell Culture
The HT1080 human fibrosarcoma cell line was grown in DMEM (Biochrom KG, Germany) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and penicillin/streptomycin at 37 °C in a humidified atmosphere of 5% (v/v) CO2. Prior to treatments, the cells were incubated in serum-free medium for 24 h at 37 °C and 5% CO2. All treatments were done in serum-free medium for 48 h prior to extraction of either RNA or protein.
RNA Isolation and Real Time PCR
Total ribonucleic acid was isolated with the use of TRIzol (Invitrogen) according to the manufacturer's instructions. Five hundred nanograms of total RNA were used for cDNA synthesis using the PrimeScript RT reagent kit (Takara, Japan) according to the manufacturer's instructions. The real time PCRs were done in an Mx3005P cycler (Stratagene). The primers were mRNA-specific to avoid misleading results from traces of DNA contamination (Table 1). For the real time PCR, we utilized the KAPA SYBR® FAST Universal qPCR kit (KAPA Biosystems) in a total volume of 20 μl. The PCR conditions used for amplification were as follows: 94 °C for 15 min and then 40 cycles at 94 °C for 20 s, 55 °C for 30 s, and 72 °C for 30 s, followed by 72 °C for 10 min. Standard curves were run in each optimized assay, which produced a linear plot of threshold cycle (Ct) against log (dilution). The amount of each target was quantified based on the concentration of the standard curve and was presented as arbitrary units. GAPDH was utilized as housekeeping gene.
TABLE 1.
Sequence of primers for the genes of interest
| Primer name | Sequence |
|---|---|
| RHAMM_F | 5′-TTC TGA ACC CTT TGG CTG G-3′ |
| RHAMM_R | 5′-ACA AGC CAA GGT GTT TTA GCC-3′ |
| GAPDH_F | 5′-GGA AGG TGA AGG TCG GAG TCA-3′ |
| GAPDH_R | 5′-GTC ATT GAT GGC AAC AAT ATC CAC T-3′ |
| CD44_F | 5′-GGT CCT ATA AGG ACA CCC CAA AT-3′ |
| CD44_R | 5′-AAT CAA AGC CAA GGC CAA GA-3′ |
Western Blot
After 48 h of respective treatments or after the completion of the in vitro adhesion assay, the cells were collected using RIPA solution, and the supernatants (culture media) were concentrated (16-fold) using 30 × 116-mm filter tubes Vivaspin, 20 ml (Biotech). The samples were electrophoresed on 8% polyacrylamide Tris/glycine gels and transferred to nitrocellulose membranes in 10 mm CAPS, pH 11, and containing 10% methanol. Membranes were blocked overnight at 4 °C with PBS containing 0.1% Tween 20 (PBS-T) and 5% (w/v) low fat milk powder. The membranes were incubated for 1 h at room temperature (RT) with the primary antibodies anti-RHAMM (1:100) in PBS containing 0.1% Tween 20 (PBS-T) and 2% (w/v) low fat milk powder, anti-FAK (1:100), anti-p-FAK (Y397) (1:100), anti-ERK1/2 (p44/42 MAPK) (1:200), anti-ERK1/2 (p44/42 MAPK) (1:200) in PBS containing 0.1% Tween 20 (PBS-T) and 2% (w/v) low fat milk powder. The immune complexes were detected after incubation with the peroxidase-conjugated secondary anti-goat antibody (1:4000), anti-mouse antibody (1:2000), anti-rabbit antibody (1:5000) in PBS-T, 1% low fat milk, with the SuperSignal West Pico chemiluminescent substrate (Pierce), according to the manufacturer's instructions.
HA Digestion
LMWHA and HMWHA were digested with Streptomyces hyaluronidase (5 units/100 μg/ml HA) for 48 h at 37 °C according to the manufacturer's instructions (Seikagaku, Japan).
LMWHA and HMWHA Purity Evaluation
As a first quality test, we examined both HA preparations for the presence of CS and HA oligosaccharides using FACE and CE. Analysis by FACE was performed before and after treatment of HA preparations with chondroitinases ABC and ACII in combination. Digestion with chondroitinases was performed in 50 mm Tris-HCl, pH 7.5, at 37 °C for 90 min using 0.01 unit/10 μg of uronic acid (44). Intact HA preparations as well as the obtained digests were freeze-dried and derivatized with 2-aminoacridone as described previously (45). FACE was performed according to protocol described by Calabro et al. (46) and Karousou et al. (47). Analysis by CE was performed on the intact preparations using a reversed polarity methodology (48). The ophthalmic preparation Viscoat®, containing both HA and CS, was used as standard.
Identification of LMWHA and HMWHA Oligomers following Digestion with Hyaluronidase
To evaluate the size of the digestion products following treatment with hyaluronidase and to examine whether both intact HA preparations contain any contaminations of HA oligomers, we performed FACE analysis. Particularly, intact HA preparations and those obtained by hyaluronidase treatment were freeze-dried, derivatized with 2-aminoacridone, and analyzed by FACE as described above. HA-derived Δ-disaccharide and HA-derived oligosaccharides (6–16-mers) were used as standards.
Transfection with siRNA
Short interfering RNA (siRNA) specific for RHAMM or CD44 and scrambled RNAi and medium GC content negative control were purchased from Invitrogen. For transfection experiments, the cells (60,000/well) were placed on a 24-well plate for 24 h. Then the DMEM containing 10% FBS was replaced by medium without serum and antibiotics. To provide for optimal transfection, siRNA (100 nm; Invitrogen) and LipofectamineTM 2000 (1 μl; Invitrogen) were first diluted separately in 50 μl of Opti-MEM® I reduced serum medium (Invitrogen). After a 5-min incubation period, 50 μl of diluted LipofectamineTM 2000 were mixed with 50 μl of diluted siRNA per well on 24-well plates and were left for 20 min at room temperature to allow siRNA-liposome complexes to form. The cells were then washed once more with medium without serum and antibiotics, and the Lipofectamine and siRNA mixture was added on top of them and shaken gently. Transfection was allowed to take place during 6 h when the medium was replaced with fresh medium containing antibiotics, and the incubation period was continued for an additional 48 h. The cells were harvested, and RNA was extracted. The optimal siRNA concentration for transfection was chosen after pilot experiments. All transfection experiments were repeated at least three times and performed in triplicate.
Cell Attachment Assay
Ninety six-well plates were used and coated with fibronectin (5 μg/cm2) for 1 h at 37 °C. The nonspecific binding sites were blocked with 1% bovine serum albumin (BSA) for 30 min at room temperature. The cells were detached with 5 mm PBS/EDTA. 5000 cells were placed onto the coated 96-well plates. The time point of 30 min was chosen after preliminary experiments as described under “Results.” HT1080 cells were serum-starved for 24 h and then treated with LMWHA (50 μg/ml), HMWHA (50 μg/ml), or ERK1/2 inhibitor (5 μm) (Cell Signaling Technology) for 48 h. Some experiments were performed by using transfected cells with siRNA specific for RHAMM (siRHAMM) or CD44 (siCD44) and control scramble (siScr). Nonadherent cells were removed with two washes using serum-free medium. All experiments were also performed on BSA-coated plates, and we calculated the cells attached specifically to fibronectin by subtracting the number of cells attached to BSA from the number of cells attached to fibronectin. The numbers of adherent cells were measured using the CyQUANT fluorometric assay (Molecular Probes, Invitrogen) according to the manufacturer's instructions. Fluorescence was measured in a Fluorometer (BioTek) using the 480/520 nm excitation and emission filters. For converting sample fluorescence values into cell numbers, a reference standard curve was created, using serial dilutions of known cell numbers. All adhesion experiments were repeated at least three times and performed in triplicate.
Immunofluorescence
HT1080 cells were seeded on round coverslips placed in 24-well plates and incubated in complete medium for 24 h. After a 24-h serum starvation, the treatments were added, and the cells were incubated for 48 h at 37 °C and 5% CO2. The cells were fixed with 5% formaldehyde and 2% sucrose in PBS for 10 min at room temperature. After three washes with PBS, 1% bovine serum solution, the permeabilizing agent Triton X-100 was added for 10 min and then washed before the addition of primary antibody anti-RHAMM for 1 h at room temperature. Coverslips not incubated with the primary antibody were utilized as negative control. The coverslips were washed again and incubated for 1 h, in the dark at RT, with anti-goat Alexa Fluor 488 (Invitrogen). TO-PRO-3 diluted 1:1000 in de-ionized H2O was applied for 10 min to stain nuclei. Actin filaments were detected using fluorescent phalloidin (Molecular Probes) diluted 1:100 in PBS for 40 min. The coverslips were then placed onto slides using glycerol as a mountant and visualized using confocal microscopy.
Statistical Analysis
The statistical significance was evaluated by Student's t test or one-way analysis of variance with Tukey's post-test, using GraphPad Prism (version 4.0) software.
RESULTS
LMWHA and HMWHA Preparations Do Not Contain CS and HA Oligosaccharides
To ensure that HMWHA and LMWHA preparations were free of chondroitin sulfate contamination and HA oligosaccharides, FACE analysis was performed. The analysis of intact LMWHA and HMWHA showed the absence of HA oligosaccharides in both preparations (supplemental Fig. S1, lanes 3 and 4) as compared with HA oligosaccharides standard (supplemental Fig. S1, lane 2). The products of digestion with chondroitinases ABC and ACII, in the case of both LMWHA (supplemental Fig. S1, lane 5) and HMWHA (supplemental Fig. S1, lane 6), were identified as Δdi-HA, and no sulfated CS-derived Δ-disaccharides were evident. The chondroitinase digest of the ophthalmic solution Viscoat® was a useful tool to identify the absence of CS. Furthermore, the absence of CS and HA oligosaccharides from LMWHA and HMWHA was also verified by CE (48). CE analysis showed that both LMWHA and HMWHA migrated as homogeneously charged populations (supplemental Fig. S2). The molecular mass range of the LMWHA preparation was 15–40 kDa; with an average mass of 16.1 kDa, as determined by gel permeation chromatography as described earlier (44).
Identification of Hyaluronidase Digestion Products
As shown in supplemental Fig. S3, lanes 5–8 and in the electropherogram of supplemental Fig. S4, treatment of both LMWHA and HMWHA with Streptomyces hyaluronidase for 48 h resulted in 4- and 6-mers of HA.
Effect of HA on Fibrosarcoma Cell Adhesion
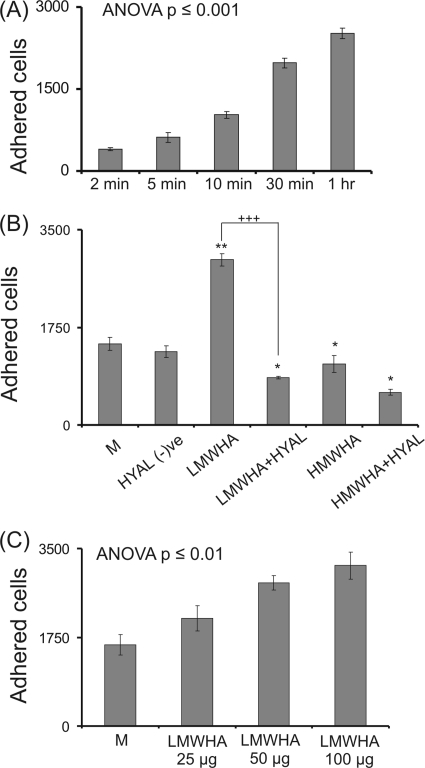
A poorly differentiated and highly metastatic human fibrosarcoma cell line consisting of rounded/elongated cells (HT1080) (49) was utilized. In our previous study the HT1080 cell migration capacity was found to be modulated through changes in HA metabolism (43). In this study, to investigate the adhesion capacity of these fibrosarcoma cells on fibronectin, an in vitro adhesion assay was performed. The HT1080 cells demonstrated specific adhesion to fibronectin, and a 30-min adhesion time was selected as a time during the near linear increase in adhesion (Fig. 1A). In continuation, the effect of exogenous addition of HMWHA (50 μg/ml) and LMWHA (50 μg/ml) was examined. LMWHA significantly increased (p ≤ 0.01) the adhesion capacity of HT1080 cells. The stimulation of cell adhesion ability by LMWHA was found to be concentration-dependent (p ≤ 0.01) (Fig. 1C), reaching a plateau at 100 μg/ml. In contrast, exogenous addition of HMWHA significantly inhibited (p ≤ 0.05) cell adhesion (Fig. 1B). To be sure that the effects are specific for HA, both LMWHA and HMWHA were digested with Streptomyces hyaluronidase according to the manufacturer's instructions. FACE analysis and capillary electrophoresis demonstrated complete digestion of both samples of HA to 4- and 6-mers as described above (supplemental Figs. S3 and S4). The hyaluronidase digests of LMWHA and HMWHA were heat-inactivated and applied to HT1080 cells for 48 h, and their adhesion capacity was assessed. The effect of LMWHA on fibrosarcoma cell adhesion was completely nullified by the digestion, which demonstrates the strict specificity of the HA chain size for the LMWHA effect (Fig. 1B). Interestingly, both HA digests had an independent inhibitory effect on fibrosarcoma cell adhesion (p ≤ 0.05). In control experiments heat-inactivated hyaluronidase did not affect HT1080 cell adhesion (Fig. 1B).
FIGURE 1.
Effect of LMWHA and HMWHA on fibronectin-dependent HT1080 cell adhesion. A, HT1080 cells harvested with PBS/EDTA were seeded, and the numbers of attached cells were measured at various time points. The results represent the average of five separate experiments in triplicate. p < 0.001 using one-way analysis of variance (ANOVA) statistical analysis. Means ± S.E. are plotted. B, HT1080 cells were treated with 0% medium (M), heat-inactivated hyaluronidase (HYAL (−)ve), LMWHA (50 μg/ml), HMWHA (50 μg/ml), and with heat-inactivated LMWHA hyaluronidase digest (LMWHA+HYAL) (50 μg/ml) or heat-inactivated HMWHA hyaluronidase digest (HMWHA+HYAL) (50 μg/ml) for 48 h before harvesting and seeding (5000 cells) for 30 min on 96-well plates coated with fibronectin (5 μg/cm2). C, HT1080 cells were incubated with different concentrations of LMWHA (25, 50, and 100 μg/ml) before harvesting and seeding (5000 cells) for 30 min on 96-well plates coated with fibronectin (5 μg/cm2). p < 0.01, using one-way analysis of variance statistical analysis. The numbers of attached cells were determined using fluorometric CyQUANT assay kit (Molecular Probes). Cells grown in serum-free medium (M) were utilized as control. The results represent the average of three separate experiments in triplicate. Means ± S.E. are plotted; statistical significance is as follows: *, p ≤ 0.05; **, p ≤ 0.01 treatment compared with control; +++, p ≤ 0.01 among samples.
Role of RHAMM in Fibrosarcoma Cell Adhesion
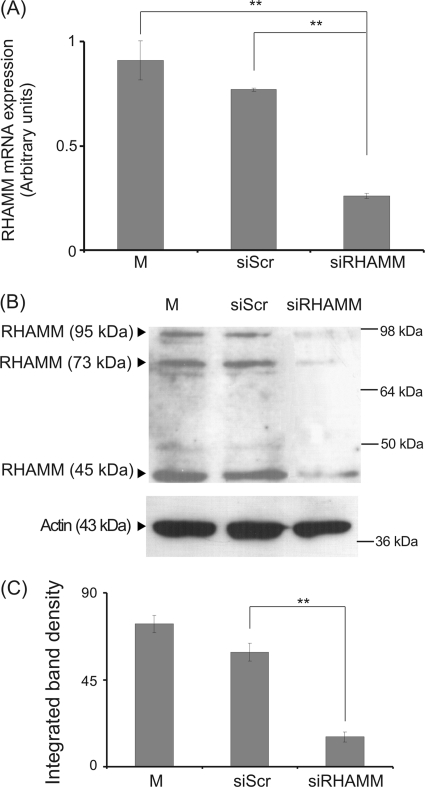
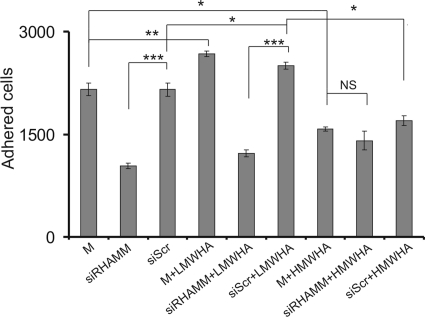
HA receptor RHAMM has been suggested to mediate HA-induced migration, adhesion, and proliferation in cancer cells (50, 51). Therefore, we investigated whether it participates in the HA-dependent fibrosarcoma cell adhesion. To determine the direct role of RHAMM on HT1080 cell adhesion, we utilized siRNA specific for the RHAMM gene. Transfection of HT1080 cells with siRHAMM resulted in a significant 70% decrease of RHAMM mRNA expression and protein expression (p ≤ 0.01) (Fig. 2). Down-regulation of RHAMM expression correlated with a strong inhibition of HT1080 cell adhesion (p ≤ 0.001) (Fig. 3). Moreover, we examined whether RHAMM mediates the HA signaling that results in altered adhesion ability of HT1080 cells. The LMWHA-induced increase in the adhesion capacity of HT1080 cells (p ≤ 0.05) was completely inhibited in RHAMM-deficient cells to the levels of the siRHAMM alone controls (p = not significant) (Fig. 3). These data demonstrate that active RHAMM signaling is obligatory for the HA effect on HT1080 cell adhesion. Interestingly, the reduction of HT1080 adhesion by HMWHA was not affected by RHAMM inhibition (p = not significant) (Fig. 3). Thus, RHAMM was shown to be an important signal mediator of HA signaling in fibrosarcoma cell adhesion in a manner that depends on HA size.
FIGURE 2.
Transfection with siRNA-specific for RHAMM. HT1080 cells were transfected with RHAMM short interfering RNA (siRHAMM) utilizing scramble interfering RNA (siScr) as a negative control. M stands for cell treated with culture medium. A, inhibition of RHAMM mRNA expression was verified by real time PCR as compared with siScr control and expressed as the ratio to siScr control at the 48-h point. Results are expressed as mean % ± S.E. Statistical significance is as follows: **, p ≤ 0.01, compared with control. B, equal amounts of protein were extracted from both siScr and siRHAMM cells, separated by PAGE, and blotted with the anti-RHAMM antibody. C, densitometric analysis of specific RHAMM protein bands. The results represent the average of three separate experiments in triplicate. Means ± S.E. are plotted; statistical significance is as follows: **, p ≤ 0.01 compared with the respective control sample.
FIGURE 3.
Effect of siRHAMM on fibronectin-dependent cell adhesion. 48 h after transfection, HT1080 siRHAMM and scramble (siScr)-transfected cells were harvested and seeded (5000 cells) for 30 min on 96-well plates coated with fibronectin (5 μg/cm2). M stands for cell treated with culture medium. The numbers of attached cells were determined using fluorometric CyQUANT assay kit. The results represent the average of three separate experiments in triplicate. Means ± S.E. are plotted; statistical significance is as follows: ***, p ≤ 0.001; **, p ≤ 0.01; *, p ≤ 0.05 compared with the respective control samples. NS, not significant.
Role of CD44 in Fibrosarcoma Cell Adhesion
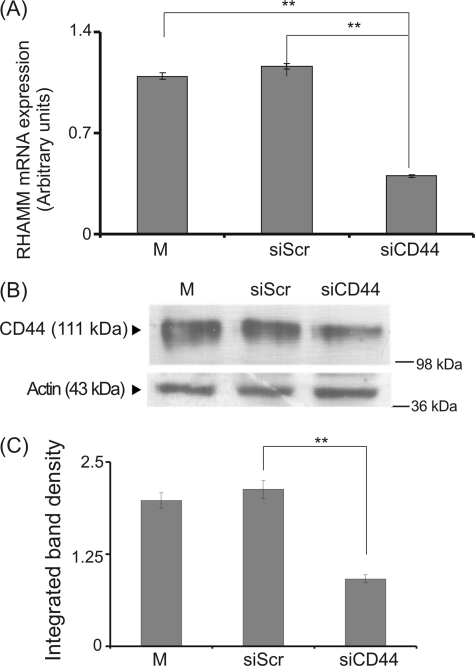
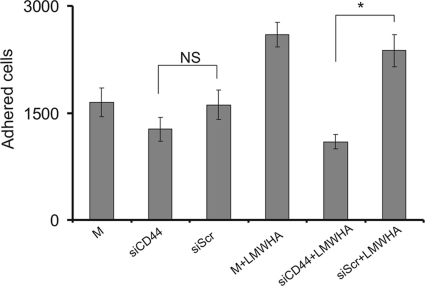
Extracellular HA receptor RHAMM interacts with CD44 to transmit its signal intracellularly (2). Thus, we investigated whether it participates in the HA-dependent fibrosarcoma cell adhesion, shown to be mediated by RHAMM. The role of CD44 was examined using RNA interference. Transfection of HT1080 cells with siCD44 resulted in a significant 60% decrease of CD44 mRNA expression and protein expression (p ≤ 0.01) (Fig. 4). CD44-deficient cells were found to have unaltered ability to adhere (Fig. 5). The down-regulation of CD44 expression, however, induced a reduction of the LMWHA-dependent increase in the adhesion capacity of HT1080 cells (p ≤ 0.05) (Fig. 5). These data demonstrate that, even though CD44 action is not obligatory in basal fibrosarcoma cell adhesive ability, it participates in the LMWHA-dependent mechanism responsible for the modulation of the adhesion of these cells.
FIGURE 4.
Transfection with siRNA specific for CD44. HT1080 cells were transfected with CD44 short interfering RNA (siCD44) utilizing siScr as a negative control. M stands for cell treated with culture medium. A, inhibition of CD44 mRNA expression was verified by real time PCR as compared with siScr control and expressed as the ratio to siScr control at the 48-h point. Results are expressed as mean % ± S.E. Statistical significance is as follows: **, p ≤ 0.01, compared with control. B, equal amounts of protein were extracted from both siScr and siCD44 cells blotted with the anti-CD44 antibody. C, densitometric analysis of specific CD44 protein bands. The results represent the average of three separate experiments in triplicate. Means ± S.E. are plotted; statistical significance is as follows: **, p ≤ 0.01 compared with the respective control sample.
FIGURE 5.
Effect of siCD44 on fibronectin-dependent cell adhesion. 48 h after transfection, HT1080 siCD44 and siScr cells were harvested and seeded (5000 cells) for 30 min on 96-well plates coated with fibronectin (5 μg/cm2). M stands for cells treated with culture medium. The numbers of attached cells were determined using fluorometric CyQUANT assay kit. The results represent the average of three separate experiments in triplicate. Means ± S.E. are plotted; statistical significance is as follows: *, p ≤ 0.05 compared with the respective control samples. NS, not significant.
Effect of HA on RHAMM mRNA and Protein Expression
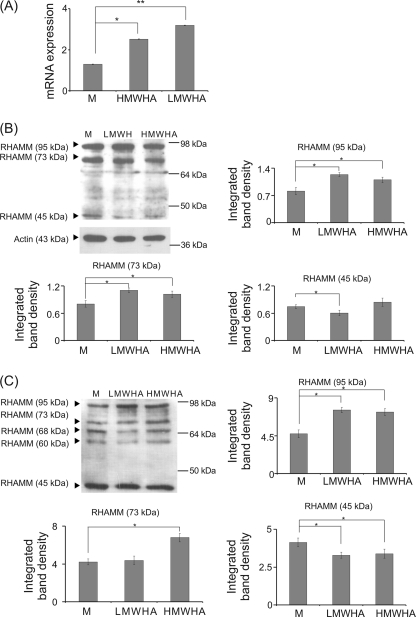
Our results demonstrated that LMWHA partly modulates fibrosarcoma cell adhesion through the RHAMM receptor. Therefore, we examined the effect of LMWHA and HMWHA on the expression of RHAMM. Real time PCR analysis showed that HT1080 cells express mRNA specific for RHAMM, whose levels were modestly increased with HMWHA treatment (p ≤ 0.05), although in the case of LMWHA the effect was more prominent (p ≤ 0.01) (Fig. 6A). In continuation, HT1080 cell extracts and concentrated media were probed for RHAMM protein. The anti-RHAMM antibody used detects a specific amino acid sequence near its carboxyl terminus, which is conserved in all RHAMM isoforms. Western blot analysis of cell extracts detected three different RHAMM isoforms of ∼95, 73, and 45 kDa. Treatment with LMWHA significantly increased expression of the 95- and 73-kDa RHAMM isoforms (53 and 37% respectively) (Fig. 6B), which correlates well with the RHAMM total transcript results. HMWHA addition likewise induced an increase of the 95- and 73-kDa isoforms (36 and 27%, respectively) (Fig. 6B). Apart from the 95, 73, and 45 RHAMM isoforms, analysis of the secreted proteins detected two additional bands of ∼68 and 60 kDa (Fig. 6C). Treatment with LMWHA gave a 60% increase of the 95-kDa isoform and a 20.5% decrease of the 45-kDa isoform. HMWHA gave a 55% increase of the 95-kDa isoform and a 18% decrease of the 45-kDa isoform (Fig. 6C). When RHAMM band density was adjusted for protein content (micrograms) of the loaded sample and then multiplied with total cell extract and media protein content in micrograms, an approximate ratio of 4:1 for cell versus secreted RHAMM protein was established. LMWHA treatment increased this ratio to 6:1 for cell versus secreted RHAMM protein.
FIGURE 6.
Effect of LMWHA and HMWHA on RHAMM expression at the mRNA and protein level. A, RHAMM mRNA expression in HT1080 cells treated with LMWHA and HMWHA (50 μg/ml) during 24 h was determined by real time PCR using primers specific for the RHAMM gene and normalized against GAPDH. B, expression of RHAMM protein isoforms of treated LMWHA, HMWHA, and control (M) HT1080 cells was determined by Western analysis. Densitometric analysis of the RHAMM isoform protein bands were normalized against actin and plotted. Representative blots are presented. C, expression of RHAMM protein isoforms in culture medium of HT1080 cells concentrated using 30 × 116 mm filter tubes. Densitometric analyses of the RHAMM isoform protein bands are plotted. Representative blots are presented. The results represent the average of three separate experiments. Means ± S.E. are plotted; statistical significance is as follows: *, p ≤ 0.05 compared with the respective control samples. and; **, p ≤ 0.01.
Effect of LMWHA Treatments on Cellular Distribution of RHAMM
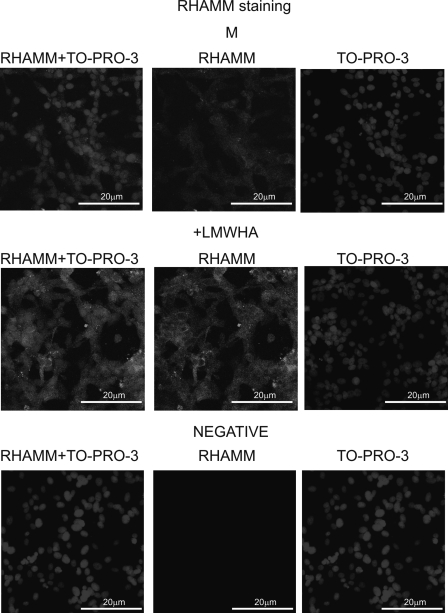
Previous studies have demonstrated that RHAMM biological functions are correlated to its cellular distribution (13–16). To clarify this point in respect to the action of LMWHA on fibrosarcoma cells, an immunofluorescence study of RHAMM distribution was conducted. When an anti-RHAMM protein was applied to HT1080 cells, a RHAMM-specific signal was found to be distributed mainly throughout the cytoplasm with a faint staining of the nuclear region (Fig. 7). LMWHA treatment strongly increased the total RHAMM-specific signal without major alterations in its distribution (Fig. 7).
FIGURE 7.
Visualization of RHAMM in HT1080 cells using immunofluorescence. RHAMM protein staining of cells and respective nuclear staining (using TO-PRO-3) were evaluated in cultures after 48 h of incubation with serum-free culture medium (M) and LMWHA (50 μg/ml). A negative control was used where the primary anti-RHAMM antibody was omitted (negative). Slides were analyzed by confocal microscopy, and pictures were taken using ×40 (with ×10 zoom).
Effect of LMWHA on Fibrosarcoma Cell Actin Organization
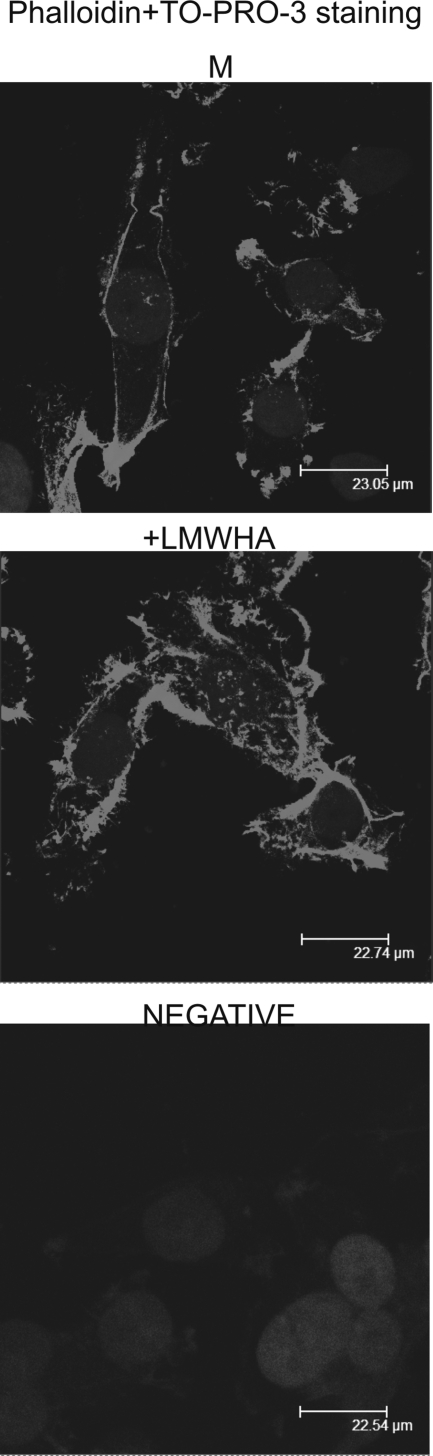
Specific cytoskeletal rearrangements of transformed tumor cells have been correlated with the efficiency of the cells to adhere and migrate (52, 53). Therefore, we examined possible effects of LMWHA on HT1080 cell actin cytoskeletal arrangement. Fibrosarcoma cells treated with LMWHA had more intense F-actin stress fibers arranged in spike-like protrusions resembling microvilli-like structures along the cell membrane (Fig. 8). This correlates well with previous studies reporting that HA chains of different sizes affect F-actin organization (54, 55).
FIGURE 8.
LMWHA affects actin fiber organization. HT1080 cells with (LMWHA) and without (M) LMWHA (50 μg/ml) treatment were seeded onto round coverslips, fixed, permeabilized, and then stained using phalloidin to visualize actin filaments. The nuclei were stained using TO-PRO-3. The signals against phalloidin and TO-PRO-3 were superimposed. Slides were analyzed by confocal microscopy, and pictures were taken using ×40.
ERK1/2 Pathway Is Obligatory for LMWHA/RHAMM-mediated HT1080 Cell Adhesion
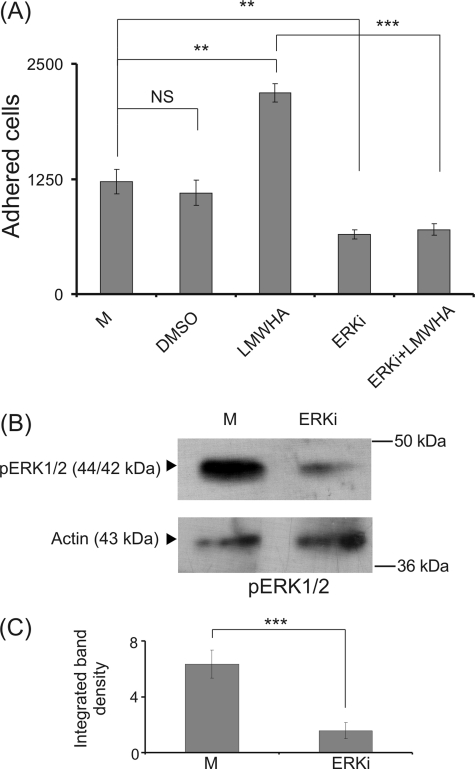
HA/RHAMM signaling has been shown to be mediated through the activation of ERK1/2, which is an important regulator of cell adhesion (56, 57). Thus, we investigated whether ERK1/2 may be a conduit through which HA regulates HT1080 cell adhesion by using a specific ERK1/2 inhibitor (ERKi). The capacity of HT1080 to adhere was diminished by the addition of the ERK inhibitor (p ≤ 0.01), therefore confirming the participation of ERK1/2 on fibrosarcoma cell adhesion (Fig. 9A). The DMSO solvent content did not affect HT1080 cell adhesion in control experiments (Fig. 9A). Noteworthy, the strong LMWHA-dependent increase in HT1080 cell adhesion was completely inhibited (p ≤ 0.001) by the ERK inhibitor (Fig. 9A). To verify the inhibitory action of ERKi on ERK1/2 phosphorylation, cells treated with ERKi were probed for pERK1/2. Western blot analysis confirmed the strong inhibitory action of ERKi (Fig. 9, B and C). These data demonstrate that ERK1/2 is a downstream effector of signaling mediated by RHAMM-LMWHA.
FIGURE 9.
Effect of ERK inhibitor (ERKi) on HT1080 fibronectin-dependent HT1080 cell adhesion. A, HT1080 cells treated with ERK inhibitor (5 μm) or LMWHA (50 μg/ml) or both for 48 h were harvested and allowed to attach (5000 cells) for 30 min on 96-well plates coated with fibronectin (5 μg/cm2). Means ± S.E. are plotted; statistical significance is as follows: **, p ≤ 0.01; ***, p ≤ 0.001 compared with the respective controls. NS, not significant. The results represent the average of three separate experiments in triplicate. B, effect of ERK inhibitor on HT1080 cell ERK1/2 phosphorylation. HT1080 cells treated with ERK inhibitor for 48 h were harvested, and equal amounts of cell extract protein were separated by PAGE and blotted with phosphorylated ERK1/2 (pERK1/2) utilizing specific antibodies. Representative blots are presented. C, densitometry analysis of the bands was normalized against actin and plotted. The results represent the average of three separate experiments. Means ± S.E. are plotted; statistical significance is as follows: ***, p ≤ 0.001 compared with the respective control samples.
LMWHA/RHAMM Signaling Enhances ERK1/2 Activation
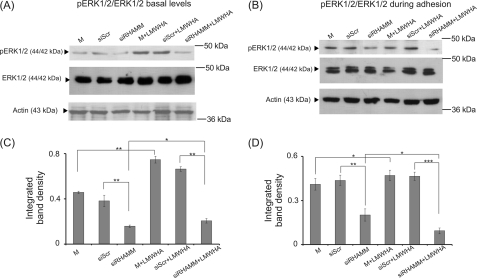
Previously, RHAMM has been reported to interact directly with ERK1/2, thereby enhancing its phosphorylation and mediating cell locomotion (15, 26, 34, 35). Therefore, we examined ERK1/2 basal and adhesion-dependent phosphorylation in si-RHAMM-treated, RHAMM-deficient HT1080 cells. Western blots revealed that both basal and adhesion-dependent phosphorylation levels of ERK1/2 were strongly decreased in RHAMM-deficient HT1080 cells (56 and 46%, respectively) (Fig. 10). In continuation, we investigated the effect of HA in combination with RHAMM signaling on ERK1/2 activation. RHAMM inhibition markedly blocked LMWHA-dependent induction of both basal and adhesion-dependent phosphorylation levels of ERK1/2 (54 and 80%, respectively) (Fig. 10). These results provide strong evidence that an LMWHA-RHAMM-ERK1/2 signaling axis is present in HT1080 fibrosarcoma cells.
FIGURE 10.
Activation of ERK1/2 in HT1080 cells. Representative Western blot analyses for phosphorylation of ERK1/2 (pERK) are shown for transfected siRHAMM, siScr cells, or nontransfected cells. A, at basal levels after 48 h of treatment with serum-free medium (M, siSRC, siRHAMM) and LMWHA (50 μg/ml); B, during adhesion onto fibronectin using the same controls and treatments. C and D, densitometry analyses of the bands are shown at basal levels (C) and during adhesion (D), respectively, with the ratio of pERK to total ERK1/2 normalized against actin. The results represent the average of three separate experiments in triplicate. Means ± S.E. are plotted; statistical significance is as follows: ***, p ≤ 0.001; **, p ≤ 0.01; *, p ≤ 0.05 compared with the respective control samples.
Role of FAK in LMWHA-RHAMM-mediated Fibrosarcoma Cell Adhesion
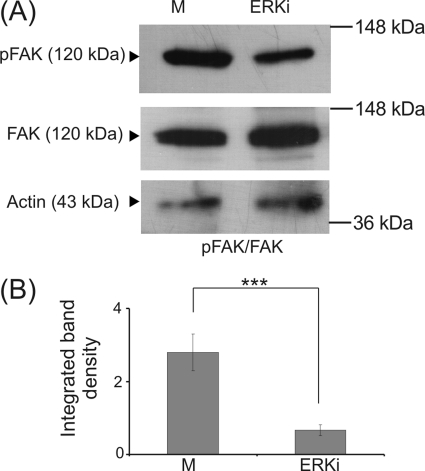
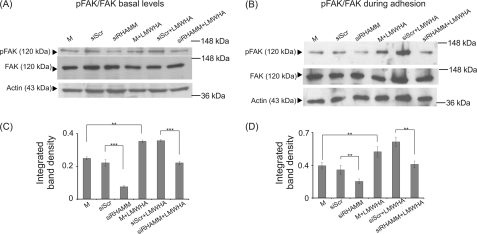
FAK, a major component of focal adhesion complexes, has been shown to be activated by ERK signaling (58). Moreover, specific interactions of RHAMM and FAK have been correlated to the regulation of cell motility (34, 35, 56). Therefore, we examined the possible participation of FAK in the proposed signaling axis. The phosphorylation of FAK, as demonstrated by Western blotting, was inhibited by treatment with the ERK inhibitor (Fig. 11A), therefore confirming a regulating role of ERK signaling on FAK activation in HT1080 cells. We also examined FAK basal and adhesion-dependent phosphorylation in RHAMM-deficient HT1080 cells. In support of our hypothesis, both basal and adhesion-dependent phosphorylation levels of FAK were strongly down-regulated (75 and 41%, respectively) in RHAMM-deficient HT1080 cells (Fig. 12). Next, we evaluated the possible effect of LMWHA-RHAMM signaling on FAK activation. Interestingly, RHAMM down-regulation completely blocked LMWHA-dependent induction of both basal and adhesion-dependent phosphorylation levels of FAK (53 and 40%, respectively) (Fig. 12). Therefore, these results show that LMWHA activates a RHAMM-ERK1/2-FAK signaling cascade to regulate fibrosarcoma cell adhesion.
FIGURE 11.
Effect of ERK inhibitor on HT1080 cell FAK (Y397) phosphorylation. HT1080 cells treated with ERK inhibitor (ERKi) for 48 h were harvested, and equal amounts of cell extract protein were separated by PAGE and blotted against phosphorylated FAK (Y397) (pFAK) and total FAK (FAK) utilizing specific antibodies. A, representative blots are shown. B, densitometry analyses of the ratio of pFAK to total FAK normalized against actin are shown. The results represent the average of three separate experiments. Means ± S.E. are plotted; statistical significance is as follows: ***, p ≤ 0.001 compared with the respective control samples.
FIGURE 12.
Activation of FAK in HT1080 cells. Representative Western blot analyses are shown for phosphorylation of FAK (pFAK) using transfected siRHAMM or siScr cells and nontransfected cells. A, at basal levels after 48 h treatment with serum-free medium (M, siSRC, siRHAMM) and LMWHA (50 μg/ml); B, during adhesion onto fibronectin, using the same controls and treatments. C and D, densitometry analyses of the ratio of pFAK to total FAK normalized against actin are shown at basal levels (C) and during adhesion (D). The results represent the average of three separate experiments. Means ± S.E. are plotted; statistical significance is as follows: ***, p ≤ 0.001, and **, p ≤ 0.01 compared with the respective control samples.
DISCUSSION
Increasing evidence indicates a crucial role of RHAMM-dependent HA signaling in tumor development and progression. In this study we show that novel LMWHA/RHAMM interactions regulate fibrosarcoma cell adhesion via ERK1/2 activation and FAK signaling.
Capable adhesion to the extracellular matrix is required for successful cancer cell invasion and metastasis. Both membrane-associated and intracellularly distributed RHAMM have been reported to affect cell motility. Specifically, HA membrane-associated RHAMM interactions have been suggested to regulate colon, breast, and prostate cancer cell adhesion ability (59–61), whereas intracellular RHAMM has been implicated in the regulation of focal adhesion turnover and in interactions with actin microfilaments and adhesion molecules like integrins (62–64). Our results demonstrate that HA specifically regulates HT1080 fibrosarcoma cell adhesion in a manner dependent on molecular weight. Thus, LMWHA (15–40 kDa) strongly increased the adhesion capacity of HT1080 cells onto fibronectin. This effect was completely abolished upon sample digestion with Streptomyces hyaluronidase verifying its strict specificity. In contrast, HMWHA (2.5 × 106 Da) significantly inhibited HT1080 cell adhesion; interestingly, HA digests had an independent inhibitory effect on fibrosarcoma cell adhesion. Differences in HA molecular weight and changes in its metabolism are well known to influence cell functions (3, 43). Recently, we have reported that HA of different molecular weights causes discriminate effects on fibrosarcoma cell migration, with exogenous HMWHA significantly decreasing HT1080 cell migration capacity, whereas treatment with LMWHA resulted in a significant stimulation of the motility of these cells (43).
The response to HA signaling, leading to changes in cell motility and invasion, was previously found to be eliminated upon inhibition of RHAMM action (15, 35, 56, 65). In our fibrosarcoma cell model system, the down-regulation of RHAMM expression led to the reduction of basal and complete inhibition of LMWHA-induced adhesion. It is worth noting that the effect of HMWHA was not attenuated in RHAMM-deficient HT1080 cells, which emphasizes the key role of HA size on RHAMM signaling. Our data demonstrated that CD44 likewise mediates the LMWHA-induced adhesion, indicating that the cooperation of both receptors modulates LMWHA signaling. The role of CD44 on tumor motility has been linked to its ability to interact with additional proteins and specifically with RHAMM (2). Although it is clear that both HA receptors can act independently, it has been suggested that their relative contributions to a certain cell action must be investigated in different cases (66). Thus, different sized HA chains have previously been shown to elucidate discriminate effects through respective RHAMM and CD44 signaling. It is noteworthy that although CD44 and RHAMM are both involved in endothelial tube formation induced by specific HA oligosaccharides, they mediate distinct angiogenic signaling pathways (54). Furthermore, in an in vitro wound healing model, HA oligosaccharides, but not native HA, were shown to stimulate endothelial cell growth and migration in a RHAMM signaling-dependent manner (67). Likewise, studies with active blocking antibodies revealed that anti-CD44 but not anti-RHAMM antibody inhibited endothelial cell adhesion to HA, whereas anti-RHAMM but not CD44 antibody blocked endothelial cell migration through the basement membrane substrate Matrigel (68).
Under monolayer culture conditions, RHAMM isoforms were distributed both in the cell layer and the medium compartment of HT1080 cells. Immunofluorescence revealed that RHAMM isoforms were mainly localized in the HT1080 cytoplasm. RHAMM was originally identified as a 56–58-kDa HA-binding protein present in the supernatant of murine fibroblasts and fibrosarcoma cell lines (69). Other isoforms recognized had molecular masses from 52 to 125 kDa (17, 36, 70–72). Both cell surface RHAMM expression and intracellular RHAMM isoforms were described to mediate HA signaling, either by interaction with CD44 or activation of intracellular signaling molecules (10).
Interestingly, RHAMM expression in HT1080 cells was greatly enhanced by addition of LMWHA at both various protein isoforms and total transcript levels. HMWHA was also, albeit modestly, found to up-regulate RHAMM expression. Immunofluorescence confirmed the marked LMWHA-stimulated up-regulation of RHAMM expression and localized the increase mainly to HT1080 cell cytoplasm regions. The exact mechanism through which LMWHA enhances RHAMM expression and distribution to the cytoplasm is currently being investigated. We propose the existence of a putative feedback regulation between HA production and respective receptor expression. Importantly, RHAMM expression and HA metabolism have been shown, through so far unknown mechanisms, to be strongly up-regulated in cancer cells as compared with their normal counterparts (2).
The oncogenic effects of RHAMM are suggested to be perpetrated through its HA-binding properties (35). Intracellular forms of RHAMM have been localized to interphase microtubules, mitotic spindles, centrosomes, and the nucleus (14, 21, 22, 71, 73, 74) suggesting that RHAMM intracellular associations have the potential to affect cell transformation and tumor progression (74, 75). Importantly, a RHAMM intracellular form has been shown to directly associate with ERK1 kinase and Src (4, 15, 36) resulting in modulations of their signaling pathways. Furthermore, HA has been found to modulate the phosphorylation of (p42/44) ERK1/2, known to mediate key cell functions, including adhesion (56, 57). In this study, the addition of the specific ERK inhibitor during HT1080 cell adhesion strongly decreased both basal and LMWHA-induced fibrosarcoma cell adhesion capacity, thereby demonstrating that ERK1/2 is a downstream effector of signals originating from RHAMM/HA interactions.
Activated ERK was recently found to be localized at focal adhesion complexes and suggested to interact with FAK (76). FAK is a cytoplasmic kinase that interacts with integrins, participates in actin polymerization, and is involved in mediating ECM signals to changes in tumor cell migration and adhesion (77–79). Furthermore, Vomastek et al. (76) report that ERK1/2 activation levels were inversely correlated to the size of focal adhesion sites, whereas a direct correlation was established to FAK activation, rate of focal adhesion disassembly, and fibroblast motility. The above data support the hypothesis that the activation of FAK promotes focal contact formation resulting in more efficient fibrosarcoma cell adhesion. Likewise, a study in prostate cancer cells showed that the regulation of FAK by ERK1/2 is required for the aggressive cell phenotype and revealed the interdependence of FAK and ERK1/2 signaling in efficient cancer cell invasion (58). In our study the utilization of the specific ERKi inhibitor resulted in a strong down-regulation of FAK activation in HT1080 cells identifying ERK1/2 as a FAK upstream effector.
Interestingly, LMWHA induced thicker F-actin stress fiber staining. Similar actin cytoskeleton reorganization was affected in dermal fibroblasts and endothelial cells by different sized HA chains (54, 55). These modifications of actin fiber organization correlate well with the proposed enhancement of focal contact formations (77).
LMWHA/RHAMM signaling was previously shown to support FAK activation and filopodia formation resulting in proliferative/migratory phenotype of esophageal cancer cells (50). In this study, basal and adhesion-dependent activation of FAK in HT1080 cells was increased with the addition of LMWHA. However, when RHAMM total expression was inhibited, HT1080 cell activation of FAK and ERK1/2 was diminished even after addition of LMWHA, therefore demonstrating the obligatory participation of RHAMM. Our results correlate well with previous studies that demonstrated that HA induced tyrosine phosphorylation of p125 (FAK), paxillin, and p42/44 ERK in human endothelial cells, which was blocked by an anti-RHAMM antibody (56). Because we have recently also demonstrated that LMWHA enhances HT1080 cell migration (43), it appears that this HA response dependent on RHAMM signaling stimulates an aggressive phenotype of fibrosarcoma cell motility.
In conclusion, a novel RHAMM-dependent but cooperative with CD44 mechanism that regulates fibrosarcoma cell adhesion has been revealed in this study. It is induced exclusively by LMWHA, involves the participation of ERK1/2/FAK downstream effectors, and may be of significance in molecularly targeted therapy of the disease.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and an additional reference.
- ECM
- extracellular matrix
- RHAMM
- receptor for hyaluronic acid-mediated motility
- LMWHA
- low molecular weight HA
- HMWHA
- high molecular weight HA
- FAK
- focal adhesion kinase
- CS
- chondroitin sulfate
- FACE
- fluorophore-assisted carbohydrate electrophoresis
- CE
- capillary electrophoresis
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid
- siScr
- scramble interfering RNA.
REFERENCES
- 1. Tammi R. H., Kultti A., Kosma V. M., Pirinen R., Auvinen P., Tammi M. I. (2008) Semin. Cancer Biol. 18, 288–295 [DOI] [PubMed] [Google Scholar]
- 2. Hamilton S. R., Fard S. F., Paiwand F. F., Tolg C., Veiseh M., Wang C., McCarthy J. B., Bissell M. J., Koropatnick J., Turley E. A. (2007) J. Biol. Chem. 282, 16667–16680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stern R., Asari A. A., Sugahara K. N. (2006) Eur. J. Cell Biol. 85, 699–715 [DOI] [PubMed] [Google Scholar]
- 4. Turley E. A., Noble P. W., Bourguignon L. Y. (2002) J. Biol. Chem. 277, 4589–4592 [DOI] [PubMed] [Google Scholar]
- 5. Toole B. P., Hascall V. C. (2002) Am. J. Pathol. 161, 745–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Price R. D., Berry M. G., Navsaria H. A. (2007) J. Plast. Reconstr. Aesthet. Surg. 60, 1110–1119 [DOI] [PubMed] [Google Scholar]
- 7. Toole B. P., Slomiany M. G. (2008) Drug Resist. Updat. 11, 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sherman L., Sleeman J., Herrlich P., Ponta H. (1994) Curr. Opin. Cell Biol. 6, 726–733 [DOI] [PubMed] [Google Scholar]
- 9. Toole B. P. (1990) Curr. Opin. Cell Biol. 2, 839–844 [DOI] [PubMed] [Google Scholar]
- 10. Cheung W. F., Cruz T. F., Turley E. A. (1999) Biochem. Soc. Trans. 27, 135–142 [DOI] [PubMed] [Google Scholar]
- 11. Aruffo A., Stamenkovic I., Melnick M., Underhill C. B., Seed B. (1990) Cell 61, 1303–1313 [DOI] [PubMed] [Google Scholar]
- 12. Lesley J., Hyman R. (1998) Front. Biosci. 3, d616–630 [DOI] [PubMed] [Google Scholar]
- 13. Assmann V., Marshall J. F., Fieber C., Hofmann M., Hart I. R. (1998) J. Cell Sci. 111, 1685–1694 [DOI] [PubMed] [Google Scholar]
- 14. Entwistle J., Hall C. L., Turley E. A. (1996) J. Cell. Biochem. 61, 569–577 [DOI] [PubMed] [Google Scholar]
- 15. Zhang S., Chang M. C., Zylka D., Turley S., Harrison R., Turley E. A. (1998) J. Biol. Chem. 273, 11342–11348 [DOI] [PubMed] [Google Scholar]
- 16. Groen A. C., Cameron L. A., Coughlin M., Miyamoto D. T., Mitchison T. J., Ohi R. (2004) Curr. Biol. 14, 1801–1811 [DOI] [PubMed] [Google Scholar]
- 17. Hardwick C., Hoare K., Owens R., Hohn H. P., Hook M., Moore D., Cripps V., Austen L., Nance D. M., Turley E. A. (1992) J. Cell Biol. 117, 1343–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Turley E. A. (1992) Cancer Metastasis Rev. 11, 21–30 [DOI] [PubMed] [Google Scholar]
- 19. Klewes L., Turley E. A., Prehm P. (1993) Biochem. J. 290, 791–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nedvetzki S., Gonen E., Assayag N., Reich R., Williams R. O., Thurmond R. L., Huang J. F., Neudecker B. A., Wang F. S., Wang F. S., Turley E. A., Naor D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 18081–18086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Assmann V., Jenkinson D., Marshall J. F., Hart I. R. (1999) J. Cell Sci. 112, 3943–3954 [DOI] [PubMed] [Google Scholar]
- 22. Maxwell C. A., Keats J. J., Belch A. R., Pilarski L. M., Reiman T. (2005) Cancer Res. 65, 850–860 [PubMed] [Google Scholar]
- 23. Maxwell C. A., McCarthy J., Turley E. (2008) J. Cell Sci. 121, 925–932 [DOI] [PubMed] [Google Scholar]
- 24. Mohapatra S., Yang X., Wright J. A., Turley E. A., Greenberg A. H. (1996) J. Exp. Med. 183, 1663–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Assmann V., Gillett C. E., Poulsom R., Ryder K., Hart I. R., Hanby A. M. (2001) J. Pathol. 195, 191–196 [DOI] [PubMed] [Google Scholar]
- 26. Wang C., Thor A. D., Moore D. H., 2nd, Zhao Y., Kerschmann R., Stern R., Watson P. H., Turley E. A. (1998) Clin. Cancer Res. 4, 567–576 [PubMed] [Google Scholar]
- 27. Hus I., Schmitt M., Tabarkiewicz J., Radej S., Wojas K., Bojarska-Junak A., Schmitt A., Giannopoulos K., Dmoszyńska A., Roliński J. (2008) Leukemia 22, 1007–1017 [DOI] [PubMed] [Google Scholar]
- 28. Zlobec I., Terracciano L., Tornillo L., Günthert U., Vuong T., Jass J. R., Lugli A. (2008) Gut 57, 1413–1419 [DOI] [PubMed] [Google Scholar]
- 29. Tolg C., Poon R., Fodde R., Turley E. A., Alman B. A. (2003) Oncogene 22, 6873–6882 [DOI] [PubMed] [Google Scholar]
- 30. Rein D. T., Roehrig K., Schöndorf T., Lazar A., Fleisch M., Niederacher D., Bender H. G., Dall P. (2003) J. Cancer Res. Clin. Oncol. 129, 161–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Akiyama Y., Jung S., Salhia B., Lee S., Hubbard S., Taylor M., Mainprize T., Akaishi K., van Furth W., Rutka J. T. (2001) J. Neurooncol. 53, 115–127 [DOI] [PubMed] [Google Scholar]
- 32. Gust K. M., Hofer M. D., Perner S. R., Kim R., Chinnaiyan A. M., Varambally S., Moller P., Rinnab L., Rubin M. A., Greiner J., Schmitt M., Kuefer R., Ringhoffer M. (2009) Neoplasia 11, 956–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ahrens T., Assmann V., Fieber C., Termeer C., Herrlich P., Hofmann M., Simon J. C. (2001) J. Invest. Dermatol. 116, 93–101 [DOI] [PubMed] [Google Scholar]
- 34. Hall C. L., Wang C., Lange L. A., Turley E. A. (1994) J. Cell Biol. 126, 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hall C. L., Yang B., Yang X., Zhang S., Turley M., Samuel S., Lange L. A., Wang C., Curpen G. D., Savani R. C., Greenberg A. H., Turley E. A. (1995) Cell 82, 19–26 [DOI] [PubMed] [Google Scholar]
- 36. Hall C. L., Lange L. A., Prober D. A., Zhang S., Turley E. A. (1996) Oncogene 13, 2213–2224 [PubMed] [Google Scholar]
- 37. Hall C. L., Collis L. A., Bo A. J., Lange L., McNicol A., Gerrard J. M., Turley E. A. (2001) Matrix Biol. 20, 183–192 [DOI] [PubMed] [Google Scholar]
- 38. Lin S. L., Chang D., Chiang A., Ying S. Y. (2008) Carcinogenesis 29, 282–290 [DOI] [PubMed] [Google Scholar]
- 39. Lin S. L., Chang D., Ying S. Y. (2007) Carcinogenesis 28, 310–320 [DOI] [PubMed] [Google Scholar]
- 40. Seger R., Krebs E. G. (1995) FASEB J. 9, 726–735 [PubMed] [Google Scholar]
- 41. Piepkorn M., Hovingh P., Linker A. (1988) J. Cell. Physiol. 135, 189–199 [DOI] [PubMed] [Google Scholar]
- 42. Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. (2003) Science 302, 1704–1709 [DOI] [PubMed] [Google Scholar]
- 43. Berdiaki A., Nikitovic D., Tsatsakis A., Katonis P., Karamanos N. K., Tzanakakis G. N. (2009) Biochim. Biophys. Acta 1790, 1258–1265 [DOI] [PubMed] [Google Scholar]
- 44. Karamanos N. K., Vanky P., Syrokou A., Hjerpe A. (1995) Anal. Biochem. 225, 220–230 [DOI] [PubMed] [Google Scholar]
- 45. Militsopoulou M., Lamari F. N., Hjerpe A., Karamanos N. K. (2002) Electrophoresis 23, 1104–1109 [DOI] [PubMed] [Google Scholar]
- 46. Calabro A., Benavides M., Tammi M., Hascall V. C., Midura R. J. (2000) Glycobiology 10, 273–281 [DOI] [PubMed] [Google Scholar]
- 47. Karousou E. G., Militsopoulou M., Porta G., De Luca G., Hascall V. C., Passi A. (2004) Electrophoresis 25, 2919–2925 [DOI] [PubMed] [Google Scholar]
- 48. Malavaki C. J., Asimakopoulou A. P., Lamari F. N., Theocharis A. D., Tzanakakis G. N., Karamanos N. K. (2008) Anal. Biochem. 374, 213–220 [DOI] [PubMed] [Google Scholar]
- 49. Rasheed S., Nelson-Rees W. A., Toth E. M., Arnstein P., Gardner M. B. (1974) Cancer 33, 1027–1033 [DOI] [PubMed] [Google Scholar]
- 50. Twarock S., Tammi M. I., Savani R. C., Fischer J. W. (2010) J. Biol. Chem. 285, 23276–23284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gares S. L., Pilarski L. M. (2000) Dev. Immunol. 7, 209–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Geiger T., Geiger B. (2010) Semin. Cancer Biol. 20, 146–152 [DOI] [PubMed] [Google Scholar]
- 53. Chalkiadaki G., Nikitovic D., Katonis P., Berdiaki A., Tsatsakis A., Kotsikogianni I., Karamanos N. K., Tzanakakis G. N. (2011) Cancer Lett., in press [DOI] [PubMed] [Google Scholar]
- 54. Matou-Nasri S., Gaffney J., Kumar S., Slevin M. (2009) Int. J. Oncol. 35, 761–773 [DOI] [PubMed] [Google Scholar]
- 55. Boraldi F., Croce M. A., Quaglino D., Sammarco R., Carnevali E., Tiozzo R., Pasquali-Ronchetti I. (2003) Tissue Cell 35, 37–45 [DOI] [PubMed] [Google Scholar]
- 56. Lokeshwar V. B., Selzer M. G. (2000) J. Biol. Chem. 275, 27641–27649 [DOI] [PubMed] [Google Scholar]
- 57. Al-Ayoubi A., Tarcsafalvi A., Zheng H., Sakati W., Eblen S. T. (2008) J. Cell. Biochem. 105, 875–884 [DOI] [PubMed] [Google Scholar]
- 58. Johnson T. R., Khandrika L., Kumar B., Venezia S., Koul S., Chandhoke R., Maroni P., Donohue R., Meacham R. B., Koul H. K. (2008) Mol. Cancer Res. 6, 1639–1648 [DOI] [PubMed] [Google Scholar]
- 59. Laurich C., Wheeler M. A., Iida J., Neudauer C. L., McCarthy J. B., Bullard K. M. (2004) J. Surg. Res. 122, 70–74 [DOI] [PubMed] [Google Scholar]
- 60. Simpson M. A., Wilson C. M., Furcht L. T., Spicer A. P., Oegema T. R., Jr., McCarthy J. B. (2002) J. Biol. Chem. 277, 10050–10057 [DOI] [PubMed] [Google Scholar]
- 61. Herrera-Gayol A., Jothy S. (2001) Int. J. Exp. Pathol. 82, 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nagy J. I., Hossain M. Z., Lynn B. D., Curpen G. E., Yang S., Turley E. A. (1996) Cell Growth Differ. 7, 745–751 [PubMed] [Google Scholar]
- 63. Gares S. L., Giannakopoulos N., MacNeil D., Faull R. J., Pilarski L. M. (1998) J. Leukocyte Biol. 64, 781–790 [DOI] [PubMed] [Google Scholar]
- 64. Gares S. L., Pilarski L. M. (1999) Scand. J. Immunol. 50, 626–634 [DOI] [PubMed] [Google Scholar]
- 65. Tolg C., Hamilton S. R., Nakrieko K. A., Kooshesh F., Walton P., McCarthy J. B., Bissell M. J., Turley E. A. (2006) J. Cell Biol. 175, 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Park J. B., Kwak H. J., Lee S. H. (2008) Cell. Adh. Migr, 2, 202–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gao F., Yang C. X., Mo W., Liu Y. W., He Y. Q. (2008) Clin. Invest. Med. 31, E106–116 [DOI] [PubMed] [Google Scholar]
- 68. Savani R. C., Cao G., Pooler P. M., Zaman A., Zhou Z., DeLisser H. M. (2001) J. Biol. Chem. 276, 36770–36778 [DOI] [PubMed] [Google Scholar]
- 69. Turley E. A., Moore D., Hayden L. J. (1987) Biochemistry 26, 2997–3005 [DOI] [PubMed] [Google Scholar]
- 70. Entwistle J., Zhang S., Yang B., Wong C., Li Q., Hall C. L., Mowat M., Greenberg A. H., Turley E. A. (1995) Gene 163, 233–238 [DOI] [PubMed] [Google Scholar]
- 71. Hofmann M., Fieber C., Assmann V., Göttlicher M., Sleeman J., Plug R., Howells N., von Stein O., Ponta H., Herrlich P. (1998) J. Cell Sci. 111, 1673–1684 [DOI] [PubMed] [Google Scholar]
- 72. Fieber C., Plug R., Sleeman J., Dall P., Ponta H., Hofmann M. (1999) Gene 226, 41–50 [DOI] [PubMed] [Google Scholar]
- 73. Maxwell C. A., Keats J. J., Crainie M., Sun X., Yen T., Shibuya E., Hendzel M., Chan G., Pilarski L. M. (2003) Mol. Biol. Cell 14, 2262–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Joukov V., Groen A. C., Prokhorova T., Gerson R., White E., Rodriguez A., Walter J. C., Livingston D. M. (2006) Cell 127, 539–552 [DOI] [PubMed] [Google Scholar]
- 75. Pujana M. A., Han J. D., Starita L. M., Stevens K. N., Tewari M., Ahn J. S., Rennert G., Moreno V., Kirchhoff T., Gold B., Assmann V., Elshamy W. M., Rual J. F., Levine D., Rozek L. S., Gelman R. S., Gunsalus K. C., Greenberg R. A., Sobhian B., Bertin N., Venkatesan K., Ayivi-Guedehoussou N., Solé X., Hernández P., Lázaro C., Nathanson K. L., Weber B. L., Cusick M. E., Hill D. E., Offit K., Livingston D. M., Gruber S. B., Parvin J. D., Vidal M. (2007) Nat. Genet. 39, 1338–1349 [DOI] [PubMed] [Google Scholar]
- 76. Vomastek T., Iwanicki M. P., Schaeffer H. J., Tarcsafalvi A., Parsons J. T., Weber M. J. (2007) Mol. Cell. Biol. 27, 8296–8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schaller M. D., Borgman C. A., Cobb B. S., Vines R. R., Reynolds A. B., Parsons J. T. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 5192–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Meng X. N., Jin Y., Yu Y., Bai J., Liu G. Y., Zhu J., Zhao Y. Z., Wang Z., Chen F., Lee K. Y., Fu S. B. (2009) Br. J. Cancer 101, 327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. McLean G. W., Carragher N. O., Avizienyte E., Evans J., Brunton V. G., Frame M. C. (2005) Nat. Rev. Cancer 5, 505–515 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.