Background: Regulator of G protein signaling 14 (RGS14) is a G protein regulatory (GPR) protein that participates in unconventional G protein signaling independent of G protein-coupled receptors (GPCRs).
Results: RGS14 forms regulated complexes with GPCRs in live cells.
Conclusion: RGS14 integrates unconventional and conventional GPCR-dependent G protein signaling pathways.
Significance: GPR proteins appear to be at the nexus of divergent G protein signaling pathways.
Keywords: G protein-coupled Receptors (GPCR), G Proteins, Guanine Nucleotide Exchange Factor (GEF), Heterotrimeric G Proteins, RGS Proteins, GPR Motif, Receptor Signaling Complex, Signal Transduction
Abstract
Regulator of G protein Signaling 14 (RGS14) is a multifunctional scaffolding protein that integrates both conventional and unconventional G protein signaling pathways. Like other RGS (regulator of G protein signaling) proteins, RGS14 acts as a GTPase accelerating protein to terminate conventional Gαi/o signaling. However, unlike other RGS proteins, RGS14 also contains a G protein regulatory/GoLoco motif that specifically binds Gαi1/3-GDP in cells and in vitro. The non-receptor guanine nucleotide exchange factor Ric-8A can bind and act on the RGS14·Gαi1-GDP complex to play a role in unconventional G protein signaling independent of G protein-coupled receptors (GPCRs). Here we demonstrate that RGS14 forms a Gαi/o-dependent complex with a Gi-linked GPCR and that this complex is regulated by receptor agonist and Ric-8A (resistance to inhibitors of cholinesterase-8A). Using live cell bioluminescence resonance energy transfer, we show that RGS14 functionally associates with the α2A-adrenergic receptor (α2A-AR) in a Gαi/o-dependent manner. This interaction is markedly disrupted after receptor stimulation by the specific agonist UK14304, suggesting complex dissociation or rearrangement. Agonist-mediated dissociation of the RGS14·α2A-AR complex occurs in the presence of Gαi/o but not Gαs or Gαq. Unexpectedly, RGS14 does not dissociate from Gαi1 in the presence of stimulated α2A-AR, suggesting preservation of RGS14·Gαi1 complexes after receptor activation. However, Ric-8A facilitates dissociation of both the RGS14·Gαi1 complex and the Gαi1-dependent RGS14·α2A-AR complex after receptor activation. Together, these findings indicate that RGS14 can form complexes with GPCRs in cells that are dependent on Gαi/o and that these RGS14·Gαi1·GPCR complexes may be substrates for other signaling partners such as Ric-8A.
Introduction
Established models of G protein signaling suggest that heterotrimeric G proteins (Gαβγ subunits) are linked to specific G protein-coupled receptors (GPCRs),3 and that these receptors act as guanine nucleotide exchange factors (GEFs) toward the Gα subunit to promote nucleotide exchange and downstream signaling events (1, 2). The regulators of G protein signaling (RGS) proteins act as GTPase accelerating proteins on the activated Gα subunit, catalyzing GTP hydrolysis to terminate G protein signaling (3–5).
Recent studies have explored novel unconventional G protein signaling pathways involved with cell division and synaptic signaling/plasticity that can operate independently of GPCRs (6–13). The hallmark of these unconventional G protein pathways are signaling complexes involving Gα-GDP bound to proteins containing one or more G protein regulatory (GPR) motifs. Resistance to inhibitors of cholinesterase 8A (Ric-8A) is a cytosolic GEF that directly promotes nucleotide exchange on Gαi, Gαo, and Gαq subunits in unconventional G protein signaling (14). Ric-8A also recognizes, binds, and regulates the formation/dissociation of some GPR·Gαi1-GDP complexes, such as AGS3·Gαi1-GDP, LGN·Gαi1-GDP, and RGS14·Gαi1-GDP (15–17).
RGS14 is a functionally and structurally complex signaling protein that is most highly expressed in the brain but also present in spleen, thymus, and lymphocytes (18–21). Within brain, RGS14 is predominately localized in the CA2 subregion of the hippocampus, where it is involved in spatial memory, learning, and synaptic plasticity (22). The unique structure of RGS14, which includes an RGS domain, two Ras/Rap binding domains, and a GPR (also known as GoLoco (23)) motif (20, 21) suggests that RGS14 functions in the brain through a variety of signaling mechanisms that may involve both G protein and MAP kinase signaling cascades (24). In addition to possessing GTPase accelerating protein activity toward activated Gαi/o-GTP subunits, RGS14 also exhibits selective guanine nucleotide dissociation inhibitor activity toward Gαi1/3-GDP subunits through direct binding of its GPR motif (18, 19, 21, 25–27). In this regard RGS14 shares similarities with the family of Group II activators of G protein signaling (AGS) proteins that are characterized by one or more GPR motifs and mediate unconventional G protein signaling (28–30). Similar to AGS3 and LGN, which form stable complexes with Gαi1-GDP via their GPR motifs (15, 16), the RGS14·Gαi1-GDP signaling complex is a substrate for Ric-8A-induced dissociation and nucleotide exchange on the resulting free Gαi1 (17).
Recent evidence suggests that unconventional pathways involving GPR·Gα-GDP complexes and conventional pathways involving GPCR·G protein complexes may be functionally linked. In particular, the GPR proteins AGS3 and AGS4 appear to interface with GPCRs in a Gαi-dependent manner (31, 32). Compelling evidence also indicates that RGS proteins directly and selectively interact with GPCRs to modulate G protein signaling (for review, see Ref. 33). Given that RGS14 is an RGS protein that interacts with Gαi/o-GTP but contains a GPR motif that binds Gαi1/3-GDP, we examined whether the RGS14·Gαi1 complex can be regulated by a Gαi/o-linked GPCR.
The non-receptor GEF Ric-8A regulates the RGS14·Gαi1 complex (17) as well as certain GPCR signaling pathways (34, 35). However, it remains unknown whether Ric-8A can modulate GPCR·Gα interactions, especially in the presence of a GPR protein such as RGS14. Therefore, we also studied the effects of Ric-8A on RGS14·Gαi1·GPCR complex formation and whether RGS14 may be at the interface between conventional and unconventional G protein signaling pathways. Here we report the first evidence that the RGS14·Gαi1-GDP complex is regulated in concert by both a Gαi/o-linked GPCR and Ric-8A in live cells. We show that RGS14 forms a stable complex with Gαi1 via its GPR motif and that this complex is proximal to GPCRs as evidenced by the presence of specific bioluminescence resonance energy transfer (BRET) signals between RGS14 and the α2A-adrenergic receptor (α2A-AR) in the presence of Gαi1. This RGS14·α2A-AR complex partially dissociates/rearranges after receptor agonist treatment and is further regulated by Ric-8A. Together, these findings illustrate that RGS14 functions together in both conventional and unconventional G protein signaling and that Ric-8A may recognize and act on GPCR·Gαi·GPR complexes to further regulate Gαi signaling.
EXPRIMENTAL PROCEDURES
Plasmids and Antibodies
The rat RGS14 cDNA used in this study (GenBankTM accession number U92279) was acquired as described (19). Rat RGS14 was used as a template in PCR reactions using TaKaRa Taq (Fisher) to generate Renilla luciferase (Luc) fusion protein constructs in the phRLucN2 vector graciously provided by Dr. Michel Bouvier (University of Montreal). The following oligonucleotides and restriction enzymes were used in the PCR amplification and subsequent digestion: RGS14 forward primer 5′-GCT CTC GAG GCC ACC ATG CCA GGG AAG CCC AAG CAC-3′, XhoI; reverse primer 5′-CGC GGT ACC TGG TGG AGC CTC CTG AGA ACC-3′, KpnI.
The RGS14-Luc GPR mutant, in which invariant glutamine and arginine residues (Gln515 and Arg516) were both mutated to alanine, was generated by site-directed mutagenesis using a Stratagene site-directed mutagenesis kit according to the manufacturer's instructions and is referred to as RGS14(GPR-null). Oligonucleotide primers used to create RGS14-Q515A/R516A-Luc (RGS14(GPR-null)) are as follows: RGS14(GPR-null) forward primer 5′-GGG GCC CAT GAC GCC GCC GGA CTT CTT CGC AAA G-3′ and reverse primer 5′-CTT TGC GAA GTC CGG CGG CGT CAT GGG CCC C-3′. The RGS14-Luc RGS domain mutant, in which invariant glutamic acid and asparagine (Glu92 and Asn93) residues were both mutated to alanine, was generated by site-directed mutagenesis using a Stratagene kit and is referred to as RGS14(RGS-null). Oligonucleotide primers used to create RGS14-E92A/N93A-Luc (RGS14(RGS-null)) are as follows: RGS14(RGS-null) forward primer 5′-AAG GAA TTC AGC GCC GCC GCC GTA ACT TTC TGG CAA GC-3′ and reverse primer 5′-GCT TGC CAG AAA GTT ACG GCG GCG GCG CTG AAT TCC TT-3′). The RGS14-Luc RGS/GPR double mutant referred to as RGS14(RGS/GPR-null) was generated by using RGS14(RGS-null) as a template and RGS14(GPR-null) primers in site-directed mutagenesis. In all cases, the plasmids were sequenced to confirm the fidelity of the PCR.
Wild-type AGS4-Luc was generated as previously described (32). Rat Gαi1-YFP (Gαi1-YFP) in pcDNA3.1 was generated by Dr. Gibson (36) and was generously provided along with pcDNA3.1::Ric-8A plasmid by Dr. Gregory Tall (University of Rochester School of Medicine and Dentistry). Gαi1-N149I-YFP (referred to as Gαi1-GPRi), Gαi1-G183S-YFP (referred to as Gαi1-RGSi), and Gαi1-G183S/N149I-YFP (referred to as Gαi1-RGSi/GPRi) were generated using the QuikChange kit (Stratagene) previously described. pcDNA3.1::Gαi1-YFP was used as a template for oligonucleotide primers Gαi1-GPRi forward primer 5′-GGG AGT ACC AGC TGC TCG ATT CGG CGG CGT A-3′ and reverse primer 5′-TAC GCC GCC GAA TCG ATC AGC TGG TAC TCC C-3′ and Gαi1-RGSi forward primer 5′-AGT GAA AAC GAC GTC AAT TGT GGA AA-3′ and reverse primer 5′-GGT TTC CAC AAT TGA CGT CGT TTT CA-3′. The Gαi1-RGSi/GPRi double mutant was constructed using the Gαi1-GPRi as a template for the Gαi1-RGSi primers. In all cases, the plasmids were sequenced to confirm the fidelity of the PCR.
Gαs-YFP and Gαq-YFP constructs were obtained from Dr. Catherine Berlot (Geisenger Institute, Danville, PA). Glu-Glu-tagged recombinant Gαi1 plasmid was purchased from UMR cDNA Resource Center (Rolla, Missouri). α2A-AR and β2-AR plasmids were generated as described and provided by Dr. Michel Bouvier (University of Montreal) (37, 38).
Anti-sera used include anti-Gαi1 (Millipore and Santa Cruz Biotechnologies, Inc.), anti-Gαi2 (Abcam), anti-Gαi3 and anti-Gαs (gifts from Dr. Thomas Gettys at Pennington Biomedical Research Center, Baton Rouge, LA), anti-FLAG (Sigma), anti-Ric-8A (provided by Dr. Gregory Tall, University of Rochester School of Medicine and Dentistry), anti-Gαq (Santa Cruz Biotechnologies, Inc.), anti-Gαo (Santa Cruz Biotechnologies, Inc.), Alexa 546 goat anti-rabbit secondary IgG (Invitrogen), Alexa 633 goat anti-mouse secondary IgG (Invitrogen), peroxidase-conjugated goat anti-mouse IgG (Rockland Immunochemicals, Inc.), and peroxidase-conjugated goat anti-rabbit IgG (Bio-Rad).
Cell Culture and Transfection
HEK293 cells were maintained in Dulbecco's minimal essential medium (without phenol red) containing 10% fetal bovine serum (5% after transfection), 2 mm glutamine, 100 units/ml penicillin, and 100 mg/ml streptomycin. Cells were incubated at 37 °C with 5% CO2 in a humidified environment. Transfections were performed using previously described protocols with polyethyleneimine (Polysciences, Inc.) (32). For immunofluorescence, cells were seeded onto glass coverslips before transfection.
BRET
BRET experiments were performed as previously described (31, 32). Briefly, HEK293 cells were transiently transfected with BRET donor and acceptor plasmids using polyethyleneimine. Forty-eight hours after transfection, the culture medium was removed, and cells were washed once with PBS and harvested with Tyrode's solution (140 mm NaCl, 5 mm KCl, 1 mm MgCl2, 1 mm CaCl2, 0.37 mm NaH2PO4, 24 mm NaHCO3, 10 mm HEPES, and 0.1% glucose (w/v), pH 7.4). Each group of cells was distributed into gray 96-well OptiPlates (PerkinElmer Life Sciences) in triplicate, with each well containing 1 × 105 cells. The acceptor (YFP/Venus-tagged) protein expression levels were evaluated by measuring total fluorescence using the TriStar LB 941 plate reader (Berthold Technologies) with excitation and emission filters at 485 and 535 nm, respectively. Data were analyzed using the MikroWin 2000 program. After fluorescence measurement, coelenterazine H (Nanolight Technology; 5 μm final concentration) was added and luminescence-detected in the 480 ± 20 and 530 ± 20 nm windows for donor (Luc) and acceptor (YFP/Venus), respectively, by the TriStar LB 941 plate reader. BRET signals were determined by calculating the ratio of the light intensity emitted by the YFP/Venus divided by the light intensity emitted by Luc. Net BRET values were corrected by subtracting the background BRET signal detected from the expression of the donor fusion protein (Luc) alone. Agonists used were UK14304 (Sigma) and isoproterenol (Sigma). Immunoblots were performed as described previously (39).
Immunofluorescence and Confocal Imaging
Transfected HEK293 cells were treated with either vehicle or 10 μm UK14304 diluted in serum-free DMEM for 5 min at 37 °C. Cells were then fixed at room temperature for 15 min in buffer containing 3.7% paraformaldehyde diluted in PBS. Cells were washed in PBS and incubated for 8 min with 0.4% Triton X-100 diluted in PBS. Cells were then blocked for 1 h at room temperature in PBS containing 10% goat serum and 3% bovine serum albumin. Next, cells were incubated in this same buffer with a 1:1000 dilution of rabbit anti- FLAG and/or mouse anti-Gαi1 antibodies at room temperature for 2 h. Cells were washed with PBS (3×) and incubated with 1:300 dilutions of Alexa 546 goat anti-rabbit and/or Alexa 633 goat anti-mouse secondary antibodies at room temperature for 1 h. Cells were washed with PBS again (3×) and mounted with ProLong Gold Antifade Reagent (Invitrogen). Confocal images were taken using a 63× oil immersion objective from a LSM510 laser scanning microscope with AxioObserver Stand (Zeiss). Images were processed using the ZEN 2009 Light Edition software and Adobe Photoshop 7.0 (Adobe Systems).
RESULTS
RGS14 Interacts Selectively with Gαi1 through Its GPR Motif
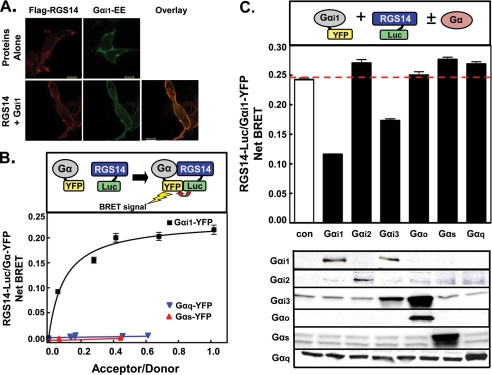
RGS14 has two distinct Gα-binding domains. The RGS domain binds activated Gαi/o subunits (18, 19, 21), whereas the GPR motif binds inactive Gαi1 and Gαi3 (19, 26, 27, 40). That RGS14 is recruited from the cytosol to the plasma membrane and co-localizes with wild-type Gαi1 (Fig. 1A, supplemental Fig. S1, and Refs. 17 and 27) suggests that RGS14 forms a stable complex with Gαi1 at the plasma membrane, which we sought to quantitatively measure using BRET. We therefore measured the strength and selectivity of a BRET signal between RGS14-Luc and various YFP-tagged Gα subunits (36, 41–43) (Fig. 1B). Of note, the YFP tag was inserted into the loop joining the αB and αC helices of each Gα (36, 41, 43), preserving nucleotide binding and hydrolysis properties similar to the wild-type protein (36). Transfection of HEK cells with increasing amounts of Gα-YFP plasmid and a fixed amount (5 ng) of RGS14-Luc plasmid showed a robust, saturable BRET signal in the presence of Gαi1-YFP, whereas no BRET signal was observed between RGS14-Luc paired with either Gαs-YFP or Gαq-YFP (Fig. 1B). This BRET signal saturation is indicative of a specific interaction between RGS14 and Gαi1 (44).
FIGURE 1.
RGS14 selectively interacts with Gαi1 and Gαi3 in the basal state of live cells as observed by BRET. A, FLAG-RGS14 and Gαi1-Glu-Glu (Gαi1-EE) plasmids were transfected into HEK cells alone and in combination. Cells were fixed, subjected to immunocytochemistry, and analyzed using confocal microscopy with a 63× objective as described under “Experimental Procedures.” Images are representative of cells observed in three separate experiments. Scale bars represent 10 μm. B, top, a diagram shows the principle of BRET using the RGS14-Luc/Gαi1-YFP pair. Non-radiative emission from the Luc tag excites the YFP if the donor/acceptor pairs are <100 Å, which then emits at 535 nm. Bottom, HEK cells were transfected with 5 ng RGS14-Luc plasmid alone or in combination with 10, 50, 100, 250, or 500 ng of either Gαi1-YFP, Gαs-YFP, or Gαq-YFP plasmid. BRET signals (luminescence measured: donor, 480 ± 20 nm; acceptor, 530 ± 20 nm) were measured, and net BRET was calculated by first calculating the 530 ± 20/480 ± 20 nm ratio and then subtracting the background BRET signal determined from cells transfected with the RGS14-Luc plasmid alone. C, top panel, HEK cells were transfected with 5 ng of RGS14-Luc and 250 ng of Gαi1-YFP plasmids alone (con) or in combination with 1 μg of untagged Gαi1, Gαi2, Gαi3, Gαo, Gαs, or Gαq plasmid. Net BRET signals are shown between RGS14-Luc and Gαi1-YFP. Bottom panel, shown is a representative immunoblot of the different untagged Gα subunits used in the BRET experiment. All BRET graphs are representative of at least three separate experiments.
To further show BRET signal selectivity for RGS14-Luc interactions with Gαi1-YFP, we performed a competition assay in cells co-expressing untagged Gα subunits (Fig. 1C) to determine which Gα subunits could displace Gαi1-YFP from RGS14-Luc and disrupt the BRET signal. As expected, the previously reported RGS14 binding partners Gαi1 and Gαi3 each disrupted the RGS14/Gαi1 BRET signal, indicative of competition with Gαi1-YFP for RGS14 binding. By contrast, Gαi2, Gαo, Gαs, and Gαq did not disrupt Gαi1-YFP binding to RGS14. This selectivity for Gαi1 and Gαi3 binding is entirely consistent with earlier reports showing RGS14 binding to only Gαi1 and Gαi3 but not other Gα through its GPR motif, further validating our BRET system (18, 19, 21, 26, 27, 40).
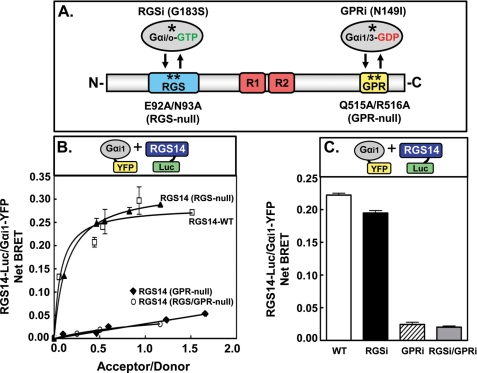
Findings in Fig. 1 suggested that the BRET signal we observed between RGS14 and Gαi1 occurs via the GPR motif. To test this hypothesis, we constructed mutants of RGS14-Luc that rendered it insensitive to binding Gαi1-YFP through either the RGS domain (RGS14-E92A/N93A-Luc; RGS-null) (18), the GPR motif (RGS14-Q515A/R516A-Luc; GPR-null) (25, 45), or both (RGS14-E92A/N93A/Q515A/R516A-Luc; RGS/GPR-null) (Fig. 2, A and B). The BRET signal between wild-type RGS14 (WT) and Gαi1 was comparable with that between RGS14(RGS-null) and Gαi1, suggesting that the majority of the observed BRET signal was not due to the RGS domain interacting with Gαi1. However, the BRET signal between RGS14(GPR-null) and Gαi1 was ∼5-fold lower than that of the RGS14-WT/Gαi1 pair. This indicates that the observed BRET signal between RGS14 and Gαi1 is primarily due to the GPR motif. As an additional approach, we generated Gαi1-YFP mutants that were insensitive to binding either the RGS domain (Gαi1-G183S-YFP; RGSi) (46), the GPR motif (Gαi1-N149I-YFP; GPRi) (47, 48), or both (Gαi1-G183S/N149I-YFP; RGSi/GPRi) (Fig. 2C). Consistent with findings in Fig. 2B, the BRET signal between RGS14 and Gαi1-GPRi was substantially (∼8-fold) lower than that generated by RGS14 paired with either wild-type Gαi1 (WT) or Gαi1-RGSi. Taken together, these findings are entirely consistent with the idea that the majority of the BRET signals observed between RGS14 and Gαi1 are due to the interaction between the RGS14 GPR motif and Gαi1.
FIGURE 2.
RGS14 BRET signals with Gαi1 in live cells are dependent on the GPR motif. A, shown is an illustration of the functional RGS14 and Gαi1 mutants, with Gαi/o-RGSi incapable of binding the RGS domain, Gαi1/3-GPRi incapable of binding the GPR motif, RGS14(RGS-null) incapable of binding active Gαi/o, and RGS14(GPR-null) incapable of binding inactive Gαi1/3. B, HEK cells were transfected with increasing amounts of Gαi1-YFP plasmid (10, 50, 100, 250, and 500 ng) in combination with 5 ng of either wild-type RGS14-Luc (RGS14-WT), RGS14(RGS-null)-Luc, RGS14(GPR-null)-Luc, or RGS14(RGS/GPR-null)-Luc plasmids. Net BRET was calculated by first calculating the 530 ± 20/480 ± 20-nm ratio and then subtracting the background BRET signal determined from cells transfected with the RGS14-Luc expression vector alone. Net BRET is shown between Gαi1-YFP and the different RGS14-Luc mutants. This figure is representative of at least three separate experiments with triplicate determinations. C, HEK cells were transfected with 5 ng of wild-type RGS14-Luc and 250 ng of either wild-type Gαi1-YFP (WT), Gαi1-RGSi-YFP, Gαi1-GPRi-YFP, or Gαi1-RGSi/GPRi-YFP plasmids. Net BRET is shown between RGS14-Luc and the different Gαi1-YFP mutants. These data are expressed as the mean of six separate experiments with triplicate determinations. *, point mutations in the proteins.
RGS14 Forms a Complex with the α2A-Adrenergic Receptor in a Gαi/o-dependent Manner
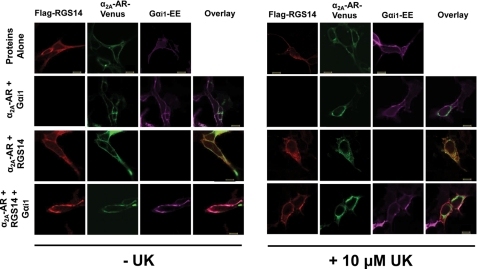
The GPR proteins AGS3 and AGS4 form Gαi-dependent complexes with GPCRs that are regulated by receptor activation (31, 32). Therefore, we sought to investigate whether the RGS14·Gαi1 complex can also be regulated by GPCRs in cells. Subcellular localization data showed that although RGS14 remained predominately cytosolic in the presence of co-expressed α2A-AR, it was recruited to the plasma membrane in the presence of both overexpressed α2A-AR and Gαi1 in the absence of agonist (Fig. 3, left panel). This suggests formation of an RGS14·Gαi1·α2A-AR complex at the plasma membrane. Although RGS14 and Gαi1 remained at the plasma membrane, the α2A-AR internalized in the presence of agonist UK14304 (Fig. 3, right panel).
FIGURE 3.
RGS14 co-localization with Gαi1 and the α2A-AR in live cells is regulated by receptor agonist. FLAG -RGS14, Gαi1- Glu-Glu (Gαi1-EE), and α2A-AR-Venus were transfected into HEK cells alone and in combination. Cells were either unstimulated (−UK) or stimulated (+UK) with 10 μm UK14304 for 5 min. Cells were fixed, subjected to immunocytochemistry, and analyzed using confocal microscopy as described under “Experimental Procedures.” Images are representative of cells observed in three separate experiments. Scale bars represent 10 μm.
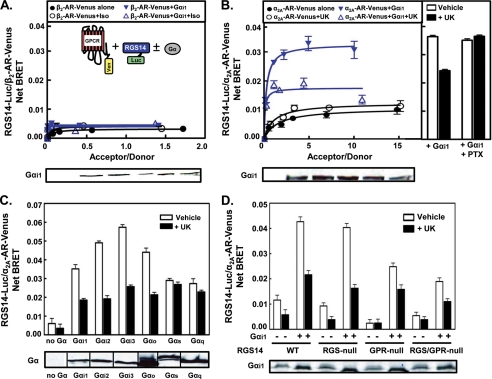
To further examine the regulatory effects of GPCRs on RGS14·Gαi1 complexes, we analyzed the BRET signals between RGS14-Luc and Venus-tagged α2A-AR or β2-AR (Fig. 4). As expected, little to no detectable BRET signal was observed between RGS14 and the Gs-linked β2-AR in the absence or presence of both Gαi1 and the receptor agonist isoproterenol (Fig. 4A). Very low specific BRET signals were observed between RGS14 and α2A-AR both in the absence and presence of receptor agonist UK14304 (Fig. 4B, filled circles and open circles, respectively). However, a 3-fold increase in BRET signal was observed between α2A-AR and RGS14 in the presence of co-expressed Gαi1 (Fig. 4B, filled triangles). This signal was reduced by ∼50% in the presence of UK14304 (Fig. 4B, open triangles). This agonist-induced reduction in BRET correlates with the lack of co-localization between RGS14 and the α2A-AR after agonist stimulation (Fig. 3, right panel). Furthermore, agonist-induced dissociation of the RGS14·α2A-AR complex was completely blocked by pretreatment with pertussis toxin (PTX) (Fig. 4B, right panel). The very low BRET signals observed between RGS14 and the β2-AR in the presence of Gαi1 (Fig. 4A) illustrate that the BRET signals observed between RGS14 and the α2A-AR are indeed specific and are not simply the result of “bystander BRET,” i.e. RGS14 localizing at the plasma membrane with Gαi1 and randomly interacting with the receptor.
FIGURE 4.
RGS14 forms a Gαi/o-dependent complex with the α2A-AR in live cells. A, Net BRET signals are shown from HEK cells transfected with 5 ng of RGS14-Luc and 0, 10, 50, 100, 250, or 500 ng of β2-AR-Venus plasmids in the presence or absence of 750 ng pcDNA3::Gαi1. Measurements were taken after treatment with either vehicle or isoproterenol (100 μm) for 5 min. A schematic representing the BRET principle used in all experiments of Fig. 4, which includes BRET measured between RGS14-Luc and a GPCR-Venus (Ven) in the presence or absence of untagged Gα, is shown within the graph. B, left panel, Net BRET signals are shown from HEK cells transfected with 5 ng of RGS14-Luc and either 0, 10, 50, 100, 250, or 500 ng of α2A-AR-Venus plasmid in the presence or absence of 750 ng of pcDNA3::Gαi1. Measurements were taken after treatment with either vehicle or α2A-AR agonist UK14304 (10 μm) for 5 min. Bottom panel, shown are representative immunoblots of untagged Gαi1 subunits used in samples with transfected Gαi1. Right panel, Net RGS14-Luc/α2A-AR-Venus BRET signals are shown from HEK cells transfected with 5 ng of RGS14-Luc, 250 ng of α2A-AR-Venus, and 750 ng of Gαi1 plasmids. Measurements were taken after treatment with UK14304 for 5 min in the absence or presence of 100 ng/ml pertussis toxin that was applied 18 h before agonist treatment, as indicated in the figure. Data are expressed as the mean of three separate experiments with triplicate determinations. C, top panel, HEK cells were transfected with 5 ng of RGS14-Luc and 100 ng of α2A-AR-Venus plasmids alone (no Gα) or in combination with 750 ng of either untagged Gαi1, Gαi2, Gαi3, Gαo, Gαs, or Gαq plasmids. Cells were either treated with vehicle or UK14304 (10 μm) for 5 min. The net BRET between RGS14-Luc and the α2A-AR-Venus under each condition is shown. Data are expressed as the mean of three separate experiments with triplicate determinations. Bottom panel, shown is a representative immunoblot of the different Gα subunits used. D, Net BRET signals for the RGS14-Luc/α2A-AR-Venus pair are shown for HEK cells transfected with 100 ng of α2A-AR-Venus and combinations of 5 ng RGS14-Luc mutant (WT, RGS-null, GPR-null, and RGS/GPR-null) plasmids in the absence or presence of 750 ng of untagged pcDNA3::Gαi1 and then treated with either vehicle or 10 μm UK14304 for 5 min. Bottom panel, shown is a representative immunoblot for Gαi1 expression. Data are expressed as the mean of four separate experiments with triplicate determinations. The net BRET between RGS14-Luc and the GPCR-Venus pairs was calculated by first calculating the 530 ± 20/480 ± 20-nm ratio and then subtracting the background BRET signal determined from cells transfected with RGS14-Luc plasmid alone.
The interaction between RGS14 and the α2A-AR was dependent on the presence of Gαi/o family members (Fig. 4C). Specific BRET signals were observed between RGS14 and the α2A-AR in the presence of Gαi1, Gαi2, Gαi3, and Gαo, with lower signals observed in the presence of Gαs and Gαq. The agonist-mediated dissociation of the RGS14·α2A-AR complex was observed in the presence of all four Gαi/o family members tested but not Gαs or Gαq (Fig. 4C).
To determine which domains of RGS14 are important for associating with the α2A-AR, we performed BRET experiments using the RGS14 constructs with mutations in the RGS domain and GPR motif as described in Fig. 2B (Fig. 4D). BRET signals observed between either RGS14-WT or RGS14(RGS-null) and the α2A-AR in the presence of co-expressed Gαi1 were comparable, with similar reductions in response to receptor agonist UK14304. This suggests that the RGS domain of RGS14 is not required for the formation of the Gαi1-dependent complex with the α2A-AR. In contrast, the BRET signals observed between the α2A-AR and RGS14(GPR-null) in the presence of Gαi1 were reduced by ∼50% in the absence of agonist compared with RGS14-WT, indicating that the GPR motif is critical to forming a complex with the α2A-AR in the presence of Gαi1. Together, these results indicate that RGS14 forms a complex with the α2A-AR in the presence of a Gαi/o protein and that the GPR motif is critical in promoting the formation of this complex (see supplemental Fig. S2A).
The RGS14·Gαi1 Complex Remains Intact after α2A-AR Stimulation
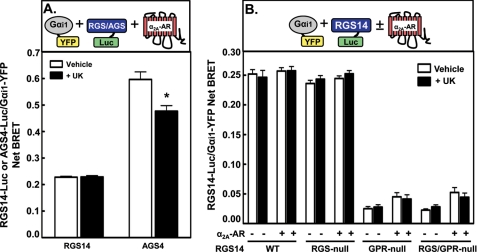
Because the presence of Gαi1 promotes the formation of an RGS14·α2A-AR complex that is regulated by agonist, we examined the effects of α2A-AR stimulation on the RGS14·Gαi1 complex (Fig. 5). To test this, we measured the BRET signal between RGS14-Luc and Gαi1-YFP in the presence of untagged α2A-AR. The RGS14·Gαi1 complex remains intact in the presence of the α2A-AR regardless of receptor stimulation (Fig. 5A). This is in marked contrast to the decrease in BRET signal observed between AGS4-Luc and Gαi1-YFP in the presence of stimulated α2A-AR (Fig. 5A and Ref. 32). Together, these findings suggest that the α2A-AR dissociates from RGS14 after agonist stimulation but that the dissociated RGS14 remains in complex with Gαi1 (supplemental Fig. S2B). This portrays a novel mechanism of GPR·Gαi complex function with GPCRs that may be unique to RGS14 compared with other Group II AGS proteins.
FIGURE 5.
RGS14 remains bound to Gαi1 after α2A-AR activation in live cells. A, HEK cells were transfected with 500 ng of untagged α2A-AR, 250 ng Gαi1-YFP, and either 5 ng of RGS14-Luc or 2 ng of AGS4-Luc plasmids. Cells were treated with either vehicle or UK14304 (10 μm) for 5 min. Net BRET generated from the RGS14-Luc/Gαi1-YFP or AGS4-Luc/Gαi1-YFP pairs was calculated by first calculating the 530 ± 20/480 ± 20-nm ratio and then subtracting the background BRET signal determined from cells transfected with RGS14-Luc or AGS4-Luc plasmid alone, respectively. Data were analyzed using paired Student's t test. *, p < 0.05 as compared with vehicle control. B, HEK cells were transfected with 250 ng of Gαi1-YFP and 5 ng of RGS14-Luc (WT, RGS-null, GPR-null, and RGS/GPR-null) plasmids with and without 500 ng of untagged α2A-AR plasmid and then treated with either vehicle or UK14304 (10 μm) for 5 min. Net BRET generated from the RGS14-Luc/Gαi1-YFP pair was calculated as in A. All data are expressed as the mean of three separate experiments with triplicate determinations.
The GPR motif is still critical for RGS14 interactions with Gαi1 in the presence of the α2A-AR (Fig. 5B), as >80% reductions in BRET signals were observed between Gαi1 and both RGS14(GPR-null) and RGS14(RGS/GPR-null) regardless of the presence of receptor. This indicates that even the presence of a GPCR cannot facilitate RGS14 interactions with Gαi1 in the absence of a functional GPR motif.
Ric-8A Promotes Dissociation of the RGS14·Gαi1 Complex
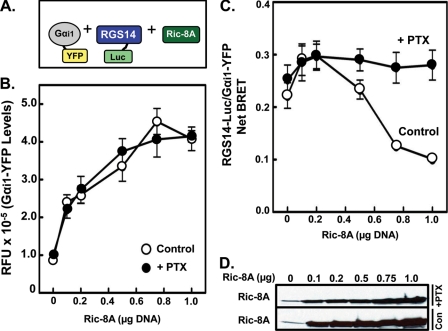
Because we observed Ric-8A regulation of RGS14·Gαi1 complexes in vitro (17), we sought to quantitatively measure Ric-8A-mediated dissociation of RGS14·Gαi1 complexes in live cells using BRET (Fig. 6A). As expected (17), increasing Ric-8A protein levels induced a decrease in BRET between RGS14-Luc and Gαi1-YFP (Fig. 6C). Ric-8A-induced reductions in RGS14/Gαi1 BRET were inhibited by pertussis toxin (+PTX) (Fig. 6C), which blocks Ric-8A binding and GEF activity toward Gαi subunits (49). Expression of Ric-8A also induces an increase in Gαi1-YFP protein expression levels (Fig. 6B), which is consistent with recent evidence showing that Ric-8A is important for the functional expression and stability of Gα subunits (50). Interestingly, the effect of Ric-8A on Gαi1-YFP expression levels was not blocked by pertussis toxin pretreatment, suggesting that the effect of Ric-8A on Gαi expression is independent from its GEF activity.
FIGURE 6.
Ric-8A facilitates dissociation of RGS14 from Gαi1 in live cells. A, shown is a diagram illustrating the BRET measured in this experiment between RGS14-Luc and Gαi1-YFP in the presence of untagged Ric-8A. B, HEK cells were transfected with 5 ng of RGS14-Luc, 250 ng of Gαi1-YFP, and increasing amounts (0, 100, 200, 500, 750, or 1000 ng) of Ric-8A plasmids. Cells were subsequently left untreated or pretreated with 100 ng/ml pertussis toxin (+PTX) for 18 h, and then the Gαi1-YFP fluorescence was measured. RFU, relative fluorescence units. C, HEK cells were transfected with 5 ng of RGS14-Luc, 250 ng of Gαi1-YFP, and increasing amounts (0, 100, 200, 500, 750, or 1000 ng) of Ric-8A plasmids. Cells were subsequently left alone or pretreated with 100 ng/ml pertussis toxin (+PTX) for 18 h, and then the net BRET between RGS14-Luc and Gαi1-YFP was measured and calculated. D, shown is a representative immunoblot of Ric-8A in each sample left alone (Con) or treated with PTX (+PTX). Measurements in B and C were taken from the exact same samples. Data are expressed as the mean of three separate experiments with triplicate determinations.
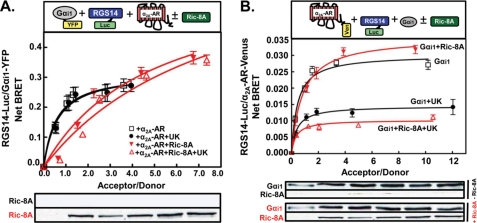
We next studied the effects of Ric-8A on RGS14·Gαi1 complexes in the presence of the α2A-AR (Fig. 7A). In the absence of Ric-8A, RGS14·Gαi1 complexes remained intact after receptor stimulation as before (see Fig. 5A). In the absence of receptor agonist, Ric-8A promoted a decrease in the RGS14/Gαi1 BRET signal. In the presence of agonist, Ric-8A induced an even greater decrease in the BRET signal (Fig. 7A). These findings suggest that Ric-8A can recognize and act on RGS14·Gαi1 complexes in the presence of GPCRs and even more so in the presence of activated receptors.
FIGURE 7.
Ric-8A induces dissociation of both Gαi1 and the α2A-AR from RGS14 after receptor stimulation. A, top panel, Net BRET signals were generated from the RGS14-Luc/Gαi1-YFP pair in HEK cells transfected with combinations of 5 ng of RGS14-Luc, 500 ng of α2A-AR, 200 ng of Ric-8A, and increasing amounts of Gαi1-YFP (0, 10, 50, 100, 250, and 500 ng) plasmids. Cells were treated with either vehicle or UK14304 (10 μm) for 5 min before BRET signals were measured. Bottom panel, shown is a representative immunoblot of Ric-8A expression for all six amounts of Gαi1-YFP plasmid transfected. Ric-8A and Ric-8A (red) represent lysates from cells without transfected Ric-8A (top immunoblot) or cells with transfected Ric-8A (bottom immunoblot), respectively. Data are expressed as the mean of three separate experiments with triplicate determinations. B, top panel, Net BRET signals generated from the RGS14-Luc/α2A-AR-Venus (Ven) pair in HEK cells transfected with combinations of 5 ng of RGS14-Luc, 100 ng of Gαi1, 200 ng of Ric-8A, and increasing amounts of α2A-AR-Venus (0, 10, 50, 100, 250, and 500 ng) plasmids. Cells were treated with either vehicle or UK14304 (10 μm) for 5 min before BRET signals were measured. Bottom panel, shown is a representative immunoblot of Ric-8A and Gαi1 expression for all six amounts of α2A-AR-Venus transfected. Data are expressed as the mean of three separate experiments with triplicate determinations.
Ric-8A Potentiates Dissociation of the RGS14· α2A-AR Complex Caused by Receptor Agonist
Because Ric-8A induced dissociation of Gαi1 from RGS14 in the presence of the α2A-AR, we next investigated the effect of Ric-8A on the RGS14·α2A-AR complex in the presence of Gαi1 (Fig. 7B). Ric-8A had little effect on the RGS14·α2A-AR complex in the presence of co-expressed Gαi1 in the absence of agonist. However, BRET signals between RGS14 and the α2A-AR in the presence of Gαi1 and receptor agonist were further reduced by ∼25% in the presence of Ric-8A (red lines) compared with the absence of Ric-8A (black lines) (Fig. 7B). These findings suggest that Ric-8A acts to facilitate dissociation of RGS14 from activated α2A-AR in the presence of Gαi1 (see supplemental Fig. S2C).
DISCUSSION
RGS14 is unusual among RGS protein family members in that it possesses two distinct Gα binding domains; that is, an RGS domain that accelerates GTP hydrolysis on activated Gαi/o subunits (18, 19, 21) and a GPR motif that forms a tight complex with inactive Gαi1/3 subunits (17, 19, 25–27). RGS14 also belongs to a second family of signaling proteins, the Group II AGS proteins, which are characterized by the presence of one or more GPR motifs that mediate newly appreciated “unconventional” G protein signaling events (28, 29). Recent studies of AGS3 and AGS4 demonstrate that these GPR domain-containing proteins interact with Gαi to form complexes with Gαi/o-linked GPCRs in cells (31, 32). Our results with RGS14 support those findings but also highlight some important differences that will be discussed. Overall, our findings indicate the following: 1) RGS14 selectively interacts with Gαi1/3 in live cells through its GPR motif, 2) RGS14 forms a Gαi/o-dependent complex with the Gi/o-linked α2A-AR in live cells, 3) RGS14 dissociates from the α2A-AR after agonist treatment but remains bound to Gαi1, 4) Ric-8A potentiates agonist-stimulated dissociation of the RGS14·α2A-AR complex, and 5) Ric-8A induces dissociation of Gαi1 and α2A-AR from RGS14, having a greater effect in the presence of stimulated α2A-AR. Taken together, these findings suggest that RGS14 integrates both unconventional Ric-8A/G protein signaling and conventional GPCR/G protein signaling. A summary and interpretation of these findings is shown in Fig. 8.
FIGURE 8.
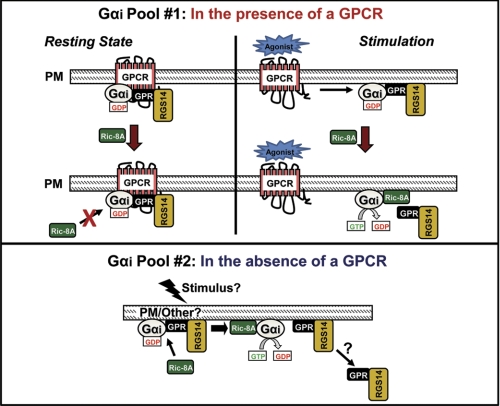
Working model depicting Ric-8A regulation of the α2A-AR·Gαi1·RGS14 complex. This visual model includes RGS14, Gαi1, α2A-AR, and Ric-8A localized at or near the plasma membrane (PM). We propose that two pools of Gαi exist in cells. Top, one pool localizes with GPCRs and Gβγ/GPR proteins at the plasma membrane to participate in conventional GPCR-dependent G protein signaling. In the resting state (left) of our model, a GPCR·Gαi·RGS14 complex forms and remains intact. Ric-8A has little effect on this complex in the absence of stimulation. Upon receptor stimulation (right), the RGS14·Gαi complex dissociates from the GPCR, where it can be further acted upon by Ric-8A. Bottom, the second Gαi pool forms complexes with GPR proteins at the plasma membrane in the absence of a GPCR to participate in unconventional GPCR-independent signaling. According to our findings, RGS14 forms a complex with Gαi through its GPR motif. Ric-8A can recognize this RGS14·Gαi complex, catalyze GTP exchange on Gαi, and induce dissociation of the complex.
RGS14 Selectively Interacts with Inactive Gαi1/3 in Live Cells through Its GPR Motif
Our BRET analysis and confocal imaging indicate that the interaction of RGS14 with inactive Gαi1/3 occurs at the plasma membrane of live cells (Fig. 1 and supplemental Fig S1). Consistent with previous studies (18, 19, 26, 27, 40), the capacity of both Gαi1 and Gαi3 (but not Gαi2, Gαo, Gαs, or Gαq) to disrupt the BRET between RGS14-Luc and Gαi1-YFP indicates that the observed BRET signal is specific for interactions between RGS14 and Gαi1/3 (Fig. 1C).
To clarify which RGS14 domains are involved in the RGS14·Gαi1 interaction, we measured the BRET signal between mutant forms of RGS14-Luc and Gαi1-YFP that specifically blocked RGS and/or GPR motif functions (Fig. 2). These studies show that the majority of the observed RGS14·Gαi1 interaction is conferred by the GPR motif of RGS14 interacting with Gαi1. The fact that the BRET signal was never completely abolished in the presence of the RGS14 and Gαi1 double mutants that ablate Gα binding to both the GPR and RGS domains (Fig. 2, B and C) is consistent with the existence of a third G protein binding site on RGS14, as has been postulated (51).
RGS14 Selectively Interacts with the α2A-AR Receptor in a Gαi/o-dependent Manner
Because RGS14 interacts with Gαi/o family members, we examined whether RGS14 can be regulated by a Gi/o-linked GPCR, specifically the α2A-AR. RGS14, Gαi1, and the α2A-AR co-localized at the plasma membrane when all three proteins were expressed together in cells (Fig. 3, left panel), consistent with the possibility that a ternary protein complex forms at the plasma membrane. After treatment with the α2A-AR agonist UK14304, RGS14 and Gαi1 remained at the plasma membrane, whereas the α2A-AR partially internalized (Fig. 3, right panel), suggesting that the ternary complex dissociates. This hypothesis was supported in our BRET experiments. Co-expression of Gαi1 resulted in an approximate 3-fold increase in RGS14/α2A-AR BRET compared with RGS14 and α2A-AR alone (Fig. 4B). The Gαi1-dependent RGS14/α2A-AR BRET signal was reduced ∼50% after receptor activation by agonist, and this agonist effect was blocked by pertussis toxin pretreatment (Fig. 4B, right panel). This implies that functional coupling of the α2A-AR to Gαi1 disrupts the RGS14·α2A-AR complex. It is possible that the interacting sites between GPCR·Gαi are different between the inactive and active states, the latter being sensitive to PTX. This is suggested by previous work on the phenomenon of guanine nucleotide-sensitive agonist binding to GPCRs and more recent work demonstrating preformed complexes of GPCRs and G proteins (52, 53).
As expected, RGS14 interaction with the α2A-AR is dependent on the presence of Gαi/o as Gαq and Gαs failed to elicit a robust RGS14/α2A-AR BRET signal. Somewhat unexpectedly, RGS14·α2A-AR association is promoted indiscriminately by the presence of any Gαi/o family member (Gαi1, Gαi2, Gαi3, and Gαo) (Fig. 4C). This is surprising given that the RGS14·α2A-AR interaction was highly dependent on the GPR motif (Fig. 4D), which only interacts with Gαi1 and Gαi3 in the absence of receptor. One possible explanation may be that RGS14 recognizes a receptor if the receptor is bound to any Gαi/o protein, reflecting the promiscuity of RGS14 GTPase accelerating protein activity toward activated Gαi/o subunits. In this regard, RGS14 is similar to RGS2. In the absence of receptor, RGS2 acts specifically on Gαq (54). However, RGS2 is capable of interacting with Gαi in the presence of a Gi/o-linked GPCR (55), albeit with 30-fold lower affinity than for Gαq (56). We note that RGS14 complexes with receptor are dependent on both the G protein and the receptor because the Gs-linked β2-AR failed to interact with RGS14 in the presence of Gαi1 (Fig. 4A).
The GPR motif interaction with Gαi1 is important in promoting formation of the RGS14·α2A-AR complex (Fig. 4D). The RGS14/α2A-AR BRET signal was greatly reduced in the presence of RGS14(GPR-null) compared with RGS14-WT, indicating that Gαi1 has a reduced capacity to bring RGS14 and the α2A-AR in close proximity when it cannot bind the GPR motif. Even when Gαi1 could no longer bind either the RGS domain or GPR motif, there was still a slight BRET signal between RGS14(RGS/GPR-null) and the α2A-AR. Several possibilities exist to explain these results; 1) there may be another (undefined) Gαi1 binding site on RGS14 (51), 2) RGS14 may be bound to Gαi1 at a distinct site on the extreme C terminus of Gαi1 (17), or 3) an unknown binding partner/scaffold may facilitate an RGS14·α2A-AR interaction.
RGS14 Remains Bound to Gαi1 after Dissociating from the α2A-AR
Although RGS14 dissociated from the α2A-AR after agonist treatment in the presence of co-expressed Gαi1 (Fig. 4), it remained in complex with Gαi1 via the GPR motif (Fig. 5). This finding is unexpected and differs from previous observations that show AGS3 and AGS4 dissociating from Gαi after receptor activation (Fig. 5A and Refs. 31 and 32)). Our result suggests that RGS14 and Gαi1 remain bound after receptor activation. This result is reminiscent of other findings showing that, in contrast to established models of G protein signaling (1), Gβγ may not necessarily always dissociate from Gα. In some cases Gβγ may rearrange relative to Gα-GTP after receptor activation (53), although in others Gβγ does appear to dissociate (Refs. 57–59 and references therein). Irrespective of the mechanism involved, our findings represent a novel mechanism of action for GPCR·Gα·RGS complexes, where the active conformation of the α2A-AR favors release of an RGS14·Gαi1 complex that may then be able to function as a signaling complex on its own or with other binding partners (such as potential MAP kinase signaling partners (24)). This complex may be regulated and function independently of the GPCR.
Ric-8A Is a Key Regulator of the GPCR·Gαi1·RGS14 Complex
Although Ric-8A has been shown to influence GPCR signaling (34, 35, 60), little is known mechanistically about if or how Ric-8A may directly interact with and regulate GPCR·G protein complexes. We recently demonstrated that Ric-8A induces dissociation of RGS14 from Gαi1 in vitro (17). In this study we sought to quantitatively measure the dissociative effects of Ric-8A on RGS14·Gαi complexes in live cells using BRET (Fig. 6). Pertussis toxin blocked Ric-8A-mediated dissociation of the RGS14·Gαi1 complex (Fig. 6, C and D), consistent with recent reports showing that pertussis toxin inhibits Ric-8A GEF activity on Gαi1 and that Ric-8A binds to Gαi1 at a region overlapping with the pertussis toxin binding site (17, 49). In the absence of pertussis toxin, Ric-8A facilitated RGS14·Gαi1 complex dissociation (Fig. 6, C and D). Ric-8A also induced dissociation of the RGS14·Gαi1 complex in the presence of the α2A-AR, even in the absence of α2A-AR stimulation (Fig. 7A). This may be explained by Ric-8A effects on Gαi1 expression levels. Because Ric-8A overexpression also induced an increase in Gαi1 expression (Fig. 6B), it may be that there is an overabundance of Gαi1 that is free to bind RGS14. The number of RGS14·Gαi1 complexes may, therefore, outnumber the number of α2A-ARs, resulting in free RGS14·Gαi1 complexes on which Ric-8A may act in the absence of receptor activation.
Ric-8A did not induce dissociation of the RGS14·α2A-AR complex in the absence of receptor stimulation (Fig. 7B). This is in contrast to its effects on the RGS14·Gαi1 complex in the presence of unstimulated receptor. It is possible that Ric-8A facilitates dissociation of RGS14·Gαi1 complexes that are not associated with receptors, accounting for the decrease in RGS14/Gαi1 BRET seen in the presence of unstimulated receptor (Fig. 7A). In a cellular signaling context, Ric-8A may function similarly to the Arr4 protein in yeast that serves a feed-forward facilitating role in pheromone receptor-G protein signaling mating responses (61). Consistent with this idea is that Ric-8A potentiates taste-receptor signaling by a potential feed-forward mechanism (34).
Taken together, these studies show that RGS14 can associate with a GPCR·Gαi/o complex in a regulated fashion and that Ric-8A is a regulatory partner in this process. Although Ric-8A potentiated dissociation of RGS14·Gαi1 complexes from the α2A-AR in both the absence and presence of receptor stimulation, it had no effect on dissociating the RGS14·α2A-AR complex itself in the absence of stimulation. We postulate that two pools of RGS14·Gαi1 complexes may exist (Fig. 8). One subset resides at membranes (plasma and others?) in the absence of a GPCR, and the other directly complexes to a cell surface receptor. Ric-8A acts differently on the RGS14·Gαi1 complex depending on whether or not the complex is coupled to a GPCR. In the absence of a GPCR (Fig. 8, bottom), Ric-8A can recognize and induce dissociation of the RGS14·Gαi1 complex. When the RGS14·Gαi1 complex is associated with a GPCR (Fig. 8, top), Ric-8A may not affect RGS14·Gαi1 complexes unless the receptor is activated. In this case Ric-8A induces dissociation of Gαi1 from RGS14 and subsequently RGS14 from receptor.
Our findings demonstrate that RGS14 functions in a unique mechanism to integrate both conventional GPCR·G protein signaling and unconventional GPCR-independent G protein signaling. These results highlight newly appreciated roles of GPR proteins at the interface of G protein signaling pathways, making them significant targets in the study of non-canonical G protein regulation and function.
Supplementary Material
Acknowledgments
We thank Dr. Gregory Tall (University of Rochester School of Medicine and Dentistry) for Ric-8A plasmids and Ric-8A antisera, Dr. Michel Bouvier (University of Montreal) for α2A-AR-Venus and β2-AR-Venus plasmids, Dr. Stephen Lanier (Medical University of South Carolina) for untagged α2A-AR plasmid, Dr. Thomas Gettys (Pennington Biomedical Research Center, Baton Rouge, LA) for Gαi3 and Gαs antisera, and Dr. Catherine Berlot (Geisenger Institute, Danville, PA) for Gαs-YFP and Gαq-YFP plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants R01NS049195 and R01NS037112 (to J. R. H.), R01GM086510 (to J. B. B.), and Pharmacological Sciences Training Grant T32 GM008602 (to C. P. V.). This work was also supported by a PhRMA Foundation Pre-doctoral Pharmacology/Toxicology Fellowship (to C. P. V.). This research was also supported in part by the Microscopy Core of the Emory Neuroscience NINDS Core Facilities Grant P30NS055077.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- GPCR
- G protein-coupled receptors
- GEF
- guanine nucleotide exchange factor
- RGS
- regulators of G protein signaling
- GPR
- G protein regulatory
- Ric-8A
- resistance to inhibitors of cholinesterase 8A
- AGS
- activators of G protein signaling
- BRET
- bioluminescence resonance energy transfer
- AR
- adrenergic receptor
- PTX
- pertussis toxin.
REFERENCES
- 1. Gilman A. G. (1987) Annu. Rev. Biochem. 56, 615–649 [DOI] [PubMed] [Google Scholar]
- 2. Hamm H. E. (1998) J. Biol. Chem. 273, 669–672 [DOI] [PubMed] [Google Scholar]
- 3. De Vries L., Zheng B., Fischer T., Elenko E., Farquhar M. G. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 235–271 [DOI] [PubMed] [Google Scholar]
- 4. Hollinger S., Hepler J. R. (2002) Pharmacol. Rev. 54, 527–559 [DOI] [PubMed] [Google Scholar]
- 5. Ross E. M., Wilkie T. M. (2000) Annu. Rev. Biochem. 69, 795–827 [DOI] [PubMed] [Google Scholar]
- 6. Colombo K., Grill S. W., Kimple R. J., Willard F. S., Siderovski D. P., Gönczy P. (2003) Science 300, 1957–1961 [DOI] [PubMed] [Google Scholar]
- 7. Groves B., Gong Q., Xu Z., Huntsman C., Nguyen C., Li D., Ma D. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 18103–18108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hampoelz B., Knoblich J. A. (2004) Cell 119, 453–456 [DOI] [PubMed] [Google Scholar]
- 9. Hess H. A., Röper J. C., Grill S. W., Koelle M. R. (2004) Cell 119, 209–218 [DOI] [PubMed] [Google Scholar]
- 10. Sans N., Wang P. Y., Du Q., Petralia R. S., Wang Y. X., Nakka S., Blumer J. B., Macara I. G., Wenthold R. J. (2005) Nat. Cell Biol. 7, 1179–1190 [DOI] [PubMed] [Google Scholar]
- 11. Sato M., Blumer J. B., Simon V., Lanier S. M. (2006) Annu. Rev. Pharmacol. Toxicol. 46, 151–187 [DOI] [PubMed] [Google Scholar]
- 12. Schade M. A., Reynolds N. K., Dollins C. M., Miller K. G. (2005) Genetics 169, 631–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Willard F. S., Kimple R. J., Siderovski D. P. (2004) Annu. Rev. Biochem. 73, 925–951 [DOI] [PubMed] [Google Scholar]
- 14. Tall G. G., Krumins A. M., Gilman A. G. (2003) J. Biol. Chem. 278, 8356–8362 [DOI] [PubMed] [Google Scholar]
- 15. Tall G. G., Gilman A. G. (2005) Proc. Natl. Acad. Sci. U. S. A. 102, 16584–16589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thomas C. J., Tall G. G., Adhikari A., Sprang S. R. (2008) J. Biol. Chem. 283, 23150–23160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vellano C. P., Shu F. J., Ramineni S., Yates C. K., Tall G. G., Hepler J. R. (2011) Biochemistry 50, 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cho H., Kozasa T., Takekoshi K., De Gunzburg J., Kehrl J. H. (2000) Mol. Pharmacol. 58, 569–576 [DOI] [PubMed] [Google Scholar]
- 19. Hollinger S., Taylor J. B., Goldman E. H., Hepler J. R. (2001) J. Neurochem. 79, 941–949 [DOI] [PubMed] [Google Scholar]
- 20. Snow B. E., Antonio L., Suggs S., Gutstein H. B., Siderovski D. P. (1997) Biochem. Biophys. Res. Commun. 233, 770–777 [DOI] [PubMed] [Google Scholar]
- 21. Traver S., Bidot C., Spassky N., Baltauss T., De Tand M. F., Thomas J. L., Zalc B., Janoueix-Lerosey I., Gunzburg J. D. (2000) Biochem. J. 350, 19–29 [PMC free article] [PubMed] [Google Scholar]
- 22. Lee S. E., Simons S. B., Heldt S. A., Zhao M., Schroeder J. P., Vellano C. P., Cowan D. P., Ramineni S., Yates C. K., Feng Y., Smith Y., Sweatt J. D., Weinshenker D., Ressler K. J., Dudek S. M., Hepler J. R. (2010) Proc. Natl. Acad. Sci. U. S. A. 107, 16994–16998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Siderovski D. P., Diversé-Pierluissi M., De Vries L. (1999) Trends Biochem. Sci. 24, 340–341 [DOI] [PubMed] [Google Scholar]
- 24. Shu F. J., Ramineni S., Hepler J. R. (2010) Cell. Signal. 22, 366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kimple R. J., Kimple M. E., Betts L., Sondek J., Siderovski D. P. (2002) Nature 416, 878–881 [DOI] [PubMed] [Google Scholar]
- 26. Mittal V., Linder M. E. (2004) J. Biol. Chem. 279, 46772–46778 [DOI] [PubMed] [Google Scholar]
- 27. Shu F. J., Ramineni S., Amyot W., Hepler J. R. (2007) Cell. Signal. 19, 163–176 [DOI] [PubMed] [Google Scholar]
- 28. Blumer J. B., Oner S. S., Lanier S. M. (2011) Acta Physiol. (Oxf), in press [DOI] [PubMed] [Google Scholar]
- 29. Blumer J. B., Smrcka A. V., Lanier S. M. (2007) Pharmacol. Ther. 113, 488–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Siderovski D. P., Willard F. S. (2005) Int. J. Biol. Sci. 1, 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oner S. S., An N., Vural A., Breton B., Bouvier M., Blumer J. B., Lanier S. M. (2010) J. Biol. Chem. 285, 33949–33958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oner S. S., Maher E. M., Breton B., Bouvier M., Blumer J. B. (2010) J. Biol. Chem. 285, 20588–20594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neitzel K. L., Hepler J. R. (2006) Semin. Cell Dev. Biol. 17, 383–389 [DOI] [PubMed] [Google Scholar]
- 34. Fenech C., Patrikainen L., Kerr D. S., Grall S., Liu Z., Laugerette F., Malnic B., Montmayeur J. P. (2009) Front. Cell. Neurosci. 3, 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nishimura A., Okamoto M., Sugawara Y., Mizuno N., Yamauchi J., Itoh H. (2006) Genes Cells 11, 487–498 [DOI] [PubMed] [Google Scholar]
- 36. Gibson S. K., Gilman A. G. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duzic E., Coupry I., Downing S., Lanier S. M. (1992) J. Biol. Chem. 267, 9844–9851 [PubMed] [Google Scholar]
- 38. Galés C., Van Durm J. J., Schaak S., Pontier S., Percherancier Y., Audet M., Paris H., Bouvier M. (2006) Nat. Struct. Mol. Biol. 13, 778–786 [DOI] [PubMed] [Google Scholar]
- 39. Blumer J. B., Chandler L. J., Lanier S. M. (2002) J. Biol. Chem. 277, 15897–15903 [DOI] [PubMed] [Google Scholar]
- 40. Kimple R. J., De Vries L., Tronchère H., Behe C. I., Morris R. A., Gist, Farquhar M., Siderovski D. P. (2001) J. Biol. Chem. 276, 29275–29281 [DOI] [PubMed] [Google Scholar]
- 41. Hein P., Rochais F., Hoffmann C., Dorsch S., Nikolaev V. O., Engelhardt S., Berlot C. H., Lohse M. J., Bünemann M. (2006) J. Biol. Chem. 281, 33345–33351 [DOI] [PubMed] [Google Scholar]
- 42. Hughes T. E., Zhang H., Logothetis D. E., Berlot C. H. (2001) J. Biol. Chem. 276, 4227–4235 [DOI] [PubMed] [Google Scholar]
- 43. Hynes T. R., Mervine S. M., Yost E. A., Sabo J. L., Berlot C. H. (2004) J. Biol. Chem. 279, 44101–44112 [DOI] [PubMed] [Google Scholar]
- 44. Mercier J. F., Salahpour A., Angers S., Breit A., Bouvier M. (2002) J. Biol. Chem. 277, 44925–44931 [DOI] [PubMed] [Google Scholar]
- 45. Khafizov K. (2009) J. Mol. Model 15, 1491–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lan K. L., Sarvazyan N. A., Taussig R., Mackenzie R. G., DiBello P. R., Dohlman H. G., Neubig R. R. (1998) J. Biol. Chem. 273, 12794–12797 [DOI] [PubMed] [Google Scholar]
- 47. Natochin M., Gasimov K. G., Artemyev N. O. (2002) Biochemistry 41, 258–265 [DOI] [PubMed] [Google Scholar]
- 48. Willard F. S., Zheng Z., Guo J., Digby G. J., Kimple A. J., Conley J. M., Johnston C. A., Bosch D., Willard M. D., Watts V. J., Lambert N. A., Ikeda S. R., Du Q., Siderovski D. P. (2008) J. Biol. Chem. 283, 36698–36710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Woodard G. E., Huang N. N., Cho H., Miki T., Tall G. G., Kehrl J. H. (2010) Mol. Cell Biol. 30, 3519–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gabay M., Tall G. G. (2011) FASEB J. 25, 804.805 [Google Scholar]
- 51. Hepler J. R., Cladman W., Ramineni S., Hollinger S., Chidiac P. (2005) Biochemistry 44, 5495–5502 [DOI] [PubMed] [Google Scholar]
- 52. Audet N., Galés C., Archer-Lahlou E., Vallières M., Schiller P. W., Bouvier M., Pineyro G. (2008) J. Biol. Chem. 283, 15078–15088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bünemann M., Frank M., Lohse M. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100, 16077–16082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Heximer S. P., Watson N., Linder M. E., Blumer K. J., Hepler J. R. (1997) Proc. Natl. Acad. Sci. U. S. A. 94, 14389–14393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ingi T., Krumins A. M., Chidiac P., Brothers G. M., Chung S., Snow B. E., Barnes C. A., Lanahan A. A., Siderovski D. P., Ross E. M., Gilman A. G., Worley P. F. (1998) J. Neurosci. 18, 7178–7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Heximer S. P., Srinivasa S. P., Bernstein L. S., Bernard J. L., Linder M. E., Hepler J. R., Blumer K. J. (1999) J. Biol. Chem. 274, 34253–34259 [DOI] [PubMed] [Google Scholar]
- 57. Digby G. J., Lober R. M., Sethi P. R., Lambert N. A. (2006) Proc. Natl. Acad. Sci. U. S. A. 103, 17789–17794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hollins B., Kuravi S., Digby G. J., Lambert N. A. (2009) Cell. Signal. 21, 1015–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lambert N. A. (2008) Sci. Signal. 1, re5 [DOI] [PubMed] [Google Scholar]
- 60. Yoshikawa K., Touhara K. (2009) Chem. Senses 34, 15–23 [DOI] [PubMed] [Google Scholar]
- 61. Lee M. J., Dohlman H. G. (2008) Current biology : CB 18, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.