Abstract
Coordinated actin remodeling is crucial for cell entry into mitosis. The WAVE regulatory complex is a key regulator of actin assembly, yet how the WAVE signaling is regulated to coordinate actin assembly with mitotic entry is not clear. Here, we have uncovered a novel mechanism that regulates the WAVE complex at the onset of mitosis. We found that the Bcr-Abl-stimulated F-actin assembly is abrogated during mitosis. This mitotic inhibition of F-actin assembly is accompanied by an attenuation of Bcr-Abl-induced tyrosine phosphorylation of the WAVE complex. We identified serine 216 of Abi1 as a target of CDK1/cyclin B kinase that is phosphorylated in cells at the onset of mitosis. The Abi1 phosphorylated on serine 216 displayed greatly reduced tyrosine phosphorylation in the hematopoietic cells transformed by Bcr-Abl. Moreover, a phosphomimetic mutation of serine 216 to aspartic acid in Abi1 was sufficient to attenuate Bcr-Abl-induced tyrosine phosphorylation of the WAVE complex and F-actin assembly. Ectopic expression of Abi1 with serine 216 mutations interfered with cell cycle progression. Together, these data show that CDK1-mediated phosphorylation of serine 216 in Abi1 serves as a regulatory mechanism that may contribute to coordinated actin cytoskeleton remodeling during mitosis.
Keywords: Actin, CDK (Cyclin-dependent Kinase), Cytoskeleton, Mitosis, Phosphorylation, Abi1, CDK1, WAVE2 Complex
Introduction
Actin cytoskeleton remodeling is a dynamic process that regulates many fundamental cellular functions such as cell adhesion, division, intracellular trafficking, morphogenesis, and motility (1, 2). This cellular process is regulated spatiotemporally not only by extracellular signals such as growth factors, chemoattractants, and extracellular matrix but also by intracellular signals that control cell cycle progression (2–4). Tight regulation by both extracellular and intracellular signals ensures that proper cytoskeleton rearrangement is achieved and coordinated with a broad range of cellular functions including directional movement and cell division.
A pivotal step in the regulation of actin assembly is actin nucleation, a process that initiates new F-actin formation from monomers. Actin nucleation is regulated by several classes of nucleation factors including actin-related protein 2/3 (Arp2/3)3 complex, formins, Spire, and cordon-bleu (COBL). Of these, the Arp2/3 complex is a key nucleation factor known to both nucleate actin and organize the branched filamentous actin (F-actin) into networks at the plasma membrane (4). The actin nucleation activity of the Arp2/3 complex requires nucleation-promoting factors (NPFs). Most mammalian NPFs belong to the Wiskott-Aldrich syndrome protein (WASP) family and are characterized structurally by containing a VCA domain (verprolin homology, central and acidic regions), which binds and activates the Arp2/3 complex (4). Among eight WASP family members identified to date, the WAVE (WASP family verprolin-homologous) subfamily proteins have been shown to play a central role in promoting Arp2/3-mediated actin nucleation and the assembly of branched F-actin networks at the plasma membrane.
WAVE proteins are present in cells as a complex with hematopoietic stem progenitor cell 300 (Hspc 300), Abl interactor (Abi), Nck-associated protein (Nap), and specifically Rac1-associated protein (Sra). In this so-called WAVE regulatory complex (WRC), Abi plays a central role in holding the complex together (5, 6) and in linking the complex to Abelson (Abl) non-receptor tyrosine kinases (7–10), whereas Sra provides a binding site for active Rac (6, 11). The formation of the WRC is believed to hold WAVE in an inactive state (11–15). Recent studies have shown that WRC can be activated by several signaling events including its binding to prenylated Rac-GTP and acidic phospholipids, as well as phosphorylation by protein kinases. The importance of these signaling events in activating WRC is supported by recent structural analysis of WRC (13). It has been shown that the interaction of WAVE with Sra sequesters its VCA domain and prevents it from interacting with actin and Arp2/3. The binding of WRC to Rac-GTP and phospholipids, or its phosphorylation by kinases, leads to a conformational change that releases the VCA domain from sequestration and makes it accessible to actin and Arp2/3 (13).
WAVE and Abi are the substrates of several protein kinases including non-receptor tyrosine kinase Abl and Src, as well as proline-directed serine/threonine kinases such as extracellular signal-regulated kinase (Erk) and cyclin-dependent kinase 5 (CDK5) (7–10, 16–21). Several lines of evidence suggest that the phosphorylation of Abi and WAVE plays an important role in regulating WRC activation. Abi proteins, which comprise three members, Abi1–3, were initially identified as a binding partner and substrate of Abl tyrosine kinases (7, 8, 22). Subsequent studies show that the Abi proteins link the WAVE complex to Abl and oncogenic Bcr-Abl tyrosine kinases, leading to its tyrosine phosphorylation and translocation to the plasma membrane (9, 10, 20). Tyrosine 150 (Tyr-150) in WAVE2 has been identified as a target of Abl tyrosine kinase; its phosphorylation by Abl is essential for the activation of WRC by growth factors (9, 10). Studies by Chen et al. (13) suggest that the tyrosine phosphorylation of WAVE induces a conformational change that releases the VCA domain from intermolecular sequestration. Remarkably, a mutation of Tyr-150 to phosphomimetic aspartic acid is sufficient to activate the NPF activity of the WAVE complex (15). Although recent studies have begun to reveal how the WAVE complex is activated, little is known about the mechanism associated with its subsequent inactivation. A rapid inactivation of the WAVE signaling is particularly important for the regulation of mitotic entry, at which point the actin assembly must be inhibited at the cell margin and Golgi apparatus to ensure proper positioning of the spindle (23) and correct disassembly/segregation of the Golgi apparatus (24). How WRC is regulated to coordinate actin cytoskeleton remodeling and cell cycle progression through mitosis remains unclear.
Previously, we and others have shown that the expression of oncogenic Bcr-Abl in hematopoietic cells stimulates actin polymerization (20, 25–27). Bcr-Abl-induced actin polymerization requires the Abi1 pathway, as the blockade of the signal transduction from Bcr-Abl to Abi1 abolishes the F-actin assembly (20). Using Bcr-Abl-transformed BaF3 cells as a model system, we set out to investigate how the Bcr-Abl-induced F-actin assembly is regulated during cell cycle progression. Here, we report that Abi1 plays a dual role in linking both Bcr-Abl and the cell cycle regulatory signals to WRC and that mitotic serine phosphorylation of Abi1 by CDK1/cyclin B serves as a cell cycle-dependent regulatory mechanism that inhibits actin assembly and tyrosine phosphorylation of WRC induced by Bcr-Abl.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Ba/F3 cells were grown in RPMI 1640 containing 10% fetal bovine serum (FBS) and 15% WEHI3-conditioned medium as a source of IL-3. The Ba/F3 cells stably expressing wild-type p185Bcr-Abl (p185wt) were cultured in RPMI 1640 containing 10% FBS. COS-7, HeLa S3 (Clontech, Mountain View, CA), and retroviral packaging cell line GP2-293 (Clontech) were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS.
The preparation of rabbit polyclonal antibodies against Abi1, Abi2, and Sra has been described previously (28, 29). The antibodies against WAVE2, cAbl (K-12), GFP, cyclin B1, Cdc2, Crkl, and phosphotyrosine-containing proteins were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and the monoclonal antibodies for Abl were obtained from BD Biosciences. Rabbit polyclonal antibodies against human Hem1 and serine 216-phosphorylated Abi1 (anti-pSer-216) were generated in conjunction with Open Biosystems (Huntsville, AL) using the peptides with sequences corresponding to human Hem1-(548–561) (KQRQAPRKGEPERD) and human Abi1-(210–222) (PNDYMTpSPARLGS), respectively, as the antigens. Protein phosphatase 1 (PP1) was purchased from New England Biolabs (Ipswich, MA). Okadaic acid and roscovitine were obtained from LC Laboratories (Woburn, MA). Nocodazole and protease inhibitor mixture, as well as the antibodies against β-actin, histone H3, and serine 10-phosphorylated histone H3, were purchased from Sigma. Alexa-conjugated phalloidin and secondary antibodies were purchased from Molecular Probes (Eugene, OR).
Retroviral Constructs and Transfection
The construction of retroviral vector MSCV-GFP has been described previously (20). This plasmid was used to construct the retroviral vectors expressing wild type and mutant forms of GFP-Abi1 proteins. The cDNAs encoding Abi1S216A and Abi1S216D were generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and human Abi1 cDNA as template. The mutated cDNAs were then subcloned into MSCV-GFP at the BglII/XhoI sites. The desired mutations were confirmed by sequencing analysis. To generate retroviruses, the GP2-293 retroviral packaging cells were transfected with plasmid DNA using the FuGENE 6 transfection reagent (Roche Applied Science) according to the manufacturer's protocols. For retrovirus-mediated gene expression, retroviral supernatants were generated as described previously (30). COS-7, p185wt, and HeLa S3 cells were infected with retroviruses and then selected with puromycin (2 μg/ml) 48 h post-transduction.
Biochemical and Cell Biology Assays
Immunoprecipitation and Western blotting analyses were performed as described previously (31). In brief, cells were lysed in lysis buffer (20 mm Hepes, pH 7.2, 150 mm NaCl, 1% Triton X-100, and 10% glycerol) containing a protease inhibitor mixture and phosphatase inhibitors and incubated with appropriate antibodies bound to Sepharose beads. The immunoprecipitates were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with the appropriate primary and secondary antibodies. To treat the immunoprecipitated Abi1 and WAVE2 with PP1 and calf intestinal alkaline phosphatase, the immunoprecipitates were incubated with 2.5 or 10 units of PP1 or calf intestinal alkaline phosphatase, respectively, in 100 μl of lysis buffer at 37 °C for 30 min. After three washes with lysis buffer, the immunoprecipitates were separated on SDS-PAGE and analyzed by Western blotting.
In vitro CDK1/cyclin B kinase assay was performed according to the manufacturer's instructions. In brief, 5 μg of GST-Abi1 and GST proteins bound on glutathione-Sepharose 4B (Amersham Biosciences) was washed three times with kinase buffer (25 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 5 mm β-glycerophosphate, 0.1 mm Na3VO4, 2 mm dithiothreitol, and 0.2 mm ATP). The proteins were then incubated with 150 ng of CDK1/cyclin B kinase (Cell Signaling Technology) in 150 μl of kinase buffer for 30 min at 30 °C. The kinase reaction was stopped by the addition of 150 μl of 50 mm EDTA (pH 8.0), and GST fusion proteins were analyzed by Western blotting using anti-pSer-216 antibody after washing three times with PBS/T (1× PBS and 0.05% Tween 20).
To deplete CDK1 from the mitotic p185wt cell extracts, 1 × 107 mitotic p185wt cells were lysed in 1 ml of lysis buffer and subjected to three rounds of immunoprecipitation using anti-CDK1 antibody (CDK1-depleted) or control IgG (mock-depleted) bound to protein A-Sepharose beads. The amount of residual CDK1 was measured by Western blotting with anti-CDK1 antibody. GFP-Abi1 was affinity-purified from asynchronous p185wt cells expressing GFP-Abi1 by anti-GFP antibody bound to protein A-Sepharose beads. The aliquots of purified GFP-Abi1 were incubated with 150 μl of mock- and CDK1-depleted extracts supplemented with 200 μm ATP. After 30 min of incubation at 37 °C, the bead-bound GFP-Abi1 was washed three times with lysis buffer and analyzed for Ser-216 phosphorylation by Western blot using anti-pSer-216 antibody. To confirm that an equal amount of GFP-Abi1 was used in the reactions, the blot was stripped and reprobed with Abi1 antibody.
A cell proliferation assay was performed in 6-well plates. Briefly, p185wt cells stably transfected with the indicated plasmids were plated in triplicate at a density of 1 × 104/well in RPMI 1640 containing 10% FBS. Cells were harvested on days 1–4, respectively, and counted.
Cell Synchronization
Synchronization of HeLa S3 cells at various stages of division cycle was performed as described previously (32). Asynchronous cells were harvested by lysing cells directly on the plate with lysis buffer after a wash with PBS. G0-synchronized cells were harvested after incubation of the cells in serum-free media for 48 h. To harvest G1-phase cells, G0-arrested cells were released into serum-containing media for 2 h and collected. To synchronize cells at the G1/S boundary, HeLa cells were incubated with media containing 2 mm thymidine for 16 h, washed with PBS, released into normal media for 12 h, and then incubated in media containing 2 mm thymidine for 15 h. S-phase cells were prepared by releasing G1/S cells into regular media for 3 h. Mitotic HeLa cells and COS-7 cells were prepared by incubating cells in media containing 100 ng/ml nocodazole for 16 h. Mitotic cells were then collected by mechanical shake-off. The Ba/F3 cells and p185wt cells were synchronized at M-phase as described (33). Briefly, cells were incubated with 200 ng/ml nocodazole for 12 h. In the last 2 h of treatment, okadaic acid was added to a final concentration of 1 μm. Under this condition, up to 75% of the p185wt cells were arrested in M-phase, as determined by examining the mitotic index of DAPI-stained cells. The treatment also resulted in the maximal inhibition of F-actin-rich structures as well as the tyrosine phosphorylation of WAVE2 complex (supplemental Fig. S1, A and C). In some experiments, as indicated in Fig. 1 and supplemental Fig. S1, treatment of p185wt cells with nocodazole (200 ng/ml for 12 h) or okadaic acid (1 μm for 4 h) alone was also performed. These treatments resulted in ∼50 and 30% of the cells, respectively, arrested in mitosis (supplemental Fig. S1A). In all treatments used for mitotic arrest of p185wt cells, cell viability was determined by a trypan blue exclusion test; no significant cell death was found in association with these methods (viability > 95%).
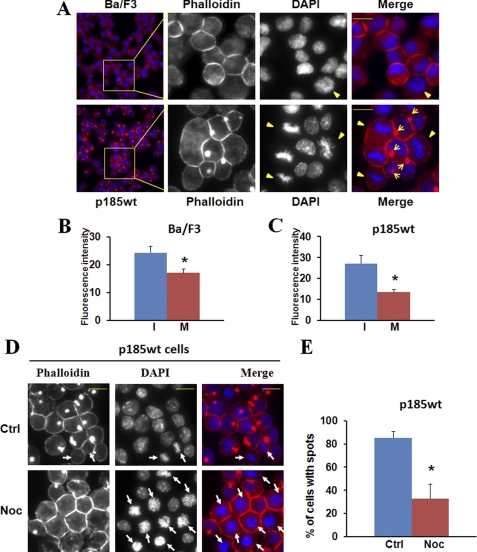
FIGURE 1.
Mitotic inhibition of F-actin assembly in Ba/F3 cells and Ba/F3 cells transformed by p185Bcr-Abl. A, Ba/F3 cells and the Ba/F3 cells transformed by p185Bcr-Abl (p185wt) were fixed, permeabilized, and stained with Alexa Fluor 546-conjugated phalloidin and DAPI to visualize F-actin (red) and nuclei (blue), respectively. The stained cells were analyzed by fluorescence microscopy at ×10 and ×40 (insets) magnification. Mitotic cells are indicated by arrowheads and F-actin-enriched spots by arrows. Bar: 10 μm. B and C, Ba/F3 cells (B) and p185wt cells (C) were fixed and stained with Alexa Fluor 546-conjugated phalloidin and DAPI. The fluorescence intensity of randomly selected interphase cells (I) and mitotic cells (M) was quantified using ImageJ software. The data are expressed as the average intensity (arbitrary units) of three independent experiments with at least 10 cells for each condition counted in each experiment. *, p < 0.01. D and E, p185wt cells were treated without (Ctrl) or with nocodazole (Noc; 200 ng/ml) for 12 h. The cells were fixed and stained with Alexa Fluor 546-conjugated phalloidin and DAPI. The cells at M-phase are indicated by arrows (D). The percentage of cells with actin-enriched spots was calculated from three randomly selected areas (E). The data are representative of three independent experiments. *, p < 0.01.
Immunocytochemistry and Fluorescence Microscopy
Cultured Ba/F3 and p185wt cell lines were fixed in 3.75% formaldehyde in PBS for 10 min, and 5–10 × 103 fixed cells were then loaded per slide by cytospin. The cells were permeabilized in 0.2% Triton X-100/PBS for 5 min, and stained with 50 μg/ml Alexa Fluor 546-conjugated phalloidin in PBS. After extensive washing with PBS and a brief staining with DAPI (Sigma) to visualize the nuclei, slides were mounted with SlowFade Gold Antifade reagent (Invitrogen). Images were captured and analyzed using Nikon TE-2000 fluorescence microscope with ImageJ software associated.
ImageJ Quantification
ImageJ software was used to quantify the fluorescence intensity of BaF3 and P185wt cells stained by Alexa Fluor 546-conjugated phalloidin. The intensity of fluorescent objects was measured after applying a fixed threshold on projected pictures, excluding objects with a size of less than 2–3 pixels. The average fluorescence intensity per spot (arbitrary units), calculated using the ”measure” function, was normalized to control. Typically, each calculated average derives from three independent experiments/condition, analyzing at least 10 cells/experiment.
Statistical Analyses
Descriptive statistics were generated for all quantitative data with the presentation of average ± S.D. The statistical significance of comparisons between experimental groups was tested using Student's t test with Microsoft Excel software.
RESULTS
Bcr-Abl-induced F-actin Assembly Is Inhibited during Mitosis
In cultured adherent cells, despite the constitutive exposure to serum and growth factors, those cells entering mitosis round up and retract the specialized adhesion and migration structures such as lamellipodia in the leading edge, suggesting a cell cycle-dependent regulatory mechanism for actin remodeling (34). To determine whether this regulatory mechanism is also at work in mitotic hematopoietic cells cultured in suspension, we stained a murine hematopoietic cell line Ba/F3 and its Bcr-Abl-transformed derivative line, Ba/F3p185Bcr-Abl (p185wt) with TRITC-conjugated phalloidin and DAPI to visualize F-actin and nuclei, respectively. We then analyzed the F-actin content in interphase and mitotic cells using fluorescence microscopy and ImageJ quantification. The F-actin content is decreased in mitotic Ba/F3 and p185wt cells compared with that in the interphase cells (Fig. 1), suggestive of a mitotic inhibition of F-actin assembly in these cells.
As reported previously (20), the expression of Bcr-Abl in Ba/F3 cells induced the assembly of an F-actin-enriched structure at the sites adjacent to membrane (Fig. 1A and supplemental Fig. S1A). This F-actin-rich structure is similar to invadopodium, a specialized adhesion/migration structure found in many tumor cells (35), in that they both appear as a dynamic, membrane-associated structure enriched with not only F-actin but also adhesion molecules such as paxillin and integrin (Ref. 20 and supplemental Fig. S2). To determine whether the Bcr-Abl-induced F-actin assembly is also inhibited during mitosis, we analyzed F-actin structures in interphase and mitotic p185wt cells by fluorescence microscopy. Remarkably, although 88% of the interphase cells formed F-actin-rich structures, none of mitotic p185wt cells displayed such structures (Fig. 1A and supplemental Fig. S1B). To confirm that the loss of F-actin-rich structures in mitotic p185wt cells is caused by cell entry into mitosis, we examined the effect of nocodazole treatment, which synchronizes cells at mitotic phase (M-phase) by interfering with the polymerization of microtubules and the ormation of mitotic spindle on Bcr-Abl-induced F-actin assembly. As expected, nocodazole treatment of p185wt cells increased the percentage of mitotic cells (the mitotic index) from 2.8 to ∼50% (Fig. 1D). Conversely, the percentage of p185wt cells with F-actin-enriched structures decreased from 85 to 30%, and none of the mitotic cells show the F-actin-rich structure (Fig. 1E). Similar results were also obtained by treating p185wt cells with okadaic acid (supplemental Fig. S1A), an inhibitor of serine/threonine phosphatases that also induces mitotic arrest of Bcr-Able-positive hematopoietic cells (36). Collectively, these data suggest that Bcr-Abl-induced F-actin assembly is inhibited when cells enter mitosis.
WAVE2 Is Tyrosine-phosphorylated by Bcr-Able, and Its Tyrosine Phosphorylation Is Attenuated when Cells Enter Mitosis
We have shown previously that Bcr-Abl-induced F-actin assembly requires Abi1 (20, 37). Blockade of the interaction between Bcr-Abl and Abi1, either by the deletion of the Src homology 3 (SH3) domain and C-terminal PXXP motif in Bcr-Abl (p185ΔSH3ΔC) or mutation of prolines 180 and 434 in the PXXP motif and the SH3 domain of Abi1 to leucine (Abi1PPLL), abolished not only the tyrosine phosphorylation of Abi1 but also the formation of F-actin-rich structures (Ref. 20 and supplemental Fig. S2). In addition, we found that Abi1 is present in Ba/F3 cells mainly as a complex with WAVE2 in the cytosol and that, upon Bcr-Abl transformation, both Abi1 and WAVE2 are translocated to membrane where they found in association with F-actin-rich structures (Ref. 20 and supplemental Fig. S2). These findings suggest that Abi1/WAVE2 signaling may play a critical role in Bcr-Abl-induced F-actin assembly.
Recent studies have shown that the tyrosine phosphorylation of WAVE by Abl tyrosine kinases is essential and sufficient for its activation (9, 10, 13). To investigate whether WAVE2 is activated by Bcr-Abl, we examined the tyrosine phosphorylation of WAVE2 in Ba/F3 cells and the Ba/F3 cells expressing p185wt and p185ΔSH3ΔC, a mutant Bcr-Abl that has constitutive tyrosine kinase activity but is defective in binding to Abi1 and inducing F-actin-rich structures (20, 30). As shown in Fig. 2A (upper panel), WAVE2 is tyrosine-phosphorylated in p185wt cells but not in parental Ba/F3 cells and the p185ΔSH3ΔC cells, suggesting that WAVE2 is activated by Bcr-Abl and that this activation is mediated by Abi1.
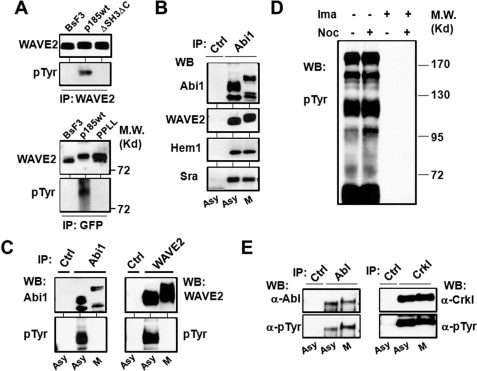
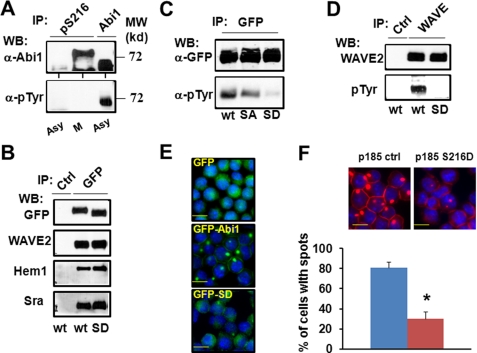
FIGURE 2.
Bcr-Abl-induced tyrosine phosphorylation of the WAVE2 complex is inhibited in mitotic cells. A, Abi1-mediated tyrosine phosphorylation of WAVE2 by Bcr-Abl. Upper panel, WAVE2 was immunoprecipitated (IP) from Ba/F3 cells and the Ba/F3 cells transformed by either wild type p185Bcr-Abl (p185wt) or mutant p185ΔSH3ΔC (ΔSH3ΔC) defective in binding to Abi1. The immunoprecipitates were analyzed by Western blotting (WB) for WAVE2 and phosphotyrosine (pTyr) as indicated. Lower panel, the lysates of Ba/F3 and p185wt cells expressing wild type GFP-Abi1 (BaF3 and p185wt), as well as p185wt cells expressing mutant GFP-Abi1PPLL (PPLL) defective in binding to Bcr-Abl, were immunoprecipitated by GFP antibody, and the immunoprecipitates were analyzed by Western blotting for phosphotyrosine. The same membrane was then stripped and reprobed with anti-WAVE2 antibody. B, Abi1 forms a complex with other components of WAVE2 complex in both interphase and mitotic cells. The p185wt cells were untreated (asynchronous cells (asy)) or synchronized at M-phase (M) as described under “Experimental Procedures.” The cell lysates were immunoprecipitated with control IgG (Ctrl) or anti-Abi1 antibody (Abi1). The immunoprecipitates were subjected to Western blot analysis using the indicated antibodies. C, Bcr-Abl-induced tyrosine phosphorylation of Abi1 and WAVE2 is inhibited during mitosis. The lysates from asynchronous or M-phase-synchronized cells were immunoprecipitated by control IgG, anti-Abi1, or anti-WAVE2 antibodies as indicated. The immunoprecipitates were subjected to Western blot analysis using the antibodies for Abi1, WAVE2, or tyrosine-phosphorylated proteins. D, profile of protein tyrosine phosphorylation in p185wt cells treated with or without nocodazole. The p185wt cells were treated with and without 200 ng/ml nocodazole in the presence or absence of 1 μm imatinib (Ima) for 12 h. The lysates containing 80 μg of total proteins were separated on 8% SDS-PAGE and analyzed by Western blotting using anti-phosphotyrosine antibody. E, the tyrosine kinase activity of Bcr-Abl is not affected during mitosis. The p185wt cells were treated with or without 200 ng/ml nocodazole for 12 h, and cell lysates were immunoprecipitated by control IgG, anti-Abl antibody (left panel), or anti-Crkl antibody (right panel). The immunoprecipitates were separated on SDS-PAGE and subjected to Western blot analysis using the indicated antibodies.
To confirm that the Bcr-Abl-induced tyrosine phosphorylation of WAVE2 is mediated by Abi1, we also examined the tyrosine phosphorylation of the WAVE2 that is associated with either wild type Abi1 or Abi1PPLL, a mutant Abi1 defective in binding to Bcr-Abl and inducing F-actin-rich structures in p185wt cells (20). GFP-tagged Abi1 and Abi1PPLL were expressed in Ba/F3 cells and p185wt cells, and the Abi1/WAVE2 complex was immunoprecipitated using GFP antibody and subsequently analyzed for tyrosine phosphorylation by Western blotting. Superimposition of blots of the same membrane probed with antibodies specific for WAVE2 and phosphotyrosine indicates that WAVE2 in complex with GFP-Abi1 is tyrosine-phosphorylated in p185wt cells but not BaF3 cells (Fig. 2A, lower panel). In contrast, the WAVE2 in complex with Abi1PPLL failed to be tyrosine-phosphorylated in p185wt cells (Fig. 2A, lower panel). Taken together, we conclude that WAVE2 is a downstream target of Bcr-Abl and that Bcr-Abl-induced tyrosine phosphorylation of WAVE2 is mediated by Abi1.
We noticed that WAVE2 in both p185wt GFP-Abi1 and p185wt GFP-Abi1PPLL cells displayed a slower mobility on SDS-PAGE compared with that in Ba/F3 cells (Fig. 2A and supplemental Fig. S3B), yet only the WAVE2 in p185wt GFP-Abi1 cells was tyrosine-phosphorylated. A possible explanation for this observation is that the mobility shift of the WAVE2 in these cells is caused mainly by serine/threonine phosphorylation. Indeed, WAVE2 has previously been shown to be phosphorylated on multiple serine/threonine residues by Erk (19), a serine/threonine kinase known to be activated by Bcr-Abl. Consistent with this, we found that the mobility shift of WAVE2 was reversible by phosphatase treatment (supplemental Fig. S3B).
To identify the mechanism associated with mitotic inhibition of Bcr-Abl-induced F-actin assembly, we examined Abi1/WAVE2 signaling in asynchronous and mitotic p185wt cells. The Abi1/WAVE2 forms a complex with Hem1 (a member of the Nap family proteins expressed in hematopoietic cells) and Sra in both interphase and mitotic p185wt cells (Fig. 2B). Both Abi1 and WAVE2 are tyrosine-phosphorylated in asynchronous p185wt cells (Fig. 2C). However, mitotic arrest of p185wt cells results in a drastic decrease in tyrosine phosphorylation of Abi1 and WAVE2 compared with asynchronous cells (Fig. 2C). Thus, our data indicate a strong correlation between the inhibition of F-actin assembly and the attenuation of Bcr-Abl-stimulated tyrosine phosphorylation of Abi1 and WAVE2 in mitotic p185wt cells.
Abi1 Is Hyperphosphorylated in Mitotic Cells
To determine whether the decrease in tyrosine phosphorylation of Abi1 and WAVE2 could be caused by a reduction of the tyrosine kinase activity of Bcr-Abl in mitotic cells, we analyzed protein tyrosine phosphorylation profiles in asynchronous and mitotic p185wt cells. Bcr-Abl stimulated robust protein tyrosine phosphorylation in both asynchronous and mitotic p185wt cells (Fig. 2D). In addition, no decrease in Bcr-Abl autophosphorylation was observed in mitotic p185wt cells compared with the asynchronous cells (Fig. 2E, left panel). Furthermore, there was no difference in the tyrosine phosphorylation of Crkl, a known substrate of Bcr-Abl tyrosine kinase, between asynchronous p185wt cells and the mitotic p185wt cells (Fig. 2E, right panel). These results suggest that it is unlikely that the decreased tyrosine phosphorylation of Abi1 and WAVE2 in mitotic p185wt cells is caused by a reduction of Bcr-Abl tyrosine kinase activity.
As Abi1 is a key component that links Bcr-Abl to the WAVE2 complex, we then asked whether the mitotic inhibition of the tyrosine phosphorylation of WAVE2 complex was attributable to a cell cycle-dependent modification of Abi1. In analyzing the complex formation and tyrosine phosphorylation of Abi1 and WAVE2 in p185wt cells, we noticed a shift of Abi1 to slower mobility on SDS-PAGE in mitotic p185wt cells compared with the asynchronous cells (Fig. 2, B and C). This suggests that either the expression of an alternative splicing variant or post-translational modification of Abi1 may occur during mitosis. To determine whether Abi1 undergoes post-translational modification such as protein phosphorylation during mitosis, we made use of BaF3 and p185wt cells expressing a GFP-tagged Abi1. These cells (named Ba/F3 GFP-Abi1 and p185wt GFP-Abi1 cells, respectively) were treated with nocodazole or nocodazole plus okadaic acid, and the expression of GFP-Abi1 was examined by Western blotting. Nocodazole treatment resulted in a partial shift of GFP-Abi1 to slower mobility on SDS-PAGE in both p185wt GFP-Abi1 (Fig. 3A, compare lane 2 with lane 1) and Ba/F3 GFP-Abi1 (Fig. 3B, compare lane 2 with lane 1) cells. This mobility shift is likely caused by serine/threonine phosphorylation, because the addition of okadaic acid in the last 2 h of nocodazole treatment led to a complete shift of Abi1 to slower mobility on SDS-PAGE (Fig. 3, A and B, compare lane 3 with lane 1 in each panel). Similar to Ba/F3 and p185wt cells, a shift of Abi1 to slower mobility on SDS-PAGE was also observed in HeLa cells synchronized at M-phase (Fig. 3, C and D). The mobility shift of Abi1 in mitotic HeLa cells was caused by serine/threonine phosphorylation, as it could be reversed by PP1, a serine/threonine phosphatase (Fig. 3E). These studies suggest that Abi1 may undergo active serine/threonine phosphorylation and dephosphorylation during mitosis.
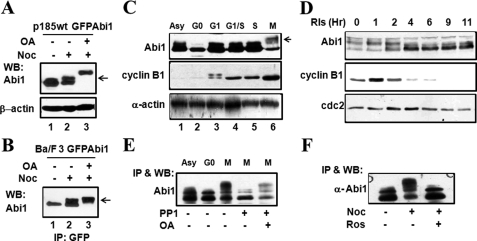
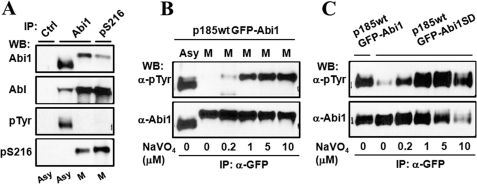
FIGURE 3.
Abi1 is hyperphosphorylated in correlation with CDK1/cyclin B activity during mitosis. A and B, mitotic arrest induces a mobility shift of Abi1 on SDS-PAGE. Ba/F3 cells (B) and p185wt cells (A) expressing GFP-Abi1 were treated with or without 200 ng/ml nocodazole (Noc) for 12 h in the presence or absence of 1 μm okadaic acid (OA) in the last 2 h of the treatment as indicated. The cell lysates containing 80 μg of total proteins (A) or immunoprecipitates by GFP-antibody (B) were separated on 8% SDS-PAGE and subjected to Western blot (WB) analysis using indicated antibodies. The arrows indicate Abi1 with slower mobility. C, cell cycle-dependent mobility shift of Abi1 in HeLa cells. Asynchronous (Asy) HeLa cells and HeLa cells synchronized at various stages of cell cycle by: serum starvation (G0), release from G0 for 2 h (G1), double thymidine block (G1/S), release from G1/S for 3 h (S), and nocodazole treatment (M) were collected. The cell lysates containing 80 μg of total proteins were separated on SDS-PAGE and subjected to Western blot analysis with the indicated antibodies. Cell synchronization was also confirmed by Western blot analysis using anti-phosphorylated histone H3 antibody, as shown in supplemental Fig. S3. The arrow indicates Abi1 with slower mobility. D, HeLa cells released from nocodazole-synchronized M-phase were collected at various time points, and the cell lysates were analyzed as described in C. E, mobility shift of Abi1 is reversed by PP1 treatment. The Abi1 immunoprecipitated from the HeLa cells synchronized at M-phase by nocodazole treatment were incubated with or without PP1 in the presence or absence of okadaic acid, as indicated. The treated immunoprecipitates, together with the Abi1 immunoprecipitates from asynchronous and G0-arrested HeLa cells as controls, were separated on SDS-PAGE and subjected to Western blotting analysis using anti-Abi1 antibody. F, mitotic mobility shift of Abi1 is inhibited by roscovitine. HeLa cells were treated with or without 100 ng/ml nocodazole for 16 h. The cells were then exposed to 50 μm roscovitine, as indicated, for an additional 2 h in the continuous presence of nocodazole. The cell lysates were immunoprecipitated (IP) by anti-Abi1 antibody followed by Western blot analysis with the indicated antibodies.
Abi proteins have been shown to interact with CDK1 (38), which is activated at the onset of mitosis (39). We found that the mitotic phosphorylation of Abi1 correlates well with the level of cyclin B, a regulatory subunit of CDK1 required for its activation at the onset of mitosis (Fig. 3, C and D). Treatment of mitotic HeLa cells with roscovitine, a pharmacologic inhibitor of CDK1, completely abrogated mitotic phosphorylation of Abi1 (Fig. 3F), suggesting an involvement of CDK1/cyclin B in mitotic phosphorylation of Abi1.
Mitotic Phosphorylation of Serine 216 in Abi1 by CDK1
Sequence analysis of Abi1 revealed that three serine/threonine (Ser/Thr) residues, Thr-198, Ser-216, and Ser-225 (Fig. 4A), are in consensus sequences for CDK1/cyclin B phosphorylation (40). Among these, Ser-216 has been shown to be a mitotic phosphorylation site in a phosphoproteomic search for mitotic phosphorylation events (41). To determine whether the Ser-216 in Abi1 is phosphorylated during mitosis, we generated a polyclonal antibody (anti-pSer-216) against the phosphopeptide that corresponds to the sequences flanking the Ser-216 in Abi1. Specificity of this antibody to Ser-216-phosphorylated Abi1 was demonstrated by three lines of evidence. First, the anti-pSer-216, but not preimmune serum (data not shown), specifically recognized the GFP-tagged Abi1 expressed in mitotic HeLa cells and p185wt cells (Fig. 4B). Second, the reaction of anti-pSer-216 to GFP-Abi1 could be competitively blocked by Ser-216 phosphopeptide but not the non-phosphopeptide in Western blotting analysis (data not shown). Third, we generated an expression vector encoding for a phospho-deficient Abi1 mutant in which Ser-216 was replaced by alanine (GFP-Abi1S216A). Using this expressing vector, we were able to show that the antibody recognizes only wild type GFP-Abi1 expressed in mitotic HeLa and p185wt cells but not GFP-Abi1S216A (Fig. 4B).
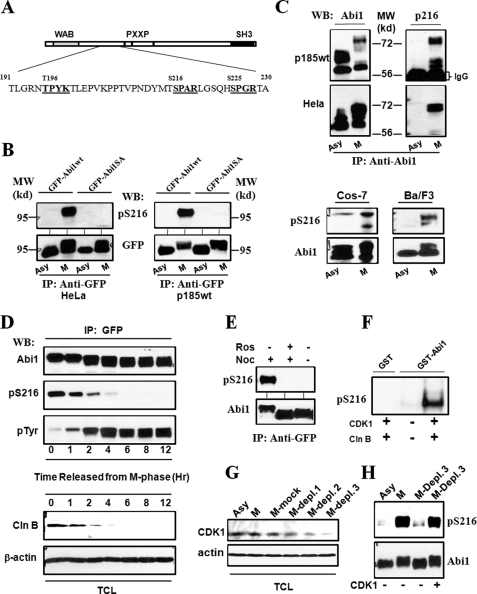
FIGURE 4.
Mitotic phosphorylation of Abi1 Ser-216 by CDK1. A, schematic representation of potential CDK1 phosphorylation sites in Abi1. The sequences that conform to consensus CDK1 phosphorylation motif ((S/T)PX(K/R)) are presented in bold. B, mitotic phosphorylation of the GFP-tagged Abi1 expressed in HeLa and p185wt cells. The HeLa (left panel) and p185wt (right panel) cells expressing GFP-Abi1 (GFP-Abi1wt) and GFP-Abi1S216A (GFP-Abi1SA) were treated with or without nocodazole for 16 and 12 h, respectively. The lysates were immunoprecipitated (IP) with anti-GFP antibody, and the immunoprecipitates were subjected to Western blot (WB) analysis using the specific antibody generated against Ser-216 phosphopeptide (anti-pSer-216) or anti-GFP antibody, as indicated. A 95-kilodalton molecular mass marker (MW) is indicated. C, mitotic phosphorylation of Ser-216 of endogenous Abi1 in Ba/F3, COS-7, HeLa, and p185wt cells. Upper panel, p185wt and HeLa cells, as indicated, were treated with or without nocodazole as described in B. The cell lysates were immunoprecipitated by anti-Abi1 antibody, and the immunoprecipitates were analyzed by Western blotting using anti-Abi1 (left panel) and anti-pSer-216 (right panel) antibodies. Lower panel, asynchronous (Asy) COS-7 and Ba/F3 cells as well as the COS-7 and Ba/F3 cells synchronized at M-phase (M), as described under “Experimental Procedures,” were lysed and analyzed by Western blotting for Ser-216 phosphorylation using anti-Abi1 and pSer-216 antibodies, as indicated. D, cell cycle-dependent phosphorylation of Abi1 in p185wt cells. p185wt GFP-Abi1 cells were synchronized at M-phase by treatment with nocodazole (200 ng/ml) and okadaic acid (1 μm) as described under “Experimental Procedures.” The cells were then washed three times with PBS and grown in normal growth medium for indicated time periods. The cell lysates were immunoprecipitated by anti-GFP antibody. The immunoprecipitates and total cell lysates (TCL) were analyzed by Western blotting using indicated antibodies. Mitotic synchronization was confirmed by examining mitotic index of DAPI-stained cells and by Western blot analysis of the cell lysates using anti-phosphorylated histone H3 antibody as shown in supplemental Figs. S1 and S3, respectively. E, inhibition of Ser-216 phosphorylation by roscovitine. p185wt cells expressing GFP-Abi1 were treated with or without 200 ng/ml nocodazole (Noc) for 12 h in the presence or absence of 50 μm roscovitine (Ros). The cell lysates were immunoprecipitated with anti-GFP antibody, and the immunoprecipitates were subjected to Western blotting analysis using anti-Abi1 and anti-pSer-216 antibodies as indicated. F, in vitro phosphorylation of Ser-216 by CDK1/cyclin B. 5 μg of GST-Abi1, or GST as a control, were incubated with or without purified CDK1/cyclin B in an in vitro kinase reaction for 30 min. The GST fusion proteins were then subjected to Western blotting analysis using anti-pSer-216 antibody. G, depletion of CDK1 from the mitotic p185wt cell lysates. The lysates from the mitotic p185wt cells (M) were subjected to three rounds of immunoprecipitation with anti-CDK1 antibody (M-Depl. 1–3) or control IgG (M-mock). These lysates, together with the lysate from asynchronous p185wt cells, were then analyzed by Western blot using the indicated antibodies. H, aliquots of the GFP-Abi1 purified from interphase p185wt GFP-Abi1 cells were incubated with the asynchronous p185wt cell lysates or the mitotic p185wt cell lysates in which CDK1 was not depleted, or they were depleted by three rounds of immunoprecipitation (M-Depl. 3) as described in G. Ser-216 phosphorylation was examined by Western blot using anti-pSer-216 antibody. The blot was then stripped and reprobed with anti-Abi1 antibody to confirm that an equal amount of GFP-Abi1 was used.
Using anti-pSer-216 antibody, we examined the phosphorylation of Abi1 Ser-216 in Ba/F3, COS-7, HeLa, and p185wt cells. When these cells were synchronized at M-phase, a robust increase in the levels of Ser-216-phosphorylated Abi1 was observed (Fig. 4C). The mitotic specific phosphorylation of Abi1 Ser-216 was also evidenced by the analysis of p185wt GFP-Abi1 cells that were synchronized at M-phase and subsequently released from mitotic arrest. As shown in Fig. 4D, the level of Ser-216-phosphorylated Abi1 peaked at M-phase and decreased rapidly as the cells exited mitosis.
To determine whether CDK1/cyclin B is responsible for the mitotic phosphorylation of Abi1 Ser-216, we compared Ser-216 phosphorylation with the expression of cyclin B during cell cycle progression through mitosis. We found that the level of Ser-216-phosphorylated Abi1 correlates well with the level of cyclin B as cells progressed through mitosis (Fig. 4D). The treatment of the mitotic p185wt cells with the CDK1 inhibitor roscovitine abrogated the Ser-216 phosphorylation of Abi1 (Fig. 4E). In addition, using an in vitro kinase assay, we were able to show that the Ser-216 of Abi1 is phosphorylated by purified CDK1/cyclin B (Fig. 4F). To further confirm that CDK1 is the specific kinase for the mitotic phosphorylation of Ser-216, we depleted CDK1 from the mitotic extracts of p185wt cell by three-round immunoprecipitation (Fig. 4G). We then examined the ability of these extracts to phosphorylate Ser-216 of the GFP-Abi1 that was purified from interphase cells. As shown in Fig. 4H, Ser-216 phosphorylation of GFP-Abi1 incubated with the mitotic p185wt cell extract was higher than that of GFP-Abi1 incubated with the asynchronous p185wt cell extract. Remarkably, depletion of CDK1 from the mitotic extracts abolished the Ser-216 phosphorylation; this could be rescued by adding CDK1/cyclin B back to the CDK1-depleted extract (Fig. 4H). Altogether, the data support the proposition that CDK1/cyclin B is the kinase responsible for the mitotic phosphorylation of Abi1 serine 216.
Phosphorylation of Ser-216 in Abi1 Inhibits Bcr-Abl-induced Tyrosine Phosphorylation of WAVE2 Complex and Attenuates F-actin Assembly
It was notable that, in p185wt GFP-Abi1 cells released from mitotic arrest, the rapid decrease of Ser-216-phosphorylated Abi1 was accompanied by an increase in tyrosine phosphorylation of Abi1 (Fig. 4D). This finding suggests that the phosphorylation of Ser-216 may attenuate the Bcr-Abl-induced tyrosine phosphorylation of Abi1. To further test this proposition, we immunoprecipitated Ser-216-phosphorylated Abi1 from mitotic p185wt cells by anti-pSer-216 antibody and compared its tyrosine phosphorylation with that of the Abi1 immunoprecipitated from interphase p185wt cells by a pan-Abi1 antibody in which little Ser-216-phosphorylated Abi1 could be detected (Fig. 4C). The endogenous Abi1 immunoprecipitated from interphase p185wt cells was heavily tyrosine-phosphorylated (Fig. 5A). However, the tyrosine phosphorylation of the Abi1 pulled down by anti-pSer-216 from mitotic p185wt cells was dramatically decreased compared with that of the Abi1 pulled down by anti-Abi1 antibody from the interphase p185wt cells, despite the fact that an approximately equivalent amount of Abi1 proteins was immunoprecipitated (Fig. 5A). These results are consistent with a role for the Ser-216 phosphorylation of Abi1 in the inhibition of its tyrosine phosphorylation by Bcr-Abl.
FIGURE 5.
Phosphorylation of Ser-216 in Abi1 inhibits Bcr-Abl-induced tyrosine phosphorylation and membrane translocation of WAVE2 complex and attenuates F-actin assembly. A, Ser-216 phosphorylation of Abi1 inhibits its tyrosine phosphorylation stimulated by Bcr-Abl. Cell lysates from asynchronous (Asy) and mitotic p185wt cells (M) were immunoprecipitated (IP) by anti-Abi1 and anti-pSer-216 antibodies. The immunoprecipitates were analyzed by Western blotting (WB) using the indicated antibodies. B, phosphomimetic mutation of Ser-216 in Abi1 does not affect its complex formation with Hem1, Sra, or WAVE2. The lysates from the p185wt cells expressing wild type GFP-Abi1 (wt) or GFP-Abi1S216D (SD) were immunoprecipitated with a control IgG or the anti-GFP antibody. The immunoprecipitates were subjected to Western blot analysis using the indicated antibodies. C, phosphomimetic mutation of the Ser-216 is sufficient to attenuate Bcr-Abl-induced tyrosine phosphorylation of Abi1. The p185wt cells were transfected by vectors expressing wild type GFP-Abi1 (wt) or the mutant forms of GFP-Abi1 in which the Ser-216 was replaced by alanine (SA) or aspartic acid (SD). The cell lysates were immunoprecipitated by a control IgG (Ctrl) or an antibody against GFP. The immunoprecipitates were subjected to Western blot analysis using anti-phosphotyrosine (pTyr) or anti-GFP antibodies as indicated. D, expression of the phosphomimetic mutant Abi1 reduced Bcr-Abl-induced tyrosine phosphorylation of WAVE2. p185wt cells expressing wild type GFP-Abi1 or GFP-Abi1 with the mutation of Ser-216 to aspartic acid were immunoprecipitated by a control IgG or anti-WAVE2 antibody (WAVE). The immunoprecipitates were subjected to Western blot analysis using the antibodies against WAVE2 and phosphotyrosine as indicated. E, phosphomimetic mutation of Ser-216 alters the subcellular distribution of Abi1 in p185wt cells. The p185wt cells expressing GFP, GFP-Abi1, and GFP-Abi1S216D (GFP-SD) were stained with DAPI. The GFP and GFP fusion proteins (green), as well as the nuclei (blue), were visualized, and images were captured by fluorescence microscopy. F, expression of Abi1S216D attenuates Bcr-Abl-stimulated actin polymerization. The p185wt (p185 ctrl) and p185wt cells expressing Abi1S216D (p185 S216D) were stained with Alexa-conjugated phalloidin and DAPI to visualize F-actin (red) and nuclei (blue), respectively. The stained cells were analyzed by fluorescence microscopy (upper panel). The cells showing F-actin-enriched spots were counted, and the percentage of these cells compared with total cells in the area was calculated and expressed as the average percentage of three randomly selected areas (lower panel). *, p < 0.01. The data are representative of three independent experiments.
To determine whether Ser-216 phosphorylation of Abi1 is sufficient to inhibit its tyrosine phosphorylation by Bcr-Abl, we generated p185wt cells expressing a GFP-tagged phosphomimetic Abi1 mutant in which Ser-216 was mutated to aspartic acid (GFP-Abi1S216D). These cells, together with the p185wt cells expressing wild type GFP-Abi1 and phospho-deficient mutant GFP-Abi1S216A, were lysed, and GFP-tagged proteins were immunoprecipitated by anti-GFP-antibodies. As shown in Fig. 5B, phosphomimetic mutation of Ser-216 does not affect the complex formation of GFP-Abi1S216D with WAVE2, Hem1, or Sra. Both GFP-Abi1wt and GFP-Abi1S216A are tyrosine-phosphorylated in p185wt cells; however, the tyrosine phosphorylation of GFP-Abi1S216D is decreased compared with GFP-Abi1wt and GFP-Abi1S216A (Fig. 5C), suggesting that the replacement of the Ser-216 by a phosphomimetic amino acid is sufficient to attenuate Bcr-Abl-induced tyrosine phosphorylation of Abi1.
We also examined the effect of the phosphomimetic mutation of Ser-216 on Bcr-Abl-induced tyrosine phosphorylation of WAVE2. As shown in Fig. 5D, the expression of GFP-Abi1S216D, but not GFP-Abi1wt, inhibited Bcr-Abl-induced tyrosine phosphorylation of WAVE2.
The tyrosine phosphorylation of WAVE2 complex is required for its activation as well as membrane translocation (9, 13). Because the phosphomimetic mutation of Ser-216 inhibited Bcr-Abl-induced tyrosine phosphorylation of Abi1 and WAVE2, we asked whether this mutation also affects the subcellular localization of GFP-Abi1S216D in p185wt cells. As we reported previously (20), GFP-Abi1wt was found concentrated in a spot adjacent to the plasma membrane in ∼80% of the p185wt cells (Fig. 5E). In contrast, GFP-Abi1S216D exhibited more diffuse cytoplasmic distribution compared with GFP-Abi1wt, suggesting that the phosphomimetic mutation of Ser-216 attenuates Bcr-Abl-induced translocation of Abi1 to membrane. To determine whether the Ser-216 phosphorylation of Abi1 plays a role in regulating WAVE2-mediated F-actin assembly in p185wt cells, we examined the effect of the expression of the phosphomimetic Abi1S216D on F-actin organization in p185wt cells. As shown in Fig. 5F, more than 80% of the p185wt cells transfected with a control vector have F-actin-enriched spots. However, the percentage of cells with F-actin-enriched structures is reduced to 30% in p185wt cells expressing Abi1S216D. Collectively, these studies strongly suggest that the phosphorylation of Abi1 at Ser-216 may attenuate, at least in part, the Abi1/WAVE2 signaling activated by Bcr-Abl tyrosine kinase.
Phosphorylation of Ser-216 Promotes Tyrosine Dephosphorylation of WAVE2 Complex
To determine how the mitotic phosphorylation of Abi1 attenuates Bcr-Abl-induced tyrosine phosphorylation of the WAVE complex, we first tested whether the mitotic phosphorylation of Abi1 affects its interaction with Bcr-Abl. Abi1 was immunoprecipitated from asynchronous and mitotic p185wt cells using anti-Abi1 antibody and anti-pSer-216 antibody, respectively. The amount of Bcr-Abl that was co-immunoprecipitated with Abi1 was then determined by Western blotting analysis. As shown in Fig. 6A, more Bcr-Abl was found in the immunoprecipitates from mitotic p185wt cells than from asynchronous p185wt cells. The increased association of Bcr-Abl with Abi1 in mitotic cells is possibly due to an increase in Bcr-Abl protein level during mitosis (data not shown). Therefore, it is unlikely that the inhibition of tyrosine phosphorylation of the WAVE complex in mitotic p185wt cells is caused by a decrease in the interaction between Abi1 and Bcr-Abl during mitosis.
FIGURE 6.
Phosphorylation of Ser-216 promotes tyrosine dephosphorylation of Abi1. A, phosphorylation of Ser-216 does not reduce the interaction between Abi1 and Bcr-Abl. Asynchronous (Asy) and M-phase-synchronized (M) p185wt cells expressing GFP-Abi1 were lysed and immunoprecipitated (IP) by anti-Abi1 (Abi1) and anti-pSer-216 (pSer-216) antibodies. The immunoprecipitates were analyzed by Western blotting using the indicated antibodies. B, treatment of mitotic p185wt cells with pervanadate rescues tyrosine phosphorylation of Abi1. p185wt cells expressing GFP-Abi1 were synchronized at M-phase as described under “Experimental Procedures.” In the last 2 h of mitotic arrest, pervanadate was added at the indicated concentrations, and cell lysates were immunoprecipitated by anti-GFP antibody. The immunoprecipitates of the treated cells, together with the immunoprecipitate from asynchronous cells as a control, were analyzed by Western blotting (WB) using the indicated antibodies. C, pervanadate treatment rescues tyrosine phosphorylation of GFP-Abi1S216D in p185wt cells. p185wt cells expressing GFP-Abi1S216D were treated with pervanadate at the indicated concentrations for 2 h. The lysates from treated cells, together with the lysate from p185wt cells expressing wild type GFP-Abi1, were immunoprecipitated by anti-GFP antibody, and the immunoprecipitates were subjected to Western blot analysis using the indicated antibodies.
Next, we examined the possibility that the mitotic serine/threonine phosphorylation of Abi1 may induce tyrosine dephosphorylation of the WAVE2 complex. To this end, p185wt GFP-Abi1 cells were arrested at mitosis, and in the last 2 h of mitotic arrest, a protein-tyrosine phosphatase inhibitor, pervanadate, was added at various concentrations. The immunoprecipitation of GFP-Abi1 followed by Western blotting analysis shows that pervanadate rescues Bcr-Abl-induced tyrosine phosphorylation of Abi1 in a dose-dependent fashion in the mitotic cells (Fig. 6B). Furthermore, the treatment of p185wt GFP-Abi1S216D cells with various concentrations of pervanadate also rescued the tyrosine phosphorylation of GFP-Abi1S216D (Fig. 6C). Taken together, these results suggest that mitotic serine/threonine phosphorylation of Abi1 may promote tyrosine dephosphorylation of WAVE complex during mitosis.
Ectopic Expression of Abi1S216A and Abi1S216D Interferes with Cell Cycle Progression
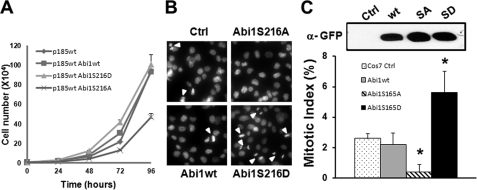
To determine whether the Ser-216 phosphorylation of Abi1 plays a role in cell growth, we examined the proliferation of the p185wt cells expressing wild type and the mutant forms of Abi1. The proliferation of p185wt cells expressing Abi1S216A was reduced compared with those of the control p185wt cells and the p185wt cells expressing Abi1wt and Abi1S216D (Fig. 7A). We then tested the effect of the overexpression of Abi1wt, Abi1S216A, and Abi1S216D on the cell cycle progression of COS-7 cells. COS-7 cells transfected with a control vector (Ctrl) show a mitotic index of 2.6% (Fig. 7, B and C). No statistically significant change in the mitotic index was observed in COS-7 cells expressing Abi1wt compared with control cells (2.2 versus 2.6%, p = 0.318). In contrast, the mitotic index of the COS-7 cells overexpressing Abi1S216D is increased by ∼1.2 fold compared with control cells (5.6 versus 2.6%, p < 0.01). On the other hand, the expression of Abi1S216A dramatically reduced the mitotic index by ∼6-fold compared with control cells (0.38 versus 2.6%, p < 0.001). Collectively, these findings provide evidence that the mitotic phosphorylation of Ser-216 in Abi1 is implicated in the regulation of cell cycle progression.
FIGURE 7.
Expression of Abi1S216A and Abi1S216D interferes with cell cycle progression. A, the p185wt cells (p185wt) and p185wt cells expressing Abi1wt (p185wt Abi1wt), Abi1S216A (p185wt Abi1S216A), or Abi1S216D (p185wt Abi1S216D) were seeded in 6-well plate (1 × 104/well) and grown in RPMI containing 10% FBS medium. Cell numbers were counted at the indicated time points. B, COS cells were stably transfected with empty retroviral vector (Ctrl) or retroviral vectors expressing GFP-Abi1wt, GFP-Abi1S216A, and GFP-Abi1S216D. The cells were stained with DAPI and examined for mitotic cells under fluorescence microscope. Arrowheads indicate the mitotic cells with condensed chromosomes. C, the mitotic cells shown in A were counted in five randomly selected areas, and the mitotic index was calculated and expressed as the average percentage of mitotic cells to total cells (lower panel). Expression of GFP-tagged Abi1 proteins was confirmed by immunoprecipitation followed by Western blot analysis using anti-GFP antibody (upper panel).
DISCUSSION
Dynamic actin assembly and the formation of membrane protrusions in the leading edge can be induced by growth factors in interphase cells; however, at the onset of mitosis, these activities appear to be inhibited as a part of the mitotic cytoskeleton remodeling process known as mitotic rounding (23, 34). These observations suggest a need for rapid activation and inactivation of WRC, a major NPF that regulates F-actin assembly in the leading edge of the cell, as cells progress through mitosis. Mitotic rounding facilitates the assembly and positioning of the spindle and therefore is essential for coordinated partitioning of the cytoplasm, intracellular organelles, and chromosomes during mitosis and cytokinesis (2). Despite its importance in the regulation of cell cycle progression, the molecular mechanism associated with mitotic regulation of actin assembly in the leading edge and the retraction of membrane protrusions is not completely understood. A major challenge in this field is the lack of an appropriate in vivo model that provides quantitative assessment of the activation and inactivation of WRC during cell cycle progression. In this study, we made use of p185wt cells, a Ba/F3-derived cell line transformed by Bcr-Abl tyrosine kinase. Several properties of this cell line make it unique for studying the mechanism associated with cell cycle-dependent regulation of the WRC signaling. First, transformation by Bcr-Abl induces the formation of an invadopodium-like structure in the leading edge that is dynamic and is enriched with not only F-actin but also adhesion molecules and extracellular matrix degradation enzymes (Refs. 20 and 42 and supplemental Fig. S2). This F-actin-rich structure is readily detected by fluorescence microscopy. Second, we found that WAVE2 is constitutively tyrosine-phosphorylated in interphase p185wt cells. Recent studies have shown that tyrosine phosphorylation of WAVE is essential and sufficient for the activation of WRC (13, 15). Therefore, it may provide an assay for the assessment of the WRC activation/inactivation during cell cycle progression. Third, we have shown that the Bcr-Abl-induced F-actin assembly and WAVE2 tyrosine phosphorylation in this cell line are dependent on Abi1. Mutations in either Abi1 or Bcr-Abl that block the interaction between Bcr-Abl and Abi1 inhibit F-actin assembly and tyrosine phosphorylation of WAVE2, suggesting an involvement of the WRC signaling in this process. Finally, similar to what is observed during mitotic rounding, we observed that the Bcr-Abl-stimulated F-actin assembly is abrogated when cells enter mitosis, suggesting that a cell cycle-dependent regulatory mechanism of the WRC signaling is at work.
Using this cell line as a model, we have uncovered a novel mechanism by which Abi1 regulates the WAVE2 complex in a cell cycle-dependent manner. We show that mitotic inhibition of F-actin assembly in p185wt cells is accompanied by an attenuation of Bcr-Abl-induced tyrosine phosphorylation of Abi1 and WAVE2. We found that Abi1 is hyperphosphorylated at the onset of mitosis and that the serine 216 of Abi1 is a mitotic phosphorylation site. More importantly, we provide evidence that mitotic serine/threonine phosphorylation of Abi1 inhibits the Bcr-Abl-induced tyrosine phosphorylation of WAVE2 complex and the assembly of F-actin.
Robust phosphorylation of serine 216 at the onset of mitosis was evidenced by three independent approaches. First, we developed an antibody that specifically recognizes Ser-216-phosphorylated Abi1. We show that this specific antibody detected endogenous Abi1 only from mitotic cells but not interphase cells. Additionally, the phosphorylation of Ser-216 was also detected in ectopically expressed GFP-Abi1, but not GFP-Abi1S216A, in HeLa and p185wt cells arrested at G2/M-phase. Moreover, the phosphoproteomic studies by other investigators have shown that Ser-216 of Abi1 is among the phosphorylation sites identified specifically in mitotic HeLa cells (41). Although these experiments clearly showed that the phosphorylation of Abi1 Ser-216 is increased at the onset of mitosis, they could not exclude the possibility that a basal phosphorylation of Ser-216 may also occur in interphase cells at a level too low to be detected by our antibody. Indeed, using a combination of affinity purification and mass spectrometry analysis, the phosphorylation of serine 216 in Abi1 has been identified in both serum-starved and EGF-stimulated A-431 cells and possibly other cells as well (15, 19), suggesting its basal phosphorylation in interphase cells.
Several lines of evidence from our studies and those of others support the conclusion that CDK1/cyclin B is responsible for mitotic phosphorylation of serine 216. First, the mitotic phosphorylation of Abi1 Ser-216 correlated well with CDK1/cyclin B activity and was inhibited by roscovitine, a pharmacological inhibitor of CDK1. Second, Ser-216 of Abi1 is an in vitro target of CDK1/cyclin B. In addition, the depletion of CDK1 from the mitotic cell extract abolished its ability to phosphorylate the Ser-216 of Abi1. Furthermore, previous studies by other investigators have shown that Abi1 interacts physically with CDK1 (38). These data, together with our findings that ectopic expression of the Abi1 with phosphomimetic and phosphodeficient mutations at Ser-216 leads to abnormal cell cycle progression, support the conclusion that CDK1-mediated phosphorylation of Ser-216 in Abi1 may serve as a regulatory mechanism for WAVE complex to coordinate actin cytoskeleton remodeling and cell cycle progression. The finding that Ser-216 of Abi1 is also phosphorylated during mitosis in adherent COS-7 and HeLa cells suggests that this cell cycle-dependent regulatory mechanism is likely common to other cells.
We propose that Abi1 may serve as a switch capable of turning on and off WRC in response to extracellular stimuli and intracellular signals. Abi1 exerts its dual role by linking the complex to either Abl tyrosine kinase, which is activated by extracellular stimuli or oncogenic mutation, or to CDK1/cyclin B, an important intracellular regulator for cell entry into mitosis (Fig. 8). The activation of Abl tyrosine kinases either by extracellular stimuli such as growth factors (9) or by oncogenic mutation (20) may result in the tyrosine phosphorylation of WRC and its translocation to the membrane, where it can be activated further by GTP-Rac and phospholipids to promote actin assembly (15). At the onset of mitosis, on the other hand, Abi1 is phosphorylated by CDK1/cyclin B, which leads to the attenuation of Abl-stimulated tyrosine phosphorylation of WAVE and inhibition of F-actin assembly in the leading edge (Fig. 8). In agreement with this notion, we show that: 1) an increase in Ser-216 phosphorylation in mitotic p185wt cells correlates well with a decrease in Bcr-Abl-stimulated tyrosine phosphorylation of Abi1 and WAVE2 (Figs. 2C and 4, C and D), an event known to be essential and sufficient for WRC activation; 2) endogenous Abi1 immunoprecipitated from mitotic p185wt cells by anti-pSer-216 displays greatly reduced tyrosine phosphorylation compared with that immunoprecipitated from interphase p185wt cells by a pan-anti-Abi1 antibody (Fig. 5A); and 3) a phosphomimetic mutation of Ser-216 in Abi1 partially attenuates Bcr-Abl-induced tyrosine phosphorylation of Abi1 and WAVE2 as well as the formation of an invadopodium-like structure (Fig. 5, C–F).
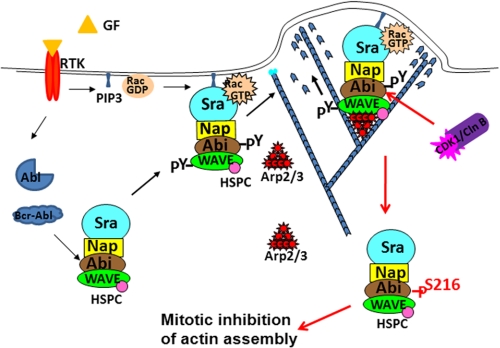
FIGURE 8.
A hypothetical model for cell cycle-dependent regulation of WRC. Stimulation by extracellular stimuli such as growth factors/receptor tyrosine kinase (RTK) may activate cAbl tyrosine kinase. Alternatively, Abl tyrosine kinase can be activated by oncogenic mutations such as fusion with Bcr sequences. Activation of Abl tyrosine kinases in turn stimulates the tyrosine phosphorylation of the WAVE complex and its translocation to membrane, where it can be further activated by active Rac and phospholipids. At the onset of mitosis, activation of CDK1/cyclin B induces serine/threonine phosphorylation of Abi1, which in turn leads to the attenuation of tyrosine phosphorylation of WRC.
It has been reported that Abl tyrosine kinases are also substrates of CDK1/cyclin B and are hyperphosphorylated during mitosis (43). This raises the question of whether the mitotic phosphorylation of Bcr-Abl reduces its tyrosine kinase activity, which might be responsible for mitotic inhibition of the tyrosine phosphorylation of WRC. A recent study by Woodring et al. (3) shows that the mitotic phosphorylation of cAbl does not reduce its tyrosine kinase activity. In agreement with that study, we found no evidence of a decrease in the tyrosine kinase activity of Bcr-Abl during mitosis. We also tested whether the mitotic phosphorylation of Abi1 would affect its interaction with Bcr-Abl, and the result does not support this hypothesis either. In an attempt to define the mechanism by which the mitotic phosphorylation of Abi1 inhibits Bcr-Abl-induced tyrosine phosphorylation of WRC, we examined whether the mitotic phosphorylation of Abi1 promotes tyrosine dephosphorylation of WRC. Our data indicate that inhibition of protein-tyrosine phosphatase activity rescued the tyrosine phosphorylation of WRC in mitosis, suggesting that tyrosine dephosphorylation by a yet to be identified pathway is likely to be responsible, at least in part, for the mitotic attenuation of tyrosine phosphorylation of WRC in p185wt cells.
In addition to serine 216, two other serine/threonine residues, Thr-196 and Ser-225, also fall within the consensus sequences for CDK1 (Fig. 4A). Therefore, it is possible that serine 216 may not be the only site in Abi1 that is phosphorylated by CDK1 during mitosis. In this regard, it is notable that, although the treatment of the mitotic p185wt cells with roscovitine (Fig. 4E) or the depletion of CDK1 from mitotic cells (Fig. 4H) significantly reversed the mitotic mobility shift of Abi1, the mutation of Ser-216 to alanine alone was not sufficient to reverse such a mobility shift (Fig. 4B). We also noticed that the phospho-deficient mutation of Ser-216 alone (S216A) failed to increase the tyrosine phosphorylation of Abi1 (Fig. 5C) and WAVE2 (data not shown) in p185wt cells. Moreover, the expression of Abi1S216A in p185wt cells did not affect the Bcr-Abl-induced assembly of F-actin-rich structures (data not shown). Together, these observations are consistent with the notion that CDK1 may phosphorylate additional serine/threonine residues in Abi1 during mitosis. Thus, a complete analysis of the mitotic phosphorylation events of the WAVE complex is needed to better understand how mitotic phosphorylation regulates the function of this complex during mitosis.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA094921 and R21 CA133597 (to Z. D.) from NCI and Grant K01 DK067191 (to Y. T.) from NIDDK. This work was also supported by a grant from the When Everyone Survives Foundation (to Z. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- Arp2/3
- actin-related protein 2/3
- Abi
- Abl interactor
- CDK
- cyclin-dependent kinase
- F-actin
- filamentous actin
- M-phase
- mitotic phase
- NPF
- nucleation-promoting factor
- PP1
- protein phosphatase 1
- SH
- Src homology
- Sra
- specifically Rac1-associated
- VCA
- verprolin homology, central and acidic regions
- WASP
- Wiskott-Aldrich syndrome protein
- WAVE
- WASP family verprolin-homologous
- WRC
- WAVE regulatory complex
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1. Pollard T. D., Cooper J. A. (2009) Science 326, 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kunda P., Baum B. (2009) Trends Cell Biol. 19, 174–179 [DOI] [PubMed] [Google Scholar]
- 3. Woodring P. J., Hunter T., Wang J. Y. (2005) J. Biol. Chem. 280, 10318–10325 [DOI] [PubMed] [Google Scholar]
- 4. Campellone K. G., Welch M. D. (2010) Nat. Rev. Mol. Cell Biol. 11, 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gautreau A., Ho H. Y., Li J., Steen H., Gygi S. P., Kirschner M. W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4379–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Innocenti M., Zucconi A., Disanza A., Frittoli E., Areces L. B., Steffen A., Stradal T. E., Di Fiore P. P., Carlier M. F., Scita G. (2004) Nat. Cell Biol. 6, 319–327 [DOI] [PubMed] [Google Scholar]
- 7. Dai Z., Pendergast A. M. (1995) Genes Dev. 9, 2569–2582 [DOI] [PubMed] [Google Scholar]
- 8. Shi Y., Alin K., Goff S. P. (1995) Genes Dev. 9, 2583–2597 [DOI] [PubMed] [Google Scholar]
- 9. Leng Y., Zhang J., Badour K., Arpaia E., Freeman S., Cheung P., Siu M., Siminovitch K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1098–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stuart J. R., Gonzalez F. H., Kawai H., Yuan Z. M. (2006) J. Biol. Chem. 281, 31290–31297 [DOI] [PubMed] [Google Scholar]
- 11. Eden S., Rohatgi R., Podtelejnikov A. V., Mann M., Kirschner M. W. (2002) Nature 418, 790–793 [DOI] [PubMed] [Google Scholar]
- 12. Ismail A. M., Padrick S. B., Chen B., Umetani J., Rosen M. K. (2009) Nat. Struct. Mol. Biol. 16, 561–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Z., Borek D., Padrick S. B., Gomez T. S., Metlagel Z., Ismail A. M., Umetani J., Billadeau D. D., Otwinowski Z., Rosen M. K. (2010) Nature 468, 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Derivery E., Lombard B., Loew D., Gautreau A. (2009) Cell Motil. Cytoskeleton 66, 777–790 [DOI] [PubMed] [Google Scholar]
- 15. Lebensohn A. M., Kirschner M. W. (2009) Mol. Cell 36, 512–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim Y., Sung J. Y., Ceglia I., Lee K. W., Ahn J. H., Halford J. M., Kim A. M., Kwak S. P., Park J. B., Ho Ryu S., Schenck A., Bardoni B., Scott J. D., Nairn A. C., Greengard P. (2006) Nature 442, 814–817 [DOI] [PubMed] [Google Scholar]
- 17. Miyamoto Y., Yamauchi J., Tanoue A. (2008) J. Neurosci. 28, 8326–8337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sossey-Alaoui K., Li X., Cowell J. K. (2007) J. Biol. Chem. 282, 26257–26265 [DOI] [PubMed] [Google Scholar]
- 19. Mendoza M. C., Er E. E., Zhang W., Ballif B. A., Elliott H. L., Danuser G., Blenis J. (2011) Mol. Cell 41, 661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Y., Clough N., Sun X., Yu W., Abbott B. L., Hogan C. J., Dai Z. (2007) J. Cell Sci. 120, 1436–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ardern H., Sandilands E., Machesky L. M., Timpson P., Frame M. C., Brunton V. G. (2006) Cell Motil. Cytoskeleton 63, 6–13 [DOI] [PubMed] [Google Scholar]
- 22. Juang J. L., Hoffmann F. M. (1999) Oncogene 18, 5138–5147 [DOI] [PubMed] [Google Scholar]
- 23. Kunda P., Pelling A. E., Liu T., Baum B. (2008) Curr. Biol. 18, 91–101 [DOI] [PubMed] [Google Scholar]
- 24. Kondylis V., van Nispen tot Pannerden H. E., Herpers B., Friggi-Grelin F., Rabouille C. (2007) Dev. Cell 12, 901–915 [DOI] [PubMed] [Google Scholar]
- 25. McWhirter J. R., Wang J. Y. (1997) Oncogene 15, 1625–1634 [DOI] [PubMed] [Google Scholar]
- 26. Skourides P. A., Perera S. A., Ren R. (1999) Oncogene 18, 1165–1176 [DOI] [PubMed] [Google Scholar]
- 27. Salgia R., Li J. L., Ewaniuk D. S., Pear W., Pisick E., Burky S. A., Ernst T., Sattler M., Chen L. B., Griffin J. D. (1997) J. Clin. Invest. 100, 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Courtney K. D., Grove M., Vandongen H., Vandongen A., LaMantia A. S., Pendergast A. M. (2000) Mol. Cell. Neurosci. 16, 244–257 [DOI] [PubMed] [Google Scholar]
- 29. Dai Z., Quackenbush R. C., Courtney K. D., Grove M., Cortez D., Reuther G. W., Pendergast A. M. (1998) Genes Dev. 12, 1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dai Z., Kerzic P., Schroeder W. G., McNiece I. K. (2001) J. Biol. Chem. 276, 28954–28960 [DOI] [PubMed] [Google Scholar]
- 31. Sun X., Li C., Zhuang C., Gilmore W. C., Cobos E., Tao Y., Dai Z. (2009) Carcinogenesis 30, 2109–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Littlepage L. E., Ruderman J. V. (2002) Genes Dev. 16, 2274–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Messenger M. M., Saulnier R. B., Gilchrist A. D., Diamond P., Gorbsky G. J., Litchfield D. W. (2002) J. Biol. Chem. 277, 23054–23064 [DOI] [PubMed] [Google Scholar]
- 34. Cramer L. P., Mitchison T. J. (1997) Mol. Biol. Cell 8, 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weaver A. M. (2006) Clin. Exp. Metastasis 23, 97–105 [DOI] [PubMed] [Google Scholar]
- 36. Mistry S. J., Atweh G. F. (2001) J. Biol. Chem. 276, 31209–31215 [DOI] [PubMed] [Google Scholar]
- 37. Yu W., Sun X., Clough N., Cobos E., Tao Y., Dai Z. (2008) Carcinogenesis 29, 1717–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin T. Y., Huang C. H., Chou W. G., Juang J. L. (2004) J. Biomed Sci. 11, 902–910 [DOI] [PubMed] [Google Scholar]
- 39. Gavet O., Pines J. (2010) Dev. Cell 18, 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moreno S., Nurse P. (1990) Cell 61, 549–551 [DOI] [PubMed] [Google Scholar]
- 41. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun X., Li Y., Yu W., Wang B., Tao Y., Dai Z. (2008) Leukemia 22, 1053–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kipreos E. T., Wang J. Y. (1990) Science 248, 217–220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.