Background: SUV3 helicase is an essential component of mitochondrial degradosome.
Results: Using specific genetic mutants, SUV3 was demonstrated to participate in mtDNA replication, which requires ATPase activity and the C-terminal conserved region.
Conclusion: SUV3 plays two distinctive roles in maintaining mtDNA stability and RNA turnover.
Significance: This study suggests a direct role of SUV3 in maintaining mtDNA stability, paving a foundation for biochemical investigation.
Keywords: SUV3, Mitochondrial DNA, Mitochondrial Metabolism, RNA Helicase, RNA Turnover, Yeast Genetics, Mitochondrial DNA (mtDNA) Replication, Mitochondrial Homeostasis, RNA Degradosome, Intron Toxicity
Abstract
Yeast SUV3 is a nuclear encoded mitochondrial RNA helicase that complexes with an exoribonuclease, DSS1, to function as an RNA degradosome. Inactivation of SUV3 leads to mitochondrial dysfunctions, such as respiratory deficiency; accumulation of aberrant RNA species, including excised group I introns; and loss of mitochondrial DNA (mtDNA). Although intron toxicity has long been speculated to be the major reason for the observed phenotypes, direct evidence to support or refute this theory is lacking. Moreover, it remains unknown whether SUV3 plays a direct role in mtDNA maintenance independently of its degradosome activity. In this paper, we address these questions by employing an inducible knockdown system in Saccharomyces cerevisiae with either normal or intronless mtDNA background. Expressing mutants defective in ATPase (K245A) or RNA binding activities (V272L or ΔCC, which carries an 8-amino acid deletion at the C-terminal conserved region) resulted in not only respiratory deficiencies but also loss of mtDNA under normal mtDNA background. Surprisingly, V272L, but not other mutants, can rescue the said deficiencies under intronless background. These results provide genetic evidence supporting the notion that the functional requirements of SUV3 for degradosome activity and maintenance of mtDNA stability are separable. Furthermore, V272L mutants and wild-type SUV3 associated with an active mtDNA replication origin and facilitated mtDNA replication, whereas K245A and ΔCC failed to support mtDNA replication. These results indicate a direct role of SUV3 in maintaining mitochondrial genome stability that is independent of intron turnover but requires the intact ATPase activity and the CC conserved region.
Introduction
SUV3 was originally identified in Saccharomyces cerevisiae as a dominant suppressor allele, SUV3-1, which suppressed a 3′ dodecamer deletion phenotypes on the VAR1 gene (1–3). Later characterization showed that the SUV3-1 allele carries a missense mutation, Val-272 to Leu (named V272L in this study) (4). Protein sequence analyses and subsequent in vitro studies revealed that SUV3 is a nuclear encoded ATP-dependent mitochondrial RNA helicase that belongs to the DExH/D-box superfamily, members of which are frequently involved in RNA processing and metabolism (5–7). SUV3 is later identified to be an essential component of the S. cerevisiae mitochondrial degradosome, along with a 3′ to 5′ exoribonuclease, DSS1 (4, 8–11). Together, the two proteins form a heterodimer in vitro, capable of degrading a double-stranded RNA (dsRNA) substrate with a 3′ overhang (7). Inactivation of either SUV3 or DSS1 in yeast led to respiratory deficiency; accumulation of aberrant RNA species, including unprocessed precursors, excised group I introns (e.g. ω intron), and transcripts with abnormal termini (8–10, 12); and loss of mitochondrial DNA (4, 6, 9, 12).
The RNA degradation assembly (also known as the RNA degradosome) has also been shown to participate in proper RNA processing and turnover in other model organisms, such as Escherichia coli and humans. The E. coli RNA degradosome is one of the best characterized; it consists of four enzymes (13–16): RNase E, an endoribonuclease (17–20); RhlB, a DEAD-box helicase (21, 22); PNPase, an ambivalent enzyme possessing both 3′ to 5′ exoribonuclease and 5′ to 3′ polyribonucleotidyltransferase activities (23, 24); and enolase, a glycolytic enzyme dispensable for in vitro reconstituted functional RNA degradosome (25, 26). The human mitochondrial degradosome, on the other hand, does not resemble the low eukaryotic counterpart because the yeast exoribonuclease, DSS1, has not been identified in humans (27–29). Instead, the 330-kDa human mitochondrial SUV3-PNPase complex of degradosome seems to function as an alternative form of E. coli degradosome lacking RNase E. The inactivation of SUV3 in the human system leads to disruption of mitochondrial homeostasis, which eventually results in cell senescence or death (30).
Because one of the most prominent phenotypes observed in yeast with null or mutated SUV3 is the accumulation of excised group I introns, which could function as endonucleases themselves, the intron toxicity theory has long been speculated as the major reason for the observed SUV3-deficient phenotypes. Initially, it was thought that the excessive nuclease activity may cause nonspecific digestion of DNA contents similar to that during ω intron conversion (8, 31–33). Later, an alternative hypothesis was proposed that excised introns may trigger exon reopening of their processed parental mRNAs through a site-specific endoribonuclease activity, based on the observation of free exons accumulated in SUV3-deleted strains (8). A more recent study further supports this hypothesis by demonstrating that SUV3 is required for recycling splicing cofactors for group I introns and removing toxic intron RNAs (34).
However, if the primary substrates for the presumably catalytically active introns are processed RNAs, this does not explain why nearly all perturbations of SUV3 in both yeast and humans result in loss of mitochondrial genome. It is even more peculiar when considering the fact that human mitochondria do not even contain introns. Whether or not there is a functional coordination of SUV3 between the proper RNA turnover and the maintenance of mitochondrial genome remains unclear. The extent to which the intron contributes to both processes also warrants a thorough investigation. Although it seems plausible that reopening of mRNAs may disrupt mitochondrial translation and affect respiratory chain function, leading to increased reactive oxygen species production that damages mtDNA5 and contributes to the loss of mitochondrial DNA (30, 35), a direct role of SUV3 in the maintenance of mitochondrial genomic stability cannot be ruled out.
To address these conundrums, we established an inducible SUV3 knockdown system in S. cerevisiae to characterize the respiratory competency, RNA degradation, and mtDNA stability of three SUV3 mutants under normal and intronless mitochondrial genome backgrounds. Our results suggest that SUV3 participates in the maintenance of mitochondrial genomic stability that is separable from its involvement in RNA degradation and processing.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
The promoter and coding region of S. cerevisiae SUV3 was subcloned into pRS415 vector to generate pRS415-ySUV3. C-terminal HA tag was generated by creating a BamHI site at the termination codon and then inserting an annealed oligonucleotide of HA tag into this BamHI site, denoted as pRS415-ySUV3HA. For generating TAP-tagged knock-in bullet, the entire SUV3 locus, including the 5′- and 3′-untranslated region, was first subcloned into pBSK as pBSK-ySUV3. Then C-terminal TAP tag was generated by creating a PacI and PmeI site at the termination codon and inserting a PacI and PmeI fragment of TAP epitope, denoted as pBSK-ySUV3TAP. Point mutations and deletion were subsequently generated by site-directed mutagenesis (QuikChange site-directed mutagenesis kit, Stratagene). For purifications of recombinant proteins, both wild-type and mutant SUV3 were subcloned into pET19b vector with the first 27 amino acids deleted. His-tagged DSS1 was constructed by subcloning the coding region of DSS1 into pQE30 vector (Novagen/EMD Biosciences) with the first 34 amino acids deleted. Purified His-tagged SUV3 recombinant protein was used as antigen to immunize mice to produce mouse polyclonal anti-ySUV3 antiserum following procedures described previously (36). ARP3 antibody was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). HA antibody was purchased from Sigma.
Yeast Strain Constructions
The S. cerevisiae strains used in this study are listed in Table 1. In brief, galactose-responsive promoter knock-in bullets, Gal1p-SUV3::KanMX6 and GAL1p-SUV3::HygroMX6, were generated by flanking Gal1 promoter between endogenous SUV3 promoter and coding sequence using standard PCR procedures. ρ0 derivatives were obtained by culturing cells in medium containing 33 μg/ml ethidium bromide for 24 h. KY81 was constructed by cytoducting mitochondria derived from Kar(167) strain (kindly provided by Dr. Gérard Faye) into Y81E strain. Δsuv3 was constructed using the d1 disruption construct described by Stepien et al. (4). STAP strains were generated by transforming Y300E with NotI and SalI-digested pBSK-ySUV3TAP bullets. The resulting strains were used to mate with either YS16 or KYS16 to generate the diploid strains. MGM101-GFP strains were constructed by knocking in a GFP bullet to the C terminus of MGM101 in either YS16 or KYS16 strains, which were then mated to STAP strains to generate diploid strains.
TABLE 1.
Summary of S. cerevisiae strains used in this study
| Strain | Nuclear genotype | Mitochondrial genotype | Source |
|---|---|---|---|
| BWG1 | MATa, his1, ade1, leu2, ura3 | ρ+, ω+ | Ref. 4 |
| BS16 | As BWG1, Gal1p-SUV3::KanMX4 | ρ+, ω+ | This study |
| Y81 | MATα, ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100 | ρ+, ω+ | Ref. 56 |
| Y81E | MATα, ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100 | ρ0 | This study |
| KY81 | MATα, ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100 | ρ+, intronless | This study |
| YS16 | As Y81, Gal1p-SUV3::HygroMX4 | ρ+, ω+ | This study |
| KYS16 | As KY81, Gal1p-SUV3::HygroMX4 | ρ+, intronless | This study |
| MGM101 YS16 | As YS16, MGM101-GFP::TRP1 | ρ+, ω+ | This study |
| MGM101 KYS16 | As KYS16, MGM101-GFP::TRP1 | ρ+, intronless | This study |
| Y300 | MATa, ade2-1 trp1-1 ura3-1 leu2-3,112 his 3-11,15 can1-100 | ρ+, ω+ | Ref. 56 |
| Y300E | MATa, ade2-1 trp1-1 ura3-1 leu2-3,112 his 3-11,15 can1-100 | ρ0 | This study |
| Δsuv3 | As Y300, suv3::URA3 | ρ0 | This study |
| STAP | As Y300E, SUV3-TAP::KanMX4 | ρ0 | This study |
| YSTAP | MATa/α, trp1-1/trp1-1 ura3-1/ura3-1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 ade2-1/ade2-1 can 1-100/can 1-100, Gal1p-SUV3::HygroMX4, SUV3-TAP::KanMX4 | ρ+, ω+ | This study |
| KYSTAP | MATa/α, trp1-1/trp1-1 ura3-1/ura3-1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 ade2-1/ade2-1 can 1-100/can 1-100, Gal1p-SUV3::HygroMX4, SUV3-TAP::KanMX4 | ρ+, intronless | This study |
| MGM101 YSTAP | As YSTAP, MGM101/MGM101-GFP::TRP1 | ρ+, ω+ | This study |
| MGM101 KYSTAP | As KYSTAP, MGM101/MGM101-GFP::TRP1 | ρ+, intronless | This study |
| DSS1TAP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, DSS1-TAP::His5MX4 | ρ+ | Open Biosystem |
| Kar(167) | MATa, Karl-I trpS cyhR [NCYC74, Δ bI, Δ bI, Δ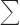 aI] aI] |
ρ+, intronless | Ref. 45 |
Reagents and Media
Culture media and standard yeast genetic methods were as described previously (37). Briefly, strains were culture in either complete medium (YEP; 1% yeast extract, 2% peptone) or synthetic medium with appropriate amino acid missing. The carbon sources were added as indicated, which could be 2% galactose, 2% glucose, or 3% glycerol. The chemicals and medium components were purchased from Sigma.
Northern Blot Analysis
Northern blot analysis was performed as described previously (38). RNAs were extracted from 20 ml of overnight yeast culture using TRIzol reagent (Invitrogen). AlkPhos with the CDP-star kit (GE Healthcare) was used for signal detection. Probes against ω intron and nuclear gene 18S were generated by PCR using the following primers: ω intron, 5′-GAA GTA AAT TGG GTG AAT TGC (forward) and 5′-TCA TTT GAG GAA TTA AAT AAG G (reverse); 18S, 5′-GAT CCT GCC AGT AGT CAT TGC TTG (forward) and 5′-CAG GAC CAA GCG GTT CTC GGT GTT (reverse).
mtDNA Copy Number Determination
DNA were extracted from 10 ml of overnight yeast culture using phenol/chloroform extraction. Quantitative PCR was performed as described previously using an iCycler (Bio-Rad) (30, 39). The relative mtDNA copy number was calculated as a ratio between the abundances of mitochondrial COX1/nuclear Actin1. Primer sequences used were as follows: COX1, 5′-CTA CAG ATA CAG CAT TTC CAA GA (forward) and 5′-GTG CCT GAA TAG ATG ATA ATG GT (reverse); Actin1, 5′-ACT ATT GGT AAC GAA AGA TTC AGA (forward) and 5′ TCA CAC TTC ATG ATG GAG TTG TAA (reverse).
Membrane Potential and ATP Production Detection
Methods were adapted from a previous publication (30). Briefly, 5 ml of yeast cultures after depletion of SUV3 in glucose for 24 h were washed with 1× PBS and incubated with 50 nm rhodamine 123 for 15 min. After washing three times with 1× PBS, flow cytometry was used to measure the uptake of rhodamine 123 by mitochondria. ATP production was determined by an ATP determination kit (Invitrogen) following the manufacturer's instructions.
Immunoprecipitation and Western Blot Analysis
10 ml of overnight yeast cultures were pelleted, and cells were resuspended in Nonidet P-40 lysis buffer (50 mm Tris-HCl (pH 7.5), 200 mm NaCl, 0.2% Nonidet P-40, 1 mm EDTA) and broken with glass beads using a FastPrep24 machine (MP Biomedicals). After removing cell debris by centrifugation at 14,000 rpm for 2 min at 4 °C, 300 μg of the lysate supernatants were then subjected to immunoprecipitation by 20 μl of IgG-Sepharose beads (GE Healthcare). Western blot was performed using rabbit IgG antibody (GeneTex) as primary antibody and rabbit IgG HRP as secondary antibody. ARP3 antibody was used for input control.
Expression and Purification of His-tagged SUV3 and DSS1
Bacteria strain Rosetta was used to express wild-type or mutant SUV3s. Subsequent purification procedures using nickel beads were based on a previous publication (40). His-tagged DSS1 was co-transformed into Rosetta with pREP4 plasmid. SUV3 and DSS1 were co-purified by mixing bacteria expressing each individual protein and purified in the same manner.
ATPase Assay, Gel Mobility Shift Assay, Helicase Assay, and Exoribonuclease Assay
Experimental procedures were adapted from a previous publication (40, 41), and detailed procedures can be found in the supplemental Experimental Procedures. All DNA and RNA oligonucleotides were purchased from Integrated DNA Technologies (San Diego, CA) Sequences included T20DNA (5′-CAA ACT CTC TCT CTC TCA AC), 3WLRNA (5′-GUU GAG AGA GAG AGA GUU UGA GAG AGA GAG GUU UGA GAG AGA), and T22RNA (5′-CUC AAA CUC UCU CUC UCU CAA C).
In Vivo Binding Assay
Cell lysates were prepared using Nonidet P-40 lysis buffer, and immunoprecipitation was performed in the same manner as described above using IgG-Sepharose beads. After SDS-PAGE, both TAP-tagged DSS1 and HA-tagged SUV3 were detected by HA antibody.
Yeast Cytology
Cells were collected at specific time points with OD < 0.5. After washing twice in water, cells were stained with 1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI) for 60 min, followed by another two washes in water. Then cells were suspended in minimal volume of water and 1 μl of cell suspensions were used for imaging. Images were obtained on an inverted Zeiss Axio Observer.Z1 microscope fluorescent system equipped with the X-Cite series 120 mercury lamps (Carl Zeiss MicroImaging, Inc.).
Mitochondrial DNA Immunoprecipitation Assay
The mitochondrial DNA immunoprecipitation (mtIP) was adapted as described (42). In brief, washed yeast cells were first treated with 2 mm ethylene glycol bis(succinimidyl succinate) for 30 min followed by 4% formaldehyde for 15 min at room temperature. The cells were lysed in mtIP lysis buffer (50 mm HEPES (pH 7.5), 140 mm NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mm EDTA) using glass beads and FastPrep24. Then DNA was sheared into small pieces using Bioruptor. IgG-Sepharose beads were used to immunoprecipitate TAP-tagged proteins at 4 °C overnight. After extensive wash, cross-link was reversed at 65 °C overnight, and the DNA was purified using a commercial PCR clean up kit (Qiagen). Specific DNA fragments were amplified and quantified by quantitative PCR. Primer sequences used were as follows: ori5, 5′-CAG AGC ACA CAT TTG TTA ATA TTT AAT AA (forward) and 5′-CCC GGA TAT CTT CTT GTT TAT C (reverse); ori6, 5′-CTC ACC CTA TTT ATT AAT CAT TAA TAA G (forward) and 5′ TTC CTA AGA ATA ATT ATT ATA ATA TTA ATT AAT TAC (reverse); 21S rRNA, 5′-TTG GTG AGA GAA AAT AAT AAA GGT C (forward) and 5′-ATA AAA TAA TCA TTT TCA TAC TTT CCC TTA CGG (reverse); COX1, 5′-CTA CAG ATA CAG CAT TTC CAA GA (forward) and 5′-GTG CCT GAA TAG ATG ATA ATG GT (reverse); ATP6, 5′-TGG TAC ACC ATT ACC ATT AGT ACC (forward) and 5′-CCA TTA ATA AAT GAC CAG CTA AGA (reverse).
RESULTS
Depletion of SUV3 Leads to Accumulation of ω Intron and Reduction of mtDNA Copy Number
To study the underlying mechanism of how SUV3 contributes to the maintenance of mitochondrial homeostasis, we developed an inducible system in a BWG1 strain, where the expression of SUV3 was regulated by a galactose-responsive promoter (the derived strain is named the BS16 strain; Fig. 1A). This inducible system allowed us to manipulate SUV3 expression at specific time points. Using this system, we performed a panel of assays to evaluate the various mitochondria functions with or without SUV3 expression.
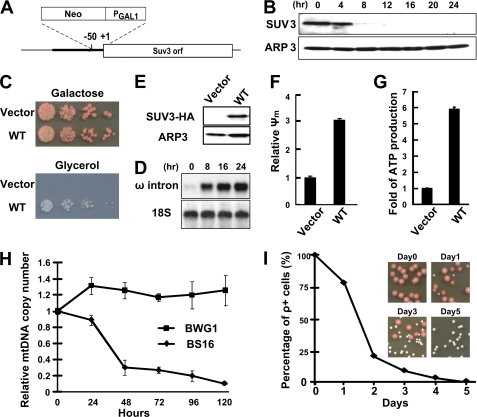
FIGURE 1.
Depletion of SUV3 leads to accumulation of ω intron and reduction of mtDNA copy number. A, a schematic representation of galactose-responsive promoter knocked in between −50 and +1 on endogenous SUV3 promoter locus. B, Western blot assay showing that endogenous SUV3 expression is reduced significantly after switching the carbon source from galactose to glucose. A nuclear encoded gene, ARP3, served as loading control. C, growth assay showing the survival of cells expressing either null (vector) or HA-tagged wild-type SUV3 on galactose- and glycerol-containing −Leu plates. D, Western blot assay showing expression of HA-tagged wild-type SUV3 in comparison with cells transformed with vector only. E, Northern blot assay demonstrating ω intron accumulation. A nuclear encoded gene, 18S, served as loading control. F and G, relative membrane potential (Ψm) and ATP production are abolished in cells expressing vector compared with cells expressing wild-type SUV3. H, qPCR assay showing a decrease of mtDNA copy number. Each point was done in triplicate for statistics. I, percentage of ρ+ cells after depletion of endogenous SUV3 for various times and representative images of yeast colonies grown on galactose plates. Error bars, S.E.
Upon switching the carbon source from galactose to glucose, the expression of SUV3 decreased dramatically in 8 h and became undetectable after 12 h, suggesting a robust and efficient SUV3 depletion (Fig. 1B). Consistent with previous studies (4), SUV3-depleted cells failed to utilize non-fermentable carbon sources like glycerol (Fig. 1C). Northern blot analysis showed a substantial accumulation of ω introns after SUV3 depletion (Fig. 1E), in contrast to the observation in the parental BWG1 strain with the same carbon source switch (supplemental Fig. 1). The SUV3 depletion in BS16 cells also severely disrupted the membrane potential and abolished the ATP production, which are typical of mitochondrial dysfunctions (Fig. 1, F and G). All of the described defects could be rescued by expressing wild-type SUV3 with an HA tag using a low copy number CEN6 vector (Fig. 1, C–G), which functionally validated our inducible system.
In S. cerevisiae, one prominent phenotype of SUV3 deficiency is the formation of petite colonies. However, the mtDNA status has never been examined directly in these cells. Therefore, we performed quantitative PCR (qPCR) to measure the mtDNA copy number, which was calculated as a ratio of mitochondrial COX1 over nuclear Actin1. Surprisingly, upon switching to glucose, mtDNA copy number declined significantly after 24 h, and less than 10% of mtDNA was retained after 5 days in the BS16 strain (Fig. 1H). In comparison, the parental BWG1 strain maintained a relatively stable mtDNA copy number throughout. To further evaluate the mtDNA integrity, endogenous wild-type SUV3 was re-expressed by plating back onto galactose plates after depleting SUV3 for various times. The pink pigmentation of BWG1 strain indicates that cells maintained their mtDNA as ρ+ status, and white petite colonies suggest that cells had been converted into ρ− or ρ0 status with mitochondrial dysfunctions. As illustrated in Fig. 1I, virtually no cells retained ρ+ status after 5 days of depletion, and re-expressing wild-type SUV3 failed to restore mtDNA. The trend of the decreasing percentages of ρ+ cells correlated well with the changes in mtDNA copy number. These observations indicate that the cells might gradually lose their mtDNA contents after SUV3 depletion, and once the mtDNA copy number falls below a certain threshold, reintroducing wild-type SUV3 will not be able to restore the ρ+ status.
Mutation in the Helicase Motifs or C-terminal Conserved Region Is Detrimental to SUV3 Functions in Maintaining Mitochondrial Homeostasis
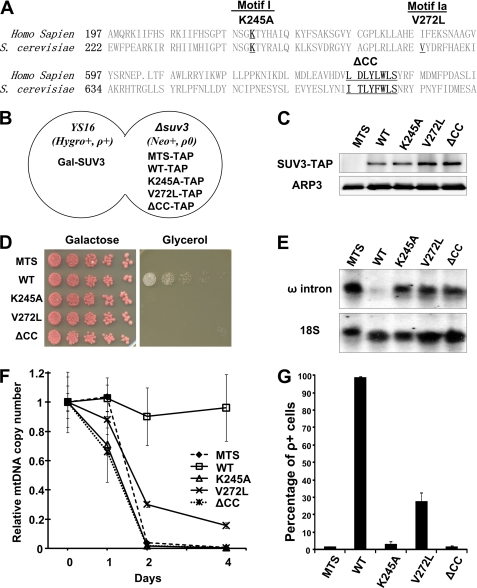
To further dissect the involvement of SUV3 in RNA turnover and mtDNA maintenance, we generated two point mutants, K245A and V272L, carrying mutations in the helicase motifs I and Ia, respectively. Mutant K245A is the yeast homologue of the previously described human SUV3 K213A mutant, which abolished the ATPase and helicase activities of SUV3 (40). Mutant V272L is the previously characterized spontaneous dominant allele, SUV3-1, that carries a missense mutation in motif Ia. Motif Ia in the DExH/D-box helicase family has been proved to be involved in RNA binding (43). In attempts to map potential novel functional motifs outside of the helicase domain, we constructed a third mutant, ΔCC, carrying an 8-amino acid deletion in the highly conserved region C-terminal to the helicase domain (Fig. 2A).
FIGURE 2.
Mutation in helicase motifs or C-terminal conserved region is detrimental to SUV3 functions in maintaining mitochondrial homeostasis. A, alignment of human and yeast SUV3 protein sequences with mutated sequences underlined. B, a schematic representation of YSTAP diploid strain. C, Western blot assay showing expression of TAP-tagged wild-type SUV3 and mutants in YSTAP strains. D, growth assay for YSTAP strains on galactose and glycerol plates. E, Northern blot assay demonstrating accumulation of ω intron after a 1-day depletion of endogenous SUV3. F, qPCR assay showing changes in mtDNA copy number of different mutants in YSTAP strain. Each point was done in triplicate for statistics. G, percentage of ρ+ cells after depleting endogenous SUV3 in glucose for 5 days. Error bars, S.E.
To characterize the physiological behaviors of these three SUV3 mutants, we adapted our inducible system into a diploid strain (schematics shown in Fig. 2B). The derived strain, YS16, showed comparable phenotypes of mitochondrial dysfunctions as the BS16 strain when depleted SUV3 (data not shown). Meanwhile, wild-type or mutant SUV3 was C-terminally TAP-tagged and knocked in to the endogenous SUV3 locus in a ρ0 Δsuv3 strain to generate YSTAP strains. A TAP tag fused to mitochondrial targeting sequence (MTS) only was similarly knocked in to serve as a negative control. After mating, the expressions of each TAP-tagged mutant SUV3 were assessed by Western blot analysis (Fig. 2C). Despite the various mutations that were introduced, each TAP-tagged mutant expressed at a level compatible to wild-type SUV3-TAP. Using this diploid inducible system with normal mtDNA contributed from the YS16 strain, we were able to examine the mutants under a relatively normal physiological background.
We first examined mitochondrial respiration status by growing the cells in different carbon sources. As shown in Fig. 2D, none of the strains expressing MTS, K245A, V272L, or ΔCC was viable on glycerol plates, suggesting that the mutations rendered the cells unable to utilize a non-fermentable carbon source. Moreover, all four strains showed significant amounts of ω intron accumulation, whereas cells expressing wild-type SUV3 had minimal level of accumulation (Fig. 2E). Cells expressing MTS, K245A, or ΔCC also displayed drastic decreases in mtDNA copy number in 5 days, whereas V272L showed a slower reduction than the other three mutant strains (Fig. 2F). To probe the integrity of mtDNA, each strain was plated back onto galactose plates after 5 days of depletion. As shown in Fig. 2G, cells expressing MTS, K245A, or ΔCC showed less than 1% of its ρ+ population, whereas the V272L mutant retained about 30% of its ρ+ cells. These results suggest that the helicase motifs and CC region are essential for normal SUV3 functions in supporting mitochondrial respiration, such as utilizing a non-fermentable carbon source, degrading introns, and maintaining mtDNA. However, V272L showed slightly higher mtDNA and ρ+ cell retention than K245A and ΔCC mutants.
Mitochondrial Homeostasis Can Be Restored by the V272L Mutant, but Not K245A or ΔCC, in an Intronless mtDNA Background
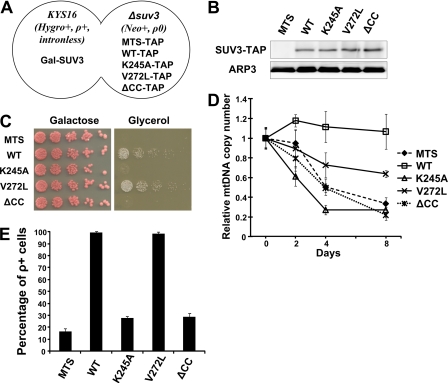
In yeast, it has been well documented that removal of all mitochondrial intron sequences has no detectable effect on mitochondrial functions under laboratory conditions (44, 45). The intronless strain may serve as an excellent tool to evaluate the alleged intron toxicity theory. For this evaluation, we first depleted mitochondrial DNA in YS16 strains by ethidium bromide treatment (the derived strain is named the Y81E strain). Intronless mitochondrial DNA genome from Kar(167) was introduced into Y81E strains by cytoduction to generate the KYS16 strain. KYS16 was then mated with STAP strains carrying different knocked in mutants, denoted as KYSTAP strains (Fig. 3A). The expression levels of wild-type and mutant SUV3 proteins are comparable (Fig. 3B). Next, we performed a growth assay to assess the survival of different intronless strains on glycerol plates (Fig. 3C). Surprisingly, in an intronless background, V272L was capable of utilizing glycerol, suggesting that the metabolic defects caused by V272L mutation can be rescued in the intronless mtDNA background. In contrast, the MTS, K245A, and ΔCC mutants failed to grow on glycerol plates even in the intronless mtDNA background. Subsequent qPCR assays showed that V272L maintained about 70% of its mtDNA after 8 days of depletion, whereas MTS, K245A, and ΔCC retained only 30% of mtDNA copies (Fig. 3D). Consistently, in the intronless context, the percentage of ρ+ cells for V272L was similar to the strain expressing the wild type, whereas the MTS, K245A, and ΔCC mutants maintained less than 30% ρ+ cells (Fig. 3E). These results implicate a role of SUV3 in mtDNA maintenance that is independent of that in intron turnover. Furthermore, this role of SUV3 in mtDNA maintenance, largely preserved in V272L, requires both intact ATPase activity and the conserved CC region.
FIGURE 3.
Mitochondrial homeostasis can be restored by the V272L mutant, but not K245A or ΔCC, in an intronless mtDNA background. A, a schematic representation of KYSTAP diploid strain. B, Western blot assay showing expression of TAP-tagged wild-type SUV3 and mutant proteins in KYSTAP strains. C, growth assay for KYSTAP strains on galactose and glycerol plates. D, qPCR assay showing changes in mtDNA copy number of different mutants in KYSTAP strains over 8 days. Each point was done in triplicate for statistics. E, percentage of ρ+ cells after depleting endogenous SUV3 for 8 days. Error bars, S.E.
The K245A Mutant Loses ATPase Activity, whereas V272L and ΔCC Mutants Have Reduced RNA Binding Activities
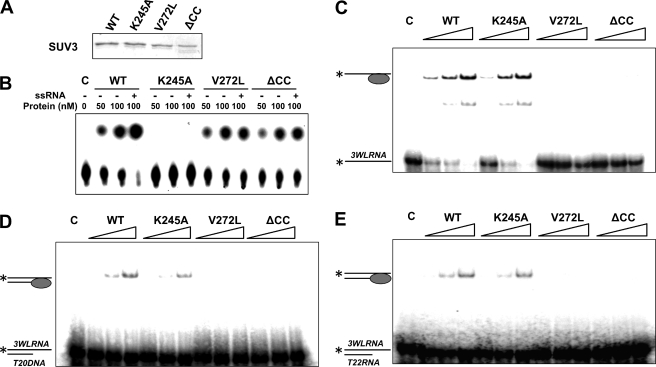
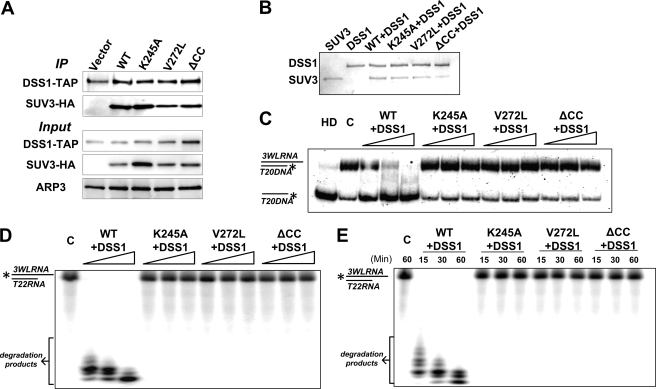
To pinpoint the defects of K245A, V272L, and ΔCC mutants, the enzymatic activities of each mutant were examined. We subcloned wild-type SUV3, K245A, V272L, and ΔCC mutants into a His-tagged E. coli expression vector, and the recombinant proteins were purified using nickel beads (Fig. 4A). As shown in Fig. 4B, purified wild-type SUV3 showed robust ATPase activity that was greatly enhanced by single-stranded RNA (ssRNA). K245A completely failed to hydrolyze ATP as expected. Both V272L and ΔCC mutants, in contrast, displayed basal levels of ATPase activity but were unable to respond to ssRNA stimulation, implying potential defects in RNA binding. To confirm this hypothesis, we performed gel mobility shift assays using various substrates (Fig. 4, C–E). Similar to their human counterparts, wild-type SUV3 and K245A had high affinities for ssRNA and double-stranded substrates (either DNA/RNA duplex or dsRNA) with a 3′ overhang. Mutants V272L and ΔCC exhibited no affinity to any of the substrates. Collectively, these biochemical analyses indicate that mutant K245A loses its ATPase activity, whereas mutants V272L and ΔCC show abolished RNA binding activities.
FIGURE 4.
The K245A mutant loses ATPase activity, whereas V272L and ΔCC mutants have reduced RNA binding activities. A, a Coomassie Blue-stained gel showing purified His-tagged recombinant wild-type SUV3 and mutants. B, increasing amounts of wild-type, K245A, V272L, and ΔCC (50 and 100 nm) were subjected to ATPase assay with or without 3WL ssRNA. Gel mobility shift assays demonstrating binding affinities of increasing amounts of wild-type SUV3 and each mutant (100, 200, and 400 nm) to single-stranded RNA (C), double-stranded DNA/RNA hybrid (D), and double-stranded RNA (E).
Helicase and RNA Degradation Activities Are Abolished in Mutants K245A, V272L, and ΔCC When Complexed with DSS1
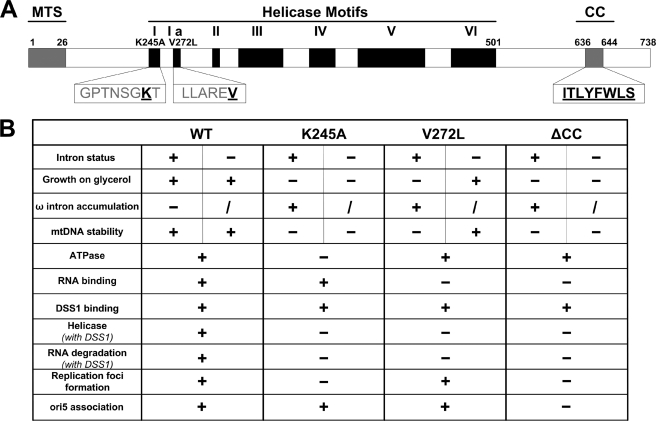
In S. cerevisiae, SUV3 has been identified as a component of the mitochondrial RNA degradosome, and the SUV3-DSS1 heterodimeric complex can efficiently degrade structured RNAs in the presence of ATP (4, 8–11). Given their physiological role together, the interactions between our SUV3 mutants and DSS1 and their activities as RNA degradosome were investigated. We tested their in vivo interactions by performing co-immunoprecipitation experiments using a DSS1-TAP strain expressing HA-tagged wild-type SUV3 and mutants. As shown in Fig. 5A, all three mutants, K245A, V272L, and ΔCC, were able to bind DSS1 to a similar extent as wild-type SUV3. Hence, SUV3 wild-type and mutants were co-purified with DSS1 (Fig. 5B), and their activities were tested by helicase and RNA degradation assays. As shown in Fig. 5C, only wild-type SUV3-DSS1 complex had the helicase activity with double-stranded DNA/RNA substrate, whereas K245A exhibited no helicase activity due to its inability to hydrolyze ATP. V272L and ΔCC failed to unwind double-stranded substrate due to defective substrate binding. Moreover, dosage and time course degradation experiments showed that only the wild-type SUV3-DSS1 complex can unwind and degrade dsRNA substrate, a task that all three mutants failed to achieve (Fig. 5, D and E). Taken together, these results indicate that none of K245A-DSS1, V272L-DSS1, or ΔCC-DSS1 complexes has a normal RNA degradosome activity.
FIGURE 5.
Helicase and RNA degradation activities are abolished in mutants K245A, V272L, and ΔCC when complexed with DSS1. A, Western blot assay showing both wild-type and mutant SUV3s retained binding ability to DSS1 in vivo. B, a Coomassie Blue-stained gel showing co-purified wild-type SUV3 and mutant with DSS1. C, increasing amounts of wild-type SUV3-DSS1, K245A-DSS1, V272L-DSS1, or ΔCC-DSS1 (100, 200, and 400 nm) were incubated with the RNA/DNA helicase substrate and assayed on a 15% polyacrylamide gel. HD, heat-denatured; C, control. D, degradation assay using increasing amounts of wild-type SUV3-DSS1, K245A-DSS1, V272L-DSS1, or ΔCC-DSS1 (100, 200, and 400 nm) to incubate with dsRNA substrate for 30 min and analyzed on a 15% denaturing gel. E, time course assay (15, 30, and 60 min) of degradation activity on dsRNA substrate from 400 nm of wild-type SUV3-DSS1, K245A-DSS1, V272L-DSS1, or ΔCC-DSS1.
Dysfunctional SUV3 Impairs mtDNA Replication in Part because SUV3 Associates with Replication Origin 5
Based on the observations that SUV3 localizes to mitochondrial nucleoids in human (46) and mtDNA copy number drops gradually after SUV3 depletion, we wondered whether or not SUV3 may contribute to mtDNA maintenance by facilitating mtDNA replication. Previous studies on yeast mtDNA replication have identified two membrane-spanning proteins, MGM101 and MMM1, that are exclusively associated with actively replicating nucleoids (47). Therefore, we modified YS16 and KYS16 strains to express C-terminally GFP-tagged MGM101, allowing us to examine the mtDNA replication status after SUV3 depletion. As illustrated in Fig. 6A, depletion of SUV3 led to drastically decreased numbers of MGM101-GFP foci (i.e. the active replication foci). Quantifications of the replication foci showed a statistically significant reduction within 8 h of SUV3 depletion (Fig. 6B). Intriguingly, similar changes were observed in intronless KYS16 strain under the same conditions (Fig. 6C). These data indicate that depletion of SUV3 is correlated with the decrease in active mtDNA replication, which is not affected by the intron status. Next, we tested the effects of SUV3 mutants using diploid strains expressing MGM101-GFP after 8 h of SUV3 depletion in glucose medium. Surprisingly, V272L showed a level of replication foci comparable with that of wild-type SUV3, whereas neither K245A nor ΔCC was able to sustain mtDNA replication (Fig. 6D). Similarly, intron status had no evident effect on replication as shown in Fig. 6E.
FIGURE 6.
Dysfunctional SUV3 impairs mtDNA replication in part because SUV3 associates with replication origin 5. A, representative images of cells expressing MGM101-GFP after depleting SUV3 for various times. Scale bar, 2 μm. B, enumeration of replication foci in MGM101-YS16 strain. C, enumeration of replication foci in MGM101-KYS16 strain. Later time points were not included in B and C because the numbers of cells with replication foci were too low to be compared with early time points. D, enumeration of replication foci for cells expressing MTS, wild-type SUV3, K245A, V272L, or ΔCC mutant in MGM101-YSTAP (D) or MGM101-KYSTAP (E) strains after 8 h of SUV3 depletion in glucose medium (images of cells were collected from two independent experiments; mean ± S.E.; n, number of cells counted; p value calculated using Student's t test, p < 0.001). F, mtIP followed by qPCR showing the selective association of wild-type SUV3 with an active mtDNA replication origin 5 while DSS1 does not bind DNA. mtIP showing association of wild type, K245A, V272L, and ΔCC with replication ori5 in YSTAP (G) and KYSTAP (H) strains. All qPCRs were done in triplicate for statistics. Error bars, S.E.
To test whether SUV3 may directly contribute to mtDNA replication, we employed the mtIP assay to evaluate the association of SUV3 with mtDNA during replication. Interestingly, wild-type SUV3 showed a significantly selective association with one of the active mtDNA replication origins, ori5, but not with ori6, an inactive replication origin (48), or the 21S rRNA locus, or the coding region for COX1 and ATP6. DSS1, on the other side, showed no evident binding to mtDNA at all (Fig. 6F), indicating that DSS1 may not be involved in regulating mtDNA replication. Following mtIP assays performed on both YSTAP and KYSTAP strains expressing wild-type and mutant SUV3s clearly showed that wild-type SUV3, K245A, and V272L but not ΔCC were able to associate with ori5 regardless of the intron status (Fig. 6, G and H). Our observations clearly suggest that SUV3 is selectively associated with ori5 through the CC domain, reflecting a potential mechanism for SUV3 to maintain mtDNA stability through regulation of mtDNA replication, which requires both intact ATPase activity and the conserved CC region.
DISCUSSION
In this paper, we delineated the roles of SUV3 in regulating proper mitochondrial RNA turnover and maintaining mtDNA stability by characterizing three mutants, K245A, V272L, and ΔCC, which carry mutations in helicase motifs I and Ia and a conserved region that is C-terminal to the helicase domain, respectively. As summarized in Fig. 7, all three mutants failed to maintain mitochondrial homeostasis under a normal mitochondrial genomic background. However, the mutant V272L can rescue the said deficiencies under an intronless mtDNA background. Biochemically, the ATPase activity is abolished in K245A, whereas V272L and ΔCC are defective in RNA binding, which all resulted in inability to unwind and degrade dsRNA substrate when complexed with DSS1. In attempts to reveal the contribution of SUV3 in mtDNA maintenance, we performed cytology assessments on actively replicating mtDNA and mtIP experiments, the results of which indicated that SUV3 disruption led to reduced mtDNA replication as SUV3 associates with an active mtDNA replication origin, ori5.
FIGURE 7.
Summary of structural and functional relationships of wild-type and mutant SUV3s. A, a schematic representation of SUV3 structure with the locations of mutants indicated in highlighted boxes. B, a table summarizing different phenotypes of wild-type SUV3 and each mutant.
The studies on yeast SUV3 are often complicated by the expression level of SUV3 and the fact that ablation of SUV3 causes a disturbance in mitochondrial homeostasis (2, 34). The inducible system that we established in this study provides advantages over other systems. First, knocking in the mutants into the endogenous SUV3 locus in one haploid strain circumvents all potential artifacts generated by ectopic expression of any given genes. Second, SUV3 level can be controlled specifically, preventing long term stress posed on the cells by dysfunctional and unstable mitochondria. Third, it allows introduction of different mitochondrial genomic backgrounds with relative ease (Figs. 2 and 3). It is the usage of such a system that permits us to manipulate SUV3 expression and analyze mutant functions in a stable but versatile mitochondrial genomic background. Using this inducible system, we confirmed the pleiotropic effects of SUV3 depletion on mitochondrial functions, including the inability to utilize non-fermentable carbon sources, accumulation of group I introns, and a decrease in mtDNA copy number, which could not be rescued by any of the three mutants analyzed (Figs. 1 and 2).
The most prominent phenotype in SUV3-depleted cells is the accumulation of ω intron and other excised group I introns, which are ribozymes that can catalyze their own splicing in vitro. Based on this, the intron toxicity theory has been the central dogma in SUV3 or mitochondrial RNA degradosome studies for decades. It is speculated that these presumably active endoribonucleases can attack ligated mRNAs, leading to the accumulation of free exons and the observed mitochondrial deficiencies (8, 34). If this conjecture were true, regardless of the status of SUV3, removing introns would eliminate the previously observed mitochondrial defects because there would be no accumulation of toxic introns. To test this hypothesis, we introduced intronless mitochondrial genome into the diploid strains expressing the mutants. The mtDNA maintenance is improved in intronless strains for all mutants compared with their counterparts with normal mtDNA backgrounds, suggesting that accumulations of introns do contribute to mtDNA instability and the disruption of mitochondrial homeostasis to a certain extent. However, only cells expressing V272L mutant are viable on a glycerol plate, whereas those of MTS, K245A, and ΔCC fail to survive (Fig. 3). The inability of K245A and ΔCC to rescue mitochondrial functions in intronless backgrounds suggests that both helicase activity and the CC region of SUV3 are essential for maintaining mitochondrial homeostasis through a mechanism independent of intron turnover.
It was noted in the previous publication (4) that V272L is able to maintain mtDNA homeostasis when expressed in BWG1 strain with a normal mitochondrial genome that contains introns. We suspected that the discrepancy was caused by genomic variations in different yeast strains. Therefore, we expressed all three mutants using CEN6 vectors in the inducible BS16 strains with normal mitochondrial genomes. As shown in supplemental Fig. 2, V272L is indeed able to survive on a glycerol plate, which K245A and ΔCC failed to achieve. Interestingly, the Y81 and Y300 strains used in this study, which are derived from the W303 strain, carry a RAD5-535 allele that inhibits postreplication repair (49) as well as an allelic variant of MIP1 that increases petite frequency (50). It is highly possible that these two mutations may aggravate V272L-associated phenotypes in a synergistic manner, implying other potential genetic interactions with SUV3. This yeast strain model allows us to reveal and characterize the phenotypes of V272L mutant under normal and intronless mitochondrial genomic backgrounds and uncouple the roles of SUV3 in the maintenance of genomic stability and RNA degradation.
In yeast, mtDNA stability can be controlled in many ways, including its replication, package, segregation, and repair (for a review, see Ref. 51). However, whether SUV3 regulates the mtDNA maintenance has never been investigated. To provide more insights, we first examined mtDNA replication by evaluating mtDNA replication foci. Meeusen and Nunnari (47) demonstrated that at any particular time point, only a subset of nucleoids undergoes active replication as indicated by BrdU labeling, and this subset of nucleoids is exclusively associated with two membrane-spanning proteins, MGM101 and MMM1. Our data showed that the numbers of MGM101-GFP-associated replication foci significantly declined within 8 h of SUV3 depletion. This suggests an immediate response of mtDNA replication to the reduction in SUV3 protein level. Thus, it is probable that SUV3 directly participates in mtDNA replication. Because mtDNA replication is often unsynchronized with mitochondrial segregation and cell division (52), it is plausible that without active mtDNA replication, the total mtDNA content will be gradually diluted out as cells divide, causing decreases in mtDNA copy number as we observed in our qPCR experiments (Figs. 1–3). In addition, this hypothesis would also provide an explanation for the other observation that when the wild-type SUV3 is reintroduced, cells are able to repopulate their mtDNA contents, although the extents of recovery depend on how much mtDNA is left after SUV3 depletion. Consistently, the ability of V272L in maintaining mtDNA replication in both normal and intronless backgrounds (Fig. 6) might be the reason why it always displays better retention of mtDNA and ρ+ cells (Figs. 2 and 3) compared with K245A and ΔCC, which fail to support mtDNA replication.
The association of SUV3 with ori5 (Fig. 6) further supports the function of SUV3 in regulating mtDNA replication and is consistent with the observation that SUV3 localizes to mitochondrial nucleoids in humans (46). According to Lecrenier and Foury (48), yeast mtDNA replication is initiated by RNA priming of mitochondrial RNA polymerase RPO41 at active replication origins, such as ori5 tested in this study. The RNA transcript synthesized by RPO41 is then processed by endoribonucleases to generate an RNA primer for DNA synthesis (53, 54). The function of SUV3 at ori5 may be involved in resolving the RNA/DNA hybrid structure formed during mtDNA replication initiation. Interestingly, the catastrophic effects caused by SUV3 deficiency could be attenuated by mutations in RPO41 as well as MTF1, another subunit of mitochondrial RNA polymerase. It is likely that such mutations severely reduce the transcription and replication initiation rate (55) and therefore relieve stress at the active replication origin.
Furthermore, it is noted that K245A and V272L, but not the ΔCC mutant, are found to associate with ori5 under both normal and intronless mtDNA backgrounds (Fig. 6, G and H). It is quite surprising to us that although both V272L and ΔCC show diminished RNA binding activities, only V272L is capable of interacting with ori5, indicating that CC domain is essential for the ori5 association. This observation could be explained by two possibilities. First, the CC region may be critical in recognizing a unique RNA/DNA hybrid structure at ori5. It is not uncommon that in DExD/D-box helicases, regions outside of the helicase core domain can contribute to substrate recognition (5). Second, the CC region may be critical for interacting with other proteins that bind specifically to ori5. Thus, although V272L fails to recognize substrate through its helicase motif Ia, it may preserve the interactions with nucleic acids or proteins through the CC region, which is sufficient to support mtDNA replication. It should also be noted that although K245A is able to bind to ori5, it fails to maintain mtDNA replication (Fig. 6, G and H), suggesting that intact ATPase activity is required at ori5 to carry out active replication. In addition, the inability of DSS1 to bind to mtDNA further dissociates the roles of SUV3 in RNA degradation and mtDNA maintenance. It is possible that SUV3 is a component of a distinct nucleoid protein complex that regulates mtDNA replication.
To summarize, our results have two implications: first, SUV3 regulates mitochondrial homeostasis at both the RNA and DNA levels; second, SUV3 harbors a distinct function that requires both ATPase activity and C-terminal conserved region for regulating mtDNA replication and maintaining mitochondrial genomic stability that is independent of its involvement in intron turnover. Although the precise biochemical role of SUV3 at the replication origin remains to be explored, our findings set a strong foundation for further endeavors.
Supplementary Material
Acknowledgments
We are grateful to Drs. Steve J. Elledge, Gérard Faye, and Ronald Butow for providing yeast strains; Dr. Haoping Liu and Shelley Lane for assistance with yeast cytology; and Dr. Guikai Wu and Brittany Ngo for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grant AG027877 (to W. H. L. and P. L. C.) and the NIH, NIGMS, Medical Scientist Training Program (University of California, Irvine) (to D.-H. W). W. H. L. serves as a member of the board of directors for GeneTex Inc. The term of this arrangement has been reviewed and approved by University of California, Irvine according to its conflict of interest policy.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. 1 and 2.
- mtDNA
- mitochondrial DNA
- mtIP
- mitochondrial DNA immunoprecipitation
- qPCR
- quantitative PCR
- MTS
- mitochondrial targeting sequence
- ySUV3
- yeast SUV3.
REFERENCES
- 1. Butow R. A., Zhu H., Perlman P., Conrad-Webb H. (1989) Genome 31, 757–760 [DOI] [PubMed] [Google Scholar]
- 2. Conrad-Webb H., Perlman P. S., Zhu H., Butow R. A. (1990) Nucleic Acids Res. 18, 1369–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu H., Conrad-Webb H., Liao X. S., Perlman P. S., Butow R. A. (1989) Mol. Cell. Biol. 9, 1507–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stepien P. P., Margossian S. P., Landsman D., Butow R. A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 6813–6817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cordin O., Banroques J., Tanner N. K., Linder P. (2006) Gene 367, 17–37 [DOI] [PubMed] [Google Scholar]
- 6. Dziembowski A., Piwowarski J., Hoser R., Minczuk M., Dmochowska A., Siep M., van der Spek H., Grivell L., Stepien P. P. (2003) J. Biol. Chem. 278, 1603–1611 [DOI] [PubMed] [Google Scholar]
- 7. Malecki M., Jedrzejczak R., Stepien P. P., Golik P. (2007) J. Mol. Biol. 372, 23–36 [DOI] [PubMed] [Google Scholar]
- 8. Margossian S. P., Li H., Zassenhaus H. P., Butow R. A. (1996) Cell 84, 199–209 [DOI] [PubMed] [Google Scholar]
- 9. Golik P., Szczepanek T., Bartnik E., Stepien P. P., Lazowska J. (1995) Curr. Genet. 28, 217–224 [DOI] [PubMed] [Google Scholar]
- 10. Dziembowski A., Malewicz M., Minczuk M., Golik P., Dmochowska A., Stepien P. P. (1998) Mol. Gen. Genet. 260, 108–114 [DOI] [PubMed] [Google Scholar]
- 11. Dmochowska A., Golik P., Stepien P. P. (1995) Curr. Genet. 28, 108–112 [DOI] [PubMed] [Google Scholar]
- 12. Stepien P. P., Kokot L., Leski T., Bartnik E. (1995) Curr. Genet. 27, 234–238 [DOI] [PubMed] [Google Scholar]
- 13. Coburn G. A., Mackie G. A. (1999) Prog. Nucleic Acid. Res. Mol. Biol. 62, 55–108 [DOI] [PubMed] [Google Scholar]
- 14. Py B., Causton H., Mudd E. A., Higgins C. F. (1994) Mol. Microbiol. 14, 717–729 [DOI] [PubMed] [Google Scholar]
- 15. Miczak A., Kaberdin V. R., Wei C. L., Lin-Chao S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 3865–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bessarab D. A., Kaberdin V. R., Wei C. L., Liou G. G., Lin-Chao S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3157–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ehretsmann C. P., Carpousis A. J., Krisch H. M. (1992) Genes Dev. 6, 149–159 [DOI] [PubMed] [Google Scholar]
- 18. Callaghan A. J., Marcaida M. J., Stead J. A., McDowall K. J., Scott W. G., Luisi B. F. (2005) Nature 437, 1187–1191 [DOI] [PubMed] [Google Scholar]
- 19. Babitzke P., Kushner S. R. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vanzo N. F., Li Y. S., Py B., Blum E., Higgins C. F., Raynal L. C., Krisch H. M., Carpousis A. J. (1998) Genes Dev. 12, 2770–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Py B., Higgins C. F., Krisch H. M., Carpousis A. J. (1996) Nature 381, 169–172 [DOI] [PubMed] [Google Scholar]
- 22. Kalman M., Murphy H., Cashel M. (1991) New Biol. 3, 886–895 [PubMed] [Google Scholar]
- 23. Carpousis A. J., Van Houwe G., Ehretsmann C., Krisch H. M. (1994) Cell 76, 889–900 [DOI] [PubMed] [Google Scholar]
- 24. Mohanty B. K., Kushner S. R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11966–11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morita T., Kawamoto H., Mizota T., Inada T., Aiba H. (2004) Mol. Microbiol. 54, 1063–1075 [DOI] [PubMed] [Google Scholar]
- 26. Chandran V., Luisi B. F. (2006) J. Mol. Biol. 358, 8–15 [DOI] [PubMed] [Google Scholar]
- 27. Briani F., Del Favero M., Capizzuto R., Consonni C., Zangrossi S., Greco C., De Gioia L., Tortora P., Dehò G. (2007) Biochimie 89, 145–157 [DOI] [PubMed] [Google Scholar]
- 28. Chen H. W., Rainey R. N., Balatoni C. E., Dawson D. W., Troke J. J., Wasiak S., Hong J. S., McBride H. M., Koehler C. M., Teitell M. A., French S. W. (2006) Mol. Cell Biol. 26, 8475–8487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sarkar D., Fisher P. B. (2006) Cell Cycle 5, 1080–1084 [DOI] [PubMed] [Google Scholar]
- 30. Khidr L., Wu G., Davila A., Procaccio V., Wallace D., Lee W. H. (2008) J. Biol. Chem. 283, 27064–27073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jacquier A., Dujon B. (1985) Cell 41, 383–394 [DOI] [PubMed] [Google Scholar]
- 32. Macreadie I. G., Scott R. M., Zinn A. R., Butow R. A. (1985) Cell 41, 395–402 [DOI] [PubMed] [Google Scholar]
- 33. Zinn A. R., Butow R. A. (1985) Cell 40, 887–895 [DOI] [PubMed] [Google Scholar]
- 34. Turk E. M., Caprara M. G. (2010) J. Biol. Chem. 285, 8585–8594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gagliardi D., Stepien P. P., Temperley R. J., Lightowlers R. N., Chrzanowska-Lightowlers Z. M. (2004) Trends Genet. 20, 260–267 [DOI] [PubMed] [Google Scholar]
- 36. Zhong Q., Chen C. F., Chen P. L., Lee W. H. (2002) J. Biol. Chem. 277, 28641–28647 [DOI] [PubMed] [Google Scholar]
- 37. Chen Y., Riley D. J., Zheng L., Chen P. L., Lee W. H. (2002) J. Biol. Chem. 277, 49408–49416 [DOI] [PubMed] [Google Scholar]
- 38. Huang H. J., Yee J. K., Shew J. Y., Chen P. L., Bookstein R., Friedmann T., Lee E. Y., Lee W. H. (1988) Science 242, 1563–1566 [DOI] [PubMed] [Google Scholar]
- 39. Ahmed K. M., Tsai C. Y., Lee W. H. (2010) J. Biol. Chem. 285, 4464–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang D. D., Shu Z., Lieser S. A., Chen P. L., Lee W. H. (2009) J. Biol. Chem. 284, 20812–20821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shu Z., Vijayakumar S., Chen C. F., Chen P. L., Lee W. H. (2004) Biochemistry 43, 4781–4790 [DOI] [PubMed] [Google Scholar]
- 42. Cheng X., Dunaway S., Ivessa A. S. (2007) Mitochondrion 7, 211–222 [DOI] [PubMed] [Google Scholar]
- 43. Büttner K., Nehring S., Hopfner K. P. (2007) Nat. Struct. Mol. Biol. 14, 647–652 [DOI] [PubMed] [Google Scholar]
- 44. Boulet A., Levra-Juillet E., Perea J., Faye G. (1990) Curr. Genet. 17, 537–541 [DOI] [PubMed] [Google Scholar]
- 45. Séraphin B., Boulet A., Simon M., Faye G. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 6810–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bogenhagen D. F., Rousseau D., Burke S. (2008) J. Biol. Chem. 283, 3665–3675 [DOI] [PubMed] [Google Scholar]
- 47. Meeusen S., Nunnari J. (2003) J. Cell Biol. 163, 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lecrenier N., Foury F. (2000) Gene 246, 37–48 [DOI] [PubMed] [Google Scholar]
- 49. Fan H. Y., Cheng K. K., Klein H. L. (1996) Genetics 142, 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mortimer R. K., Johnston J. R. (1986) Genetics 113, 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen X. J., Butow R. A. (2005) Nat. Rev. Genet. 6, 815–825 [DOI] [PubMed] [Google Scholar]
- 52. Spelbrink J. N. (2010) IUBMB Life 62, 19–32 [DOI] [PubMed] [Google Scholar]
- 53. Masters B. S., Stohl L. L., Clayton D. A. (1987) Cell 51, 89–99 [DOI] [PubMed] [Google Scholar]
- 54. Xu B., Clayton D. A. (1995) Mol. Cell Biol. 15, 580–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rogowska A. T., Puchta O., Czarnecka A. M., Kaniak A., Stepien P. P., Golik P. (2006) Mol. Biol. Cell 17, 1184–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Allen J. B., Zhou Z., Siede W., Friedberg E. C., Elledge S. J. (1994) Genes Dev. 8, 2401–2415 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.