Abstract
The normal microbial occupants of the mammalian intestine are crucial for maintaining gut homeostasis, yet the mechanisms by which intestinal cells perceive and respond to the microbiota are largely unknown. Intestinal epithelial contact with commensal bacteria and/or their products has been shown to activate noninflammatory signaling pathways, such as extracellular signal-related kinase (ERK), thus influencing homeostatic processes. We previously demonstrated that commensal bacteria stimulate ERK pathway activity via interaction with formyl peptide receptors (FPRs). In the current study, we expand on these findings and show that commensal bacteria initiate ERK signaling through rapid FPR-dependent reactive oxygen species (ROS) generation and subsequent modulation of MAP kinase phosphatase redox status. ROS generation induced by the commensal bacteria Lactobacillus rhamnosus GG and the FPR peptide ligand, N-formyl-Met-Leu-Phe, was abolished in the presence of selective inhibitors for G protein-coupled signaling and FPR ligand interaction. In addition, pretreatment of cells with inhibitors of ROS generation attenuated commensal bacteria-induced ERK signaling, indicating that ROS generation is required for ERK pathway activation. Bacterial colonization also led to oxidative inactivation of the redox-sensitive and ERK-specific phosphatase, DUSP3/VHR, and consequent stimulation of ERK pathway signaling. Together, these data demonstrate that commensal bacteria and their products activate ROS signaling in an FPR-dependent manner and define a mechanism by which cellular ROS influences the ERK pathway through a redox-sensitive regulatory circuit.
Keywords: Cell Proliferation, Epithelial Cell, ERK, Mouse, Reactive Oxygen Species (ROS), Commensal Bacteria, Dual Specific Phosphatase 3, Formyl Peptide Receptor, Lactobacillus rhamnosus GG, fMLF
Introduction
The mammalian intestinal epithelium coexists in intimate contact with up to 1014 prokaryotic organisms that comprise the gut microbiota (1, 2). This prokaryotic community thrives in a generally symbiotic fashion with the host, contributing to vitamin and micronutrient synthesis, simulation of immune development and function, extraction of calories from otherwise indigestible complex carbohydrates, and competitive exclusion of pathogens (3, 4). Recently, direct microbial effects on intrinsic epithelial processes have been described. For example, commensal bacteria were shown to augment intestinal barrier function and stimulate reparative responses (5–7). Studies in germ-free mice reveal a slower turnover of intestinal epithelial cells with a marked decrease in crypt-to-villus transit time (8). Due to their therapeutic potential, commensal bacteria have also recently been used as oral supplements (probiotics) for the treatment of inflammatory and developmental disorders in the intestinal tract (9). However, the mechanism by which the microbiota engage in cross-talk with the intestinal epithelia to mediate gut homeostatic events is not well understood.
Bacteria signal to the intestinal epithelia through transmembrane or intracellular pattern recognition receptors, which recognize conserved bacterial structural motifs, known as microbial associated molecular patterns. A class of newly characterized epithelial pattern recognition receptors is the formyl peptide receptors (FPRs).3 FPRs are membrane-bound receptors that bind prokaryotic translation products modified with the bacteria-specific amino acid N-formyl methionine, N-formyl peptides, or fMLF (10–12). Ligand-bound FPRs undergo phosphorylation resulting in conformational changes that recruits the Gi family of G proteins. FPR activation within professional phagocytes initiates signaling pathways that (i) influence actin dynamics including chemotaxis, (ii) induce the transcription of inflammatory effectors and cytokines, and (iii) lead to the activation of NADPH oxidase enzymes that result in ROS production (respiratory burst). Recently, FPRs have been characterized on nonphagocyte cell types, including intestinal epithelia (13). Our research group has shown that commensal bacteria signal directly through FPRs to activate the MAPK ERK signaling pathway without promoting the proinflammatory IκB or proapoptotic JNK pathways (14). Additionally, we have shown that commensal bacteria can down-regulate intestinal epithelial proinflammatory signaling through cellular ROS generation (6). However, it is unknown whether epithelial cells generate FPR-dependent ROS production in response to fMLF recognition (analogous to events well known in phagocytes) or whether ROS plays a role in homeostatic signaling.
The physiological generation of ROS by nonphagocyte NADPH oxidase enzymes (NOXs) regulates diverse cellular processes through transient oxidative inactivation of catalytic cysteine residues on a spectrum of regulatory enzymes (15–17). A well studied class of redox-sensitive regulatory enzymes is the dual specific phosphatases (DUSPs) (18). DUSPs are a subset of protein-tyrosine phosphatases that dephosphorylate threonine and tyrosine residues in the consensus motif Thr-Xaa-Tyr (where Xaa is Glu, Gly, or Pro) on MAPKs ERK, JNK, and p38. Once MAPKs are dephosphorylated, the signaling cascade becomes inactive. Conversely, when the DUSP catalytic cysteines are oxidatively inactivated through localized ROS production, DUSP phosphatase activity is reversibly inhibited, leading to sustained activation of MAPKs.
Here, we show that a member of the commensal microbiota, and a bacterial product, activate FPR-dependent ROS generation in the intestinal epithelia and that generated ROS is sufficient to modulate ERK pathway signaling through the oxidative inactivation of DUSP3. These findings indicate that commensal bacteria induce ROS generation through intestinal FPRs and identify a mechanism by which the microbiota influence intestinal epithelial homeostatic signaling.
EXPERIMENTAL PROCEDURES
Reagents
H2O2 and N-acetylcysteine (NAC) were purchased from Sigma-Aldrich. MG-262 was purchased from Biomol (Plymouth Meeting, PA). Pertussis toxin (PTx) was purchased from Calbiochem, and the FPR antagonist N-tert-butoxycarbonyl-Met-Leu-Phe (Boc2) was purchased through MP Biomedicals (Aurora, OH).
Cell Culture
Human intestinal epithelial cell line SK-CO15 was grown in high glucose (4.5 g/liter) DMEM supplemented with 10% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, 15 mm HEPES (pH 7.4), 2 mm l-glutamine, and 1% nonessential amino acids as described previously (13) at 37 °C in a 4% CO2 incubator.
ROS Detection
Epithelial cells were treated with Lactobacillus rhamnosus GG (LGG; 5 × 107 cfu/ml) (ATCC 53103) or fMLF (500 nm) (Sigma-Aldrich) for the indicated times were washed with HBSS and incubated in the dark with 5 mm CM-H2DCF-DA (Molecular Probes) for an additional 5 min as described previously (6). To evaluate whether inhibitors of G protein-coupled receptors or FPRs could block observed ROS generation, cells were pretreated with NAC (20 mm), diphenyliodinium (DPI) (40 mm), PTx (1 μg/ml), or Boc2 (100 ng/ml) 30 min prior to LGG or fMLF treatment. All images were acquired using a confocal laser scanning microscope (Zeiss LSM 510) at ×20 magnification. Images were captured using 488 nm laser for excitation and a 515–540 nm emission filter. Quantification of fluorescence intensity was determined using ImageJ (National Institutes of Health, Bethesda, MD) software with treatments at 30 min.
For detection of ROS in mice, food and water of 6–8-week-old C57BL/6 mice were removed, then mice were anesthetized and injected intrarectally with hydrocyanine3 (7.5 μm) 1 h prior to administration of PBS, LGG, or fMLF for 7 min. Mice were euthanized, and tissues were removed for analysis. The colon was opened along the mesenteric border, placed in 4% paraformaldehyde for 20 min, and processed for confocal laser scanning microscopy at ×20 magnification. Images were captured using a 535 nm laser for excitation and 560 nm emission filter. Quantification of fluorescence intensity was determined using ImageJ software.
Immunoblotting
Antibodies were obtained as follows: anti DUSP3, anti-myc, and phospho-ERK (Cell Signaling, Danvers, MA); β-actin (Sigma-Aldrich); and HRP-conjugated donkey anti-rabbit or sheep anti-mouse secondary antibody (GE Healthcare). Immunoblotting was performed as described previously (19). DNA was stained with To-Pro-3 iodide (Molecular Probes). Fluorescent images were acquired by laser confocal microscopy at ×63 magnification. Prior to the detection of endogenous DUSP3, cells were incubated with MG-262, a proteosome inhibitor.
Reporter Gene Assays
SK-CO15 cells were transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For luciferase reporter assays, cells were transfected with ERK-dependent Elk1 reporter plasmids (Luciferase Trans-Reporting Systems; Stratagene) according to the manufacturer's instructions. Following cell treatment, cells were lysed in reporter lysis buffer (Promega), and luciferase activity was determined using the Dual Luciferase Reporter Assay System (Promega).
Quantitative RT-PCR Analysis
Total RNA was extracted from either cultured SK-CO15 cells or colonic epithelial scrapings from three mice using TRIzol reagent (Invitrogen). Reverse transcription reactions were performed using the QuantiTect Reverse Transcription kit (Qiagen) and PCRs undertaken using QuantiTect SYBR Green PCR kit (Qiagen). Primers used for PCRs include 18 S (forward) 5′-gtaacccgttgaaccccatt-3′, 18 S (reverse) 5′-ccatccaatcggtagtagcg-3′, hDUSP3 (forward) 5′-taaaaaccccaccatttgga-3′, and hDUSP3 (reverse) 5′-cttcctgcttgtcttctgg-3′. DUSP3 gene expression was standardized against 18 S transcript levels in the same sample, and experimental results were recorded as -fold increase relative to measurements in PBS-treated samples. PCRs were performed in triplicate using two separate RNA preparations for each data point.
Plasmids and Constructs
The DUSP3 open reading frame was cloned into pCMV-myc to create pCMV-myc-DUSP3. A catalytically inactive form of DUSP3 was created (mDUSP3) where the cysteine residue at position 124 is replaced by an alanine which is reported to render DUSP3 catalytically inactive.
Analysis of Protein Oxidation
DUSP3 redox state was monitored by electrophoretic mobility shift on 10% SDS-PAGE under nonreducing conditions. Cell lysates were prepared using a buffer containing 20 mm Tris-HCl (pH 7.4), 1% Triton X-100, 10 mm N-ethylmaleimide (NEM), and Complete Mini protease inhibitor mixture (Roche Applied Science). Lysates were separated by SDS-PAGE in the presence or absence of β-mercaptoethanol and immunoblotted using myc or DUSP3 antibodies (Cell Signaling).
Mice
All murine experimental procedures were undertaken according to the Emory University guidelines for ethical treatment of animals. For analysis of colonic tissue by intrarectal treatment, the food and water of 6–8-week-old B6 mice were removed 4 h prior to anesthetization, then intrarectal administration of PBS, LGG, or fMLF for 7 or 30 min. Mice were euthanized, and tissues were removed for analysis. The colon was opened along the mesenteric border and placed in 4% paraformaldehyde for 20 min, and subsequent colon whole mounts were prepared as described below. For control experiments, mice were systemically administered 1 μg/ml PTx via intraperitoneal injection for 18 h prior to LGG treatment. For the fMLF peptidomimetic control, mice were intrarectally administered 100 μg/ml Boc2 through a soft catheter 30 min prior to LGG treatment.
Colon Whole Mount Preparation
Dissected murine tissues were fixed for 20 min in 4% paraformaldehyde, washed in PBS, permeabilized with 0.1% Triton X-100 for 5 min, and washed again. Samples were blocked in 5% normal goat serum for 1 h before incubation with rabbit anti-phospho-ERK (Cell Signaling) for 1 h at 37 °C (or overnight at 4 °C) and then with FITC-conjugated goat anti-rabbit IgG (Invitrogen). Cellular actin was stained with Alexa Fluor Phallodin-633 (Molecular Probes). Tissue was cut into 2-mm to 5-mm pieces, mounted on slides, and visualized by laser confocal microscopy at ×63 magnification.
Reproducibility and Data Presentation
p values ≤ 0.05 using Student's t test were considered significant. The results of statistical analyses are given in the figure legends.
RESULTS
fMLF and Commensal Bacteria Induce FPR-dependent ROS Generation in Cultured Human Intestinal Cells or in Vivo in Murine Colonic Epithelia
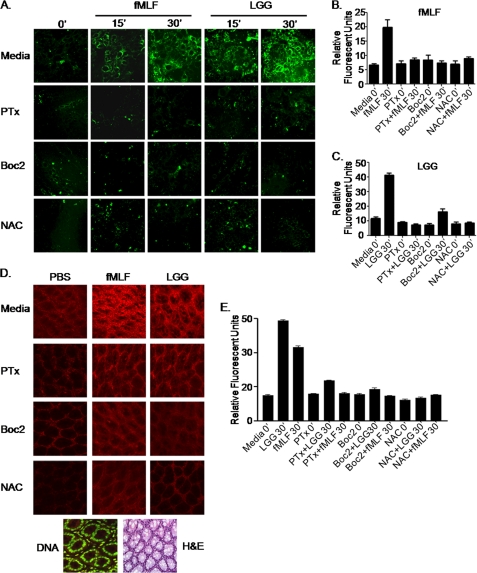
fMLF is a classic inducer of FPR-dependent ROS generation in neutrophils (10–12, 20). We investigated the extent to which commensal bacteria or fMLF activated FPRs situated on the apical surface of epithelial cells and mediated an analogous ROS response. As a candidate commensal bacterium, we used LGG, a member of the human microbiota and commonly used probiotic agent. Polarized cultured SK-CO15 colonic epithelial cells were treated with either fMLF or LGG for up to 30 min, and ROS generation was detected by oxidation of the redox sensitive CM-H2DCF-DA dye as reported previously (6). ROS was rapidly generated in fMLF- or LGG-treated cells within 15 min of contact and peaked at 30 min (data past 30 min not shown) (Fig. 1A). We confirmed these results using another oxidation-sensitive dye, dihydroethidine and show that ROS generation is inhibited by the flavoprotein inhibitor DPI (supplemental Fig. 1). ROS was detected in the cytoplasm of contacted cells, whereas no ROS was evident in the nucleus. The levels of ROS generated by application of fMLF or LGG were comparable with those generated in response to contact with 250 μm H2O2 (supplemental Fig. 2). Importantly, both fMLF- and LGG-induced ROS generation was abrogated in cells pretreated with Boc2, a competitive inhibitor of fMLF binding to FPRs (21, 22), and by PTx, an inhibitor the Gαi subunit of G protein-coupled receptors (23) (Fig. 1A). Furthermore, pretreatment of the stimulated cells with NAC, a glutathione (GSH) precursor and ROS sink, also abrogated fMLF- and LGG-induced ROS generation. ROS was quantified by densitometry of panels in Fig. 1A (Fig. 1, B and C). To investigate whether commensal bacteria-induced ROS generation occurs in vivo, we pretreated mice with a newly developed redox-sensitive hydrocyanine dye that fluoresces when oxidized (24). Intrarectal administration of either LGG or fMLF rapidly induced ROS generation in colonic enterocytes (Fig. 1D). H&E or DNA staining of tissues similar to those analyzed for ROS generation show the colonic architecture of the examined areas (Fig. 1D). ROS generation detected in tissues shown in Fig. 1D was also quantified by densitometry (Fig. 1E). Consistent with responses seen in vitro, pretreatment of the murine colon with PTx, Boc, or NAC also inhibited LGG- or fMLF-induced ROS generation in vivo (Fig. 1E). Together, these data demonstrate that fMLF and LGG induce comparable physiological levels of FPR-dependent ROS generation in contacted cells.
FIGURE 1.
LGG and fMLF induce the generation of ROS in cultured epithelial cells in an FPR-dependent manner. A, CM-H2DCF-DA (5 μm)-mediated detection of ROS in SK-CO15 cells treated with fMLF (500 nm) or LGG (5 × 107 cfu/ml) over 30 min. B, quantitative representation of ROS production in A for fMLF at 30 min. C, quantitative representation of ROS production in A for LGG at 30 min. D, hydrocyanine3 (7.5 μm)-mediated detection of ROS in murine colonic enterocytes treated with fMLF (500 nm) or LGG (5 × 107 cfu/ml) for 7 min. Fluorescence was measured at ×40 magnification by confocal laser scanning microscopy (Zeiss). DNA was stained with Syto 24 and H&E sections for tissue orientation. E, quantitative representation of ROS production in D. For B, C, and E, data are representative of three independent assays quantified with ImageJ software and are expressed in units of fluorescence. Error bars, S.E.
Dampening of Cellular ROS Levels Attenuates LGG- and fMLF-induced ERK Pathway Activation and Cellular Proliferation
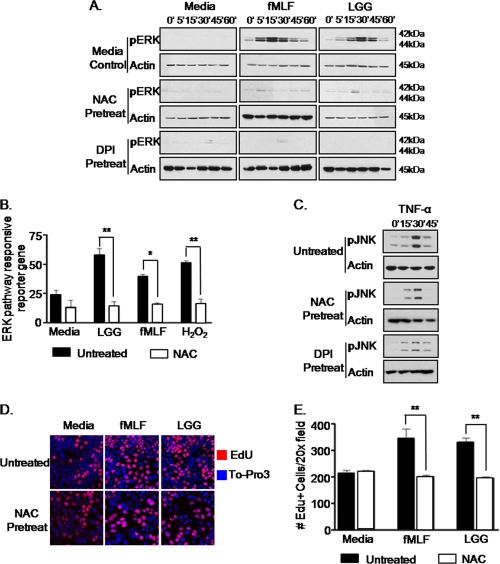
We reported previously that contact of the apical surface of enterocytes by commensal bacteria or fMLF specifically activated the ERK signaling pathway (14), and here we confirm that L. rhamnosus or fMLF does not induce inflammatory cytokine production, whereas treatment of cultured cells with Salmonella or TNF potently induces high levels of IL-8 (supplemental Fig. 3). Having shown that enterocytes also generate ROS in response to contact by fMLF and LGG, we next investigated the extent to which physiological levels of ROS influenced ERK signaling. Cultured SK-CO15 cells were treated with NAC or the NADP(H) oxidase inhibitor, DPI, prior to treatment with LGG or fMLF. Strikingly, whereas ERK is rapidly phosphorylated in response to LGG of fMLF, no ERK phosphorylation was detected in NAC- or DPI-treated cells, indicating a function for ROS in LGG- or fMLF-induced ERK signaling (Fig. 2A). We also investigated ERK pathway activation in the presence of NAC using a transfection-based ERK pathway-dependent reporter assay. Consistent with the results from immunoblot analysis, pretreatment of cells with NAC inhibited LGG- or fMLF-induced ERK pathway-responsive transcriptional activation (Fig. 2B). Importantly, neither NAC nor DPI treatment inhibited TNF-α-induced JNK phosphorylation (Fig. 2C), demonstrating that TNF-α-induced signaling is not ROS-dependent. We also reported previously that apical contact of polarized SK-CO15 cultured cells by LGG or fMLF induced cellular proliferation (14). To determine whether ROS generated in response to contact by LGG and fMLF mediates these proliferative events, we assayed for EdU incorporation in cultured SK-CO15 cells pretreated with the antioxidant NAC before stimulation with either LGG or fMLF. We observed that antioxidant pretreatment significantly reduced the number of EdU-positive cells induced by LGG and fMLF cellular contact (Fig. 2, D and E), thus implicating a role for commensal bacteria-induced ROS generation in mediating cellular proliferation and/or homeostatic events in the gut.
FIGURE 2.
Dampening of cellular ROS levels attenuates LGG- or fMLF-induced ERK pathway activation and cellular proliferation. A, immunoblot analysis for phospho-ERK in cultured SK-CO15 cells treated with NAC (20 μm) or DPI (40 μm) 30 min prior to stimulation with LGG (5 × 107 cfu/ml) or fMLF (500 nm) up to 1 h. B, ERK pathway-specific luciferase reporter gene assay from transfected SK-CO15 cells treated with NAC (20 μm) 30 min prior to LGG (5 × 107 cfu/ml), fMLF (500 nm), or H2O2 (1 mm) stimulation.*, p < 0.05; **, p < 0.001. C, immunoblot analysis for phospho-JNK in cultured SK-CO15 cells treated with NAC (20 μm) or DPI (40 μm) 30 min prior to stimulation with TNF-α (1 ng/ml) up to 30 min. D, EdU incorporation into cultured SK-CO15 cells treated with NAC (20 μm) prior to incubation for 12 h with LGG (5 × 107 cfu/ml) or fMLF (500 nm). Blue, To-Pro-3 for DNA; red, EdU for proliferation. Confocal microscope images were recorded at ×63 magnification. E, quantitative representation of EdU-positive cells in C. Shown is the number of EdU-positive cells/10 fields of view at ×20 for three replicates/treatment. *, p < 0.05. Error bars, S.E.
LGG and fMLF Up-regulate DUSP3 mRNA and Protein Levels
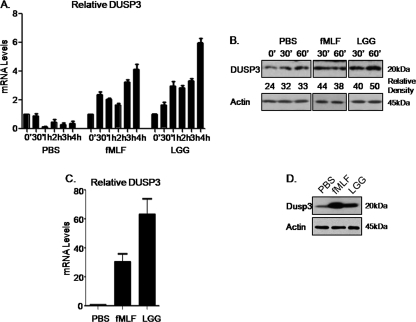
ROS has been shown to modulate cellular signaling events via the oxidation of hyperreactive cysteine residues within a subset of regulatory enzymes (15). A well studied example is the family of MAPK phosphatases or DUSPs, which dephosphorylate MAPK tyrosine and threonine residues, thus rendering MAPKs inactive (18). Using microarray analysis, our research group showed previously that LGG treatment in the 2-week-old murine intestine induced the transcriptional up-regulation of DUSP3, which is known to specifically dephosphorylate ERK and thus inhibit ERK pathway signaling (25). As a first step toward understanding the relationship between LGG/fMLF and DUSP3, we confirmed DUSP3 up-regulation in treated SK-CO15 cells by both quantitative PCR analysis (Fig. 3A) and immunoblot detection for endogenous DUSP3 (Fig. 3B). We also confirmed these results in vivo by quantitative PCR and immunoblot analyses of intestinal scrapings from mice treated intrarectally with LGG or fMLF. Results confirm increased DUSP3 mRNA and total protein levels in LGG- and fMLF-treated murine colonic epithelium (Fig. 3, C and D). Together, these data indicate that intestinal epithelial DUSP3 is up-regulated in response to whole commensal bacteria or formyl peptides, concurrent with ERK phosphorylation and pathway stimulation, plausibly acting as a negative feedback loop for the down-regulation of ERK pathway signaling. Importantly, because DUSP proteins are sensitive to cellular redox levels, DUSP3 may be a target for ROS modulation of ERK signaling.
FIGURE 3.
LGG and fMLF up-regulate DUSP3 mRNA and protein levels. A, quantitative RT-PCR analysis of DUSP3 mRNA levels in cultured SK-CO15 cells stimulated with LGG (5 × 107 cfu/ml) or fMLF (500 nm) for 30 min. PCRs were performed in triplicate using two separate RNA preparations for each data point. Error bars represent S.E. B, immunoblot analysis for total DUSP3 in cultured SK-CO15 cells stimulated with LGG (5 × 107 cfu/ml) or fMLF (500 nm) up to 1 h. C, quantitative RT-PCR analysis of DUSP3 mRNA levels in mouse colonic epithelial scrapings treated in vivo with 100 μl LGG (107 cfu/ml) or fMLF (500 nm) for 30 min. PCRs were performed in triplicate using two separate RNA preparations for each data point. Error bars represent S.E. D, immunoblot analysis for total DUSP3 in mouse colonic epithelial cell scrapings treated in vivo with 100 μl of LGG (107 cfu/ml) or fMLF (500 nm) for 30 min.
LGG- and fMLF-induced Cellular ROS Oxidizes DUSP3
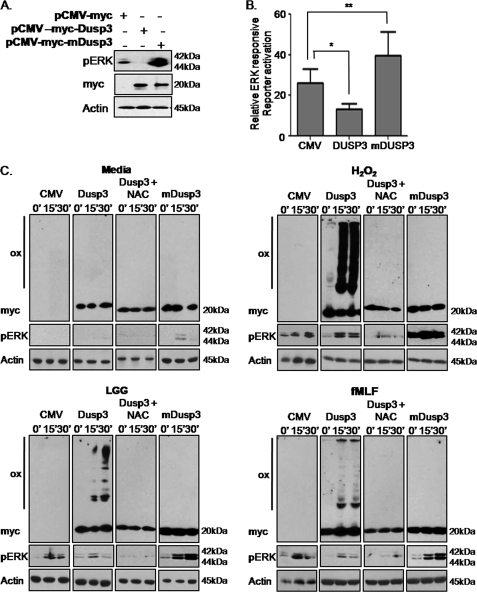
We then sought to establish the mechanism by which LGG- or fMLF-induced cellular ROS influences ERK signaling circuitry. Thus far, we have established that ROS is generated within minutes of LGG or fMLF contact with epithelial cells and that ensuing events include the phosphorylation of the ERK and the up-regulation of ERK-responsive gene, DUSP3. Following pathway activation, DUSP3 is known to catalyze the dephosphorylation of phosphorylated ERK, serving as a negative feedback loop (18). Absent inducing stimuli, DUSP3 is expressed at basal levels that are sufficient to suppress overt ERK pathway activation. Importantly, the phosphatase activity of DUSP3 is dependent on a critical cysteine at position 124. This conserved residue within the DUSP family is readily oxidized in response to elevated cellular redox conditions, rendering DUSP proteins, including DUSP3, catalytically inactive and unable to suppress MAPK pathway activity (26). We thus hypothesized that the rapid generation of ROS in response to LGG and fMLF contact leads to the oxidation and transient inactivation of DUSP3, thus facilitating ERK phosphorylation and pathway activation. To test this hypothesis, we transfected cultured SK-CO15 cells with plasmids harboring either wild-type DUSP3 or a catalytically inactive mutant form of DUSP3 (mDUSP3), where the critical cysteine residue at position 124 was replaced by serine, and assessed its ability to dephosphorylate ERK. Whereas wild-type DUSP3 potently repressed ERK phosphorylation markedly below basal levels, introduction of mDUSP3, presumably acting as a dominant negative, resulted in dramatically elevated levels of phosphorylated ERK (Fig. 4A). Consistent with the immunoblot analysis, wild-type DUSP3 was effective in suppressing basal ERK pathway activity, whereas the mutant form served to augment ERK-dependent gene transcription (Fig. 4B). Next, we evaluated DUSP3 or mDUSP3 oxidation in response to elevated cellular ROS levels following treatment with LGG, fMLF, or H2O2 (control). The oxidation status of DUSP3 was examined by immunoblot analysis using nonreducing SDS-PAGE as described previously (26). These conditions allow detection of higher molecular mass disulfide dimer and aggregate forms. Untreated DUSP3 and mDUSP3 migrate at 20 kDa (Fig. 4C), whereas LGG, fMLF, H2O2 treatment resulted in a characteristic band shift indicative of rapid DUSP3 oxidation and formation of higher order aggregates (Fig. 4C). Importantly, no band shift was detected in lysates prepared from cells transfected with plasmids expressing mDUSP3, where the redox-sensitive cysteine 124 residue is mutated to serine. This confirms that the critical cysteine residue on DUSP3 required for its phosphatase activity is responsive to cellular ROS induced by bacterial products (Fig. 4C). These findings are further supported in studies where pretreatment of transfected cultured cells with NAC before treatment with LGG or fMLF markedly reduced the levels of oxidized DUSP3, thus indicating that DUSP3 oxidation is a direct result of LGG- and fMLF-induced ROS generation (Fig. 4C). In all cases, the appearance of oxidized forms of DUSP3 correlated with ERK phosphorylation, whereas mDUSP3 exhibited superbasal levels of ERK phosphorylation. Importantly, when the reducing agent β-mercaptoethanol was added to treated cell lysates, all high molecular mass bands were abolished, indicating that the slow migrating forms are likely mixed disulfide bonds between DUSP3 and other cellular proteins (supplemental Fig. 4). Together, these data show that LGG- or fMLF-induced ROS generation modulates the dynamic interaction between DUSP3 and its target substrate, phosphorylated ERK.
FIGURE 4.
LGG- or fMLF-induced generation of ROS oxidizes DUSP3. A, SK-CO15 cultured cells transfected with vector control or plasmids expressing DUSP3 or mDUSP3 assayed by immunoblotting for basal levels of phospho-ERK. B, ERK-responsive luciferase reporter gene assay for basal levels of ERK stimulation from SK-CO15 cells transfected with vector control or plasmids expressing DUSP3 or mDUSP3.*, p < 0.05; **, p < 0.001. Error bars, S.E. C, SK-CO15 cultured cells transfected with vector control or plasmids expressing DUSP3 or mDUSP3 or DUSP3 treated with NAC (20 μm) 30 min prior to stimulation with LGG (5 × 107 cfu/ml) or fMLF (500 nm) up to 30 min. Lysates were then assayed for DUSP3 oxidation status by immunoblotting for myc in nonreducing conditions or phospho-ERK by immunoblotting in reducing conditions. All cells were lysed in a buffer containing 10 mm NEM to prevent oxidation of cysteines during sample preparation. DUSP3 oxidation was monitored by changes in electrophoretic mobility. Ox, oxidized DUSP3. myc, reduced DUSP3.
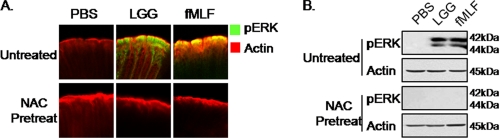
Antioxidant Pretreatment Inhibits LGG- and fMLF-induced Phosphorylation of ERK in Murine Enterocytes
We previously reported that LGG or fMLF induced the phosphorylation of ERK in murine enterocytes (14). To show that this activity is dependent on LGG- or fMLF-induced ROS generation in the murine colon, we pretreated mice intrarectally with NAC before stimulation with LGG or fMLF for 7 min. Strikingly, LGG- or fMLF-induced ERK phosphorylation was completely abrogated in the colonic epithelium of NAC-pretreated mice, compared with untreated control mice where ERK is phosphorylated within minutes of contact by LGG or fMLF (Fig. 5A). To corroborate these observations, colonic epithelial scrapings from these mice were examined by immunoblot analysis using an antibody specific for phosphorylated ERK. Consistent with the immunofluorescent analysis, NAC pretreatment markedly reduced LGG- and fMLF-induced phosphorylation of ERK compared with nonpretreated samples (Fig. 5B). Collectively, these data show that fMLF and LGG induce ERK via FPR-dependent redox modulation of DUSP3 in the murine colon.
FIGURE 5.
Antioxidant pretreatment inhibits LGG- or fMLF-induced phosphorylation of ERK in murine enterocytes. A, immunofluorescence of phospho-ERK within intestinal whole mount preparations (as described under “Experimental Procedures”) in either media- or NAC (20 μm)-pretreated intestinal mucosa 30 min prior to treatment with 100 μl of LGG (107 cfu/ml) or fMLF (500 nm) for 7 min. B, immunoblot analysis for phospho-ERK in mouse colonic epithelial cell scrapings pretreated with NAC (20 μm) 30 min prior to in vivo treatment with 100 μl LGG (107 cfu/ml) or fMLF (500 nm) for 7 min.
DISCUSSION
The intestinal mucosa has evolved mechanisms to perceive and respond to bacteria, including the resident microbiota and the occasional enteric pathogen. For example, microbe-induced signals from epithelial Toll-like receptors and intracytoplasmic Nod proteins are well known to mediate proinflammatory and cytoprotective cellular responses and are vital in the host defense against pathogens. In this work, we describe a mechanism by which commensal bacteria can stimulate important non-proinflammatory eukaryotic signaling pathways that influence proliferative or homeostatic pathways, rather than defense responses. We recently reported that epithelial cells can perceive commensal bacteria via FPRs located on the surface of epithelial cells (14), a function that has been reported extensively in professional phagocytic cells. We also reported that ligand binding to FPRs in intestinal enterocytes induced the specific activation of ERK pathway signaling and stimulation of cellular proliferation (14). Here, we assess whether other aspects of FPR-mediated signaling reported in phagocytes are also functionally important in epithelial cells.
Previous reports by our research group showed that commensal bacteria and their soluble fermentation products stimulated ROS generation within intestinal epithelial cells (6, 27). This process may be analogous to the activation of high levels of ROS in professional phagocytes stimulated with FPR ligands (respiratory burst). The FPR-dependent respiratory burst in professional phagocytes is mediated via the gp91phox complex (NOX2) in response to microorganisms or formylated peptides. Colonic epithelial cells also express NOX1 and both DUOX proteins (28). We thus speculated that ROS within epithelial cells may be generated via the same mechanism. Here, we demonstrate that commensal bacterial contact, or fMLF contact with cultured cells or in vivo epithelium, results in the FPR-dependent generation of ROS and that pharmacological inhibition of NADP(H) oxidase by DPI attenuates this response. These data identify NOX1 and/or DUOX as the enzymes that generate commensal bacteria-induced ROS in colonic epithelial cells. Additionally, we show that inhibition of ROS generation also inhibits commensal bacteria- or fMLF-induced phosphorylation of ERK. Together, these data implicate ROS in the signaling events that mediate commensal bacteria-induced ERK pathway signaling.
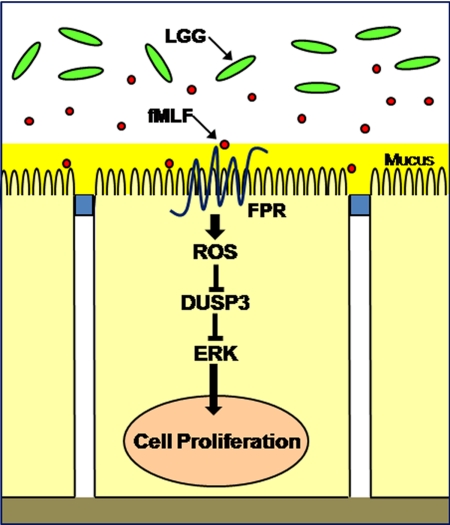
The generation of ROS as a response to bacteria and their products is highly conserved and is widely represented in plants and lower metazoans (29–32). For example, plants purposefully generate intracellular ROS as signaling molecules to control various processes, including pathogen defense, programmed cell death, and stomatal behavior. Interestingly, in both plants and lower metazoans, ROS-producing enzymes and their regulation by GTPases appear to be evolutionarily conserved mechanisms for ROS generation (30). It is also thought that many ROS are short lived, highly localized molecules, with a small radius of action, which may allow for specific targeting of particular signaling pathways. ROS signaling is transduced by a subset of sensor enzymes that are transiently inactivated by reversible oxidation of catalytic cysteine residues within the active sites (33). Such enzymes include a variety of tyrosine phosphatases such as PTEN, antioxidants such as thioredoxins and peroxiredoxins, and members of the Ubc family of proteins (6). Our laboratory established that bacterially elicited ROS transiently inactivate Ubc12, which normally neddylates the Cul1 subunit of the ubiquitin ligase complex targeting IκB-α for proteosomal degradation. With IκB-α no longer targeted for degradation, NF-κB remains trapped in the cytosol, unable to translocate to the nucleus to activate transcription of inflammatory mediators (6), resulting in inflammatory suppression. Additionally, we have shown commensal generated ROS augment cell motility by oxidative inactivation of the tyrosine phosphatases LMW-PTP and SHP-2, which are responsible for desphosphorylation of FAK, thus allowing sustained activation of FAK, formation of focal adhesions, and subsequent enhancement of in vitro and in vivo wound healing (34). These studies represent a body of data directly implicating ROS as signaling molecules in intestinal homeostasis. Here, we show commensal-induced ROS is also crucial for sustaining ERK signaling and subsequent cellular proliferative events by modulating DUSP3 activity. DUSP3, also known as the human Vaccinia H1-related (VHR) phosphatase (35, 36), is a 185-amino acid (20-kDa) protein identified based on its homology with the Vaccinia virus H1 open reading frame. Recently, DUSP3 was shown to be important in the control of cell growth and differentiation (37). We show that commensal bacteria-induced generation of ROS can lead to the direct oxidative inactivation of DUSP3 enzymatic activity, thus relieving DUSP3-mediated suppression of ERK pathway signaling (Fig. 6). Together, our data identify a molecular mechanism by which commensal bacteria directly activate a homeostatic signaling pathway in the mammalian intestine.
FIGURE 6.
Model for commensal bacteria-induced ERK pathway signaling via FPR-dependent redox modulation of DUSP3.
Characterization of the molecular mechanisms by which commensal bacteria mediate beneficial effects on health and disease is important for advancing the application of these microbes in preventative or therapeutic medical approaches. In fact, supplementation of the normal microbiota with exogenous bacteria, “probiotics,” has already been shown to result in promising therapeutic benefits. Specifically, probiotic therapy decreases inflammatory responses, augments barrier function, and increases epithelial proliferation (7). Probiotic preparations are clinically indicated for the treatment of ulcerative colitis and postsurgical pouchitis (38, 39) and the prevention of necrotizing enterocolitis (25). Additionally, abnormal composition of the microbiota, “dysbiotic flora,” is implicated in the pathogenesis of inflammatory bowel disease and potentially other systemic immune disorders. We show that some of the beneficial effects attributed to probiotics, and of a normal microbiota, are potentiated by FPR-dependent and ROS-mediated modulation of cellular signaling pathways. Characterization of FPRs as receptors that mediate commensal bacterial signaling to intestinal epithelial cells, as well as identifying the signal pathways within intestinal epithelial cells that are modified by commensal bacteria, will contribute to a better understanding of the symbiotic role of the microbiota and the potential benefits of probiotics.
Acknowledgment
We thank Jeffrey Mercante for advice during manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK071604, DK089763, and AI064462.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- FPR
- formyl peptide receptor
- Boc2
- N-tert-butoxycarbonyl-Met-Leu-Phe
- DPI
- diphenyliodinium
- DUSP
- dual-specific phosphatase
- fMLF
- N-formyl-Met-Leu-Phe
- LGG
- Lactobacillus rhamnosus GG
- NAC
- N-acetylcysteine
- NOX
- NADPH oxidase enzyme
- PTx
- pertussis toxin
- ROS
- reactive oxygen species
- EdU
- 5-ethynyl-2′-deoxyuridine.
REFERENCES
- 1. Gill S. R., Pop M., Deboy R. T., Eckburg P. B., Turnbaugh P. J., Samuel B. S., Gordon J. I., Relman D. A., Fraser-Liggett C. M., Nelson K. E. (2006) Science 312, 1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu J., Mahowald M. A., Ley R. E., Lozupone C. A., Hamady M., Martens E. C., Henrissat B., Coutinho P. M., Minx P., Latreille P., Cordum H., Van Brunt A., Kim K., Fulton R. S., Fulton L. A., Clifton S. W., Wilson R. K., Knight R. D., Gordon J. I. (2007) PLoS Biol. 5, e156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hooper L. V., Bry L., Falk P. G., Gordon J. I. (1998) Bioessays 20, 336–343 [DOI] [PubMed] [Google Scholar]
- 4. Neish A. S. (2009) Gastroenterology 136, 65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park J., Floch M. H. (2007) Gastroenterol. Clin. North Am. 36, 47–63, v [DOI] [PubMed] [Google Scholar]
- 6. Kumar A., Wu H., Collier-Hyams L. S., Hansen J. M., Li T., Yamoah K., Pan Z. Q., Jones D. P., Neish A. S. (2007) EMBO J. 26, 4457–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin P. W., Myers L. E., Ray L., Song S. C., Nasr T. R., Berardinelli A. J., Kundu K., Murthy N., Hansen J. M., Neish A. S. (2009) Free Radic. Biol. Med. 47, 1205–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hooper L. V., Gordon J. I. (2001) Science 292, 1115–1118 [DOI] [PubMed] [Google Scholar]
- 9. Pull S. L., Doherty J. M., Mills J. C., Gordon J. I., Stappenbeck T. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Migeotte I., Communi D., Parmentier M. (2006) Cytokine Growth Factor Rev. 17, 501–519 [DOI] [PubMed] [Google Scholar]
- 11. Rabiet M. J., Huet E., Boulay F. (2007) Biochimie 89, 1089–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ye R. D., Boulay F., Wang J. M., Dahlgren C., Gerard C., Parmentier M., Serhan C. N., Murphy P. M. (2009) Pharmacol. Rev. 61, 119–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Babbin B. A., Jesaitis A. J., Ivanov A. I., Kelly D., Laukoetter M., Nava P., Parkos C. A., Nusrat A. (2007) J. Immunol. 179, 8112–8121 [DOI] [PubMed] [Google Scholar]
- 14. Wentworth C. C., Jones R. M., Kwon Y. M., Nusrat A., Neish A. S. (2010) Am. J. Pathol. 177, 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Forman H. J., Maiorino M., Ursini F. (2010) Biochemistry 49, 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Janssen-Heininger Y. M., Mossman B. T., Heintz N. H., Forman H. J., Kalyanaraman B., Finkel T., Stamler J. S., Rhee S. G., van der Vliet A. (2008) Free Radic. Biol. Med. 45, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Foreman J., Demidchik V., Bothwell J. H., Mylona P., Miedema H., Torres M. A., Linstead P., Costa S., Brownlee C., Jones J. D., Davies J. M., Dolan L. (2003) Nature 422, 442–446 [DOI] [PubMed] [Google Scholar]
- 18. Cerignoli F., Rahmouni S., Ronai Z., Mustelin T. (2006) Cell Cycle 5, 2210–2215 [DOI] [PubMed] [Google Scholar]
- 19. Collier-Hyams L. S., Zeng H., Sun J., Tomlinson A. D., Bao Z. Q., Chen H., Madara J. L., Orth K., Neish A. S. (2002) J. Immunol. 169, 2846–2850 [DOI] [PubMed] [Google Scholar]
- 20. Torres M., Hall F. L., O'Neill K. (1993) J. Immunol. 150, 1563–1577 [PubMed] [Google Scholar]
- 21. Gavins F. N., Yona S., Kamal A. M., Flower R. J., Perretti M. (2003) Blood 101, 4140–4147 [DOI] [PubMed] [Google Scholar]
- 22. Babbin B. A., Lee W. Y., Parkos C. A., Winfree L. M., Akyildiz A., Perretti M., Nusrat A. (2006) J. Biol. Chem. 281, 19588–19599 [DOI] [PubMed] [Google Scholar]
- 23. Bokoch G. M., Gilman A. G. (1984) Cell 39, 301–308 [DOI] [PubMed] [Google Scholar]
- 24. Kundu K., Knight S. F., Willett N., Lee S., Taylor W. R., Murthy N. (2009) Angew Chem. Int. Ed. Engl. 48, 299–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin P. W., Nasr T. R., Berardinelli A. J., Kumar A., Neish A. S. (2008) Pediatr. Res. 64, 511–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. (2005) Cell 120, 649–661 [DOI] [PubMed] [Google Scholar]
- 27. Kumar A., Wu H., Collier-Hyams L. S., Kwon Y. M., Hanson J. M., Neish A. S. (2009) J. Immunol. 182, 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lambeth J. D. (2004) Nat. Rev. Immunol. 4, 181–189 [DOI] [PubMed] [Google Scholar]
- 29. Ha E. M., Oh C. T., Ryu J. H., Bae Y. S., Kang S. W., Jang I. H., Brey P. T., Lee W. J. (2005) Dev. Cell 8, 125–132 [DOI] [PubMed] [Google Scholar]
- 30. Kotchoni S. O., Gachomo E. W. (2006) J. Biosci. 31, 389–404 [DOI] [PubMed] [Google Scholar]
- 31. Pauly N., Pucciariello C., Mandon K., Innocenti G., Jamet A., Baudouin E., Hérouart D., Frendo P., Puppo A. (2006) J. Exp. Bot. 57, 1769–1776 [DOI] [PubMed] [Google Scholar]
- 32. Tanaka A., Christensen M. J., Takemoto D., Park P., Scott B. (2006) Plant Cell 18, 1052–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reddie K. G., Carroll K. S. (2008) Curr. Opin. Chem. Biol. 12, 746–754 [DOI] [PubMed] [Google Scholar]
- 34. Swanson P. A., 2nd, Kumar A., Samarin S., Vijay-Kumar M., Kundu K., Murthy N., Hansen J., Nusrat A., Neish A. S. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 8803–8808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guan K. L., Broyles S. S., Dixon J. E. (1991) Nature 350, 359–362 [DOI] [PubMed] [Google Scholar]
- 36. Liu K., Lemon B., Traktman P. (1995) J. Virol. 69, 7823–7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rahmouni S., Cerignoli F., Alonso A., Tsutji T., Henkens R., Zhu C., Louis-dit-Sully C., Moutschen M., Jiang W., Mustelin T. (2006) Nat. Cell Biol. 8, 524–531 [DOI] [PubMed] [Google Scholar]
- 38. Hart A. L., Stagg A. J., Kamm M. A. (2003) J. Clin. Gastroenterol. 36, 111–119 [DOI] [PubMed] [Google Scholar]
- 39. Kruis W., Fric P., Pokrotnieks J., Lukás M., Fixa B., Kascák M., Kamm M. A., Weismueller J., Beglinger C., Stolte M., Wolff C., Schulze J. (2004) Gut 53, 1617–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]