Abstract
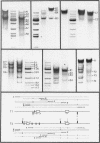
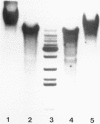
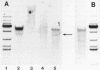
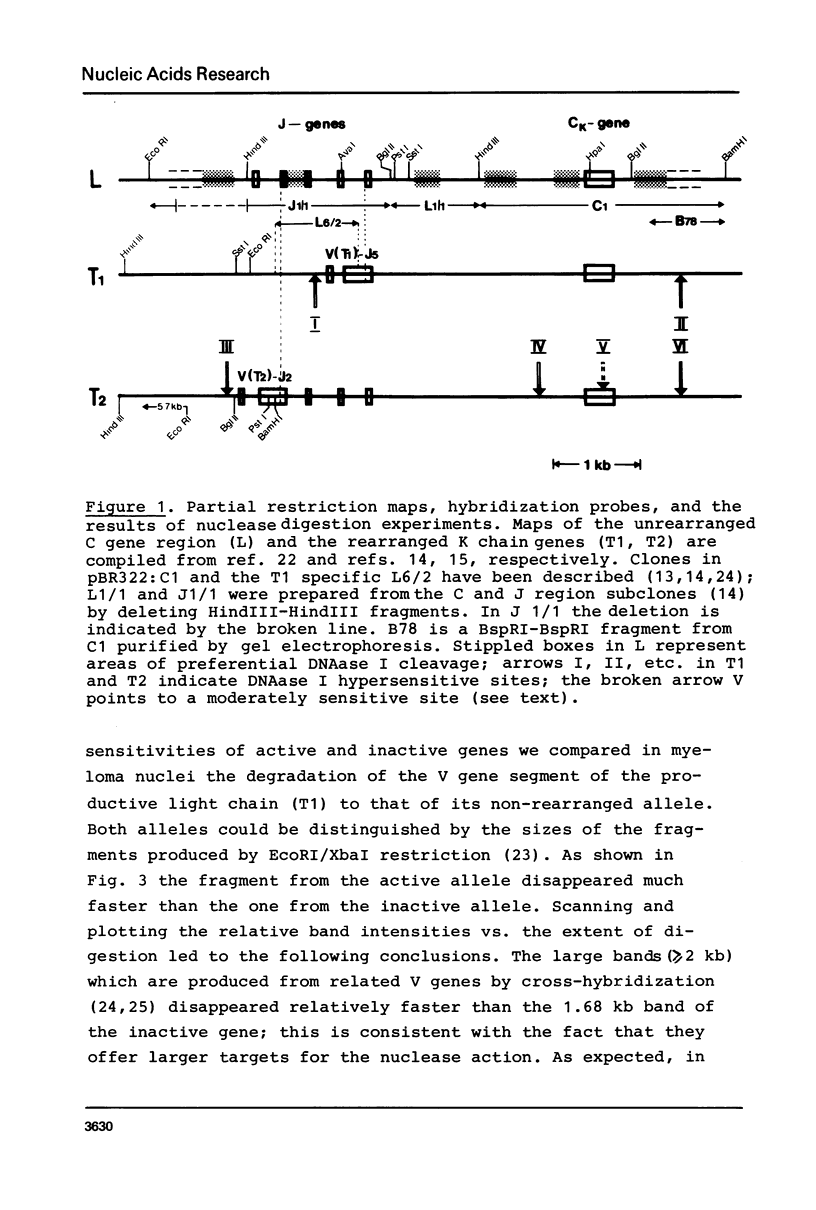
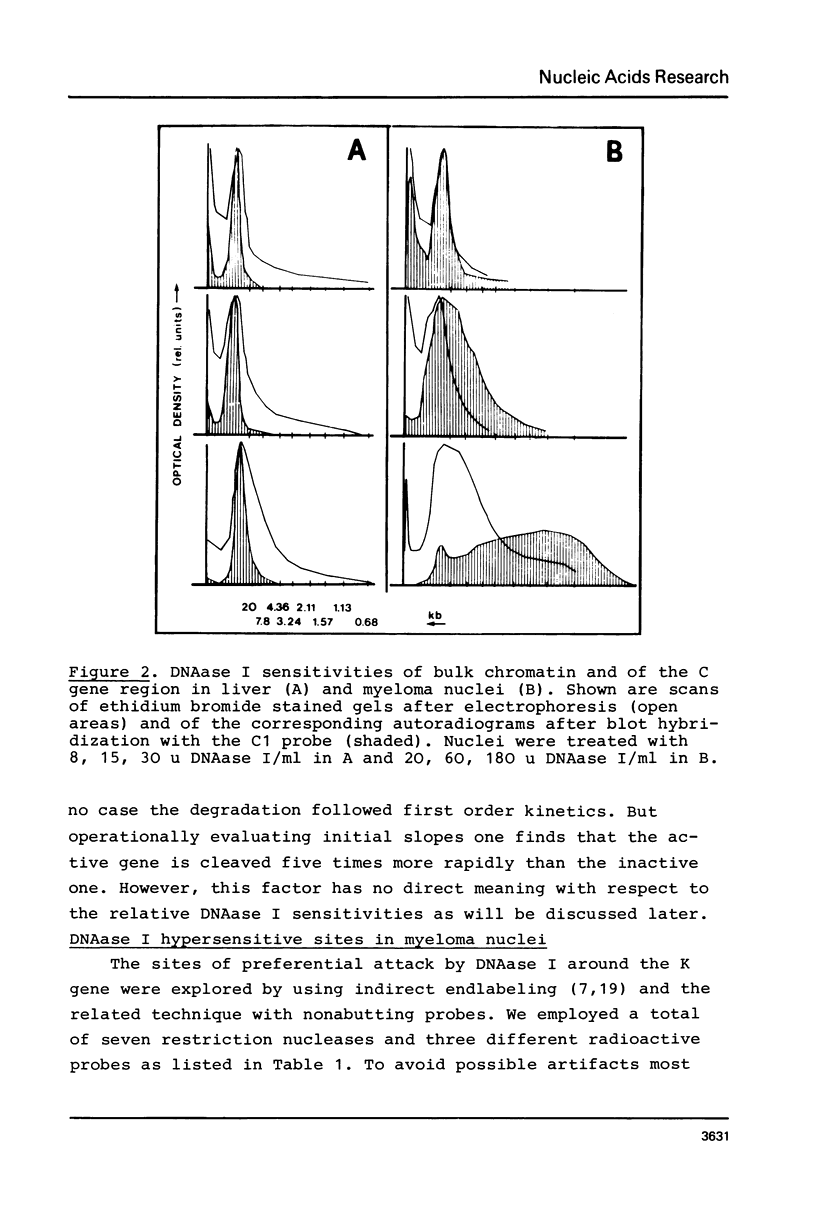
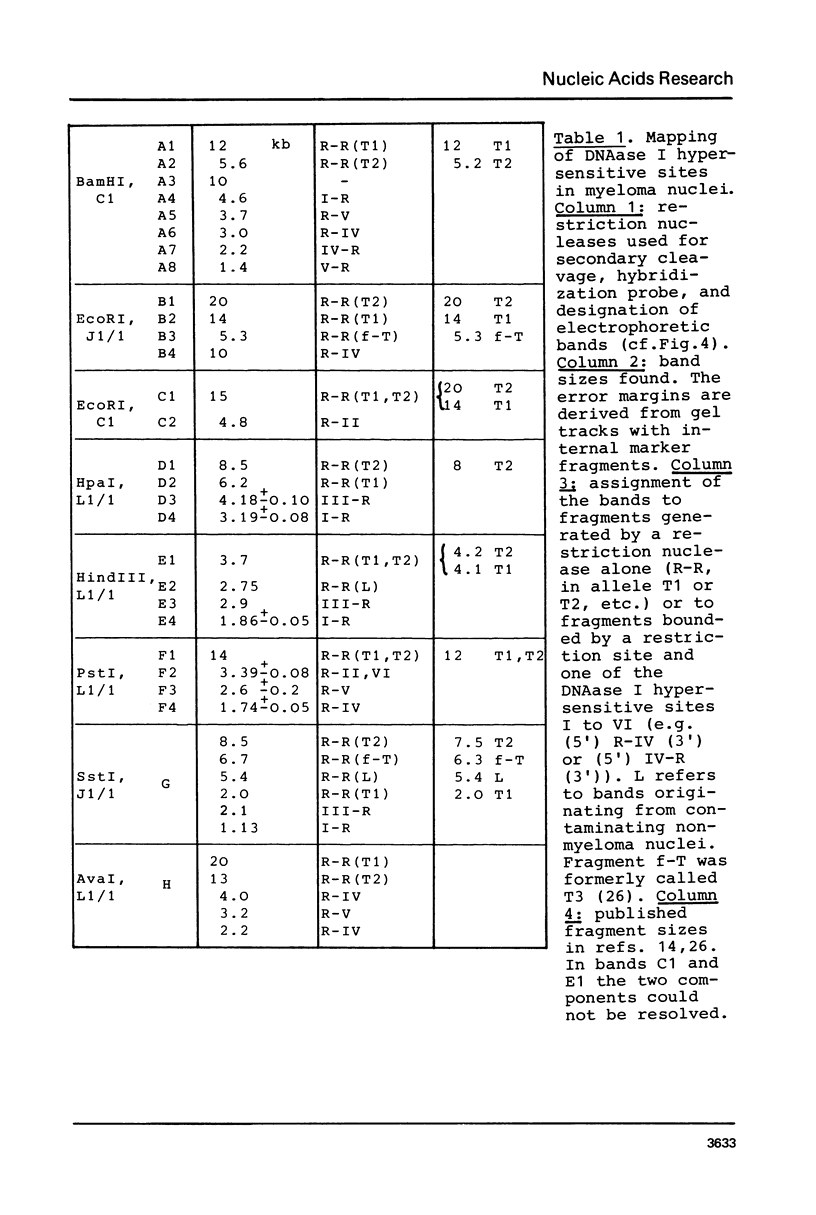
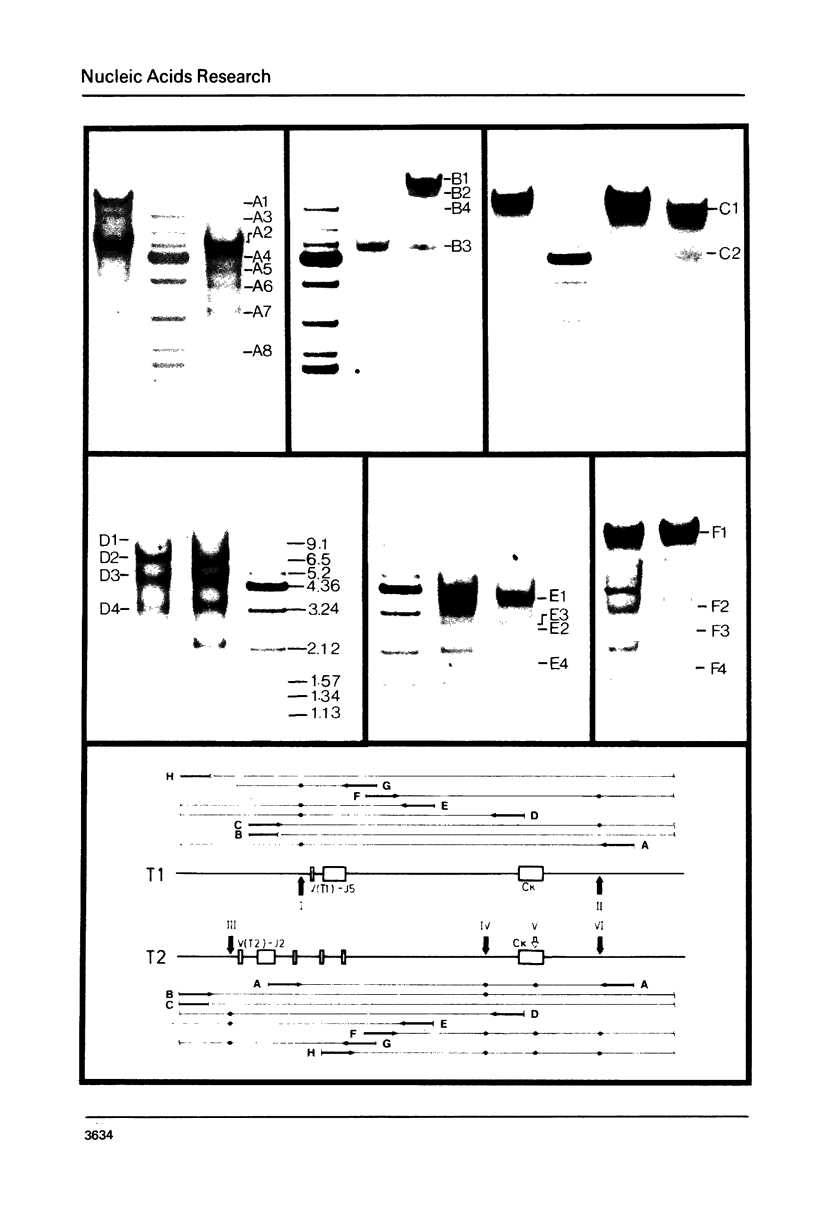
In mouse myeloma T the productive kappa light chain gene differs from its aberrantly rearranged allele in the patterns of DNAase I hypersensitive sites. In the region of the alleles where they are identical in sequence they have one site in common which lies 0.8 kb downstream of the coding region; but two sites upstream of and within the C gene segment (2) are found only on the non-productive allele. Within the region of different sequences both alleles have analogously located DNAase I hypersensitive sites; they lie 0.15 kb upstream of the respective leader segments and cover putative promoter sequences. Only one of the six DNAase I hypersensitive sites is also very sensitive towards micrococcal nuclease due to its particular DNA sequence. The non-rearranged gene studied in liver nuclei has no DNAase I hypersensitive sites but is preferentially cleaved in A/T rich regions.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenburger W., Neumaier P. S., Steinmetz M., Zachau H. G. DNA sequence of the constant gene region of the mouse immunoglobulin kappa chain. Nucleic Acids Res. 1981 Feb 25;9(4):971–981. doi: 10.1093/nar/9.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenburger W., Steinmetz M., Zachau H. G. Functional and non-functional joining in immunoglobulin light chain genes of a mouse myeloma. Nature. 1980 Oct 16;287(5783):603–607. doi: 10.1038/287603a0. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Bryan P. N., Hofstetter H., Birnstiel M. L. Nucleosome arrangement on tRNA genes of Xenopus laevis. Cell. 1981 Dec;27(3 Pt 2):459–466. doi: 10.1016/0092-8674(81)90387-1. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Lomonossoff G. P., Laskey R. A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Hood L. Allelic exclusion and nonproductive immunoglobulin gene rearrangements. Cell. 1981 Apr;24(1):1–3. doi: 10.1016/0092-8674(81)90492-x. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7102–7106. doi: 10.1073/pnas.77.12.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junowicz E., Spencer J. H. Studies on bovine pancreatic deoxyribonuclease A. II. The effect of different bivalent metals on the specificity of degradation of DNA. Biochim Biophys Acta. 1973 Jun 8;312(1):85–102. [PubMed] [Google Scholar]
- Loening U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem J. 1967 Jan;102(1):251–257. doi: 10.1042/bj1020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Max E. E., Maizel J. V., Jr, Leder P. The nucleotide sequence of a 5.5-kilobase DNA segment containing the mouse kappa immunoglobulin J and C region genes. J Biol Chem. 1981 May 25;256(10):5116–5120. [PubMed] [Google Scholar]
- Max E. E., Seidman J. G., Leder P. Sequences of five potential recombination sites encoded close to an immunoglobulin kappa constant region gene. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3450–3454. doi: 10.1073/pnas.76.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Pech M., Höchtl J., Schnell H., Zachau H. G. Differences between germ-line and rearranged immunoglobulin V kappa coding sequences suggest a localized mutation mechanism. Nature. 1981 Jun 25;291(5817):668–670. doi: 10.1038/291668a0. [DOI] [PubMed] [Google Scholar]
- Pfeiffer W., Zachau H. G. Accessibility of expressed and non-expressed genes to a restriction nuclease. Nucleic Acids Res. 1980 Oct 24;8(20):4621–4638. doi: 10.1093/nar/8.20.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H., Hüppi K., Heinrich G., Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979 Jul 26;280(5720):288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Altenburger W., Zachau H. G. A rearranged DNA sequence possibly related to the translocation of immunoglobulin gene segments. Nucleic Acids Res. 1980 Apr 25;8(8):1709–1720. doi: 10.1093/nar/8.8.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Höchtl J., Schnell H., Gebhard W., Zachau H. G. Cloning of V region fragments from mouse liver DNA and localization of repetitive DNA sequences in the vicinity of immunoglobulin gene segments. Nucleic Acids Res. 1980 Apr 25;8(8):1721–1729. doi: 10.1093/nar/8.8.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Zachau H. G. Two rearranged immunoglobulin kappa light chain genes in one mouse myeloma. Nucleic Acids Res. 1980 Apr 25;8(8):1693–1707. doi: 10.1093/nar/8.8.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storb U., Wilson R., Selsing E., Walfield A. Rearranged and germline immunoglobulin kappa genes: different states of DNase I sensitivity of constant kappa genes in immunocompetent and nonimmune cells. Biochemistry. 1981 Feb 17;20(4):990–996. doi: 10.1021/bi00507a053. [DOI] [PubMed] [Google Scholar]
- Van Ness B. G., Weigert M., Coleclough C., Mather E. L., Kelley D. E., Perry R. P. Transcription of the unrearranged mouse C kappa locus: sequence of the initiation region and comparison of activity with a rearranged V kappa-C kappa gene. Cell. 1981 Dec;27(3 Pt 2):593–602. doi: 10.1016/0092-8674(81)90401-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weischet W. O., Allen J. R., Riedel G., Van Holde K. E. The effects of salt concentration and H-1 depletion on the digestion of calf thymus chromatin by micrococcal nuclease. Nucleic Acids Res. 1979;6(5):1843–1862. doi: 10.1093/nar/6.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]