Background: Murine models suggest differences between cellular actions of Gαs and XLαs, but these are unknown.
Results: XLαs, unlike Gαs, remains in the plasma membrane and can generate sustained cAMP signaling.
Conclusion: The unique actions of XLαs likely stem from its strong affinity for plasma membrane.
Significance: Cellular actions of XLαs have implications in cAMP signaling and diseases caused by mutations in this protein.
Keywords: Cell Differentiation, Cholera Toxin, Cyclic AMP (cAMP), Fluorescence Resonance Energy Transfer (FRET), G Protein-coupled Receptors (GPCR), Heterotrimeric G Proteins, Intracellular Trafficking, Parathyroid Hormone Receptor, gsp Oncogene, Osteoblast Differentiation
Abstract
Murine models indicate that Gαs and its extra-long variant XLαs, both of which are derived from GNAS, markedly differ regarding their cellular actions, but these differences are unknown. Here we investigated activation-induced trafficking of Gαs and XLαs, using immunofluorescence microscopy, cell fractionation, and total internal reflection fluorescence microscopy. In transfected cells, XLαs remained localized to the plasma membrane, whereas Gαs redistributed to the cytosol after activation by GTPase-inhibiting mutations, cholera toxin treatment, or G protein-coupled receptor agonists (isoproterenol or parathyroid hormone (PTH)(1–34)). Cholera toxin treatment or agonist (isoproterenol or pituitary adenylate cyclase activating peptide-27) stimulation of PC12 cells expressing Gαs and XLαs endogenously led to an increased abundance of Gαs, but not XLαs, in the soluble fraction. Mutational analyses revealed two conserved cysteines and the highly charged domain as being critically involved in the plasma membrane anchoring of XLαs. The cAMP response induced by M-PTH(1–14), a parathyroid hormone analog, terminated quickly in HEK293 cells stably expressing the type 1 PTH/PTH-related peptide receptor, whereas the response remained maximal for at least 6 min in cells that co-expressed the PTH receptor and XLαs. Although isoproterenol-induced cAMP response was not prolonged by XLαs expression, a GTPase-deficient XLαs mutant found in certain tumors and patients with fibrous dysplasia of bone and McCune-Albright syndrome generated more basal cAMP accumulation in HEK293 cells and caused more severe impairment of osteoblastic differentiation of MC3T3-E1 cells than the cognate Gαs mutant (gsp oncogene). Thus, activated XLαs and Gαs traffic differently, and this may form the basis for the differences in their cellular actions.
Introduction
GNAS is a complex locus giving rise to multiple translated and nontranslated gene products (1–4). Gene association and copy number variation studies have associated GNAS with the pathogenesis of multiple different complex diseases and cancers (5–15). Inherited mutations within or nearby GNAS that directly impair the functions and/or the expression of its gene products are responsible for several different human diseases, including, but not limited to, pseudohypoparathyroidism, various endocrine and nonendocrine tumors, and McCune-Albright syndrome (16, 17). One of the products of GNAS is the α-subunit of the heterotrimeric stimulatory GTP-binding protein (Gαs), a ubiquitous protein essential for the actions of many hormones, neurotransmitters, and paracrine factors (1). The GTP-bound form of the Gαs subunit transduces the activation of cell surface G protein-coupled receptors (GPCRs)3 into intracellular signaling by stimulating a number of effectors, such as adenylyl cyclases, which catalyze the synthesis of cAMP.
GNAS also encodes a long Gαs variant, termed XLαs (Fig. 1A) (18), whose cellular actions remain uncertain. XLαs uses a promoter that is distinct from the promoter of Gαs and is active on the paternal allele only, i.e. XLαs expression is limited to a single parental allele (19). Although Gαs is encoded by GNAS exons 1–13, XLαs uses an alternative first exon (exon XL) that splices onto exons 2–13 (19, 20). Thus, Gαs and XLαs differ in their N-terminal regions but are otherwise identical. A variant of XLαs, termed XXLαs, is also derived from GNAS (Fig. 1A) (21, 22). The latter includes the entire coding sequence of XLαs but extends in the N terminus, thus having about 300 additional amino acids. Like XLαs, the cellular actions of XXLαs are unclear.
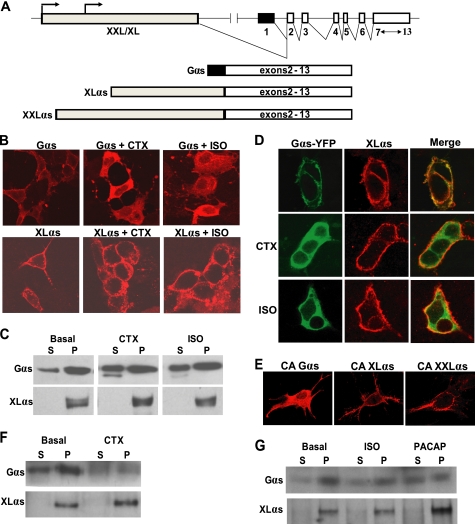
FIGURE 1.
XLαs is targeted differently from wild-type Gαs upon activation. A, depiction of the GNAS locus and the transcripts encoding Gαs, XLαs, and XXLαs. Exons and introns are indicated by boxes and connecting lines, respectively. Splicing pattern is shown by angled lines. Arrows indicate the origin of transcription. B and C, HEK293 cells were transiently transfected with cDNA encoding either HA-tagged Gαs or XLαs. Forty eight hours after transfection, cells were treated with 10−5 m isoproterenol for 20 min or with 1 μg/ml CTX for 4 h, and subcellular localizations of Gαs and XLαs were examined by immunocytochemical analysis by using the anti-HA antibody (B), and Western blot analysis by using either the anti-HA antibody or a polyclonal antibody against the common C terminus of Gαs and XLαs. (C). D, HEK293 cells were transiently co-transfected with cDNA encoding HA-tagged XLαs and Gαs-YFP. Forty eight hours after transfection, cells were treated with 10−5 m isoproterenol (ISO) for 20 min or with 1 μg/ml CTX for 4 h. XLαs and Gαs-YFP were detected in the same cells by immunocytochemical analysis using the anti-HA antibody and by confocal fluorescence microscopy. E, GnasE2−/E2− cells were transiently transfected with cDNA encoding HA-tagged Gαs-R201H, XLαs-R543H, or XXLαs-R844H. Two days later, immunofluorescence confocal microscopy using anti-HA antibody was employed to determine the subcellular localization of those mutants. F and G, subcellular localizations of endogenous Gαs and XLαs in PC12 cells stimulated by 1 μg/ml CTX (F), 10 μm isoproterenol (ISO; G), or 100 μm PACAP (G). Endogenous Gαs and XLαs in soluble (S) and particulate (P) fractions were subjected to immunoblotting with the polyclonal antibody against Gαs and XLαs.
Studies with transfected cell lines have shown that XLαs can mimic the action of Gαs regarding the stimulation of cAMP generation in response to receptor activation or upon treatment with cholera toxin (CTX), which ADP-ribosylates the α-subunit and thus inhibits the intrinsic GTPase activity (23–25). When expressed ectopically in transgenic mice, XLαs can also enhance cellular responses that are typically mediated by Gαs (26). However, data obtained from gene knock-out studies do not readily support a “Gαs-like” role for XLαs at the cellular level. Mice in which XLαs (together with XXLαs) is ablated show poor adaptation to feeding, early postnatal lethality, and a hypermetabolic phenotype (27–30), but these findings are strikingly different from and, in terms of energy and lipid metabolism, the opposite of the findings observed in Gαs knock-out mice (31, 32). Thus, although XLαs can seemingly contribute to cAMP signaling, its cellular actions are predicted to differ significantly from the cellular actions of Gαs.
Like the Gαs subunit, the XLαs subunit is localized to the plasma membrane at the basal state (25, 33). Gαs redistributes to the cytoplasmic compartments following activation (34, 35), and this regulatory process is critical for limiting the activation of Gαs (36, 37). However, recent studies have also shown that, in response to certain receptor agonists, the internalized Gαs protein can continue to stimulate cAMP production within endosomes (38, 39). The fate of XLαs upon activation, however, has remained unknown. A previous study using subcellular fractionation detected differences between the distributions of ADP-ribosylated forms of XLαs and Gαs (33), and in another study, a GTPase-deficient XLαs mutant was localized differently from the cognate Gαs mutant by immunostaining (22). Based on these findings, we hypothesized that XLαs traffics differently from Gαs upon activation. We herein investigated this hypothesis by examining the subcellular localization of XLαs before and after activation. Our findings revealed that XLαs remain localized to the plasma membrane even upon activation and can thereby generate sustained signaling.
EXPERIMENTAL PROCEDURES
Expression Constructs and Mutagenesis
Construction of cDNA plasmids encoding hemagglutinin (HA) epitope-tagged Gαs, XLαs, and XXLαs, all in pcDNA3.1, were described previously (22, 34). The truncation and point mutation constructs of Gαs, XLαs, and XXLαs were generated using circular wild-type cDNA plasmid as a template and specific mutagenic oligonucleotides as primers by using the QuikChange mutagenesis kit (Stratagene). cDNA encoding the XLαs-Gαs chimera in which residues 247–318 of human XLαs replaced Gly-2 and Cys-3 of Gαs was generated by standard methods and cloned into pcDNA3.1. All plasmid DNAs were sequenced to verify the presence of desired mutations and that they were free of aberrant random mutations. Restriction endonucleases and other enzymes for making constructs were obtained from New England Biolabs (Beverley, MA). Plasmids encoding Gαs-YFP, -Gβ1, and -Gγ2 were kindly provided by Dr. Matthew Mahon (Massachusetts General Hospital and Harvard Medical School). The plasmid encoding Gαs-GFP was kindly provided by Dr. Catherine Berlot (Geisinger Health, Weis Center for Research). The plasmid encoding XLαs-GFP was constructed by substituting, in the Gαs-GFP construct, the sequences encoding the XL domain for the sequences of Gαs derived from exon 1. The plasmid encoding the PTHR-DsRed fusion protein (PTHR-DsRed) was constructed by inserting cDNA encoding DsRed into the exon 2 encoded portion of PTHR.
Cell Culture and Transient Transfection
Cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2. HEK293, PC12, and MC3T3-E1 cells were obtained from American Type Culture Collection (ATCC; Manassas, VA). HEK293 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum. PC12 cells were grown on collagen type IV-coated culture plates in ATCC-formulated F-12K medium supplemented with 15% fetal horse serum and 2.5% fetal bovine serum. Fibroblastic GnasE2−/E2− cells null for Gαs, XLαs, and XXLαs, which have been described previously (24), were cultured in DMEM/F-12 medium supplemented with 5% FBS. MC3T3-E1 cells were maintained in α-minimal essential medium.
For determining and comparing subcellular localizations of Gαs, XLαs, and XXLαs and mutants thereof, and for experiments involving the quantification of cAMP levels, cells were additionally transfected with plasmids encoding Gβ1 and Gγ2. HEK293 cells were transfected by using FuGENE 6 (Roche Applied Science) and GnasE2−/E2− cells by using JetPEI DNA transfection reagent (PolyPlus transfectionTM, Illkirch, France) following the protocols supplied by the manufacturer. Stimulation of cells with isoproterenol or pituitary adenylate cyclase activating peptide-27 (PACAP) was done for 20 min at 37 °C. CTX stimulation was performed for 4 h at 37 °C. Isoproterenol, PACAP, and CTX were purchased from Sigma. Preosteoblastic MC3T3-E1 cells were transfected by using the PolyJet transfection reagent (SignaGen Laboratories).
Cell Lysis and Subcellular Fractionation
Whole cell lysates were prepared by either 2× SDS-polyacrylamide gel loading buffer or 1% Triton X-100 in a Tris-HCl (pH 7.8)-buffered solution containing protease inhibitors. For preparation of total cell membranes, cells were lysed in isotonic buffer without detergent (10 mm Tris-HCl (pH 7.8), 4 mm EDTA, and protease inhibitors) by passing 10–15 times through a 28-gauge syringe tip on ice. After 10 min of centrifugation at 1,000 × g at 4 °C, the resulting supernatant was further centrifuged at 100,000 × g (Beckman TL-100 Tabletop Ultracentrifuge, Beckman, Palo Alto, CA) for 1 h at 4 °C. The supernatant after the second centrifugation was designated as the soluble fraction. The pellet was resuspended in a buffer containing 20 mm Hepes (pH 7.4), 0.1 m NaCl, 3 mm MgSO4, and 20% glycerol. The pellet obtained after the ultracentrifugation was designated as the total membrane fraction (particulate fraction). Protein concentration was determined by BCA protein assay kit (Pierce) using bovine serum albumin as standard.
Western Blot Analysis
The particulate and soluble fractions of cell lysates were subjected to electrophoresis for separation of proteins by either 10% (Gαs) or 6% (for XLαs and XXLαs) SDS-PAGE. Whole cell lysates were separated by 4–15% gradient SDS-PAGE. Separated proteins were transferred onto Immobilon PVDF membranes (Millipore, Temecula, CA). After blocking with 5% nonfat milk in Tris-buffered saline, 0.1% Tween 20 for 1 h at room temperature, the blots were probed with primary antibodies, either rabbit anti-HA antibody (AbCam, Cambridge, MA) or a rabbit antibody against the C-terminal decapeptide common to Gαs, XLαs, and XXLαs (Millipore). The immunoblots were then reacted to goat anti-rabbit IgG secondary antibody conjugated to horseradish peroxidase (HRP) (Santa Cruz Biotechnology), and immunoreactive proteins were visualized using Western Lightning Plus-ECL enhanced chemiluminescence detection kit (PerkinElmer Life Sciences). Blots were stripped by using the Re-Blot Plus solution (Millipore), and subsequently immunoreacted to a polyclonal antibody against β-actin (Santa Cruz Biotechnology) as a gel loading control. Densitometric analysis of blots was carried out by using FluorChem SP imaging system and AlphaeaseFC software version 4.1.0 (Alpha Innotech, San Leandro, CA).
Immunocytochemistry and Confocal Microscopy
Cells were grown and transfected in collagen-coated, four-well chamber slides with cover (Nunc, Naperville, IL). Cells were washed three times with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde in PBS for 20 min. After permeabilization with 0.1% saponin in PBS, 0.5% BSA for 15 min and subsequently blocking for 30 min with 0.1% saponin and 0.5% BSA in PBS, cells were incubated with a rabbit anti-HA antibody (Santa Cruz Biotechnology) and then incubated with Cy3-conjugated anti-rabbit IgG (Amersham Biosciences). The immunoreactivity was visualized and analyzed by using a laser scanning confocal fluorescent microscope (Nikon, Tokyo, Japan).
Analysis of Protein Palmitoylation
To inhibit palmitoylation, HEK293 cells were cotransfected with expression constructs encoding XLαs-GFP, Gβ1, and Gγ2, and immediately following transfection, these cells were treated with 25 μm 2-bromo-hexadecanoic acid (2BP) or vehicle (DMSO) alone. The subcellular localization of the protein was evaluated by confocal fluorescent microscopy 24 h after transfection or by subcellular fractionation followed by Western blot 48 h after transfection. For direct determination of XLαs palmitoylation, cells transiently expressing either wild-type XLαs or the XLαs-C287S,C318S mutant were metabolically labeled with 0.16 mCi/ml [3H]palmitic acid ([9,10-3H]palmitic acid (PerkinElmer Life Sciences)) in Dulbecco's modified Eagle's medium supplemented with 0.2% fatty acid-free bovine serum albumin at 37 °C for 5 h. After washing with PBS, cells were lysed in PBS containing 5 mm EDTA, 1% Triton X-100, and a protease inhibitor mixture (Sigma). Wild-type and the mutant XLαs were immunoprecipitated by a mouse monoclonal anti-HA antibody (Abcam). Following separation by 10% SDS-PAGE, proteins were analyzed by fluorography to detect the emission of 3H. Briefly, gels were fixed and processed by using the EN3HANCE reagent (PerkinElmer Life Sciences) according to the manufacturer's instructions. Dried gels were exposed to a Kodak BioMax MS x-ray film for at least 6 weeks at −80 °C. In some experiments, immunoprecipitated proteins were separated on two parallel 7.5% SDS-polyacrylamide gels. One of the gels was blotted onto an Immobilon PVDF membrane and immunoreacted to the polyclonal antibody against the Gαs/XLαs/XXLαs C terminus. The other gel was sliced according to the position of the immunoreactive bands in the Western blot. Radioactivity in each gel slice was counted in a Beckman scintillation counter. Background was determined by counting the radioactivity in blank gel slices.
TIRF Microscopy
HEK293 cells were transiently transfected with cDNA encoding Gαs-GFP or XLαs-GFP, as well as PTHR-DsRed, Gβ1, and Gγ2, and 48 h later, cells were plated on FluoroDish (World Precision Instruments, Inc.) coated with poly-l-lysine. The following day, cells were washed two times with Hanks' balanced salt solution (Invitrogen) and stimulated with 10 nm [Nle8,Nle21,Tyr34]rPTH(1–34) (PTH(1–34)). Fluorescence measurements of single cells were performed using a total internal reflection (TIRF) objective, a motorized laser TIRF illumination unit as the TIRF microscopy condenser (Nikon), and argon laser (Nikon). GFP was excited with 488 nm laser line using 530 nm emission filter. The fluorescent images were recorded for 30 min after PTH(1–34) stimulation, and the intensity of green emission fluorescence was recorded. The intensity of green fluorescence was measured and analyzed by NIS Elements software (Nikon). To minimize photobleaching during the experiment, recording was performed at 5-s intervals in the first 5 min, 10-s intervals from 5 to 20 min, and 30-s intervals from 20 to 30 min.
Determination of cAMP Response
Basal cAMP accumulation was determined 72 h after transfection of HEK293 cells. Cells were lysed after incubation in a buffer containing 2 mm isobutyl methylxanthine (Sigma) for 15 min at 37 °C. The medium was removed, and cells were lysed with 50 mm HCl. Radioimmunoassay was performed to determine the amount of cAMP. For determining the time course of cAMP generation in live cells, a FRET-based assay was used, as described previously (38). HEK293 cells stably expressing PTHR and transiently expressing a cAMP biosensor, Epac-CFP/YFP with or without XLαs were continuously perfused with control buffer, 100 nm M-PTH(1–14) or 10 μm isoproterenol; the details of the “M” substitution and the signaling properties of this PTH analog has been described previously (40, 41).
Osteoblastic Differentiation
Preosteoblastic murine MC3T3-E1 cells grown in 24-well plates were transfected with various expression plasmids. Forty eight hours after transfection, growth medium was replaced (day 0) with osteogenic differentiation medium containing ascorbic acid (50 μg/ml). Differentiation of cells into osteoblasts was assessed on days 0, 2, and 5 by staining for alkaline phosphatase activity, which was performed on cells fixed with 10% formalin. After washing with PBS, cells were incubated for 30 min at room temperature with a 0.1 m Tris-HCl (pH 8.5)-buffered solution containing 0.01% naphthol AS-MX phosphate (Sigma) and 0.06% Fast Blue BB salt (Sigma). RNA was also isolated on the same day by using the RNeasy mini kit (Qiagen), and first strand cDNA was synthesized by using Superscript III reverse transcriptase (Invitrogen) and random hexamer primers. To quantify the level of alkaline phosphatase transcript, real time RT-PCR using the SYBR Green reagent (Qiagen) was performed. The β-actin transcript was also amplified as an internal control. Forward and reverse PCR primers for amplification of the alkaline phosphatase transcript were 5′-AACCCAGACACAAGCATTCC-3′ and 5′-CGAAGGGTCAGTCAGGTTGT-3′, respectively. Primers for amplification of β-actin were described previously (42).
Statistical Analyses
Statistical significance of difference between two sample means was determined by paired Student's t test. Statistical significance of difference among multiple sample means was determined by one-way analysis of variance. A p value of less than 0.05 was considered to be significant.
RESULTS
XLαs Is Targeted Differently from Gαs upon Activation
To determine whether XLαs mimics Gαs regarding activation-induced subcellular redistribution, we transfected HEK293 cells with cDNA encoding HA-tagged XLαs or Gαs. Immunofluorescence confocal microscopy using an anti-HA antibody demonstrated that both proteins were localized to the plasma membrane at the basal state, although the staining for XLαs appeared to be punctate (Fig. 1B). In cells treated with a saturating concentration of isoproterenol (10 μm), a selective full agonist for the endogenous β2AR (a class A GPCR; β2AR), or with 1 μg/ml cholera toxin, which directly stimulates Gαs by inhibiting its intrinsic GTPase activity, Gαs was detected at intracellular localizations, whereas XLαs staining remained in the plasma membrane (Fig. 1B). In the same assays, the extended XLαs variant XXLαs behaved similarly to XLαs with respect to subcellular localization before and after activation (supplemental Fig. S1). Using either anti-HA antibody (for Gαs) or a polyclonal antibody directed against the C terminus of XLαs and Gαs (for XLαs), Western blot analysis of particulate and soluble fractions of these transfected HEK293 cells also showed that at the basal state Gαs exists mostly within the particulate fraction and that stimulation of these cells with isoproterenol or cholera toxin results in a modest redistribution to the soluble fraction (Fig. 1C). In contrast, XLαs immunoreactivity was detected entirely in the particulate fraction both at the basal state and upon stimulation with isoproterenol or cholera toxin (Fig. 1C). Findings similar to those observed for XLαs were also obtained when using cells transfected with XXLαs (supplemental Fig. S1).
We then co-transfected HEK293 cells with cDNA encoding HA-tagged XLαs or XXLαs and a YFP-labeled Gαs subunit (Gαs-YFP). At the basal state, immunocytochemical analysis using the anti-HA antibody and fluorescence confocal microscopy revealed both Gαs-YFP and XLαs at the plasma membrane, but after stimulation by isoproterenol or cholera toxin, only Gαs-YFP became cytoplasmic (Fig. 1D). In the same cells, immunostaining for XLαs remained at the plasma membrane (Fig. 1D). The subcellular localization of XXLαs relative to that of Gαs-YFP was identical to that of XLαs. It remained plasma membrane bound upon activation as opposed to Gαs-YFP, which internalized when stimulated by isoproterenol or cholera toxin (supplemental Fig. S1).
To rule out that the activation-induced differences between the subcellular localizations of Gαs and XLαs reflect excessive amounts of total Gαs expression in transfected HEK293 cells, which endogenously express Gαs (but not XLαs) at readily detectable levels, we employed mouse embryonic fibroblasts that genetically lack endogenous Gαs, XLαs, and XXLαs (GnasE2−/E2− cells) expression (24). We transfected these cells with cDNA encoding a GTPase-deficient, constitutively active mutant of either Gαs (R201H) or XLαs (R543H), which was used as a substitute for receptor- or cholera toxin-induced activation. Immunostaining of these cells with the anti-HA antibody also showed that Gαs-R201H is localized to the cytoplasm, whereas constitutively active XLαs-R543H, as well as the cognate XXLαs mutant (R844H), is localized to the plasma membrane (Fig. 1E). Moreover, we compared the subcellular distribution of Gαs and XLαs in PC12 cells, a rat pheochromocytoma cell line that expresses both XLαs and Gαs endogenously. Western blot analysis of nonstimulated PC12 cells using the C-terminal Gαs/XLαs antibody showed that whereas Gαs is localized mostly, but not exclusively, to the particulate fraction, XLαs is entirely confined to the particulate fraction (Fig. 1F). Following treatment of PC12 cells with cholera toxin, the abundance of Gαs shifted partly toward the soluble fraction, whereas XLαs continued to be localized exclusively in the particulate fraction (Fig. 1F). In addition, stimulation of PC12 cells with either isoproterenol or PACAP, which bind their respective endogenously expressed Gαs-coupled receptors, also resulted in a modest shift of Gαs immunoreactivity from the particulate to the soluble fraction, whereas these receptor agonists failed to alter the association of XLαs with the particulate fraction (Fig. 1G). These findings indicated that XLαs (as well as XXLαs), unlike Gαs, is not subject to internalization upon activation.
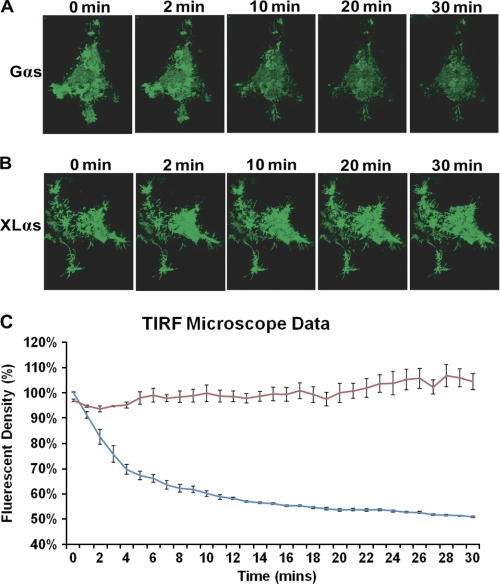
XLαs can functionally couple to PTHR (24, 25), which belongs to the family of class B GPCRs. To determine whether the differences observed between the subcellular localizations of Gαs and XLαs also exist following activation of PTHR, we transiently coexpressed PTHR and either Gαs-GFP or XLαs-GFP in HEK293 cells, which were then stimulated with PTH(1–34) (10 nm) and monitored for 30 min by TIRFM. The changes in the intensity of green fluorescence were strikingly different between cells expressing Gαs-GFP and those expressing XLαs-GFP. The fluorescence intensity diminished by about 50% at the end of the 30-min time course, with a half-life of ∼5 min, in cells expressing Gαs-GFP (Fig. 2, A and C), whereas it remained constant in cells expressing XLαs-GFP (Fig. 2, B and C) (supplemental videos). These findings confirmed that XLαs remains attached to the plasma membrane despite being activated.
FIGURE 2.
TIRFM distinguishes the plasma membrane localizations of Gαs and XLαs upon activation of PTHR in live cells. HEK293 cells were cotransfected with cDNAs encoding either Gαs-GFP and PTHR (A) or XLαs-GFP and PTHR (B), and live imaging was performed using TIRFM. The cells were treated with 10−8 m PTH(1–34) for 30 min and monitored over time. Images show fluorescence intensity at 0, 2, 10, 20, and 30 min after ligand addition. C, intensity of GFP fluorescence was measured to study the localization of Gαs (blue) and XLαs (red) at the plasma membrane and shown as the time course. Values are normalized to fluorescence at t = 0 s, and data represent the mean ± S.D. of n = 3 experiments. The complete image sequences of the experiments shown in A and B are shown in supplemental videos 1 and 2.
Conserved Cysteine Residues and a Region Comprising HCD (but not PRR) Confer Strong Plasma Membrane Avidity to XLαs and XXLαs
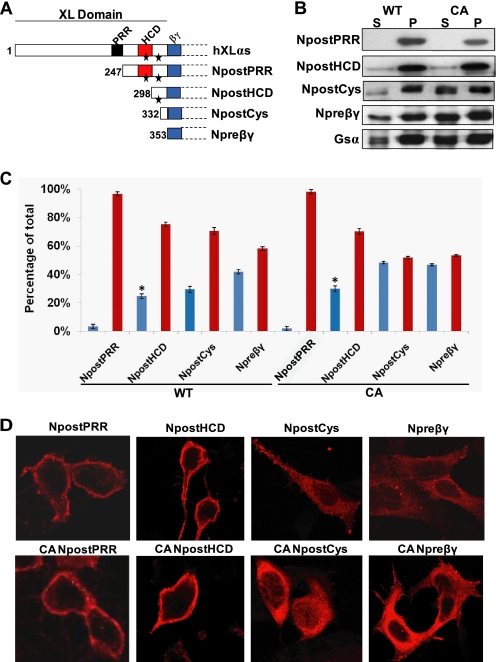
To determine the structural features of XLαs and XXLαs that prevent activation-induced internalization, we generated a series of N-terminally truncated mutants, and we compared the subcellular localization of each mutant. The truncations started with deletion of the N-terminal residues, including the PRR, and gradually omitted the HCD and the residues before the putative Gβγ interaction domain (corresponding to the exon 1-encoded portion of Gαs) (Fig. 3A). The subcellular localization of each truncated mutant was determined in transiently transfected HEK293 cells by using Western blot analyses using the anti-HA antibody following subcellular fractionation and confocal immunofluorescence microscopy. Truncation of the N-terminal residues, including PRR, but not HCD did not affect subcellular localization, as this mutant (NpostPRR) was detected almost exclusively in the particulate fraction (Fig. 3, B and C). However, further removal of the region extending from the C-terminal end of PRR to the C-terminal end of HCD augmented the percentage of soluble protein from 3 ± 0.5 to 25 ± 0.5% of total (NpostPRR versus NpostHCD; p < 0.001) (Fig. 3, B and C). With more truncation of the XL domain sequences, increasingly more recombinant proteins were detected in the soluble fraction. Comparing these results with those obtained from the truncation mutants carrying point mutations that are analogous to Gαs-R201H (i.e. GTPase inhibiting), we observed a significant difference in the localization of the mutant missing not only PRR and HCD but also both of the conserved cysteines (NpostCys). Whereas 29 ± 0.8% of total NpostCys was detected in the soluble fraction, 48 ± 0.3% of its GTPase-deficient form was in the same fraction (p < 0.001; Fig. 3, B and C). This finding, however, was not confirmed by confocal immunofluorescence microscopy, which detected both of these mutants in the cytoplasm (Fig. 3D). This discrepancy likely reflected localization of NpostCys in intracellular vesicles. Overall, these studies did not reveal clear differences between the native and GTPase-deficient forms of the truncation mutants and thus prevented us from identifying a distinct domain(s) responsible solely for keeping activated XLαs and XXLαs proteins in the plasma membrane.
FIGURE 3.
Region of XLαs comprising HCD and two conserved cysteines is important for plasma membrane targeting. HEK293 cells were transiently transfected with N-terminally truncated XLαs, and the subcellular localization of these HA-tagged truncation mutants was analyzed by immunocytochemistry and subcellular fractionation followed by Western blot after 2 days of transfection. A, schematic diagram of Gαs, XLαs domain structure, and XLαs truncated mutants used in this study. Wild-type XLαs consists of a conserved XL domain, which contains three conserved domains as follows: PRR, proline-rich region; HCD, highly charged domain; βγ, putative Gβγ interaction domain. Asterisks depict the two conserved cysteines in the XL domain. B, Western blot (anti-HA antibody) showing the subcellular distribution of N-terminal XLαs truncations in both wild-type and constitutively active (CA) forms. S, soluble fraction; P, particulate fraction. C, percentage of soluble (blue bars) or particulate (red bars) fraction in total protein, as calculated by densitometry of Western blots. Data represent means ± S.D. of three to five independent experiments. *, p < 0.01 compared with NpostPRR according to Student's t test. D, immunocytochemical analysis of N-terminal truncations of XLαs in HEK293 cells by using the anti-HA antibody.
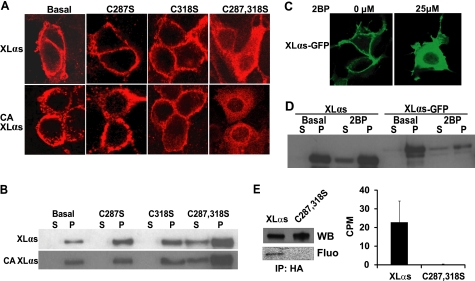
These data, however, made it clear that the region comprising the two conserved cysteines has an important role in the plasma membrane targeting of XLαs and XXLαs at the basal state. Cysteines are targets for palmitoylation, and palmitoylation is a critical post-translational modification for membrane attachment (43). To determine whether these cysteine residues were required for the membrane localization of XLαs, the two cysteine residues were mutated into serines individually or together (i.e. XLαs-C287S, XLαs-C318S, and XLαs-C287S,C318S)). Immunofluorescence confocal microscopy demonstrated that, although mutation of each cysteine alone did not affect the membrane targeting, mutations of both cysteines to serines resulted in a marked reduction of plasma membrane attachment (Fig. 4A). When analogous mutations were introduced into full-length XXLαs (i.e. XXLαs-C589S, XXLαs-C619S, XXLαs-C589S,C619S), similar results were obtained (supplemental Fig. S2). Additional introduction of the GTPase-inhibiting mutation analogous to Gαs-R201H into these Cys-to-Ser mutants also yielded similar results (Fig. 4A; supplemental Fig. S2). Western blots using the anti-Gαs C-terminal antibody verified these findings when analyzing soluble and particulate fractions of cell lysates (Fig. 4B; supplemental Fig. S2). These findings indicated that at least one of these conserved cysteines is necessary for the plasma membrane targeting of XLαs and XXLαs. We then tested the effect of 2BP, an inhibitor of protein palmitoylation, on the subcellular localization of XLαs. Treatment of cells with 2BP resulted in retardation of XLαs in the cytoplasm, as determined by fluorescence microscopy of fixed HEK293 cells transiently expressing XLαs-GFP (Fig. 4C) and Western blot analysis of lysates from HEK293 cells transiently expressing either native XLαs or XLαs-GFP (Fig. 4D). In addition, based on metabolic labeling experiments using radiolabeled palmitic acid, wild-type XLαs, but not the XLαs-C287S,C318S mutant, was palmitoylated in HEK293 cells transiently expressing these proteins (Fig. 4E). Together, these results indicated that protein palmitoylation plays a critical role in the plasma membrane targeting of XLαs (and XXLαs) at the steady state.
FIGURE 4.
Substitution of conserved cysteine residues in the XL domain and inhibition of protein palmitoylation disrupts plasma membrane targeting of XLαs at steady state. A, immunocytochemical analysis of subcellular distribution for wild-type and Cys-to-Ser mutants of XLαs in HEK293 cells by using the anti-HA antibody. HEK293 cells were transiently transfected with expression constructs encoding HA-tagged wild-type or Cys-to-Ser mutants of XLαs (Cys-287 and Cys-318). Forty eight hours after transfection, subcellular localizations of these XLαs mutants were investigated. CA, GTPase deficient, constitutively active form analogous to R201H. B, Western blots using the anti-Gαs and -XLαs C-terminal antibody was performed for determining the subcellular localizations of Cys-to-Ser mutants of XLαs in transfected HEK293 cells. C287S,C318S (C287,318S). C, subcellular localization of XLαs-GFP transiently expressed in HEK293 cells treated with either the vehicle or 25 μm 2BP immediately after transfection. D, Western blot analysis (anti-Gαs C-terminal antibody) of HEK293 cell lysates transiently expressing native XLαs or XLαs-GFP. Cells were treated with either the vehicle or 25 μm 2BP immediately after transfection. S, soluble fraction; P, particulate fraction. E, palmitoylation of XLαs, but not the XLαs-C287S,C318S mutant, transiently expressed in HEK293 cells. Following metabolic labeling with [3H]palmitic acid, transfected cells were lysed, and the HA-tagged recombinant proteins were immunoprecipitated (IP). Proteins were separated by SDS-PAGE and analyzed either by Western blot (WB) analysis using the anti-Gαs C-terminal antibody or by fluorography (Fluo), for which the exposure time was at least 6 weeks (representative of two independent experiments). In some experiments, gel slices corresponding to the immunoreactivity of XLαs and the mutant were counted to determine palmitoylation (right). Data are counts per min (CPM) above the background and represent mean ± S.E. of three independent experiments.
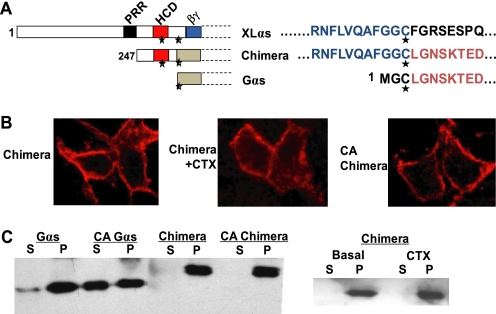
Our truncation experiments indicated that a 72-amino acid segment of XLαs extending from the C-terminal end of PRR to the second conserved cysteine (residues 247–318 according to XLαs) is critical with respect to plasma membrane targeting. Cys-318 is homologous to Gαs Cys-3, which has been shown to undergo palmitoylation and to be important for subcellular targeting (44, 45). We therefore generated an XLαs-Gαs chimera in which the 72-amino acid region of XLαs replaced Gly-2 and Cys-3 of Gαs (Fig. 5A). In transfected HEK293 cells, the XLαs-Gαs chimera was localized to the plasma membrane. Importantly, it remained localized to the plasma membrane upon activation by cholera toxin treatment or by introduction of a GTPase inhibiting mutation analogous to Gαs-R201H (Fig. 5, B and C), thus indicating that the structural features sufficient to anchor XLαs (and XXLαs) in the plasma membrane, even upon activation, are included in this 72-amino acid segment including HCD and the conserved cysteine residues.
FIGURE 5.
A 72-amino acid segment of XLαs containing HCD and the two conserved cysteines is sufficient to anchor activated Gαs to the plasma membrane. A, diagram of the XLαs-Gαs chimera protein, with the 72-amino acid segment of XLαs spanning the two cysteines to replace the N-terminal 2 residues of Gαs. PRR, proline-rich region; HCD, highly charged domain; βγ, putative Gβγ interaction domain. Asterisks indicate the conserved cysteine residues. B, immunocytochemical analysis using the anti-HA antibody was performed to study the subcellular distribution of the XLαs-Gαs chimera in HEK293 cells transfected with cDNA encoding HA-tagged XLαs-Gαs chimera or a constitutively active (CA) form of the chimera carrying a GTPase inhibiting mutant analogous to R201H. CTX was also used to stimulate the native form of the chimera. C, Western blot analysis using the anti-HA antibody for comparing the subcellular localizations of Gαs, the XLαs-Gαs chimera, and the GTPase-deficient form of the chimera transiently expressed in HEK293 cells. S, soluble fraction; P, particular fraction.
XLαs Extends the Duration of cAMP Response Induced by PTHR Activation
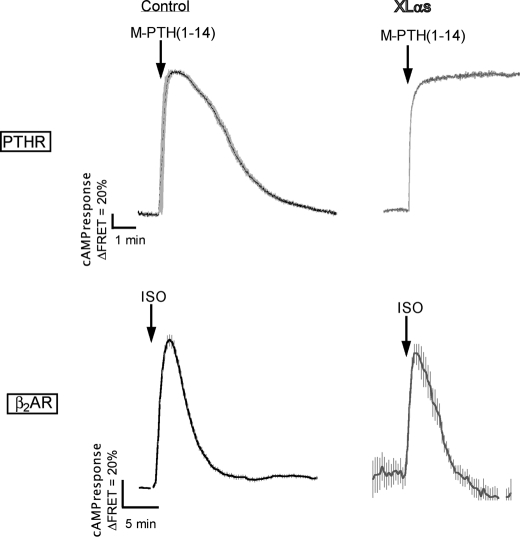
Activation-induced subcellular redistribution of Gαs serves as a regulatory mechanism that limits the time-course of cAMP generation at the plasma membrane (36, 38). We therefore reasoned that the absence of XLαs internalization could prolong the duration of cAMP generation in response to receptor activation. We addressed this hypothesis by using a Förster resonance energy transfer (FRET)-based reporter that permits real time recording of cAMP production in live cells (46). As an agonist, we chose a PTH analog, M-PTH(1–14), which has been shown to induce a short lived cAMP response (41). As expected, a rapid change in the FRET signal was observed upon the addition of 100 nm M-PTH(1–14) onto HEK293 cells expressing the PTHR alone or in combination with XLαs and Gβ1γ2 (Fig. 6). After ligand washout, the FRET signal returned back to its initial level in control cells, whereas it remained at the maximal level at least for 6 min after washout in XLαs-expressing cells (Fig. 6). Thus, XLαs extended the duration of the cAMP response induced by the typically short acting agonist M-PTH(1–14). Although this finding was consistent with the absence of XLαs internalization upon activation, a similar effect was not observed when the same cells were stimulated by isoproterenol. The kinetic profile of the cAMP response elicited by isoproterenol in cells expressing XLαs appeared indistinguishable from that in control cells (Fig. 6), indicating that the ability of XLαs to prolong agonist-induced cAMP response is receptor-specific.
FIGURE 6.
In response to PTHR, but not β2AR, activation, XLαs expression results in sustained cAMP production at the plasma membrane. cAMP response measured by FRET changes from HEK293 cells stably expressing PTHR and transiently expressing a cAMP-biosensor, Epac-CFP/YFP with or without XLαs. Cells transfected with XLαs cDNA were co-transfected with plasmids encoding Gβ1 and Gγ2. During the experiment, cells were continuously perfused with a control buffer and stimulated with either 100 nm M-PTH(1–14) or 10 μm isoproterenol (ISO). Data are mean ± S.E. of five independent experiments; cell number, n = 80.
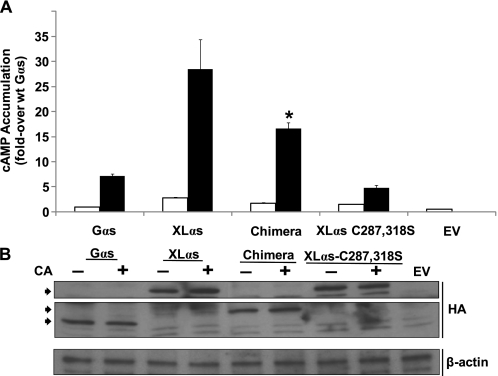
Constitutive XLαs Activity Is Markedly More Effective Than Constitutive Gαs Activity
GTPase inhibiting mutations of GNAS cause certain endocrine and nonendocrine tumors and McCune-Albright syndrome (16, 17). Because these mutations can affect both Gαs and XLαs and lead to constitutive cAMP production (23, 47), we tested whether a GTPase-deficient XLαs mutant (R543H) would be more effective in mediating basal cAMP accumulation than the cognate Gαs mutant (R201H). In transfected HEK293 cells, XLαs-R543H showed significantly (p < 0.001) higher basal cAMP accumulation compared with Gαs-R201H (Fig. 7A). Basal cAMP accumulation was also significantly (p < 0.05) higher in cells transiently expressing the GTPase-deficient version of the XLαs-Gαs chimera than Gαs-R201H, consistent with the plasma membrane localization of the former; however, the amount of cAMP accumulated in cells expressing XLαs-R543H was still significantly higher (p < 0.05) than that in cells expressing the GTPase- deficient XLαs-Gαs chimera (Fig. 7A). In contrast, the XLαs mutant carrying both C287S,C318S substitutions and an analogous GTPase-deficient mutation, which showed poor plasma membrane localization (see Fig. 4), displayed much lower basal activity than any of the other constitutively active proteins (Fig. 7A). Western blot analyses using whole cell lysates and anti-HA antibody showed that the immunoreactivity for each of these proteins was comparable with one another, indicating that the observed differences in cAMP accumulation were unlikely to reflect total expression levels (Fig. 7B).
FIGURE 7.
Basal cAMP accumulation through a GTPase-deficient XLαs mutant is significantly higher than that through the cognate Gαs mutant. A, basal cAMP accumulation in HEK293 cells transfected with plasmids encoding wild-type (white bars) or GTPase-deficient (black bars) versions of Gαs, XLαs, the XLαs-Gαs chimera (Chimera), or the XLαs-C287S,C318S mutant; cells were transfected with empty vector (EV) as control. Seventy two hours after transfection, cells were incubated with 2 mm isobutyl methylxanthine for 15 min at 37 °C. Amount of cAMP was determined by a radioimmunoassay. Values were normalized to the level obtained in cells transfected with the plasmid encoding wild-type Gαs. Data represent mean ± S.E. of seven independent experiments. *, p < 0.05 compared with constitutively active (CA) Gαs and with constitutively active XLαs according to one-way analysis of variance. B, Western blot analysis of whole cell lysates obtained from the transfected HEK293 cells analyzed in A. Specific anti-HA immunoreactivity is indicated by arrowheads. β-Actin immunoreactivity was used as a control for loading. The image is representative of three experiments with similar results.
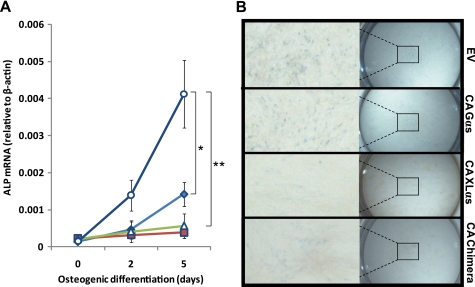
Gαs mutants carrying GTPase inhibiting mutations inhibit differentiation of osteoblasts, as seen in patients with fibrous dysplasia of bone (48, 49). We thus compared the effects of Gαs-R201H and XLαs-R543H expression on preosteoblastic MC3T3-E1 cells, in which the cAMP signaling pathway inhibits osteoblastic differentiation (50). When transfected MC3T3-E1 cells were grown under osteogenic conditions for 5 days, transient expression of Gαs-R201H significantly impaired osteoblastic differentiation, as judged by significantly lower alkaline phosphatase mRNA levels in these cells compared with control cells transfected with empty vector (Fig. 8A). XLαs-R543H expression also impaired osteoblastic differentiation of MC3T3-E1 cells, and in fact, almost no increase in alkaline phosphatase mRNA expression was detected in cells transfected with cDNA encoding XLαs-R543H (Fig. 8A). Similarly, cells transiently expressing the GTPase-deficient XLαs-Gαs chimera failed to show a significant increase in alkaline phosphatase mRNA levels under osteogenic conditions (Fig. 8A). These findings regarding alkaline phosphatase mRNA levels were corroborated by experiments measuring alkaline phosphatase activity. At day 5, although all transfected cells displayed weaker staining than control cells transfected with the empty vector, the staining of cells transiently expressing XLαs-R543H or the GTPase-deficient XLαs-Gαs chimera appeared to be nearly absent (Fig. 8B). These findings are consistent with the above results showing that activated XLαs can continue to signal, and thus, its cellular effects can be more prolonged than the cellular effects of activated Gαs.
FIGURE 8.
XLαs-R543H expression more severely inhibits osteoblastic differentiation of MC3T3-E1 cells than Gαs-R201H. A, alkaline phosphatase (ALP) mRNA levels in MC3T3-E1 cells transfected with empty vector (EV) (○), Gαs-R201H (♦), XLαs-R543H (■), or the cognate GTPase-deficient mutant of the XLαs-Gαs chimera (Δ). Osteogenic medium was introduced 2 days after transfection (day 0), and alkaline phosphatase mRNA was measured by using real time RT-PCR. Values were normalized to β-actin mRNA. Data represent mean ± S.E. of three to five independent experiments. *, p < 0.05, and **, p < 0.01 compared with empty vector-transfected cells. CA, constitutively active. B, alkaline phosphatase staining of MC3T3-E1 cells at day 5 of osteogenic differentiation. Cells grown in 24-well plates were transfected with the plasmids, as indicated; CA, GTPase-deficient form. Staining was performed after fixation, as described under “Experimental Procedures.” Images were captured at 2 or 6.3× magnification.
DISCUSSION
XLαs is a variant of Gαs that can mediate receptor-activated stimulation of adenylyl cyclases, but the study of mouse models in which XLαs is ablated indicate that the cellular actions of this protein differ importantly from those of Gαs. In this study we revealed a marked difference between these proteins, which entailed their subcellular localization following activation. Our results showed that XLαs and its N-terminally extended variant XXLαs, unlike Gαs, are not subject to activation-induced subcellular internalization. In response to the activation of most GPCRs, Gαs traffics away from the plasma membrane via an endocytic pathway, and this mechanism limits continuous generation of cAMP at the plasma membrane (36–38, 51, 52). Because XLαs lacks activation-induced internalization, it induces cAMP generation in a sustained manner, and mutational inhibition of GTPase activity results in markedly stronger constitutive activity for XLαs than for Gαs.
Recent studies have shown that isoproterenol-stimulated cAMP accumulation mediated by XLαs is higher than that mediated by Gαs in the presence of phosphodiesterase inhibitors (47, 53). These findings can now be explained, at least in part, by the sustained association of activated XLαs with the plasma membrane. However, additional mechanisms may also be involved, because in our study the degree of basal cAMP accumulation mediated by the constitutively active XLαs mutant was significantly higher than that mediated through the constitutively active XLαs-Gαs chimera (see Fig. 7), which was indistinguishable from XLαs regarding the degree of plasma membrane association. These additional mechanisms are likely to be at the level of adenylyl cyclase stimulation rather than involving nucleotide exchange or GTPase activity, given that the differences between Gαs and XLαs regarding cAMP production were also observed in a GTPase-deficient state. Studies have shown that Gαs is associated with lipid rafts and that this localization hinders its ability to fully stimulate adenylyl cyclase (52, 54). It is thus possible that XLαs is excluded from lipid rafts, which would be consistent with the potent adenylyl cyclase stimulation by this protein. Localization of XLαs within different membrane microdomains needs to be investigated in future studies.
Expression of XLαs resulted in a prolonged cAMP response after PTHR but not the β2AR stimulation (see Fig. 6). This finding could reflect the differences between the mechanisms underlying the desensitization of these two receptors. The inability of activated XLαs to extend isoproterenol-induced cAMP response, despite remaining in the plasma membrane (see Fig. 1), could reflect receptor phosphorylation and arrestin binding, which are well characterized mechanisms desensitizing β2AR (55–57). PTHR, however, appears to utilize distinct mechanisms of activation and desensitization. Unlike β2AR, PTHR is able to generate prolonged cAMP signaling in response to certain agonists (38, 41), and arrestins, contrary to their actions on β2AR, enhance the cAMP signaling induced by PTH (58). These differences may thus explain why the strong plasma membrane localization of XLαs yields a prolonged cAMP response to PTH but not to isoproterenol. Alternatively, PTHR and β2AR might interact differently with XLαs, such that the mechanisms desensitizing PTHR, regardless of being different from those desensitizing β2AR, fail to overcome the effect of activated XLαs localization in the plasma membrane. These possibilities, which may have significant implications in GPCR signaling and the relative roles of Gαs and XLαs, remain to be addressed.
Previous studies had originally indicated that rat XLαs is localized to the trans-Golgi network (18), and six cysteine residues within the XL domain have been previously identified as being critical for this subcellular localization at the basal state (59). More recent studies have established that both rat and human XLαs are localized to the plasma membrane (25, 33). Only two of the six cysteines shown to be important for the subcellular localization of rat XLαs are conserved in human XLαs, which is used in this study, and our findings show that at least one of those conserved cysteines is absolutely required for plasma membrane targeting. Cysteine residues are predicted to be substrates for palmitoylation. Indeed, rat XLαs has been shown to be palmitoylated (33), and the cysteine residues within the cysteine-rich region appeared to be required for this modification (59). Our present results demonstrate that human XLαs is also palmitoylated. Because this lipid modification is required for the insertion of Gαs to the plasma membrane (60), it is likely to be required for the plasma membrane targeting of XLαs as well. Our results obtained with the Cys-to-Ser mutants and the palmitoylation inhibitor 2BP are consistent with this prediction.
According to our results obtained from truncation mutants, plasma membrane targeting of XLαs requires not only one of the two conserved cysteines in the XL domain but also the region between PRR and the C-terminal end of HCD (see Fig. 4), which consists of a highly acidic N-terminal portion and a highly basic C-terminal portion. Polybasic amino acid regions are important for membrane targeting of other proteins, such as Ras and Rho family of small GTPases (61), and indeed, a recent study identified an N-terminal polybasic region within Gαs as another signal for plasma membrane targeting (62). Similarly, the corresponding region in XLαs, i.e. the C-terminal end of the XL domain, includes multiple basic residues, and these residues, in addition to those within HCD, may contribute to the plasma membrane targeting of XLαs at the steady state. The polybasic regions within HCD could also play a role in preventing the activation-induced subcellular redistribution of XLαs, because replacing several N-terminal residues of Gαs with a polybasic region from G protein-coupled receptor kinase 5 has inhibited the cytosolic redistribution of Gαs upon activation (36).
It is clear that XLαs can mimic Gαs regarding cAMP production both in transfected cells (23–25) and in transgenic mice (26). In fact, our results, together with recent data (47, 53), show that it can even be more effective than Gαs. Then, why do data from Gαs knock-out mouse models (31, 32) indicate that endogenous XLαs is unable to compensate for Gαs ablation? There could be two possible reasons. First, because activated Gαs can also act on effector molecules that are localized intracellularly (63, 64), and because it can apparently signal from internalized vesicles (38, 39), it is conceivable that the defects caused by Gαs ablation result primarily from the loss of its actions that occur through intracellular effectors/mechanisms. This hypothesis is certainly consistent with the inability of XLαs to substitute for Gαs despite being a strong stimulator of cAMP production at the plasma membrane. Second, it is possible that the internalization of Gαs upon activation is required for effective cAMP signaling. For example, the Gαs-specific regulator of G protein signaling RGS-PX1 is localized to endosomes (65), and it has been suggested that the termination of Gαs activation and thereby reassembly of the G protein heterotrimer is achieved at this subcellular site (36). Conversely, it is important to note that the presence of endogenous Gαs expression in XLαs knock-out mice is also insufficient to prevent the phenotypes observed in this model (27, 28). Therefore, it appears that XLαs and Gαs have nonredundant contributions to cAMP signaling, perhaps by having spatially and temporally different expression profiles or by mediating cAMP signaling in response to different types of stimuli. In addition, it is likely that XLαs interacts with unique effectors at the plasma membrane and thereby trigger signaling pathways that differ entirely from the cAMP signaling pathway.
GTPase inhibiting mutations that lead to constitutive XLαs activity are found in patients with certain endocrine and nonendocrine tumors, fibrous dysplasia of bone, and McCune-Albright syndrome (47, 66). Our findings predict that by enhancing the basal levels of cAMP and/or as-yet-undefined second messengers, constitutive XLαs signaling at the plasma membrane is likely to contribute to the pathogenesis of these disorders. For example, XLαs is normally expressed in undifferentiated skeletal progenitors (47, 67), and GTPase-deficient XLαs mutants may thus play a role in inhibiting the differentiation of these cells into normal bone.
Recent studies have identified the GNAS locus as one of few genes whose expression and/or copy numbers are increased in various cancers (14, 15, 68). Given that XLαs activity is regulated less rigorously than Gαs activity, at least with respect to cAMP production, it is tempting to speculate that elevation of XLαs levels plays an important role in the development of some of these cancers. Consistent with that hypothesis, XLαs expression is normally limited to the paternal GNAS allele, and it is known that overexpression of paternally expressed gene products, such as insulin-like growth factor 2 (69), can lead to neoplasia.
In summary, our results show that XLαs traffics differently from Gαs upon activation and thereby is able to extend cAMP signaling at the plasma membrane. The unique cellular actions of XLαs and its variant XXLαs remain unknown, and their strong association with the plasma membrane may form the basis for these unique actions.
Supplementary Material
Acknowledgments
We thank Matthew James Webber and Richard Bouley (Massachusetts General Hospital, Program in Membrane Biology) for providing technical advice and support for the TIRFM experiments. We also thank Thomas Gardella (Massachusetts General Hospital) for the critical review of this manuscript and Harald Jüppner (Massachusetts General Hospital) for helpful discussions throughout the study.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK073911 (to M. B.) and R01DK087688 (to J.-P. V.) from NIDDK. This work was also supported in part by March of Dimes Foundation Research Grant 6-FY10-336 (to M. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and videos.
- GPCR
- G protein-coupled receptor
- Gαs
- α-subunit of the stimulatory G protein
- XLαs
- extra-large αs
- XXLαs
- N-terminally extended XLαs
- PTH
- PTHR, type 1 PTH/PTHrP receptor
- β2AR
- β2-adrenergic receptor
- TIRF
- total internal reflection fluorescence
- TIRFM
- total internal reflection fluorescence microscopy
- PACAP
- pituitary adenylate cyclase activating peptide-27
- HCD
- highly charged domain
- PRR
- proline-rich region
- 2BP
- 2-bromo-hexadecanoic acid
- CTX
- cholera toxin.
REFERENCES
- 1. Kozasa T., Itoh H., Tsukamoto T., Kaziro Y. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 2081–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weinstein L. S., Liu J., Sakamoto A., Xie T., Chen M. (2004) Endocrinology 145, 5459–5464 [DOI] [PubMed] [Google Scholar]
- 3. Peters J., Williamson C. M. (2008) Adv. Exp. Med. Biol. 626, 16–26 [DOI] [PubMed] [Google Scholar]
- 4. Plagge A., Kelsey G., Germain-Lee E. L. (2008) J. Endocrinol. 196, 193–214 [DOI] [PubMed] [Google Scholar]
- 5. Chen Y., Nakura J., Jin J. J., Wu Z., Yamamoto M., Abe M., Tabara Y., Yamamoto Y., Igase M., Bo X., Kohara K., Miki T. (2003) Hypertens Res. 26, 439–444 [DOI] [PubMed] [Google Scholar]
- 6. Yamamoto M., Abe M., Jin J. J., Wu Z., Tabara Y., Mogi M., Kohara K., Miki T., Nakura J. (2004) Hypertens. Res. 27, 919–924 [DOI] [PubMed] [Google Scholar]
- 7. Frey U. H., Alakus H., Wohlschlaeger J., Schmitz K. J., Winde G., van Calker H. G., Jöckel K. H., Siffert W., Schmid K. W. (2005) Clin. Cancer Res. 11, 5071–5077 [DOI] [PubMed] [Google Scholar]
- 8. Frey U. H., Eisenhardt A., Lümmen G., Rübben H., Jöckel K. H., Schmid K. W., Siffert W. (2005) Cancer Epidemiol. Biomarkers Prev. 14, 871–877 [DOI] [PubMed] [Google Scholar]
- 9. Frey U. H., Lümmen G., Jäger T., Jöckel K. H., Schmid K. W., Rübben H., Müller N., Siffert W., Eisenhardt A. (2006) Clin. Cancer Res. 12, 759–763 [DOI] [PubMed] [Google Scholar]
- 10. Hahn S., Frey U. H., Siffert W., Tan S., Mann K., Janssen O. E. (2006) Eur. J. Endocrinol. 155, 763–770 [DOI] [PubMed] [Google Scholar]
- 11. Otterbach F., Callies R., Frey U. H., Schmitz K. J., Wreczycki C., Kimmig R., Siffert W., Schmid K. W. (2007) Breast Cancer Res. Treat. 105, 311–317 [DOI] [PubMed] [Google Scholar]
- 12. Schmitz K. J., Lang H., Frey U. H., Sotiropoulos G. C., Wohlschlaeger J., Reis H., Takeda A., Siffert W., Schmid K. W., Baba H. A. (2007) Neoplasia 9, 159–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lehnerdt G. F., Franz P., Zaqoul A., Schmitz K. J., Grehl S., Lang S., Schmid K. W., Siffert W., Jahnke K., Frey U. H. (2008) Clin. Cancer Res. 14, 1753–1758 [DOI] [PubMed] [Google Scholar]
- 14. Kan Z., Jaiswal B. S., Stinson J., Janakiraman V., Bhatt D., Stern H. M., Yue P., Haverty P. M., Bourgon R., Zheng J., Moorhead M., Chaudhuri S., Tomsho L. P., Peters B. A., Pujara K., Cordes S., Davis D. P., Carlton V. E., Yuan W., Li L., Wang W., Eigenbrot C., Kaminker J. S., Eberhard D. A., Waring P., Schuster S. C., Modrusan Z., Zhang Z., Stokoe D., de Sauvage F. J., Faham M., Seshagiri S. (2010) Nature 466, 869–873 [DOI] [PubMed] [Google Scholar]
- 15. Sandgren J., Andersson R., Rada-Iglesias A., Enroth S., Akerstrom G., Dumanski J. P., Komorowski J., Westin G., Wadelius C. (2010) Exp. Mol. Med. 42, 484–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weinstein L. S., Chen M., Xie T., Liu J. (2006) Trends Pharmacol. Sci. 27, 260–266 [DOI] [PubMed] [Google Scholar]
- 17. Spiegel A. M., Weinstein L. S. (2004) Annu. Rev. Med. 55, 27–39 [DOI] [PubMed] [Google Scholar]
- 18. Kehlenbach R. H., Matthey J., Huttner W. B. (1994) Nature 372, 804–809 [DOI] [PubMed] [Google Scholar]
- 19. Hayward B. E., Kamiya M., Strain L., Moran V., Campbell R., Hayashizaki Y., Bonthron D. T. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10038–10043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peters J., Wroe S. F., Wells C. A., Miller H. J., Bodle D., Beechey C. V., Williamson C. M., Kelsey G. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3830–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abramowitz J., Grenet D., Birnbaumer M., Torres H. N., Birnbaumer L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8366–8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aydin C., Aytan N., Mahon M. J., Tawfeek H. A., Kowall N. W., Dedeoglu A., Bastepe M. (2009) Endocrinology 150, 3567–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klemke M., Pasolli H. A., Kehlenbach R. H., Offermanns S., Schultz G., Huttner W. B. (2000) J. Biol. Chem. 275, 33633–33640 [DOI] [PubMed] [Google Scholar]
- 24. Bastepe M., Gunes Y., Perez-Villamil B., Hunzelman J., Weinstein L. S., Jüppner H. (2002) Mol. Endocrinol. 16, 1912–1919 [DOI] [PubMed] [Google Scholar]
- 25. Linglart A., Mahon M. J., Kerachian M. A., Berlach D. M., Hendy G. N., Jüppner H., Bastepe M. (2006) Endocrinology 147, 2253–2262 [DOI] [PubMed] [Google Scholar]
- 26. Liu Z., Segawa H., Aydin C., Reyes M., Erben R. G., Weinstein L. S., Chen M., Marshansky V., Frohlich L. F., Bastepe M. (2011) Endocrinology 152, 1222–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Plagge A., Gordon E., Dean W., Boiani R., Cinti S., Peters J., Kelsey G. (2004) Nat. Genet. 36, 818–826 [DOI] [PubMed] [Google Scholar]
- 28. Xie T., Plagge A., Gavrilova O., Pack S., Jou W., Lai E. W., Frontera M., Kelsey G., Weinstein L. S. (2006) J. Biol. Chem. 281, 18989–18999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu S., Gavrilova O., Chen H., Lee R., Liu J., Pacak K., Parlow A. F., Quon M. J., Reitman M. L., Weinstein L. S. (2000) J. Clin. Invest. 105, 615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Skinner J. A., Cattanach B. M., Peters J. (2002) Genomics 80, 373–375 [DOI] [PubMed] [Google Scholar]
- 31. Chen M., Gavrilova O., Liu J., Xie T., Deng C., Nguyen A. T., Nackers L. M., Lorenzo J., Shen L., Weinstein L. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7386–7391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Germain-Lee E. L., Schwindinger W., Crane J. L., Zewdu R., Zweifel L. S., Wand G., Huso D. L., Saji M., Ringel M. D., Levine M. A. (2005) Endocrinology 146, 4697–4709 [DOI] [PubMed] [Google Scholar]
- 33. Pasolli H. A., Klemke M., Kehlenbach R. H., Wang Y., Huttner W. B. (2000) J. Biol. Chem. 275, 33622–33632 [DOI] [PubMed] [Google Scholar]
- 34. Levis M. J., Bourne H. R. (1992) J. Cell Biol. 119, 1297–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wedegaertner P. B., Bourne H. R., von Zastrow M. (1996) Mol. Biol. Cell 7, 1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thiyagarajan M. M., Bigras E., Van Tol H. H., Hébert T. E., Evanko D. S., Wedegaertner P. B. (2002) Biochemistry 41, 9470–9484 [DOI] [PubMed] [Google Scholar]
- 37. Makita N., Sato J., Rondard P., Fukamachi H., Yuasa Y., Aldred M. A., Hashimoto M., Fujita T., Iiri T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17424–17429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ferrandon S., Feinstein T. N., Castro M., Wang B., Bouley R., Potts J. T., Gardella T. J., Vilardaga J. P. (2009) Nat. Chem. Biol. 5, 734–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Calebiro D., Nikolaev V. O., Gagliani M. C., de Filippis T., Dees C., Tacchetti C., Persani L., Lohse M. J. (2009) PLoS Biol. 7, e1000172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shimizu N., Dean T., Tsang J. C., Khatri A., Potts J. T., Jr., Gardella T. J. (2005) J. Biol. Chem. 280, 1797–1807 [DOI] [PubMed] [Google Scholar]
- 41. Okazaki M., Ferrandon S., Vilardaga J. P., Bouxsein M. L., Potts J. T., Jr., Gardella T. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16525–16530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bastepe M., Weinstein L. S., Ogata N., Kawaguchi H., Jüppner H., Kronenberg H. M., Chung U. I. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14794–14799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smotrys J. E., Linder M. E. (2004) Annu. Rev. Biochem. 73, 559–587 [DOI] [PubMed] [Google Scholar]
- 44. Linder M. E., Middleton P., Hepler J. R., Taussig R., Gilman A. G., Mumby S. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3675–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Parenti M., Viganó M. A., Newman C. M., Milligan G., Magee A. I. (1993) Biochem. J. 291, 349–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nikolaev V. O., Bünemann M., Hein L., Hannawacker A., Lohse M. J. (2004) J. Biol. Chem. 279, 37215–37218 [DOI] [PubMed] [Google Scholar]
- 47. Mariot V., Wu J. Y., Aydin C., Mantovani G., Mahon M. J., Linglart A., Bastepe M. (2011) Bone 48, 312–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weinstein L. S., Shenker A., Gejman P. V., Merino M. J., Friedman E., Spiegel A. M. (1991) N. Engl. J. Med. 325, 1688–1695 [DOI] [PubMed] [Google Scholar]
- 49. Schwindinger W. F., Francomano C. A., Levine M. A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 5152–5156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tintut Y., Parhami F., Le V., Karsenty G., Demer L. L. (1999) J. Biol. Chem. 274, 28875–28879 [DOI] [PubMed] [Google Scholar]
- 51. Hynes T. R., Mervine S. M., Yost E. A., Sabo J. L., Berlot C. H. (2004) J. Biol. Chem. 279, 44101–44112 [DOI] [PubMed] [Google Scholar]
- 52. Allen J. A., Yu J. Z., Donati R. J., Rasenick M. M. (2005) Mol. Pharmacol. 67, 1493–1504 [DOI] [PubMed] [Google Scholar]
- 53. Kaya A. I., Ugur O., Oner S. S., Bastepe M., Onaran H. O. (2009) J. Pharmacol. Exp. Ther. 329, 350–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Allen J. A., Yu J. Z., Dave R. H., Bhatnagar A., Roth B. L., Rasenick M. M. (2009) Mol. Pharmacol. 76, 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kohout T. A., Lefkowitz R. J. (2003) Mol. Pharmacol. 63, 9–18 [DOI] [PubMed] [Google Scholar]
- 56. Ferguson S. S., Zhang J., Barak L. S., Caron M. G. (1998) Life Sci. 62, 1561–1565 [DOI] [PubMed] [Google Scholar]
- 57. Bünemann M., Lee K. B., Pals-Rylaarsdam R., Roseberry A. G., Hosey M. M. (1999) Annu. Rev. Physiol. 61, 169–192 [DOI] [PubMed] [Google Scholar]
- 58. Feinstein T. N., Wehbi V. L., Ardura J. A., Wheeler D. S., Ferrandon S., Gardella T. J., Vilardaga J. P. (2011) Nat. Chem. Biol. 7, 278–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ugur O., Jones T. L. (2000) Mol. Biol. Cell 11, 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wedegaertner P. B., Chu D. H., Wilson P. T., Levis M. J., Bourne H. R. (1993) J. Biol. Chem. 268, 25001–25008 [PubMed] [Google Scholar]
- 61. Williams C. L. (2003) Cell. Signal. 15, 1071–1080 [DOI] [PubMed] [Google Scholar]
- 62. Crouthamel M., Thiyagarajan M. M., Evanko D. S., Wedegaertner P. B. (2008) Cell. Signal. 20, 1900–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Castellone M. D., Teramoto H., Williams B. O., Druey K. M., Gutkind J. S. (2005) Science 310, 1504–1510 [DOI] [PubMed] [Google Scholar]
- 64. Yu J. Z., Dave R. H., Allen J. A., Sarma T., Rasenick M. M. (2009) J. Biol. Chem. 284, 10462–10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zheng B., Ma Y. C., Ostrom R. S., Lavoie C., Gill G. N., Insel P. A., Huang X. Y., Farquhar M. G. (2001) Science 294, 1939–1942 [DOI] [PubMed] [Google Scholar]
- 66. Mantovani G., Bondioni S., Lania A. G., Corbetta S., de Sanctis L., Cappa M., Di Battista E., Chanson P., Beck-Peccoz P., Spada A. (2004) J. Clin. Endocrinol. Metab. 89, 3007–3009 [DOI] [PubMed] [Google Scholar]
- 67. Michienzi S., Cherman N., Holmbeck K., Funari A., Collins M. T., Bianco P., Robey P. G., Riminucci M. (2007) Hum. Mol. Genet. 16, 1921–1930 [DOI] [PubMed] [Google Scholar]
- 68. Tominaga E., Tsuda H., Arao T., Nishimura S., Takano M., Kataoka F., Nomura H., Hirasawa A., Aoki D., Nishio K. (2010) Gynecol. Oncol. 118, 160–166 [DOI] [PubMed] [Google Scholar]
- 69. Reik W., Constancia M., Dean W., Davies K., Bowden L., Murrell A., Feil R., Walter J., Kelsey G. (2000) Int. J. Dev. Biol. 44, 145–150 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.