Background: The structure of SNAT amino acid transporter is unknown.
Results: Using chemical labeling, N-glycosylation mapping, immunofluorescence, and molecular modeling, we resolved topological structure of SNAT4.
Conclusion: SNAT4 contains 10 transmembrane helices with extracellular N and C termini and a large N-glycosylated extracellular loop domain.
Significance: This is the first study providing topological structural information of a member of mammalian SNAT neutral amino acid transporter family.
Keywords: Amino Acid Transport, Computer Modeling, Membrane, Membrane Proteins, Transport Amino Acids, Membrane Topology
Abstract
Members of system N/A amino acid transporter (SNAT) family mediate transport of neutral amino acids, including l-alanine, l-glutamine, and l-histidine, across the plasma membrane and are involved in a variety of cellular functions. By using chemical labeling, glycosylation, immunofluorescence combined with molecular modeling approaches, we resolved the membrane topological structure of SNAT4, a transporter expressed predominantly in liver. To analyze the orientation using the chemical labeling and biotinylation approach, the “Cys-null” mutant of SNAT4 was first generated by mutating all five endogenous cysteine residues. Based on predicted topological structures, a single cysteine residue was introduced individually into all possible nontransmembrane domains of the Cys-null mutant. The cells expressing these mutants were labeled with N-biotinylaminoethyl methanethiosulfonate, a membrane-impermeable cysteine-directed reagent. We mapped the orientations of N- and C-terminal domains. There are three extracellular loop domains, and among them, the second loop domain is the largest that spans from amino acid residue ∼242 to ∼335. The orientation of this domain was further confirmed by the identification of two N-glycosylated residues, Asn-260 and Asn-264. Together, we showed that SNAT4 contains 10 transmembrane domains with extracellular N and C termini and a large N-glycosylated, extracellular loop domain. This is the first report concerning membrane topological structure of mammalian SNAT transporters, which will provide important implications for our understanding of structure-function of the members in this amino acid transporter family.
Introduction
Amino acid transporters are a group of membrane proteins transporting various amino acids across the membrane and providing substrates for protein biosynthesis and metabolism, precursors for neurotransmitters and hormones, etc. (1). Mammalian amino acid transporters are categorized into various protein transporter families, including cationic amino acid transporter, system N/A transporter (SNAT),2 excitatory amino acid transporters, 4F2hc and rBAT (2–4). SNATs are Na+-dependent transporters that transport zwitterionic amino acids, such as l-alanine, l-glutamine, l-histidine, and l-serine (5). Molecular identification of system N/A transporters has recently revealed that there are seven highly homologous isoforms of this amino acid transporter family named as SNAT1–7, which are encoded by 7 members of the SLC38 gene family (SLC38A1–7), respectively (6, 7). Previous studies suggest that SNAT1, SNAT2, and SNAT4 are highly homologous; SNAT1 and SNAT2 share 52% homology, and SNAT4 shares 48 and 57% homology with SNAT1 and SNAT2, respectively. However, there are some functional differences among the different members of SNATs. SNAT1 and SNAT2 are voltage- and Na+-dependent transporters and also transport substrates for oxidative and nitrogen metabolism (8–11), whereas SNAT4 has a lower substrate affinity for neutral amino acid and transports cationic amino acids in a Na+-independent manner (12). Despite the previous physiological and functional studies of SNATs, little is known regarding the molecular structures of the members of this transporter family.
Compared with soluble proteins, there are very limited high resolution structures available for membrane proteins due to technical difficulties (only 180 membrane protein structures solved thus far) (13). Recently, several high resolution structures of amino acid transporters have been resolved (14–16); however, none of them is a mammalian transporter. To understand the function of SNAT proteins at the molecular level, it is important to obtain the structural information of these proteins, especially their native conformations on the membrane. Given the high degree of similarity between SNAT family members, it is of great interest to study the topological property of one of the SNAT transporters. By using homology modeling with the high resolution structure of LeuTAa, a bacterial leucine transporter (14), a recent study shows that there are 11 transmembrane domains on SNAT2 (17), which is similar to the hypothetical structure estimated by using the secondary structure prediction analyses (18). This study also indicates that computer simulation can be used as a guiding tool for the experimental determination of membrane topological structure.
In this study, we experimentally resolved the membrane topological structure of a SNAT amino acid transporter, SNAT4. SNAT4 is found to predominantly express in liver hepatocytes surrounding the central vein (19) and is thought to be a liver-specific isoform of SNATs (12). Other groups have also documented its expression in placenta and placental cell lines (20, 21). Here, we utilized chemical surface labeling approach with a membrane-impermeable Cys-directed reagent, MTSEA-biotin, which has been used for extracellular cross-linking of cysteine residues (22). In addition, other approaches, including indirect immunofluorescence labeling and N-linked glycosylation mapping in conjunction with sequence analysis and computer simulation, were used. This is the first study experimentally defining the membrane topological structure of SNAT amino acid transporters.
EXPERIMENTAL PROCEDURES
Materials
Anti-SNAT4 antibody was raised against the N-terminal fragment of SNAT4 in rabbit and affinity purified as described previously (19). A QuikChange site-directed mutagenesis kit was purchased from Stratagene (La Jolla, CA). Anti-myc antibody was obtained from Invitrogen. MicroBCA kit, Ultralink immobilized NeutrAvidin, and Super Signal West Pico Chemiluminescence Substrate kit were purchased from Pierce. MTSEA-biotin was purchased from Toronto Research Chemicals (Toronto, ON, Canada) and dissolved in 0.1% dimethyl sulfoxide to 2 mm. All restriction enzymes and PNGase-F were purchased from New England Biolabs. All other reagents were purchased either from Invitrogen or Sigma.
Computer Prediction of Topological Models and Molecular Modeling
Mouse SNAT4 sequence was retrieved from the NCBI data base (GenBank AY027919.1; Swiss-Prot Q8R1S9.1). The sequence was analyzed by topology prediction servers, including HMMTOP and MEMSAT-SVM (Table 1). Molecular modeling servers, such as HHpred and Swiss Model, were also adopted for constructing the hypothetical three-dimensional structure of SNAT4. The sequence of SNAT4 was submitted to SWISS-MODEL automated module server, and the hypothetical model was constructed by using Arg-bound AdiC protein (Protein Data Bank code 3L1L) as a template. In HHpred, the pairwise query-template sequence alignment between SNAT4 and AdiC protein was first generated by performing hidden Markov method, and the predicted structure of SNAT4 was obtained by MODELLER program based on sequence alignment.
TABLE 1.
Summary of SNAT4 topology prediction results using various methods
| Prediction Servers | Number of TMH | N/C terminus | Methods |
|---|---|---|---|
| HMMTOP | 10 | Same side | Hidden Markov model |
| MEMSAT-SVM | 13 | Opposite side | SVM |
| Phobius | 10 | Same side | Consensus |
| SOSUI | 10 | Same side | Hydrophobicity-based |
| ConPred II | 10 | Same side | Consensus |
| TMpred | 10 | Same side | Rule-based |
| Philius | 9 | Opposite side | Neural network |
| TopPred2 | 10 | Same side | Hydrophobicity-based |
| TOPTMH | 9 | Opposite side | Consensus |
| TMHMM | 11 | Opposite side | Consensus |
Preparation of DNA Constructs Containing SNAT4 Mutants
Mouse SNAT4 was cloned and identified as described previously (19). The entire open reading frame of SNAT4 was amplified by using a pair of DNA primers: sense, 5′-GACTGGATCCAAATGGACCCCATGGAACTGAAC-3′ and antisense, 3′-GACTTCTAGAGTGGTGATTGGGATTCG-5′. PCR products were purified and digested with XbaI and BamHI before cloning into a pcDNA 3.1 expression vector. Mutants of SNAT4 were generated by using a QuikChange site-directed mutagenesis kit, and DNA primers used are listed in Table 2. The correctness of all the sequences was verified by a sequencing facility at the University of Texas Health Science Center at San Antonio DNA Core. The “Cys-null” mutant was first made by mutating all five cysteines in WT SNAT4 to alanines. Using Cys-null mutant as a backbone, a single cysteine was introduced individually into predicted nontransmembrane domains based on the topological models generated by computer simulation.
TABLE 2.
DNA primers used for site-directed mutagenesis
| Mutation sites | Sense | Antisense |
|---|---|---|
| C18A | ACGGAGACAGCGCCAGCGGGGACAGT | ACTGTCCCCGCTGGCGCTGTCTCCGT |
| C232A | GATTTTCTCTCTCCGCCATGGTGTTTTTCG | CGAAAAACACCATGGCGGAGAGAGAAAATC |
| C249A | ATTCCAAATTCCCGCCCCTCTGCCTG | CAGGCAGAGGGGCGGGAATTTGGAAT |
| C321A | GTGCTGAGAAAGCCCAACCAAAATACT | AGTATTTTGGTTGGGCTTTCTCAGCAC |
| C345A | CTTTTGCTTTTGTCGCCCACCCTGAGGT | ACCTCAGGGTGGGCGACAAAAGCAAAAG |
| T54C | CGAAAGTCAGAAGTTCCTGTGCAATGGGTTTTTAGGGAAG | CTTCCCTAAAAACCCATTGCACAGGAACTTCTGACTTTCG |
| A129C | CCTTTTGCTGAAGACATGCAAGGAAGGAGGGTC | GACCCTCCTTCCTTGCATGTCTTCAGCAAAAGG |
| A183C | GAACTGCCTGAAGTAATCAGATGCTTCATGGGACTTGAAGAAAAC | GTTTTCTTCAAGTCCCATGAAGCATCTGATTACTTCAGGCAGTTC |
| L219C | CTCTCTCCTTAAAAATTGCGGCTACCTTGGCTACAC | GTGTAGCCAAGGTAGCCGCAATTTTTAAGGAGAGAG |
| I474C | GTCATCCTCGTGCCTACCTGCAAATACATCTTTGGATTC | GAATCCAAAGATGTATTTGCAGGTAGGCACGAGGATGAC |
| T366C | CCCGCAGAAAGATGCAGTGCGTGTCCAACATTTCCATC | GATGGAAATGTTGGACACGCACTGCATCTTTCTGCGGG |
| A401C | GAAGACGAGCTGCTGCATTGCTACAGCAAGGTCTACAC | GTGTAGACCTTGCTGTAGCAATGCAGCAGCTCGTCTTC |
| T440C | CCGTACTTCGGTGATCTGCCTGCTGTTTCCAAGG | CCTTGGAAACAGCAGGCAGATCACCGAAGTACGG |
| L507C | CTCGTCAAGAAAGAACCTTGCAGATCACCCCAGAAGATTG | CAATCTTCTGGGGTGATCTGCAAGGTTCTTTCTTGACGAG |
| N543C | AGTGGTGATTGGGACACGGCGGGTTGTAGA | TCTACAACCCGCCGTGTCCCAATCACCACT |
| N260Q | GGATCACAACAACGGACAGCTGACGTTCAACAACAC | GTGTTGTTGAACGTCAGCTGTCCGTTGTTGTGATCC |
| N264Q | CAACGGAAATCTGACGTTCCAGAACACACTTCCGATTCAC | GTGAATCGGAAGTGTGTTCTGGAACGTCAGATTTCCGTTG |
| N260,264Q | CAACGGACAACTGACGTTCCAGAACACACTTCCGATTCAC | GTGAATCGGAAGTGTGTTCTGGAACGTCAGTTGTCCGTTG |
Cell Culture, Transfection, and Immunofluorescence
HepIR cells were obtained from Dr. Feng Liu's laboratory (University of Texas Health Science Center at San Antonio) and cultured in humid 5% CO2, 33 °C incubator with DMEM containing 4% fetal bovine serum (FBS). Chinese hamster ovary (CHO) cells were cultured in humid 5% CO2, 37 °C incubator in Ham's F-12 medium containing 10% FBS and 1% penicillin/streptomycin. Two micrograms of DNA constructs were transiently transfected into CHO cells using Lipofectamine 24 h after plating of the cells on 60-mm dishes. Immunofluorescence staining was performed on glass coverslips using anti-SNAT4 antibody or anti-c-myc antibody, and followed by the incubation with FITC-conjugated anti-rabbit or anti-mouse IgG secondary antibody, respectively. DAPI was used to label the nuclei. The cells were then visualized and analyzed using confocal laser scanning microscopy (Fluoview; Olympus Optical, Tokyo, Japan) at the Imaging Core facility (University of Texas Health Science Center at San Antonio).
Chemical Labeling of SNAT4 Mutants with MTSEA-Biotin and Western Blotting
MTSEA-biotin was first dissolved in 0.1% dimethyl sulfoxide at a concentration of 0.2 m in PBS. Twenty-four hours after transfection, CHO cells were washed three times with PBS (with Ca2+ and Mg2+) and then treated with 2 mm MTSEA-biotin/PBS for 30 min at 4 °C with or without 0.25% Triton X-100. The cells were then collected in chilled RIPA buffer (25 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1% Nonidet P-40, 0.1% sodium deoxycholate) containing protease inhibitors and homogenized with a 26½-gauge needle 20 times. The total protein concentration was determined by MicroBCA kit (Pierce), and equal amounts of total protein were mixed with Ultralink-immobilized NeutrAvidin for 30 min at 4 °C. Biotinylated proteins were eluted from NeutrAvidin beads using 40 μl of 2× Laemmli sample buffer. The sample was then separated by 10% SDS-PAGE, transferred to a nitrocellulose membrane, immunoblotted by anti-SNAT4 (1:500 dilutions) or anti-GAPDH/β-actin (1:5000 dilutions) antibody, and subsequently, incubated with HRP-conjugated goat anti-rabbit IgG secondary antibody or HRP-conjugated donkey anti-mouse (1:5000 dilutions), respectively. The signals were detected by Super West Pico Chemiluminescence Substrate kit.
Deglycosylation Assay
CHO cells were collected 24 h after transfection in chilled RIPA buffer and homogenized with a 26½-gauge needle 20 times. The total protein concentration was determined by MicroBCA. Twenty micrograms of total protein was first denatured in glycoprotein denaturing buffer at 100 °C for 10 min and later incubated with a reaction mix (New England Biolabs) containing G7 reaction buffer, Nonidet P-40, and PNGase-F at 37 °C for 2 h, followed by Western blotting assay as described above.
RESULTS
Membrane Topology Predictions of SNAT4
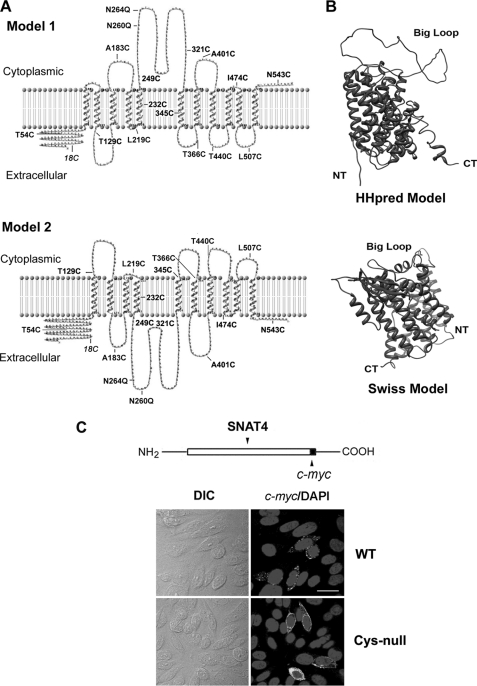
To identify the membrane topology of SNAT4, we first used sequence-based prediction methods, referred to as “hydropathy index,” which was previously used to identify transmembrane helix (TMH) in membrane proteins (23, 24). Meanwhile, topology or side prediction was also accomplished by the approach that incorporates the charge biased distribution between the inner and outer loops, also known as the “positive-inside” rule (25). Some commonly used approaches, such as hidden Markov modeling and neuron network, extract information from various regions of a TM protein (26, 27). More advanced methods were recently developed by combining hidden Markov models, rule-based scheme, and support vector machines (28). As suggested by a previous paper (29), topological predictions would be more accurate when individual methods show consistency in their prediction rather than when they are inconsistent. Recently, several consensus prediction methods based on multiple predictors have also been developed (Phobius, ConPred II, and TOPTMH), which yield more reliable and accurate results (see review, see Ref. 30). These analyses result in two predicted models (Table 1 and Fig. 1A); model 1 has 11 TMHs with opposite orientations of N and C termini, and model 2 consists of 10 TMHs with both N and C termini located at the same side of plasma membrane. The major difference between these two models is the configuration of first through fourth helical motifs, which results in different orientations of N terminus and the largest loop domain (between the third and fourth TMHs). The locations of the mutations used in MTSEA-biotin chemical labeling are indicated on the illustration of these two models (Fig. 1A).
FIGURE 1.
Predicted membrane topological models of SNAT4. A, two putative topological models of SNAT4 were constructed using TMpred and TopPred and confirmed by other methods listed in Table 1. The major difference between these two models is the orientation of the largest loop domain, which spans from amino acid residue ∼242 to ∼335. This results in the different membrane orientations of the last four extra- or intracellular loops. The mutations as indicated were generated based on these two models. B, three-dimensional structure models of SNAT4 were generated by HHpred or SWISS-MODEL, using Arg-bound AdiC protein (Protein Data Bank code 3L1L) as a template. NT, N terminus; CT, C terminus. C, SNAT4-c-myc construct was generated by fusing c-myc tag with SNAT4 in pcDNA 3.1 vector. The Cys-null mutant was generated by replacing all endogenous cysteines with alanines. Expression of wild-type and the Cys-null mutant of SNAT4 in CHO cells was examined by immunolabeling with anti-c-myc antibody. Scale bar, 20 μm.
Two most commonly used molecular modeling methods, HHpred and SWISS-MODEL, were also employed to reconstruct hypothetical three-dimensional model of proteins, using a known three-dimensional structure of bacterial AdiC transporter as a template. The three-dimensional structural models were re-constructed by the MODELLER software using HHPre (Fig. 1B, upper) or SWISS-MODEL modeling method (Fig. 1B, lower). Both programs predicted the similar three-dimensional structure of SNAT4, which harbors 10 TMHs and the same membrane orientations of N and C termini.
Extracellular Orientations of N terminus and C terminus of SNAT4 Protein
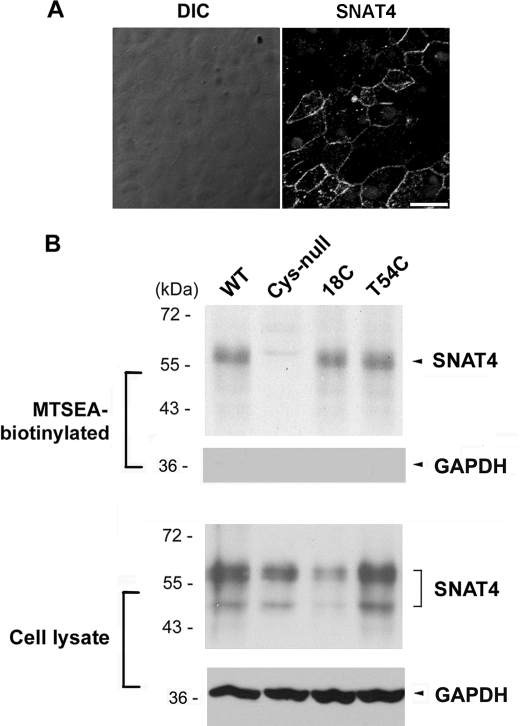
We first investigated the orientation of SNAT4 N terminus by using a hepatocyte-like cell line, HepIR. The level of SNAT4 in HepIR cells was enhanced by insulin treatment (16 μg/ml for 24 h) as reported previously (19). SNAT4 protein expressed on the cell surface was immunolabeled with affinity-purified anti-SNAT4 antibody against the N terminus of SNAT4 in the absence of detergent. Immunofluorescence shows the positive signals outlining the cell (Fig. 2A), suggesting that N-terminal domain of SNAT4 is localized to the extracellular side of the cell.
FIGURE 2.
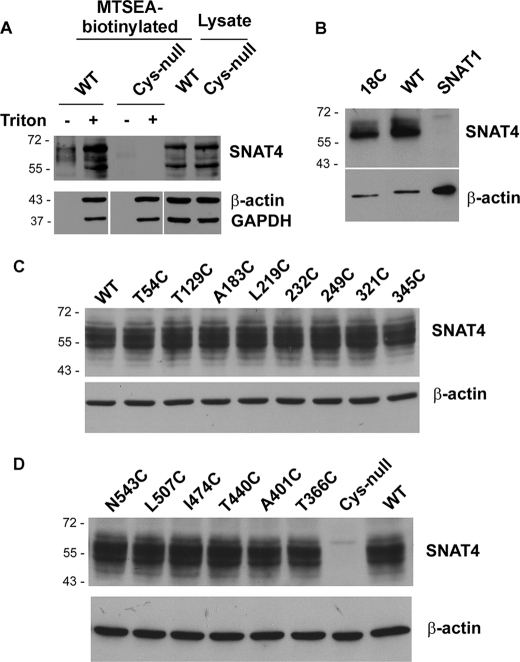
N terminus of SNAT4 is oriented at the extracellular side of the cell determined by immunofluorescence and MTSEA-biotin chemical labeling. A, SNAT4 protein was immunolabeled by an antibody against the N terminus of SNAT4 expressed in HepIR cells in the absence of Triton X-100. The SNAT4 signals were detected by FITC-conjugated goat anti-rabbit IgG secondary antibody. Nucleus was counterstained with DAPI. Scale bar, 20 μm. B, using the Cys-null mutant as a template, cysteine was individually introduced back to generate 18C and T54C mutants. MTSEA-biotin labeling was performed in CHO cells expressing exogenous WT, Cys-null, 18C, and T54C mutants of SNAT4. The lysates of the cell labeled with MTSEA-biotin were loaded on NeutrAvidin beads. Biotinylated proteins (upper) and the preloaded cell lysates (lower) were immunoblotted with affinity-purified anti-SNAT4 antibody or anti-GAPDH antibody.
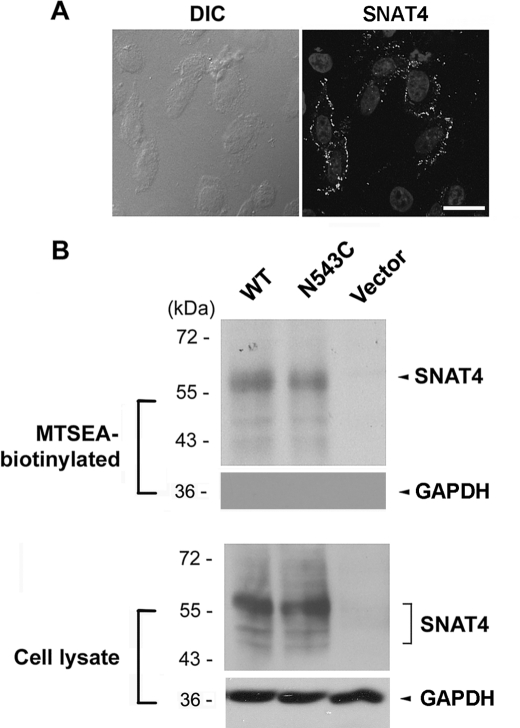
To determine the orientation of C terminus, c-myc-tagged SNAT4 was expressed in CHO cells. Immunofluorescence labeling with anti-c-myc antibody showed that in the absence of the detergent, the signal was also visible on the cell surface (Fig. 3A), suggesting that the C terminus, like N terminus, also faces the extracellular side of the cell. However, in neighboring untransfected cells, the fluorescence signal was absent.
FIGURE 3.
C terminus of SNAT4 faces the extracellular side of the cell. A, CHO cells expressing exogenous c-myc-tagged SNAT4 were immunolabeled with anti-c-myc antibody in the absence of Triton X-100. Nuclei were counterstained with DAPI. Scale bar, 20 μm. B, Cys-null mutant was generated by replacing all endogenous cysteines with alanines. The N543C mutant was generated using the Cys-null mutant as a template. Cells expressing WT, N543C, and vector control were labeled with MTSEA-biotin. The isolated biotinylated proteins (upper) and the preloaded cell lysates (lower) were immunoblotted with anti-SNAT4 antibody or anti-GAPDH antibody.
MTSEA-biotin chemical labeling was employed to confirm the membrane orientation of N and C termini of SNAT4 through a cross-linking reaction. The Cys-null mutant was generated by mutating all endogenous cysteine resides to alanines. Like wild-type SNAT4, Cys-null mutants were expressed and localized to the surface of CHO cells (Fig. 1C). Site mutants of SNAT4 containing single cysteines, 18C (endogenous), T54C and N543C expressed in CHO cells were reactive with MTSEA-biotin and retained by NeutrAvidin (Figs. 2B and 3B). GAPDH, an intracellular protein, was only detected in cell lysates, but not in biotinylated samples, suggesting that MTSEA-biotin is not assessable to intracellular proteins (Figs. 2B and 3B). Immunofluorescence and chemical labeling results demonstrated the extracellular orientation of both N and C termini.
Membrane Topological Structure of SNAT4 Determined by MTSEA-biotin Chemical Labeling
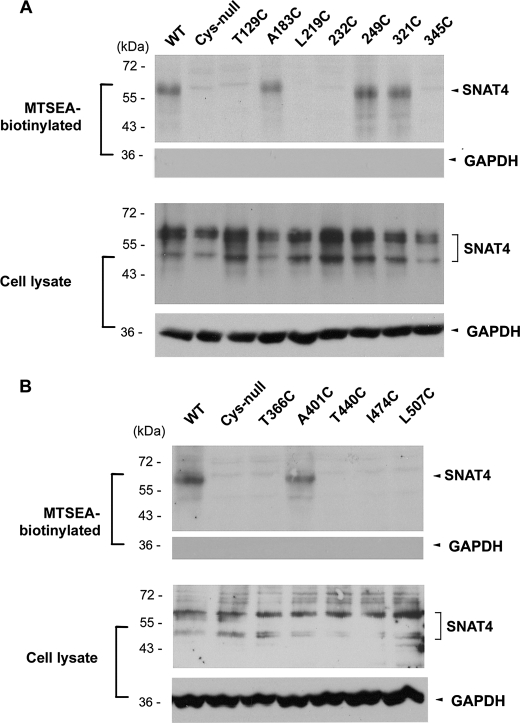
Using Cys-null as a template, cysteines were introduced individually to all hypothetical loop domains predicted by computational prediction (Fig. 1, A and B). CHO cells expressing these mutants were labeled with MTSEA-biotin. Among all mutants containing single cysteines, positive biotinylation results in cells expressing 249C and 321C mutants suggest that the largest loop domain faces the extracellular side of the cell (Fig. 4A). 345C is predicted to be located in the fifth TMH, and as expected, it was not labeled with MTSEA-biotin.
FIGURE 4.
Identification of extracellular loop domains of SNAT4 using MTSEA-biotin chemical labeling. DNA constructs containing single cysteine (T129C, A183C, L219C, 232C, 249C, 321C, 345C, T366C, A401C, T440C, I474C, and L507C) on the Cys-null template of SNAT4 were generated and transfected into CHO cells. The cells were labeled with MTSEA-biotin, and the isolated biotinylated proteins (upper) and the preloaded cell lysates (lower) were immunoblotted with anti-SNAT4 antibody or anti-GAPDH antibody. A, amino acid residues at positions 183, 249, and 321 are located at the extracellular loop domains. B, amino acid residue at 401 is located at an extracellular loop domain.
Based on the Cys-null template, three mutants, A129C, A183C, and L219C located on the first three loop domains, respectively, were generated. MTSEA-biotin labeling suggests that only the second loop domain containing Ala-183, but not the first and third loop domains containing Ala-129 and Leu-219, respectively, faces outside (Fig. 4A). Five other mutants containing single cysteines, T366C, A401C, T440C, I474C, and L507C, were generated to determine the orientation of the last five hypothetical loop domains. Only the loop domain containing A401C was labeled by MTSEA-biotin, suggesting its extracellular orientation (Fig. 4B). Biotinylated samples were also probed by anti-GAPDH antibody, and the results showed the inaccessibility of MTSEA-biotin labeling on intracellular GAPDH protein (Fig. 4). We showed that only in membrane-permeabilized cells treated with Triton X-100, GAPDH and β-actin were labeled by MTSEA-biotin (Fig. 5A). These control experiments with intracellular proteins further confirm that MTSEA-biotin is membrane-impermeable, primarily reacting with extracellular cysteine residues.
FIGURE 5.
MTSEA-biotin reacts with all single cysteine mutants of SNAT4 in the presence of detergent. A, CHO cells expressing exogenous WT or Cys-null mutant were pretreated with or without 0.25% Triton X-100 prior to MTSEA-biotin labeling. The isolated biotinylated proteins or the preloaded cell lysates were immunoblotted with anti-SNAT4 antibody (upper) or anti-β-actin/anti-GAPDH antibody (lower). B–D, CHO cells expressing exogenous WT, single cysteine mutants generated based on the Cys-null template, the Cys-null mutant or SNAT1, another member of SNAT family, were pretreated with or without 0.25% Triton X-100 prior to MTSEA-biotin labeling. The isolated biotinylated proteins were immunoblotted with anti-SNAT4 antibody (upper) or anti-β-actin antibody as control (lower).
To exclude the possibility that the lack of the labeling is due to the nonreactivity of certain cysteine residues, we permeabilized the cells with Triton X-100 and found that MTSEA-biotin labeled all mutants containing single cysteine residues as well as wild type SNAT4 at comparable levels, whereas the Cys-null mutant was not labeled (Fig. 5, B–D). These results suggest that the lack of reactivity is not due to the problem associated with MTSEA-biotin labeling, but instead, is representative of the intracellular membrane orientation.
Identification of Two N-Glycosylation Sites at the Largest Extracellular Loop Domain of SNAT4
Extracellular orientation of N-linked glycosylation modification provides a unique way for determining topology of the membrane proteins. N-Glycosylated sites of SNAT4 protein were predicted by the NetNGlyc 1.0 server (Fig. 6A) (43). Among six possible asparagine residues, Asn-8 is located at the N terminus and Asn-83 is at the predicted second TMH. Amino acid residues Asn-260 and Asn-264 are the two most likely N-glycosylated residues located on the largest hypothetical loop domain. These two Asn residues were mutated into Gln, and their expression in CHO cells was confirmed by Western blotting (Fig. 6B). Compared with the wild-type control, both N260Q and N264Q mutant proteins showed a mobility shift from around 60 kDa to 53 kDa (Fig. 6B). The double mutant containing both N260Q and N264Q showed a dramatic band shift to around 35 kDa. Deglycosylation by PNGase-F further increased the mobility of SNAT4 on SDS-PAGE. These results suggest that both Asn-260 and Asn-264 sites are modified by N-glycosylation and further confirm the extracellular orientation of the largest loop domain.
FIGURE 6.
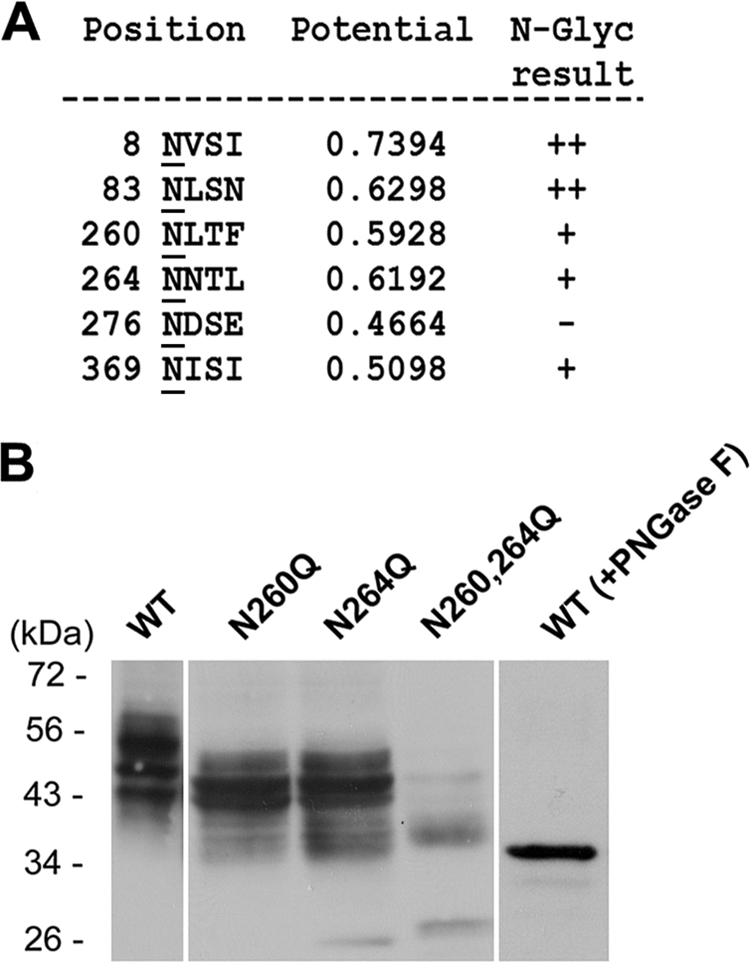
Asn-260 and Asn-264 located in the largest loop domain are modified by N-linked glycosylation. A, the N-linked glycosylated sites of SNAT4 protein were predicted by NetNGlyc 1.0 server. B, two predicted N-glycosylated sites, Asn-260 and Asn-264, were mutated to Gln. WT and single and double mutants of SNAT4 were expressed in CHO cells. Isolated membrane samples were immunoblotted with anti-SNAT4 antibody. Membrane protein samples isolated from the cell expressing WT SNAT4 were treated with PNGase-F and immunoblotted with anti-SNAT4 antibody.
DISCUSSION
In this paper, we determined the membrane topological structure of SNAT4 protein using biochemical approaches in conjunction with computational means. Based on two models predicted by various prediction and molecular modeling methods, we generated a Cys-null mutant and other mutants containing only a single cysteine. MTSEA-biotin used here specifically labels cysteines located on the extracellular side of the membrane protein. Immunofluorescence was used to confirm the orientation of N and C termini, and a deglycosylation assay was employed to define the orientation of the largest loop domain. Our data suggest that SNAT4 contains 10 TMHs with both extracellular N and C termini, and a large, extracellular N-glycosylated loop domain spanning from ∼242 to ∼335 amino acids.
Most of the integral membrane proteins consist of one or more tightly packed transmembrane α-helices, which form independently stable folding units in a membrane environment. Based on amino acid propensities of TMHs and hydrophobicity scales, several methods have been developed, which are used to determine potential membrane-spanning segments (23). Based on all of the prediction outcomes, two topological models for SNAT4 were generated: 11 transmembrane segments with opposite orientation of C or N terminus (model 1) and 10 TMHs with the identical orientations of C and N termini (model 2). These two models differed primarily in the orientation of N terminus and the largest loop domain. The difference in the N-terminal orientation results in changes in the number of transmembrane segments and the orientation of the biggest loop. To further confirm the topological prediction at detailed molecular structural level, we utilized two homology modeling approaches, HHpred and SWISS-MODEL. The HHpred method uses de novo repeat identification in protein sequence by building pairwise query-template alignments. SWISS-MODEL modeling, on the other hand, requires at least one experimentally determined three-dimensional structure of a protein as a template. Although neither of these two homology modeling programs is generally designed for study of the membrane proteins, the results from both modeling systems predict that the three-dimensional structure of SNAT4 has a high degree of homology with a major facilitator superfamily, APC (amino acid/polyamine/organocation) family (AdiC; Protein Data Bank code 3L1L) (15). This analysis is consistent with the previous studies showing that SNAT family shares the sequence homology with bacterial AdiC protein, a member of APC family (31, 32). Here, homological modeling again suggests that SNAT4 shares the same features of transmembrane segments with AdiC protein with 10 TMHs and the same orientation of N and C termini.
We used MTSEA-biotin surface chemical labeling to experimentally define membrane topology of SNAT4. MTSEA-biotin reagent, a derivative of methanethiosulfonate (MTS), forms mixed disulfides with free sulfhydryls, covalently linking the -SCH2CH2X group to the cysteine sulfhydryl. MTSEA is predominantly positively charged (33) and membrane-permeable (34). Conjugation with biotin moiety renders MTSEA membrane- impermeable under mild condition (34). To determine the topology of membrane proteins by using MTS chemicals or biotinylated derivatives, one must consider carefully the bulkiness of the compound and the accessibility of substituted residues to the compound. MTSEA-biotin has a relatively large size (14.5 Å), which could potentially make it less accessible to certain, buried cysteine residues. However, in our study, the MTSEA-biotin was able to label all single cysteines, but not cysteine-null mutant in Triton X-100-permeabilized cells, demonstrating the effectiveness of this chemical labeling. We first generated the Cys-null mutant by mutating all five endogenous cysteines to alanines. Using this mutant as a backbone, we made single cysteine site mutants of SNAT4 on the predicted loop domains. The mutated amino acid residues were relatively conserved nonpolar ones, including Leu, Thr, or Ile, which presumably would not affect the overall structure of SNAT4. By using immunofluorescence as well as MTSEA-biotin labeling, we show that both N and C termini of SNAT4 are extracellular.
The extracellular orientation of C termini of SNAT family proteins have previously been determined both experimentally (35) and by computer simulation (9, 17, 36), which is consistent with our results. However, based primarily on theoretical prediction, but not on experimental data, previous published studies, including those from our laboratory, suggest an intracellular orientation of N termini for SNAT family proteins (10, 11, 17, 36, 37). Based on the prediction by HHpred, we noticed that SNAT family proteins share a certain degree of sequential homologies with plant auxin permease, auxin1, and bacterial transporters, AdiC and Mhp1. In addition, Bröer et al. (38) shows a similar predicted hydropathicity plot profile between SNAT3 and Mhp1, suggesting the same number of transmembrane segments between Mhp1 and SNAT3, and the identical membrane orientation of N and C termini. Despite the unrelated amino acid sequences, above mentioned transporters have been shown to have similar core membrane topological structures, with identical orientation of both N and C termini. However, contrary to that of SNAT4, the intracellular orientation of N terminus of a plant auxin permease, auxin1, which shares low degree of sequential similarity with SNAT family members, is reported determined by immunofluorescence method (39). The crystal structure of AdiC shows identical intracellular orientation of both C and N termini, with transmembrane segments similar to those of SNAT4 (15). These results are consistent with our model showing the identical orientation of both termini, but disagree with the N and C termini being extracellular. This could be an example of topology inversion during evolution for homologous proteins with the same number of transmembrane segments (40).
SNAT4 contains a total of six potential N-linked glycosylation sites. Because N-glycosylation normally occurs at the extracellular side of the membrane proteins, determination of N-glycosylation sites offers an alternative approach for the determination of membrane topology (41, 42). The mutant with an ablation of the two glycosylation sites located in the largest loop domain increased mobility on SDS-PAGE compared with wild-type SNAT4, suggesting extracellular orientation of this big loop domain. In addition to these two glycosylation sites, a slight slower mobility of this mutant compared with deglycosylated SNAT4 implies the existence of additional N-glycosylation site(s). The most likely one is the Asn-8 residue at the N terminus, which is consistent with extracellular orientation of the N terminus.
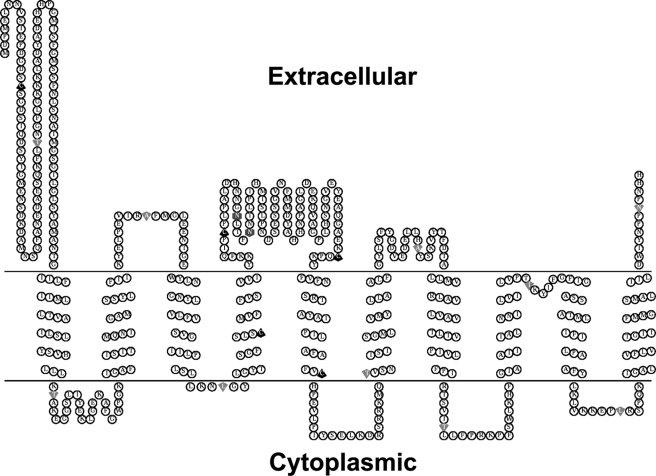
Other extracellular and intracellular loop domains were similarly determined by MTSEA-biotin labeling. For the last five loop domains, only the one containing Ala-401 is an extracellular loop domain, and all the others are intracellular. These results suggest that the model 2 closely represents the topological structure of SNAT4 with one modification; the fourth loop harboring Ile-474 is not extracellular. Interestingly, the fifth extracellular loop domain contains four amino acid residues. Due to its size, this domain is likely to be buried in the membrane instead of being exposed at the cell surface, thereby making it inaccessible to MTSEA-biotin chemical labeling. Based on all of our experimental data, the membrane topology of a SNAT amino acid transporter, for the first time, has been resolved experimentally (Fig. 7). Determination of the membrane topological structure will help to dissect the structure-function of SNAT amino acid transporters.
FIGURE 7.
Schematic illustration of SNAT4 topological structure and the location of mutation sites used. Based on the data we obtained, the diagram of topological structure of SNAT4 was generated using the TOPO2 server and modified by Photoshop software (Adobe Systems, San Jose, CA). Five endogenous cysteines are labeled in black up arrows; other single cysteine mutation sites are represented as down arrows; two glycosylation sites, Asn-260 and Asn-264, are indicated in hexagons.
Acknowledgments
We thank members of Dr. Jiang's laboratory for critical reading of the manuscript.
The work was supported, in whole or in part, by National Institutes of Health Grant EY12085. This work was also supported by Welch Foundation Grant AQ-1507.
- SNAT
- system N/A amino acid transporter
- MTS
- methanethiosulfonate
- MTSEA
- N-biotinylaminoethyl methanethiosulfonate
- TMH
- transmembrane helix
- PNGase-F
- peptide: N-glycosidase F.
REFERENCES
- 1. Christensen H. N. (1990) Physiol. Rev. 70, 43–77 [DOI] [PubMed] [Google Scholar]
- 2. Palacín M., Estévez R., Bertran J., Zorzano A. (1998) Physiol. Rev. 78, 969–1054 [DOI] [PubMed] [Google Scholar]
- 3. Castagna M., Shayakul C., Trotti D., Sacchi V. F., Harvey W. R., Hediger M. A. (1997) J. Exp. Biol. 200, 269–286 [DOI] [PubMed] [Google Scholar]
- 4. Malandro M. S., Kilberg M. S. (1996) Annu. Rev. Biochem. 65, 305–336 [DOI] [PubMed] [Google Scholar]
- 5. Mackenzie B., Erickson J. D. (2004) Pflügers Arch. 447, 784–795 [DOI] [PubMed] [Google Scholar]
- 6. Sundberg B. E., Wååg E., Jacobsson J. A., Stephansson O., Rumaks J., Svirskis S., Alsiö J., Roman E., Ebendal T., Klusa V., Fredriksson R. (2008) J. Mol. Neurosci. 35, 179–193 [DOI] [PubMed] [Google Scholar]
- 7. Hägglund M. G., Sreedharan S., Nilsson V. C., Shaik J. H., Almkvist I. M., Bäcklin S., Wrange O., Fredriksson R. (2011) J. Biol. Chem. 286, 20500–20511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang H., Huang W., Sugawara M., Devoe L. D., Leibach F. H., Prasad P. D., Ganapathy V. (2000) Biochem. Biophys. Res. Commun. 273, 1175–1179 [DOI] [PubMed] [Google Scholar]
- 9. Yao D., Mackenzie B., Ming H., Varoqui H., Zhu H., Hediger M. A., Erickson J. D. (2000) J. Biol. Chem. 275, 22790–22797 [DOI] [PubMed] [Google Scholar]
- 10. Gu S., Roderick H. L., Camacho P., Jiang J. X. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3230–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gu S., Roderick H. L., Camacho P., Jiang J. X. (2001) J. Biol. Chem. 276, 24137–24144 [DOI] [PubMed] [Google Scholar]
- 12. Hatanaka T., Huang W., Ling R., Prasad P. D., Sugawara M., Leibach F. H., Ganapathy V. (2001) Biochim. Biophys. Acta 1510, 10–17 [DOI] [PubMed] [Google Scholar]
- 13. White S. H. (2009) Nature 459, 344–346 [DOI] [PubMed] [Google Scholar]
- 14. Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. (2005) Nature 437, 215–223 [DOI] [PubMed] [Google Scholar]
- 15. Gao X., Lu F., Zhou L., Dang S., Sun L., Li X., Wang J., Shi Y. (2009) Science 324, 1565–1568 [DOI] [PubMed] [Google Scholar]
- 16. Shaffer P. L., Goehring A., Shankaranarayanan A., Gouaux E. (2009) Science 325, 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Z., Albers T., Fiumera H. L., Gameiro A., Grewer C. (2009) J. Biol. Chem. 284, 25314–25323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R. (2005) Nucleic Acids Res. 33, W116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gu S., Langlais P., Liu F., Jiang J. X. (2003) Biochem. J. 371, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Desforges M., Lacey H. A., Glazier J. D., Greenwood S. L., Mynett K. J., Speake P. F., Sibley C. P. (2006) Am. J. Physiol. Cell Physiol. 290, C305–312 [DOI] [PubMed] [Google Scholar]
- 21. Novak D., Quiggle F., Haafiz A. (2006) Biochimie 88, 39–44 [DOI] [PubMed] [Google Scholar]
- 22. Loo T. W., Ho C., Clarke D. M. (1995) J. Biol. Chem. 270, 19345–19350 [DOI] [PubMed] [Google Scholar]
- 23. Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 24. Snider C., Jayasinghe S., Hristova K., White S. H. (2009) Protein Sci. 18, 2624–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. von Heijne G., Gavel Y. (1988) Eur. J. Biochem. 174, 671–678 [DOI] [PubMed] [Google Scholar]
- 26. Zheng W. J., Spassov V. Z., Yan L., Flook P. K., Szalma S. (2004) Comput. Biol. Chem. 28, 265–274 [DOI] [PubMed] [Google Scholar]
- 27. Viklund H., Elofsson A. (2004) Protein Sci. 13, 1908–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ahmed R., Rangwala H., Karypis G. (2010) J. Bioinform. Comput. Biol. 8, 39–57 [DOI] [PubMed] [Google Scholar]
- 29. Nilsson J., Persson B., von Heijne G. (2000) FEBS Lett. 486, 267–269 [DOI] [PubMed] [Google Scholar]
- 30. Punta M., Forrest L. R., Bigelow H., Kernytsky A., Liu J., Rost B. (2007) Methods 41, 460–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jack D. L., Paulsen I. T., Saier M. H. (2000) Microbiology 146, 1797–1814 [DOI] [PubMed] [Google Scholar]
- 32. Lolkema J. S., Slotboom D. J. (2008) Mol. Membr. Biol. 25, 567–570 [DOI] [PubMed] [Google Scholar]
- 33. Zhang H., Karlin A. (1997) Biochemistry 36, 15856–15864 [DOI] [PubMed] [Google Scholar]
- 34. Holmgren M., Liu Y., Xu Y., Yellen G. (1996) Neuropharmacology 35, 797–804 [DOI] [PubMed] [Google Scholar]
- 35. Hyde R., Cwiklinski E. L., MacAulay K., Taylor P. M., Hundal H. S. (2007) J. Biol. Chem. 282, 19788–19798 [DOI] [PubMed] [Google Scholar]
- 36. Zhang Z. I., Zander C. B., Grewer C. (2011) Biochem. J. 434, 287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gu S., Adan-Rice D., Leach R. J., Jiang J. X. (2001) Genomics 74, 262–272 [DOI] [PubMed] [Google Scholar]
- 38. Bröer S., Schneider H. P., Bröer A., Deitmer J. W. (2009) J. Biol. Chem. 284, 25823–25831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blakeslee J. J., Bandyopadhyay A., Lee O. R., Mravec J., Titapiwatanakun B., Sauer M., Makam S. N., Cheng Y., Bouchard R., Adamec J., Geisler M., Nagashima A., Sakai T., Martinoia E., Friml J., Peer W. A., Murphy A. S. (2007) Plant Cell 19, 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. von Heijne G. (2006) Nat. Rev. Mol. Cell Biol. 7, 909–918 [DOI] [PubMed] [Google Scholar]
- 41. Devoto A., Piffanelli P., Nilsson I., Wallin E., Panstruga R., von Heijne G., Schulze-Lefert P. (1999) J. Biol. Chem. 274, 34993–35004 [DOI] [PubMed] [Google Scholar]
- 42. van Geest M., Lolkema J. S. (2000) Microbiol. Mol. Biol. Rev. 64, 13–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gupta R., Brunak S. (2002) Pac. Symp. Biocomput. 7, 310–322 [PubMed] [Google Scholar]