Background: Classical C2H2 zinc finger proteins generally bind DNA via a three-finger motif.
Results: We have identified the DNA site recognized by ZNF217 and defined its mechanism of binding.
Conclusion: Two classical C2H2 zinc fingers, rather than the typical three, are sufficient to bind an eight base pair sequence.
Significance: This work broadens our understanding of DNA binding by classical zinc fingers.
Keywords: Breast Cancer, DNA-binding Protein, Gene Regulation, Gene Transcription, Transcription Factors, DNA-Protein Interaction, ZNF217, Zinc Finger
Abstract
Classical C2H2 zinc finger proteins are among the most abundant transcription factors found in eukaryotes, and the mechanisms through which they recognize their target genes have been extensively investigated. In general, a tandem array of three fingers separated by characteristic TGERP links is required for sequence-specific DNA recognition. Nevertheless, a significant number of zinc finger proteins do not contain a hallmark three-finger array of this type, raising the question of whether and how they contact DNA. We have examined the multi-finger protein ZNF217, which contains eight classical zinc fingers. ZNF217 is implicated as an oncogene and in repressing the E-cadherin gene. We show that two of its zinc fingers, 6 and 7, can mediate contacts with DNA. We examine its putative recognition site in the E-cadherin promoter and demonstrate that this is a suboptimal site. NMR analysis and mutagenesis is used to define the DNA binding surface of ZNF217, and we examine the specificity of the DNA binding activity using fluorescence anisotropy titrations. Finally, sequence analysis reveals that a variety of multi-finger proteins also contain two-finger units, and our data support the idea that these may constitute a distinct subclass of DNA recognition motif.
Introduction
Transcription factors are sequence-specific DNA-binding proteins that localize to promoters and enhancers/silencers and recruit co-regulatory factors, such as histone modifying enzymes, to turn genes on or off (1). Sequence-specific DNA binding can be achieved through a number of different structural domains, and most known transcription factors are categorized by the nature of their DNA recognition domains. The most prominent classes of transcription factors are classical (or C2H2) zinc fingers (2), homeodomains (3), basic leucine zipper (4), basic helix-loop-helix (5), nuclear receptor domains (6), and MADS boxes (7). The relative abundance of the different domains varies somewhat between organisms, with nuclear receptors, for instance, being more abundant in the worm Caenorhabditis elegans than in other organisms (8) and MADS boxes being particularly common in plants (9), but overall classical zinc fingers are the major class of sequence-specific DNA binding domains across Eukarya.
There are several reasons why zinc fingers might have become so abundant during evolutionary history. First, they are small structures that tend to be thermodynamically stable (in part due no doubt to the cross-linking effect of the zinc coordination) and are dependent on only a small number of residues for proper folding, perhaps allowing rapid evolution. Indeed, a zinc binding module has been recorded to be among the first domains to arise during in vitro evolution experiments (10, 11). Second, these domains bind DNA in a modular fashion, which allows mixing and matching to create proteins with novel DNA binding specificities through the addition of extra zinc fingers. It is also for these reasons that zinc fingers have been successfully used for the generation of artificial transcription factors and nucleases with novel specificities (12–14). Finally, zinc fingers can mediate interactions with RNA or with other proteins (15–17), and this functional diversity, although currently not well understood, may also have led to their proliferation in the genome.
Although classical zinc finger proteins have been extensively studied, the vast majority of the work has been centered on arrays of three or four classical zinc fingers separated by canonical TGE(K/R)P linkers. The three-dimensional structures of several such arrays bound to DNA target sites have been determined (for example, Zif268 (18), Gli5 (19), TFIIIA (20), and the designed zinc finger protein Aart (21)), revealing a shared recognition mode in which residues on one surface of the α-helix of each finger make base-specific contacts in the major groove of the DNA. Additional stabilizing interactions involving other residues in the helix and residues in the TGE(K/R)P linkers are also commonly observed.
It is notable, however, that a large number of zinc finger proteins do not have three or four tandem-arranged zinc fingers separated by the canonical linker sequences, raising the question of how these proteins identify their target elements and indeed whether or not they are DNA-binding proteins at all.
We have studied one large multi-zinc finger protein, namely ZNF217, and investigated the mechanisms by which it regulates gene expression (22). ZNF217 has been recognized as an important oncogene, with overexpression of ZNF217 associated with breast, ovarian, and numerous other cancers (23–25). The mechanisms through which it operates are complex (26–28), but one proposal has been that ZNF217 directly binds to and represses the E-cadherin gene promoter via a two-zinc finger domain that recognizes the consensus sequence CAGAAY (29). E-cadherin is an important cell adhesion molecule, and repression of E-cadherin, which presumably reduces cell-cell contacts, has been associated with cancers with increased metastatic potential (30, 31).
We began by investigating how ZNF217 might localize to the E-cadherin promoter and confirmed the previous observation that only fingers 6 and 7 detectably contact DNA (29). However, our results suggested that the E-cadherin site was bound with low affinity, prompting further mapping and mutagenesis experiments to search for higher affinity recognition sites. This work suggested a preferred site of (T/A)(G/A)CAGAA(T/G/C), which is related to but distinct from the previously proposed CAGAAY site. We then showed that the integrity of the double-finger domain is required for functional regulation through this DNA element. We went on to examine the affinity with which the two-finger domain contacted the preferred site and to determine whether it used similar molecular contacts to those found in three and four zinc finger units. Our results demonstrate that a single two-finger motif rather than the usual three or four finger units is sufficient to allow ZNF217 and potentially other multi-zinc finger proteins to contact DNA.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Residues 469–525 of human ZNF217 were subcloned into the pGEX-2T vector, and the construct was overexpressed in E. coli BL21 cells overnight at 18 °C by the addition of 0.4 mm isopropyl 1-thio-β-d-galactopyranoside to a log phase culture. Expressed proteins were purified by glutathione affinity chromatography. For NMR analysis, the ZNF217-F67 was cleaved from GST with thrombin, and the resulting protein was purified by gel filtration chromatography (S75) carried out in 50 mm Tris (pH 7.4) containing 300 mm NaCl and 1 mm DTT. Point mutants were handled similarly, except that gel filtration was not carried out.
Electrophoretic Mobility Shift Assays (EMSA)
Nuclear extracts were prepared, and EMSAs were carried out as described previously (32). The modified binding buffer used in EMSAs consisted of 20 mm HEPES (pH 7.9), 100 mm KCl, 0.2 mm EDTA, 0.2 mm EGTA, 0.1% Nonidet P-40, 5% glycerol, 0.5 mm PMSF, 0.5 mm DTT, 1 μg of BSA, and 0.5 μof g poly(dI·dC), and 1 mm ZnSO4 per reaction. The following probe sequences were used: human E-cadherin promoter (−230 to −159), CCAGTGGAATCAGAACCGTGCAGGTCCCATAACCCACCTAGACCCTAGCAACTCCAGGCTAGAGGGTCACCG (Fig. 2A), and ZNF217 site selection consensus (29), GGATCCATTGCAGAATTGTGGTTGCTGTAGGAATTC (Fig. 2B). Probes used to define the core ZNF217-F67 binding consensus (Fig. 3) are given in Tables 1 and 2.
FIGURE 2.
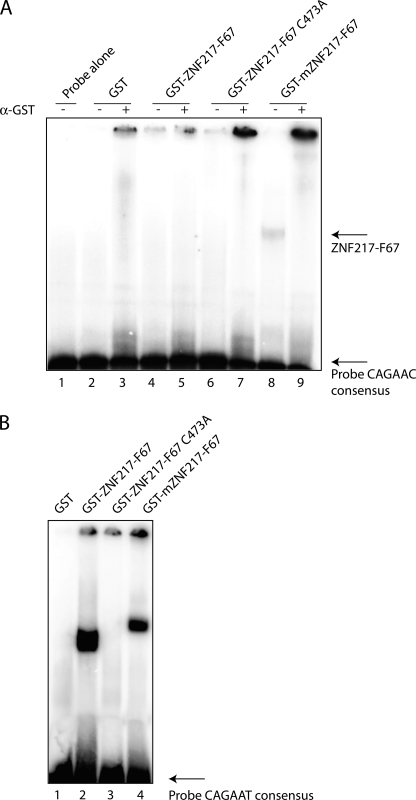
ZNF217-F67 binds weakly to the human E-cadherin promoter. A, various GST-ZNF217-F67 constructs were used in EMSA to assess binding to the −230 to −159 region of the human E-cadherin promoter containing the consensus sequence CAGAAC. 1 μg of recombinant protein was used in each lane, and αGST antibody was used to supershift any complexes containing GST fusions. Lanes are: probe only (lane 1), GST only (lanes 2 and 3), human ZNF217-F67 (lanes 4 and 5), human ZNF217-F67 containing the C473A mutation of F6 (lanes 6 and 7), murine ZNF217-F67 (lanes 8 and 9). B, 2 μg of recombinant GST (lane 1), GST-hZNF217-F67 (lane 2), GST-hZNF217-F67 C473A (lane 3), and GST-mZNF217-F67 (lane 4) were used in EMSA with a probe based on a previously identified CAST sequence, containing the consensus sequence CAGAAT (29).
FIGURE 3.
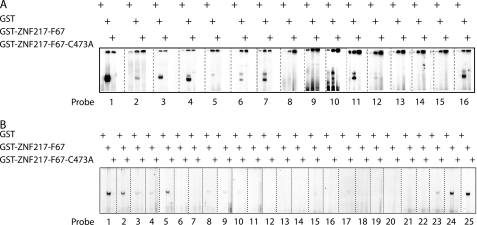
ZNF217-F67 binds to a consensus sequence of (T/A)(G/A)CAGAA(T/G/C). A, 2 μg of recombinant GST, GST-ZNF217-F67, and GST-ZNF217-F67 C473A were tested in EMSA for their ability to bind to probes containing either mutations or deletions in and around the core binding consensus of TGCAGAAT. Mutated bases were altered to G residues. The sequences of the probes and the relative strength of binding are given in Table 1. B, 2 μg of recombinant GST, GST-ZNF217-F67, and GST-ZNF217-F67 C473A were tested in EMSA for their ability to bind to probes containing individual site-specific mutations of the TGCAGAAT core DNA binding sequence. The sequences of the probes and the relative strengths of binding are given in Table 2.
TABLE 1.
ZNF217-F67 binds to a core sequence of TGCAGAAT
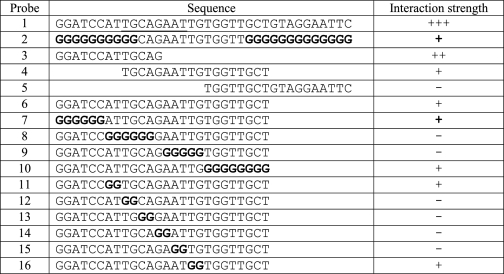
Shown are the probe sequences used in Fig. 3A to determine the core binding sequence for ZNF217-F67. Sequences are aligned to indicate deletions, and mutated bases are shown in bold. The eight-base pair core is underlined in the probe for gel 1. Also shown is the relative strength of binding of ZNF217-F67 to each of the probes.
TABLE 2.
ZNF217-F67 binds to a consensus sequence of (T/A)(G/A)CAGAA(T/G/C)
Shown are the probe sequences used in Fig. 3B to define the ZNF217-F67 binding consensus sequence. Mutated bases are shown in bold. Also shown is the relative strength of binding of ZNF217-F67 to each of the probes.
| Probe | Sequence | Interaction strength |
|---|---|---|
| 1 | TCCATTGCAGAATTGTGG | +++ |
| 2 | TCCATAGCAGAATTGTGG | ++ |
| 3 | TCCATGGCAGAATTGTGG | + |
| 4 | TCCATCGCAGAATTGTGG | + |
| 5 | TCCATTACAGAATTGTGG | ++ |
| 6 | TCCATTTCAGAATTGTGG | − |
| 7 | TCCATTCCAGAATTGTGG | − |
| 8 | TCCATTGTAGAATTGTGG | + |
| 9 | TCCATTGAAGAATTGTGG | + |
| 10 | TCCATTGGAGAATTGTGG | + |
| 11 | TCCATTGCTGAATTGTGG | − |
| 12 | TCCATTGCGGAATTGTGG | − |
| 13 | TCCATTGCCGAATTGTGG | − |
| 14 | TCCATTGCAAAATTGTGG | − |
| 15 | TCCATTGCATAATTGTGG | + |
| 16 | TCCATTGCACAATTGTGG | − |
| 17 | TCCATTGCAGTATTGTGG | + |
| 18 | TCCATTGCAGGATTGTGG | − |
| 19 | TCCATTGCAGCATTGTGG | − |
| 20 | TCCATTGCAGATTTGTGG | − |
| 21 | TCCATTGCAGAGTTGTGG | − |
| 22 | TCCATTGCAGACTTGTGG | − |
| 23 | TCCATTGCAGAAATGTGG | − |
| 24 | TCCATTGCAGAAGTGTGG | +++ |
| 25 | TCCATTGCAGAACTGTGG | +++ |
Luciferase Repression Assays
HEK-293 cells were transfected using FuGENE 6 (Roche Applied Science) in 6-well plates with between 0.25 and 2 μg of pMT3-FLAG-ZNF217 or point mutations of pMT3-FLAG-ZNF217. Also included was 3 μg of reporter plasmid (either pGL2-(TGCAGAAT)3-LexA-Luc or pGL2-(CAGAAT)3-LexA-Luc or pGL2-(CTGGAGTA)3-LexA-Luc) and 1 μg of LexA-VP16 expression plasmid. pMT3-FLAG empty vector was added to make the amount of DNA in each transfection equal. 10 μg of a plasmid expressing Renilla Luciferase (pRL-Luc, Promega) was used in each transfection to control for transfection efficiency. Cells were incubated for 48 h after transfection, and luciferase assays were then performed using the Promega Luciferase Assay System.
NMR Spectroscopy
Samples of ZNF217-F67 or DNA were exchanged into a buffer comprising 10 mm Na2HPO4 (pH 7.0), 50 mm NaCl, and 1 mm DTT by dialysis and concentrated using centrifugal concentrators. NMR spectra were recorded on Bruker Avance 600 or 800 MHz NMR spectrometers equipped with cryoprobes. Backbone resonance assignments were made using HNCACB, CBCA(CO)NH, HNCO, HBHA(CO)NH, and HNCA spectra. Spectra were processed using Topspin 2.5 (Bruker) and visualized using Sparky 3.11.
Data-driven Three-dimensional Structure Prediction Using CS-Rosetta
The 13Cα, 13Cβ, 13C′, 15N, Hα, and HN chemical shift assignments of ZNF217-F67 were used as input for the CS-Rosetta software (33) to generate data-driven models of ZNF217-F6 (residues 469–494) and ZNF217-F7 (residues 495–525). For each domain, 10,000 models were generated, and the top 500 models ranked by energy were chosen for further analysis. The 10 lowest-energy structures for ZNF217-F6 and ZNF217-F7 were accepted according to the published criteria (33); 1) a “funneling” distribution, indicating a convergence of the structure prediction, was observed in the plot of Rosetta all-atom energy against Cα root mean square deviation relative to the model with the lowest energy, and 2) the low energy models clustered within 2 Å from the model with the lowest energy. The structures were visualized by using PyMOL.
Fluorescence Anisotropy Titrations
GST-tagged ZnF217-F67 and fluorescein-labeled dsDNA oligonucleotides (WT sequence, 5′-fluorescein-TCCATTGCAGAATTGTGG-3′; mutated sequence, 5′-fluorescein-TCCATCTGGAGTATGTGG-3′) were dialyzed into a 10 mm phosphate buffer (pH 6.5) containing 50 mm NaCl and 1 mm DTT. Fluorescence anisotropy titrations were performed at 25 °C on a Cary Eclipse fluorescence spectrophotometer with a slit width of 10 nm, and data were averaged over 15 s. The excitation and detection wavelengths were 495 and 520 nm, respectively. In each titration the fluorescence anisotropy of a solution of 50 nm fluorescein-tagged dsDNA was measured as a function of the added protein concentration. Binding data were fitted to a simple 1:1 binding model by nonlinear least squares regression. Each titration was performed three times, and the final affinity was taken as the mean of these measurements.
RESULTS
Zinc Fingers 6 and 7 Are Capable of Binding DNA
ZNF217 contains eight classical C2H2 zinc fingers arranged in two major clusters (Fig. 1). To identify the domain(s) responsible for DNA recognition, we first expressed the various zinc finger clusters as GST fusion proteins in bacteria and purified them by glutathione affinity chromatography. Recent work has suggested that ZNF217 binds a particular cis-element, CAGAAC, in the human E-cadherin promoter (29). We tested various zinc finger combinations of both mouse and human ZNF217 with this element in an EMSA. We detected weak binding with murine fingers 6 and 7 (mZNF217-F67, Fig. 2A, lanes 8 and 9). Supershift experiments with an anti-GST serum eliminated the retarded band, consistent with the view that the binding was due to the GST-mZNF217-F67 recombinant protein. However, we were not able to detect any binding of the human protein (ZNF217-F67) to the E-cadherin promoter (Fig. 2A, lanes 6 and 7). Furthermore, no binding was observed with either mouse or human constructs encompassing the other zinc fingers (data not shown).
FIGURE 1.
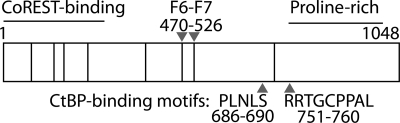
The domain topology of human ZNF217. Shown are the positions of the eight C2H2 zinc fingers (black bars), the ZNF217-F67 domain (F6-F7), the CtBP-binding sites, the CoREST interaction domain, and a proline-rich domain (22, 23, 29).
We also tested the DNA binding ability of various ZNF217 zinc finger constructs with Pentaprobe, a collection of DNA fragments that together contain all possible five-base pair sequences (34). Again, no binding was detected with individual fingers 1, 5, and 8 and a domain containing fingers 1–4, whereas fingers 6 and 7 were able to bind to the Pentaprobe sequence (data not shown). These data suggest that fingers 6 and 7 are the main determinants of DNA recognition by ZNF217.
ZNF217 Recognizes an 8-bp Sequence with the Consensus ((T/A)(G/A)CAGAA(T/G/C)
While carrying out the DNA binding experiments described above, we noted that binding of ZNF217 to the E-cadherin promoter sequence, although reproducible, was barely detectable. We then repeated recently published EMSA experiments using a consensus ZNF217 binding sequence identified by site selection (CASTing) (29). This site, CAGAAT, is related to but distinct from the actual site identified in the E-cadherin promoter. This sequence gave rise to robust DNA binding with both the murine and human GST fusion proteins encompassing zinc fingers 6 and 7 of ZNF217 (Fig. 2B). To confirm that the observed bands arose from sequence-specific binding to the intact zinc finger domain, we mutated a key residue that is required for zinc finger structure. The first cysteine residue in zinc finger 6 of ZNF217 (C473) was mutated to alanine (ZNF217-F67C473A); this change eliminated binding to both the E-cadherin promoter and the CAST-derived oligonucleotide, confirming that DNA recognition was dependent on zinc finger integrity (Fig. 2).
To investigate why the site-selected probe was robustly recognized by mouse and human GST-ZNF-F67, whereas the E-cadherin promoter site was not, we carried out a series of mutagenesis experiments on the CAST-selected probe to define the site most strongly bound by ZNF217-F67 in EMSA assays. We generated and tested both 5′ and 3′ deletions and site-specific mutations of the probe and identified a core sequence of TGCAGAAT (Fig. 3A, Table 1). This site is related to the previously identified essential CAGAAC core present in the human E-cadherin promoter. However, it also includes two additional 5′-flanking residues, TG, that are not found in the E-cadherin site but that appear to be important for binding.
To assess the site in more detail, we carried out EMSAs using a series of mutant probes in which each nucleotide in the extended core TGCAGAAT was mutated to every other residue (Fig. 3B, Table 2). The results confirmed that all eight residues can influence DNA binding and identified a consensus site of (T/A)(G/A)CAGAA(T/G/C). This site differs from the E-cadherin promoter site (ATCAGAAC) at two critical residues, explaining the difference in binding properties observed between Fig. 2, A and B.
ZNF217 Can Repress Transcription through Its Consensus DNA-binding Site in Cellular Assays
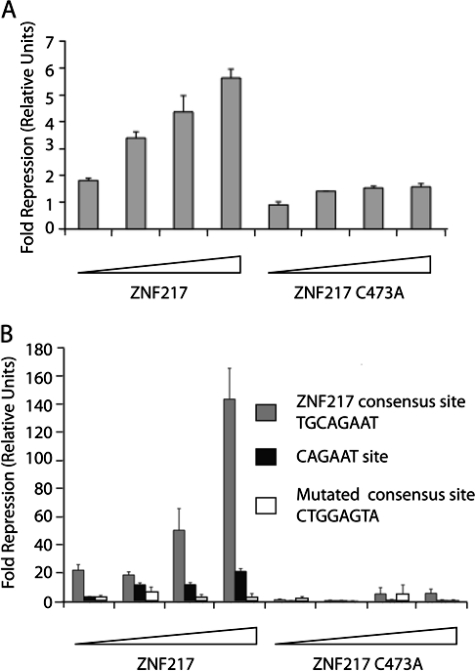
ZNF217 has been shown to function as a transcriptional repressor. It recruits co-repressors of the C-terminal-binding protein (CtBP)3 family via Pro-Ile-Asp-Leu-Ser (PIDLS) and Arg-Arg-Thr (RRT) motifs (22, 35). CtBPs then recruit other co-regulatory molecules such as histone deacetylases, histone methyltransferases, and histone demethylases to repress gene expression (36–38). To determine whether ZNF217 was able to repress transcription in cellular assays via the consensus target site we had identified, we constructed a simple reporter system. We used a luciferase reporter gene, driven by a minimal adenovirus 1B promoter containing a LexA binding site and three copies of the ZNF217 consensus element. Transcription was activated by the expression of a LexA-VP16 fusion protein, and repression was measured when full-length ZNF217 was co-expressed in HEK-293 cells. As shown in Fig. 4A, dose-dependent repression was observed when increasing amounts of a ZNF217-encoding expression vector were added. In contrast, a similar vector encoding the ZNF217C473A mutant produced only minimal repression. These results confirm the ability of ZNF217 to function in gene repression and indicate that an intact ZNF217-F67 domain is required for binding to target promoters containing the (T/A)(G/A)CAGAA(T/G/C) consensus.
FIGURE 4.
ZNF217 represses gene expression by sequence specific binding to the TGCAGAAT consensus. A, HEK-293 cells were transfected with 0.25–2 μg of pMT3-FLAG-ZNF217 or pMT3-FLAG-ZNF217 C473A as well as 3 μg of pGL2-(TGCAGAAT)3-LexA-Luc reporter plasmid and 1 μg of LexA-VP16 expression plasmid. Shown are -fold repressions of LexA-VP16 transactivation by ZNF217 or ZNF217 C473A compared with empty vector control (n = 3, error bars show S.D.). B, HEK-293 cells were transfected with 0.25–2 μg of pMT3-FLAG-ZNF217or pMT3-FLAG-ZNF217 C473A as well as 3 μg of either pGL2-(TGCAGAAT)3-LexA-Luc, pGL2-(CAGAAT)3-LexA-Luc, or pGL2-(CTGGAGTA)3-LexA-Luc reporter plasmids and 1 μg of LexA-VP16 expression plasmid. Shown are -fold repressions of LexA-VP16 transactivation by ZNF217 compared with empty vector control (n = 3, error bars show S.D.).
As a further control for the specificity of the sequence element recognized by ZNF217, we constructed two additional reporter genes. One carried 3 copies of the CAGAAT element related to that found in the E-cadherin promoter instead of the 8-bp core we identified above, whereas the other contained mutations chosen to severely disrupt our core element. We again observed strong dose-dependent repression by ZNF217 with the new core element (TGCAGAAT) and observed minor but detectable repression using the shorter sequence related to the E-cadherin core (CAGAAT) (Fig. 4B), consistent with the weak binding observed in vitro to this latter site (Fig. 2A). In contrast, we observed no significant repression with the mutated version of the new core (CTGGAGTA) (Fig. 4B). Finally, mutation of the critical cysteine 473 to alanine in finger 6 essentially abrogated repression (Fig. 4B). Taken together these results confirm that ZNF217 can function as a transcriptional repressor and demonstrate that it can be localized to its target promoter in a sequence specific manner via a DNA binding domain that comprises zinc fingers 6 and 7.
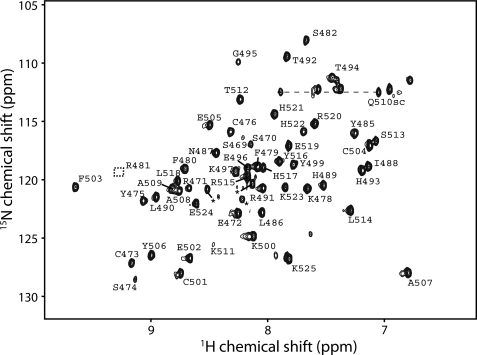
Identification of Residues That Are Important for DNA Recognition
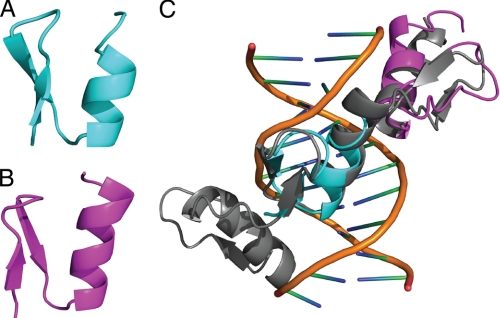
Given that the binding of two zinc fingers to an eight-base recognition site was somewhat unexpected (typically three zinc fingers are required for binding to sites of around nine bases), we next probed the nature of the ZNF217-DNA interaction at a molecular level using NMR spectroscopy. A 15N HSQC spectrum of uniformly 15N-labeled ZNF217-F67 (residues 469–525) showed excellent dispersion, indicating that the construct formed a stable structure in solution (Fig. 5). We used standard triple resonance approaches to assign the signals in the 15N HSQC to specific residues in the protein and then used the program CS-Rosetta (33) to calculate three-dimensional structures for each domain based on the chemical shifts. These structures (Fig. 6, A and B) have folds that closely resemble each other (the backbone root mean square deviation over the structured regions of the lowest energy models of F6 and F7 is 0.9 Å) and other classical zinc fingers (the backbone root mean square deviation of finger 3 of Zif268 to F6 is 1.1 Å and to F7 is 1.0 Å). Fig. 6C shows an overlay of F6 and F7 with fingers 2 and 3 from Zif268, showing the high structural similarity.
FIGURE 5.
15N HSQC spectrum and calculated three-dimensional structures of ZNF217 fingers 6 and 7. Shown is an 15N HSQC spectrum of 200 μm hZNF217-F67, recorded at 25 °C in 10 mm Na2HPO4 (pH 7.0) containing 50 mm NaCl and 1 mm DTT. Residue assignments are given. Arg-481 lies below the plotted contour level.
FIGURE 6.
A and B, shown are ribbon representations of the lowest energy models of finger 6 (A) and finger 7 (B) of ZNF217-F67, calculated using CS-ROSETTA, and backbone chemical shifts obtained from the analysis of triple resonance NMR data. C, shown is an overlay (backbone atoms) of F6 (cyan) and F7 (magenta) with fingers 2 and 3 from Zif268 using the structure for the latter determined in the presence of DNA (PDB 1ZAA (18)).
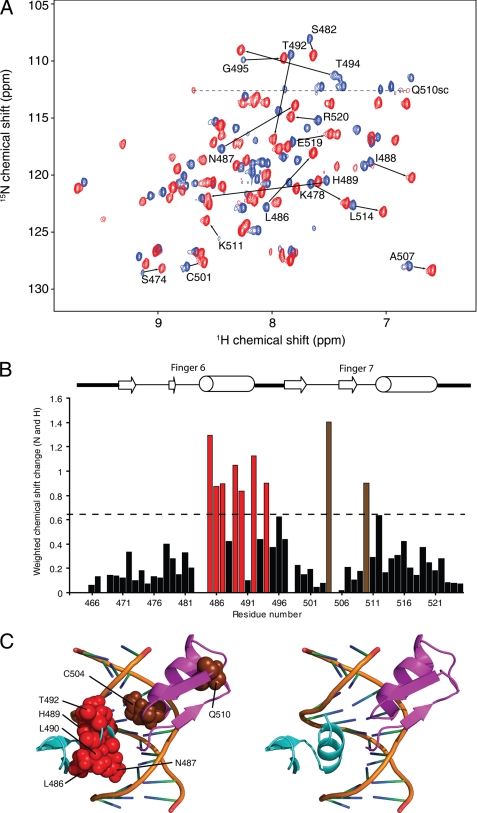
We assessed DNA binding by ZNF217-F67 by titrating into a sample of 15N-labeled ZNF217-F67 a 14-bp double-stranded oligonucleotide bearing the extended recognition sequence deduced above (5′-CATTGCAGAATTGT-3′). As shown in Fig. 7A, many signals shifted after the addition of DNA, and the good quality of the 15N HSQC spectrum after saturation with DNA indicates the formation of a well defined and well ordered complex. We recorded triple resonance data for the protein-DNA complex and again made assignments of the backbone atoms. Fig. 7B shows the magnitude of the chemical shift changes for the backbone nuclei (HN, N) of each residue after the addition of DNA. A larger number of significant changes was observed in finger 6 compared with finger 7. These changes are shown mapped onto the structural models of each ZF in Fig. 7C.
FIGURE 7.
15N HSQC titration of ZNF217-F67 with DNA. A, shown is an overlay of 15N HSQC spectra of ZNF217-F67 in the absence (blue) and presence (red) of two molar equivalents of a dsDNA oligonucleotide with the sequence 5′-CATTGCAGAATTGT-3′. Assignments are marked for selected residues that underwent substantial chemical shift changes upon the addition of DNA. B, shown are weighted average chemical shift changes for backbone nuclei of ZNF217-F67 after the addition of DNA. The dotted line indicates a change 1 S.D. greater than the mean change. Residues undergoing changes larger than 1 S.D. are shown in red (F6) or brown (F7). C, mapping of residues that underwent substantial chemical shift changes are shown. Residues from F6 and F7 are indicated in red and brown, respectively. The zinc fingers are shown in the same orientation as in Fig. 6C together with the DNA from the Zif268 structure (PDB 1ZAA).
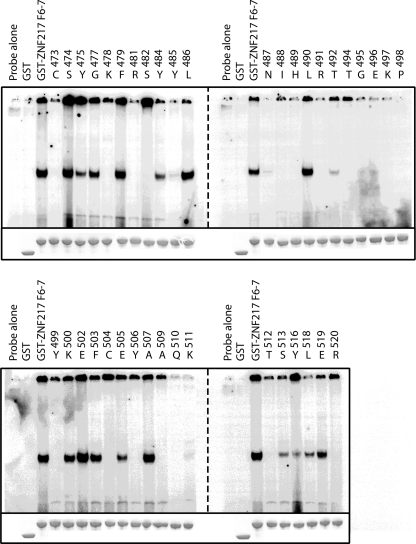
Chemical shift changes may be indicative of direct DNA contacts or may result from local perturbations in the structure arising from the binding event. To assist in distinguishing between these two possibilities and to corroborate the NMR data, we mutated finger 6 and 7 residues to alanine or, where alanines were present, to glutamine. We then tested the ability of these mutants to bind to DNA in EMSAs (Fig. 8 and supplemental Table 1). One-dimensional 1H NMR spectroscopy was used to confirm that all mutants, apart from Y506A, folded correctly. DNA binding was found to be severely compromised for a number of mutants in both zinc fingers. As expected, mutations that interfered with zinc binding, such as C473A, H489A, and C504A, disrupted DNA binding, and it was also notable that mutations in the TGEKP linker between the two zinc fingers abrogated binding.
FIGURE 8.
Mutagenic scan of the ZNF217-F67-DNA interaction. Residues were mutated to alanine (or glutamine where alanines were already present) and were expressed as GST fusion proteins. 1 μg of each mutant was used in EMSA to assess binding to an oligonucleotide containing the consensus sequence TGCAGAAT (See also supplemental Table 1).
Affinity and Specificity
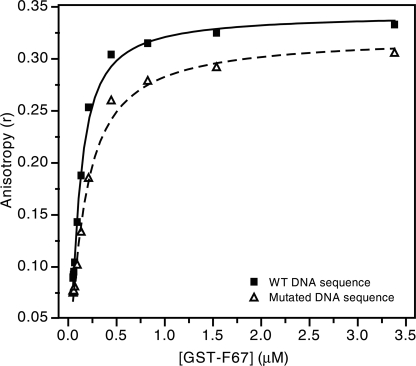
To quantify the DNA binding activity of ZNF217, we used fluorescence anisotropy to measure the affinity of the interaction between ZNF217-F67 and a dsDNA oligonucleotide containing the TGCAGAAT sequence (Fig. 9). The data fit well to a simple 1:1 binding model and gave a dissociation constant of 80 ± 25 nm. To assess the specificity of binding, we carried out a fluorescence anisotropy titration using an oligonucleotide in which the core recognition sequence was mutated to CTGGAGTA. The affinity we measured was ∼2.5 times lower (190 ± 50 nm) than the affinity measured for the optimized sequence.
FIGURE 9.
Fluorescence anisotropy data measuring the binding of ZNF217-F67 to DNA. Purified recombinant ZNF217-F67 was titrated into dsDNA carrying a 5′-fluorescein tag, and fluorescence anisotropy was measured as a function of protein concentration. Filled squares and the solid line show binding to an oligonucleotide containing the consensus sequence identified in the current study, and open triangles and the dashed line show binding to an oligonucleotide designed to eliminate this consensus sequence. Fits are shown for a simple 1:1 binding model.
DISCUSSION
DNA Recognition by ZNF217
The mechanisms by which proteins can be targeted to particular DNA sequences in the genome are of considerable interest. Zinc finger proteins are the most abundant DNA-binding proteins found in eukaryotic genomes, and there have been recent successes with targeting artificial zinc finger proteins to chosen genomic sites (12–14). Although the mechanisms by which some naturally occurring zinc finger proteins bind DNA have been identified, the vast majority of work has focused on constructs comprising three tandem-arranged classical zinc fingers. However, an examination of the human genome reveals that many hundreds of classical zinc finger domains do not lie within closely spaced three-finger modules of this type, and although several have been shown to function as protein recognition modules (39–42), the functions of the vast majority of these domains remain undefined.
Here we have investigated the eight-zinc finger protein ZNF217 and shown that a construct comprising fingers 6 and 7 of this protein is able to bind an eight base pair double-stranded DNA site in a sequence-specific manner. Furthermore, this double finger domain is able, in the context of the full-length ZNF217 protein, to mediate transcriptional repression in a cellular reporter gene assay. The conventional view has been that three classical zinc fingers are required for physiologically relevant DNA binding, with a few exceptions such as the single GAGA zinc finger (43) and the Tramtrack pair of zinc fingers (44); our work strengthens the argument that shorter two-finger units can be functionally relevant for DNA recognition.
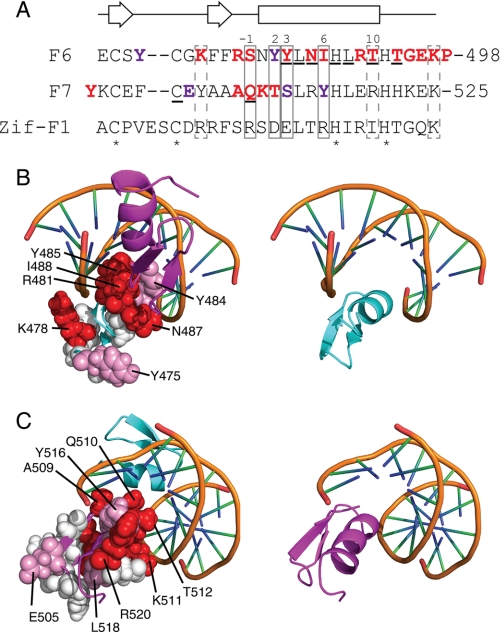
Typically, classical zinc fingers use residues at positions −1, 2, 3, and 6 of their α-helix to make sequence-specific contacts with DNA (Fig. 10A) and can make additional interactions with the phosphodiester backbone via other residues in the α-helix. Our NMR data show that the helix of F6 is strongly involved in binding DNA. Fewer large chemical shift changes were observed in F7, although a substantial change is observed for Gln-510, which lies at the −1 position in the F7 helix. Because 15N HSQC titration data are most sensitive to changes in the conformation or environment of nuclei in the backbone of the protein, we also carried out alanine-scanning mutagenesis across the F67 polypeptide to more directly assess the involvement of side chains in contacting the DNA. In Fig. 10, B and C, the structures of F6 and F7 are overlaid in the positions of F2 and F3 of Zif268 (as shown in Fig. 6) with the residues that were mutated, shown in space-filling representation. Red indicates residues that were essential or very important for DNA binding, pink indicates those that make some contribution to binding, and white designates residues that do not contribute measurably to binding.
FIGURE 10.
Residues important for ZNF217-F67 DNA recognition. A, the amino acid sequences of F6 and F7 of ZNF217 are shown together with the sequence of finger 1 from the prototypical DNA binding zinc finger protein Zif268. Residues that typically make sequence-specific contacts with DNA in classical zinc fingers are boxed with solid lines, residues that often make nonspecific interactions with the DNA backbone are shown in dashed boxes, residues that underwent substantial chemical shift changes upon the addition of DNA to ZNF217-F67 are underlined, and zinc-ligating residues are indicated with asterisks. Those residues shown by site-directed mutagenesis to mostly or completely eliminate DNA binding are in bold and colored red; residues that reduced but did not abolish DNA binding are in bold and colored purple. Numbering of the α-helix is that typically used for classical zinc fingers. The secondary structure for F6 and F7, as predicted from an analysis of F67 chemical shifts, is shown above the sequences. B and C, the structures of F6 and F7 were overlaid onto F2 and F3 of Zif268 in the x-ray crystal structure of this protein bound to DNA (PDB 1ZAA). Residues that were mutated in Fig. 8 are shown in space-filling representation. Those residues shown by site-directed mutagenesis to mostly or completely eliminate DNA binding are colored red, residues that reduced but did not abolish DNA binding are colored pink, and residues that had little or no effect on DNA binding are shown in white. Red residues are more prominent on the surface of the domains used in Zif268 to contact DNA, whereas white residues are concentrated on the opposite face of each domain. B shows residues in F6, whereas C shows residues in F7. The right-hand panels show F6 or F7 in the same orientation as the corresponding left-hand panel for reference.
It is clear from Fig. 10 that the majority of residues that make substantial contributions to DNA binding lie on a surface that, in Zif268, forms the DNA contact surface. Many of these residues are in canonical DNA binding positions in the α-helix (as indicated in Fig. 10A), and it is also notable that Lys-478 was identified as making contact with DNA. A basic residue in this position has been shown to contact the DNA backbone in other zinc finger-DNA complexes, such as Zif268 (18) and TFIIIA (45).
Two of the residues for which significant chemical shift changes were observed, namely Leu-486 and Leu-490, proved to be dispensable for binding, according to our mutagenesis data, suggesting that the chemical shift changes observed for these residues might be caused by local conformational rearrangements; consistent with this idea, we have observed significant changes in the dynamics of the helices of classical zinc fingers from other proteins after DNA binding.4 Two further residues that were identified in our NMR analysis are zinc binding residues (His-489 and Cys-504), and their direct involvement in DNA binding is difficult to ascertain via mutagenesis because of their structural importance. The equivalent histidine in other zinc finger-DNA complexes does lie near the DNA backbone and commonly makes an electrostatic interaction with a phosphate group (2). Finally, mutation of any residue in the linker between F6 and F7 abrogated binding, indicating that this sequence is important for DNA recognition, as observed for other zinc finger-DNA complexes. These data together point to the conclusion that the overall topology of the ZNF217-DNA complex resembles previously characterized complexes containing more than two zinc fingers.
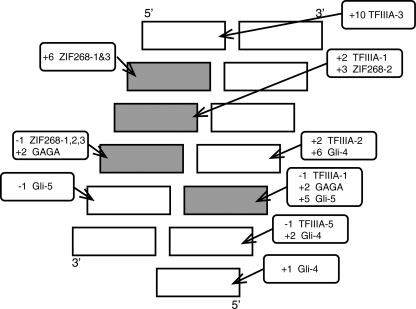
A notable feature of the ZNF217-DNA interaction is the observation that a two-zinc finger unit appears to recognize an eight-base pair DNA sequence. Although classical zinc fingers are sometimes thought of as contacting a three-base pair unit, there are a number of examples of contacts being made by residues in the α-helix to bases that lie 3′ to the canonical recognition sequence (Fig. 11). Interactions between residues at the +2 position in the helix and the base pair immediately 3′ to the core three-base pair target site are common (46), and interactions to bases as far as three nucleotides away have also been observed. Furthermore, an arginine at the +10 position in finger 3 of TFIIIA makes a base-specific contact with a guanine (47) that lies outside the three-base recognition site in the 5′ direction. Thus, it is possible in principle for a two-finger unit to exhibit sequence preferences across a recognition site of up to 12–14 base pairs, and mechanisms of this type are most likely used by ZNF217 to confer selectivity across its 8-base pair target site.
FIGURE 11.
Summary of sequence-specific interactions that have been observed between classical zinc fingers and DNA (adapted from Wolfe et al. (46)). Rectangles represent nucleotides in dsDNA. The four nucleotides that have been shown most frequently to be contacted by residues in classical zinc fingers are shaded gray. Rounded rectangles show the position of residues in the recognition helix that have been shown to contact the indicated bases, and an example of a zinc finger that displays each type of interaction is given (Zif268–1 indicates finger 1 of Zif268) (18, 19, 45, 60).
DNA Recognition by ZNF217 in Vivo
The consensus sequence (T/A)(G/A)CAGAA(T/G/C) that we have identified differs from two previously proposed recognition sequences. It is highly related to the core sequence CAGAAY identified by PCR site selection experiments (29) but differs in that it contains two additional 5′-flanking residues. Our data show that these two residues are important for binding. Surprisingly, a quite different consensus (ATTCC(G/A)AC) was proposed after a bioinformatics analysis of ZNF217-regulated genes identified from ChIP-ChIP data (27). This may reflect the ability of in vivo assays to identify biological sites where binding is weaker than those selected by in vitro assays. Alternatively, the bioinformatics-derived consensus sequence may represent the site of binding of a different transcription factor onto which ZNF217 associates, via protein-protein interactions (piggy-backing).
The question of how ZNF217 in particular localizes to the E-cadherin promoter remains unresolved. The previously identified CAGAAY core element in the E-cadherin promoter may be involved, but our work suggests that this is a low affinity site, because it lacks the critical 5′-flanking residues that are present in the extended core element. There is some good evidence that ZNF217 does localize to and repress E-cadherin, including chromatin immunoprecipitation experiments (29), but on the other hand, E-cadherin did not emerge from the microarray experiments (27), perhaps suggesting it is a ZNF217 target in some but not all tissues. Recent experiments have indicated very clearly that transcription factors identify different subsets of target genes in different cell types either as a result of chromatin accessibility or the availability of different partner proteins (48). Our work showing relatively weak binding to the E-cadherin promoter suggests that ZNF217 might not localize to this sequence solely via the zinc finger 6 and 7 domain. It is possible that it uses additional mechanisms such as protein-protein interactions. ZNF217 binds to CtBP (22), as do several other proteins implicated in the regulation of E-cadherin, such as Zeb (49) and Klf8 (50, 51). The fact that CtBP can multimerize (37, 52) raises the possibility that it forms a nucleus that helps recruit multiple transcription factors and in doing so stabilizes the interaction of these factors at the target promoter. We have also tested for direct protein-protein interactions between ZNF217 and Klf8 and between ZNF217 and Zeb but have found no clear evidence for direct contact.5
How Much Affinity and Specificity Are Enough?
The majority of sequence-specific DNA-binding proteins for which quantitative studies have been carried out exhibit dissociation constants in the nanomolar range or lower. For example, Zif268 has been shown to bind a target sequence with a dissociation constant of 6 nm (18), and the HoxD9 and Antp homeodomains bind to their target sites with a KD of ∼1.5 nm (53, 54). It has consequently become commonly accepted that affinities of this magnitude represent “biologically significant” interactions and that weaker interactions are less likely to be relevant. However, a number of weaker interactions have been measured that have clear biological relevance. The DNA binding domain of THAP1 contacts its target site with an affinity of only 8 μm (55), and the double zinc finger domain of Tramtrack recognizes DNA with a dissociation constant of 400 nm (56). The affinity measured in the current study for the ZNF217-DNA interaction (80 nm) thus falls well within the range of known affinities and further emphasizes the idea that biologically relevant protein-DNA interactions are not exclusively associated with nanomolar affinities.
These data lead to the question of whether weaker interactions hint at a mechanism of action that is distinct from that employed by “strong binders.” Weaker interactions will almost certainly be associated with faster off-rates for the protein-DNA complexes, and so one possibility is that regulatory complexes involving such proteins are found more commonly in systems that must respond more rapidly to signals that are intended to alter the expression of target genes. It is also possible that weak interactors are generally found in large multiprotein complexes where DNA contacts are made by more than one protein simultaneously and that additional affinity is provided in that way. A system of this type could have the advantage of being able to “mix and match” DNA binding domains from different proteins and thereby target different genes depending on which regulatory proteins were available.
The other question raised by the DNA binding data for ZNF217 is that of what constitutes specificity for a sequence-specific DNA-binding protein. Our data showed that ZNF217-F67 did display a higher affinity for its selected target sequence than for an unrelated sequence, but the degree of selectivity was only a factor of 2.5. In contrast, the HoxD9 and Antp homeodomains bind to unrelated DNA sequences with KD values of 100–300 nm (53, 54), representing 60–200-fold selectivity for their cognate targets, and the specificity of the GATA1 C-terminal zinc finger for its target site has been estimated to be ∼1000-fold (57). However, quantitative assessments of DNA binding specificity are relatively rare, and it is possible that many other DNA-binding proteins do not display this high level of selectivity.
The question of how a DNA-binding protein finds its cognate targets in the “noise” of three billion base pairs of genomic DNA is largely an unanswered one, but it is often thought that such a protein must have high specificity to act effectively. Surprisingly, despite the selectivity of ZNF217 being apparently very low, our transient transfection data demonstrate that the protein can effectively find targets and generate functional outcomes in a cellular context (albeit on naked transfected DNA rather than a chromatinized target). This ability is significantly compromised when even small changes are made in the DNA recognition site (Fig. 4). The selectivity of ZNF217 is thus apparently substantially greater in vivo than in vitro. This phenomenon might reflect the recruitment of partner proteins that enhance specificity, although it is notable that the sequence preferences of ZNF217 in transient transfections closely match those observed in vitro with recombinant protein. That is, binding is strongest to the extended TGCAGAAT sequence and weaker to CAGAAT or CTGGAGTA oligonucleotides; it is simply the magnitude of the difference in specificity that is apparently magnified in cells. We have recently observed similar results during the analysis of an unrelated DNA-binding domain,6 and so we speculate that small apparent specificities might well be functionally significant, although the mechanism underlying this phenomenon remains to be elucidated.
Implications for Other Zinc Finger Proteins
The structure of the two-zinc finger domain of Tramtrack bound to DNA (44) established the idea that two tandem classical zinc finger domains could mediate sequence-specific DNA binding, and it was later noted that a number of zinc finger proteins contained two-finger units (58), although at that time little was known about the properties of proteins other than Tramtrack. GATA-family transcription factors also use two zinc fingers to bind to DNA, but these domains are structurally distinct to the classical zinc fingers (59). Likewise, Drosophila GAGA factor (GAF) interacts with DNA via a single classical C2H2 finger, but in this case additional contacts to the minor groove are made by an N-terminal basic region (60). Despite these reports and although yeast zinc finger proteins have been predicted to bind DNA using two-finger motifs (61), it is generally accepted that in mammals classical zinc finger proteins tend to use three zinc fingers to contact their target DNA sequences (2).
An inspection of the UniProtKB protein sequence data base revealed that of the 838 confirmed human C2H2 zinc finger proteins, 18 contain only a single two-zinc finger unit. There are also >100 additional proteins containing multiple zinc fingers in which a discrete double-finger unit is present, and it is possible that these proteins can interact with DNA via a mechanism similar to that of ZNF217. Indeed, certain mammalian transcriptional regulators such as BCL11a and ZNF219 contain double zinc finger domains that are known to contact DNA (62, 63). There are a number of other proteins, such as ZNF536, that may also have DNA binding activity via their double zinc finger domains (64). ZNF536, like ZNF217, also binds CtBP. On the other hand, zinc finger proteins such as FOG, which also contain double finger domains but lack the typical TGERP linker and appropriate spacing, may well not be capable of sequence-specific DNA binding.
Supplementary Material
Acknowledgments
We thank Gottfried Otting for access to the 800-MHz NMR spectrometer at the Australian National University and Bill Bubb and Ann Kwan for maintenance of the NMR facility at the University of Sydney. We thank David Segal (UC Davis) for useful discussions.
This work was supported by grants from the National Health and Medical Research Council, Australian Research Council, and the Prostate Cancer Foundation of Australia (to M. C. and J. P. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
J. Font and J. P. Mackay, unpublished data.
J. P. Mackay and M. Crossley, unpublished data.
S. Setiyaputra and J. P. Mackay, unpublished results.
- CtBP
- C-terminal-binding protein
- HSQC
- heteronuclear single quantum correlation.
REFERENCES
- 1. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 2. Wolfe S. A., Nekludova L., Pabo C. O. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 183–212 [DOI] [PubMed] [Google Scholar]
- 3. Scott M. P., Tamkun J. W., Hartzell G. W., 3rd. (1989) Biochim. Biophys. Acta 989, 25–48 [DOI] [PubMed] [Google Scholar]
- 4. Amoutzias G. D., Veron A. S., Weiner J., 3rd, Robinson-Rechavi M., Bornberg-Bauer E., Oliver S. G., Robertson D. L. (2007) Mol. Biol. Evol. 24, 827–835 [DOI] [PubMed] [Google Scholar]
- 5. Jones S. (2004) Genome Biol. 5, 226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krizek B. A., Meyerowitz E. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4063–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Magner D. B., Antebi A. (2008) Trends Endocrinol. Metab. 19, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Immink R. G., Kaufmann K., Angenent G. C. (2010) Semin. Cell Dev. Biol. 21, 87–93 [DOI] [PubMed] [Google Scholar]
- 10. Lo Surdo P., Walsh M. A., Sollazzo M. (2004) Nat. Struct. Mol. Biol. 11, 382–383 [DOI] [PubMed] [Google Scholar]
- 11. Keefe A. D., Szostak J. W. (2001) Nature 410, 715–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bibikova M., Carroll D., Segal D. J., Trautman J. K., Smith J., Kim Y. G., Chandrasegaran S. (2001) Mol. Cell. Biol. 21, 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Papworth M., Kolasinska P., Minczuk M. (2006) Gene 366, 27–38 [DOI] [PubMed] [Google Scholar]
- 14. Klug A. (2010) Q. Rev. Biophys. 43, 1–21 [DOI] [PubMed] [Google Scholar]
- 15. Gamsjaeger R., Liew C. K., Loughlin F. E., Crossley M., Mackay J. P. (2007) Trends Biochem. Sci. 32, 63–70 [DOI] [PubMed] [Google Scholar]
- 16. Brown R. S. (2005) Curr. Opin. Struct. Biol. 15, 94–98 [DOI] [PubMed] [Google Scholar]
- 17. Mackay J. P., Font J., Segal D. J. (2011) Nat. Struct. Mol. Biol. 18, 256–261 [DOI] [PubMed] [Google Scholar]
- 18. Pavletich N. P., Pabo C. O. (1991) Science 252, 809–817 [DOI] [PubMed] [Google Scholar]
- 19. Pavletich N. P., Pabo C. O. (1993) Science 261, 1701–1707 [DOI] [PubMed] [Google Scholar]
- 20. Foster M. P., Wuttke D. S., Radhakrishnan I., Case D. A., Gottesfeld J. M., Wright P. E. (1997) Nat. Struct. Biol. 4, 605–608 [DOI] [PubMed] [Google Scholar]
- 21. Segal D. J., Crotty J. W., Bhakta M. S., Barbas C. F., 3rd, Horton N. C. (2006) J. Mol. Biol. 363, 405–421 [DOI] [PubMed] [Google Scholar]
- 22. Quinlan K. G., Nardini M., Verger A., Francescato P., Yaswen P., Corda D., Bolognesi M., Crossley M. (2006) Mol. Cell. Biol. 26, 8159–8172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Collins C., Rommens J. M., Kowbel D., Godfrey T., Tanner M., Hwang S. I., Polikoff D., Nonet G., Cochran J., Myambo K., Jay K. E., Froula J., Cloutier T., Kuo W. L., Yaswen P., Dairkee S., Giovanola J., Hutchinson G. B., Isola J., Kallioniemi O. P., Palazzolo M., Martin C., Ericsson C., Pinkel D., Albertson D., Li W. B., Gray J. W. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8703–8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang G., Krig S., Kowbel D., Xu H., Hyun B., Volik S., Feuerstein B., Mills G. B., Stokoe D., Yaswen P., Collins C. (2005) Hum. Mol. Genet. 14, 3219–3225 [DOI] [PubMed] [Google Scholar]
- 25. Nonet G. H., Stampfer M. R., Chin K., Gray J. W., Collins C. C., Yaswen P. (2001) Cancer Res. 61, 1250–1254 [PubMed] [Google Scholar]
- 26. Banck M. S., Li S., Nishio H., Wang C., Beutler A. S., Walsh M. J. (2009) Epigenetics 4, 100–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krig S. R., Jin V. X., Bieda M. C., O'Geen H., Yaswen P., Green R., Farnham P. J. (2007) J. Biol. Chem. 282, 9703–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thillainadesan G., Isovic M., Loney E., Andrews J., Tini M., Torchia J. (2008) Mol. Cell. Biol. 28, 6066–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cowger J. J., Zhao Q., Isovic M., Torchia J. (2007) Oncogene 26, 3378–3386 [DOI] [PubMed] [Google Scholar]
- 30. Gumbiner B. M. (2005) Nat. Rev. Mol. Cell Biol. 6, 622–634 [DOI] [PubMed] [Google Scholar]
- 31. Jeanes A., Gottardi C. J., Yap A. S. (2008) Oncogene 27, 6920–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Crossley M., Whitelaw E., Perkins A., Williams G., Fujiwara Y., Orkin S. H. (1996) Mol. Cell. Biol. 16, 1695–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shen Y., Lange O., Delaglio F., Rossi P., Aramini J. M., Liu G., Eletsky A., Wu Y., Singarapu K. K., Lemak A., Ignatchenko A., Arrowsmith C. H., Szyperski T., Montelione G. T., Baker D., Bax A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4685–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kwan A. H., Czolij R., Mackay J. P., Crossley M. (2003) Nucleic Acids Res. 31, e124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quinlan K. G., Verger A., Kwok A., Lee S. H., Perdomo J., Nardini M., Bolognesi M., Crossley M. (2006) Mol. Cell. Biol. 26, 8202–8213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., Shi Y. (2004) Cell 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 37. Shi Y., Sawada J., Sui G., Affar, el B., Whetstine J. R., Lan F., Ogawa H., Luke M. P., Nakatani Y., Shi Y. (2003) Nature 422, 735–738 [DOI] [PubMed] [Google Scholar]
- 38. Ueda J., Tachibana M., Ikura T., Shinkai Y. (2006) J. Biol. Chem. 281, 20120–20128 [DOI] [PubMed] [Google Scholar]
- 39. Fox A. H., Liew C., Holmes M., Kowalski K., Mackay J., Crossley M. (1999) EMBO J. 18, 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liew C. K., Kowalski K., Fox A. H., Newton A., Sharpe B. K., Crossley M., Mackay J. P. (2000) Structure 8, 1157–1166 [DOI] [PubMed] [Google Scholar]
- 41. Liew C. K., Simpson R. J., Kwan A. H., Crofts L. A., Loughlin F. E., Matthews J. M., Crossley M., Mackay J. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matthews J. M., Kowalski K., Liew C. K., Sharpe B. K., Fox A. H., Crossley M., MacKay J. P. (2000) Eur. J. Biochem. 267, 1030–1038 [DOI] [PubMed] [Google Scholar]
- 43. Matharu N. K., Hussain T., Sankaranarayanan R., Mishra R. K. (2010) J. Mol. Biol. 400, 434–447 [DOI] [PubMed] [Google Scholar]
- 44. Fairall L., Schwabe J. W., Chapman L., Finch J. T., Rhodes D. (1993) Nature 366, 483–487 [DOI] [PubMed] [Google Scholar]
- 45. Wuttke D. S., Foster M. P., Case D. A., Gottesfeld J. M., Wright P. E. (1997) J. Mol. Biol. 273, 183–206 [DOI] [PubMed] [Google Scholar]
- 46. Wolfe S. A., Grant R. A., Elrod-Erickson M., Pabo C. O. (2001) Structure 9, 717–723 [DOI] [PubMed] [Google Scholar]
- 47. Nolte R. T., Conlin R. M., Harrison S. C., Brown R. S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kajimura S., Seale P., Tomaru T., Erdjument-Bromage H., Cooper M. P., Ruas J. L., Chin S., Tempst P., Lazar M. A., Spiegelman B. M. (2008) Genes. Dev. 22, 1397–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Postigo A. A., Dean D. C. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 6683–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Vliet J., Turner J., Crossley M. (2000) Nucleic Acids Res. 28, 1955–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang X., Zheng M., Liu G., Xia W., McKeown-Longo P. J., Hung M. C., Zhao J. (2007) Cancer Res. 67, 7184–7193 [DOI] [PubMed] [Google Scholar]
- 52. Kumar V., Carlson J. E., Ohgi K. A., Edwards T. A., Rose D. W., Escalante C. R., Rosenfeld M. G., Aggarwal A. K. (2002) Mol. Cell 10, 857–869 [DOI] [PubMed] [Google Scholar]
- 53. Iwahara J., Zweckstetter M., Clore G. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15062–15067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Affolter M., Percival-Smith A., Müller M., Leupin W., Gehring W. J. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 4093–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bessière D., Lacroix C., Campagne S., Ecochard V., Guillet V., Mourey L., Lopez F., Czaplicki J., Demange P., Milon A., Girard J. P., Gervais V. (2008) J. Biol. Chem. 283, 4352–4363 [DOI] [PubMed] [Google Scholar]
- 56. Fairall L., Harrison S. D., Travers A. A., Rhodes D. (1992) J. Mol. Biol. 226, 349–366 [DOI] [PubMed] [Google Scholar]
- 57. Omichinski J. G., Trainor C., Evans T., Gronenborn A. M., Clore G. M., Felsenfeld G. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 1676–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Iuchi S. (2001) Cell. Mol. Life Sci. 58, 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Trainor C. D., Ghirlando R., Simpson M. A. (2000) J. Biol. Chem. 275, 28157–28166 [DOI] [PubMed] [Google Scholar]
- 60. Omichinski J. G., Pedone P. V., Felsenfeld G., Gronenborn A. M., Clore G. M. (1997) Nat. Struct. Biol. 4, 122–132 [DOI] [PubMed] [Google Scholar]
- 61. Böhm S., Frishman D., Mewes H. W. (1997) Nucleic Acids Res. 25, 2464–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Avram D., Fields A., Senawong T., Topark-Ngarm A., Leid M. (2002) Biochem. J. 368, 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sakai T., Hino K., Wada S., Maeda H. (2003) DNA Res. 10, 155–165 [DOI] [PubMed] [Google Scholar]
- 64. Qin Z., Ren F., Xu X., Ren Y., Li H., Wang Y., Zhai Y., Chang Z. (2009) Mol. Cell. Biol. 29, 3633–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.