Background: DUSP9/MKP-4 dephosphorylates and inactivates MAP kinases.
Results: The kinase interaction motif of DUSP9/MKP-4 is phosphorylated by protein kinase A, and this reduces its ability to interact with and inactivate MAP kinase substrates.
Conclusion: DUSP9/MKP-4 represents a point of cross-talk between PKA and MAP kinase signaling.
Significance: Cross-talk between distinct signal transduction pathways may be important in determining physiological response.
Keywords: MAP Kinases (MAPKs), p38 MAPK, Protein Kinase A (PKA), Protein Phosphatase, Protein Phosphorylation, Signal Transduction
Abstract
MAP kinase phosphatase 4 (DUSP9/MKP-4) plays an essential role during placental development and is one of a subfamily of three closely related cytoplasmic dual-specificity MAPK phosphatases, which includes the ERK-specific enzymes DUSP6/MKP-3 and DUSP7/MKP-X. However, unlike DUSP6/MKP-3, DUSP9/MKP-4 also inactivates the p38α MAP kinase both in vitro and in vivo. Here we demonstrate that inactivation of both ERK1/2 and p38α by DUSP9/MKP-4 is mediated by a conserved arginine-rich kinase interaction motif located within the amino-terminal non-catalytic domain of the protein. Furthermore, DUSP9/MKP-4 is unique among these cytoplasmic MKPs in containing a conserved PKA consensus phosphorylation site 55RRXSer-58 immediately adjacent to the kinase interaction motif. DUSP9/MKP-4 is phosphorylated on Ser-58 by PKA in vitro, and phosphorylation abrogates the binding of DUSP9/MKP-4 to both ERK2 and p38α MAP kinases. In addition, although mutation of Ser-58 to either alanine or glutamic acid does not affect the intrinsic catalytic activity of DUSP9/MKP-4, phospho-mimetic (Ser-58 to Glu) substitution inhibits both the interaction of DUSP9/MKP-4 with ERK2 and p38α in vivo and its ability to dephosphorylate and inactivate these MAP kinases. Finally, the use of a phospho-specific antibody demonstrates that endogenous DUSP9/MKP-4 is phosphorylated on Ser-58 in response to the PKA agonist forskolin and is also modified in placental tissue. We conclude that DUSP9/MKP-4 is a bona fide target of PKA signaling and that attenuation of DUSP9/MKP-4 function can mediate cross-talk between the PKA pathway and MAPK signaling through both ERK1/2 and p38α in vivo.
Introduction
DUSP9/MKP-44 is one of a group of three cytoplasmic dual-specificity MAP kinase phosphatases that includes the prototypic ERK-specific phosphatase DUSP6/MKP-3 and DUSP7/MKP-X (1–3). However, unlike DUSP6/MKP-3, which is highly specific in its ability to interact with and inactivate the classical ERK1/2 MAPKs (4, 5), DUSP9/MKP-4 can also interact with and inactivate p38α MAPK both in vitro and in vivo, indicating that this phosphatase regulates both mitogen and stress-activated MAPK signaling (6, 7). Deletion of the murine Dusp9 gene causes embryonic lethality at mid-gestation. This is caused by placental insufficiency due to a failure in labyrinth development, and this end point correlates exactly with the spatial and temporal pattern of DUSP9/MKP-4 expression in the normal placenta (8). Despite a failure to observe gross changes in the levels of either ERK1/2 or p38α phosphorylation in placental tissues lacking DUSP9/MKP-4, disruption of either p38α or MEK1, an upstream activator of ERK1/2, gives rise to placental defects almost identical to those seen on loss of DUSP9/MKP-4 (9, 10). This indicates that the abnormal regulation of either or both of these MAPK pathways likely contributes to placental failure in animals lacking DUSP9/MKP-4.
Tetraploid rescue experiments, which bypass the lethality caused by deletion of DUSP9/MKP-4, give rise to animals that develop normally with no obvious phenotype and are fertile. Furthermore, despite high levels of DUSP9/MKP-4 expression in the developing liver, adult kidney, and testis, these tissues develop normally in embryos lacking DUSP9/MKP-4, indicating that the essential function of this phosphatase is restricted to the extraembryonic tissues (8). More recently, DUSP9/MKP-4 has been implicated in the regulation of insulin signaling in murine models of obesity and stress-induced insulin resistance (11, 12). A possible link between this gene and susceptibility to type 2 diabetes in humans is also suggested by the recent identification of a type 2 diabetes risk locus near DUSP9 in a genome-wide association study, the first such locus to be identified on the X chromosome (13).
MKPs recognize and bind their cognate MAPK substrates through an arginine-rich kinase interaction motif (KIM) located in the amino-terminal non-catalytic domain of the protein. Furthermore, MAPK binding via this motif causes conformational changes at the active site of the enzyme leading to catalytic activation of MKPs in vitro (1, 2). Consistent with its activity toward both ERK2 and p38α, DUSP9/MKP-4 interacts with both of these MAPKs in yeast two-hybrid assays, and the catalytic activity of DUSP9/MKP-4 toward para-nitrophenyl phosphate (p-NPP) in vitro is increased on incubation with either recombinant ERK2 or p38α (6, 14). Here we demonstrate that the ability of DUSP9/MKP-4 to recognize both ERK1/2 and p38α is mediated by a conserved KIM comprising essential arginine residues at positions 52 and 53. Furthermore, DUSP9/MKP-4 is unique among the cytoplasmic MKPs in containing a conserved cAMP-dependent protein kinase (PKA) consensus phosphorylation site (Ser-58) immediately carboxyl-terminal to the KIM. This site can be modified by PKA in vitro, and phosphorylation abolishes the interaction of the amino-terminal domain of DUSP9/MKP-4 with both ERK2 and p38α in glutathione S-transferase (GST) pulldown assays. Consistent with a role for this modification in regulating the activity of DUSP9/MKP-4 toward these MAPKs in vivo, phospho-mimetic (glutamic acid) substitution of Ser-58 also reduces the ability of DUSP9/MKP-4 to inactivate both ERK1/2 and p38α MAPKs when expressed in mammalian cells. Finally, using a phospho-specific antibody raised against this site, we demonstrate that endogenous DUSP9/MKP-4 is modified on Ser-58 in response to the PKA agonist forskolin and also in murine placental tissue in vivo. We conclude that DUSP9/MKP-4 is a bona fide target of PKA signaling and that attenuation of DUSP9/MKP-4 function can mediate cross-talk between the PKA pathway and both mitogen- and stress-activated MAPK signaling.
EXPERIMENTAL PROCEDURES
Reagents
Forskolin was purchased from Sigma. Okadaic acid was obtained from Calbiochem. Monoclonal antibodies against myc and hemagglutinin (HA) were from Cancer Research UK. Antibodies against phospho-ERK, ERK, phospho-p38, p38, protein kinase A catalytic subunit (PKAc), and phospho-cyclic AMP response element-binding protein (CREB)/activating transcription factor 1 (ATF1) were purchased from Cell Signaling Technology. The anti-tubulin antibody was purchased from Santa Cruz. The sheep polyclonal antiserum (#302) raised against murine DUSP9/MKP-4 and the anti-GST rabbit polyclonal antibody have been described previously (6, 15). All cell culture reagents were from Invitrogen.
Bacterial and Yeast Strains
Escherichia coli DH5α and Rosetta DE3 were from Novagen. Saccharomyces cerevisiae strains PJ69-4A and PJ69-4α (16) were used for two-hybrid assays. Maintenance, propagation, and transformation were all performed according to standard methods (17).
DNA Constructs
The plasmids pGADT7.ERK2, pGADT7.JNK1, pGADT7.p38α, pSG5.ERK2-HA, pSG5.p38α-HA, pSG5.mDUSP9/MKP-4-myc, and pGEX5X constructs encoding GST alone and GST-PTP-SL-(147–288) have been described previously (6, 18, 19). Lenti-UBC-PKA-CQR encoding a constitutively active mutant of PKA under the control of the ubiquitin C promoter (20) was kindly provided by Anthony Zeleznik (University of Pittsburgh). The human DUSP9/MKP-4 cDNA was cloned by PCR amplification using a human kidney cDNA library (Clontech) as template using primers MKP-4 forward (5′-ATGAATTCATATGGAGGGTCTGGGCCGCTCG-3′) and MKP-4 reverse (5′-ATCTCGAGCTAGGTGGG GGCCAGCTCGAAGG-3′). To clone the human DUSP9/MKP-4 ORF into a modified pSG5 (Stratagene) vector allowing in-frame fusion to a carboxyl-terminal myc epitope tag, the reverse primer MKP-4-SG5 5′-ATCTCGAGGGTGGGGGCCAGCTCGAAGGCG-3′ was used. This replaces the stop codon with an in-frame XhoI site. Reading frames were then subcloned as NdeI-XhoI fragments into pET15B (Novagen) and pGBKT7/pGADT7 (Clontech) vectors and as EcoRI-XhoI fragments into pSG5. DUSP9/MKP-4 ORFs encoding either S58A, S58E, or KIM mutants were generated using the DUSP9/MKP-4 cDNA as template by overlap extension PCR using the following overlapping primer pairs in conjunction with the MKP-4 forward and either the MKP-4 reverse or MKP-4-SG5 external flanking primers: S58A forward (5′-CGCCTGCGGAGGGGCGCCCTGTCGGTGGCG-3′) and reverse (5′-CGCCACCGACAGGGCGCCCCTCCGCAGGCG-3′); S58E forward (5′-CGCCTGCGGAGGGGCGAGCTGTCGGTGGCG-3′) and reverse (5′-CGCCACCGACAGCTCGCCCCTCCGCAGGCG-3′); KIM forward (5′-GCGCTCCTGCTGGCCGCCCTGGCGAGGGGCAGCCTG-3′) and reverse (5′-CAGGCTGCCCCTCGCCAGGGCGGCCAGGAGCGC-3′. Mutant cDNAs were then digested with XmaI and XhoI and subcloned into the appropriate expression vector containing the wild-type DUSP9/MKP-4 ORF predigested with XmaI and XhoI, thus replacing the wild-type DUSP9/MKP-4 sequence with a cDNA fragment containing the appropriate mutation. The murine DUSP9/MKP-4 S58A mutant was made by PCR amplification using the mouse DUSP9/MKP-4 cDNA as a template and the following external flanking primers: MF5 (5′-ATGGATCCATGGAGAGTCTGAGTCGGTCATGC-3′) and MR3 (5′-ATTCTAGATTATGTGGGGTCCAGCTCAAAGAC-3′) in conjunction with primers S58A forward (5′-GCTATGTCGGTGCGGTCGCTCTTG-3′) and S58A reverse (5′-CCCCCTCCGCAGGCGGCGCAGCAT-3′). The two PCR products were ligated and amplified using the flanking external primers. The resulting fragment was then digested with BamHI and XbaI and cloned into pCDNA3-myc predigested with BamHI and XbaI. All DNA constructs were verified by DNA sequencing.
Expression and Purification of Recombinant Proteins
Recombinant histidine-tagged proteins were expressed in the E. coli strain Rosetta DE3. All subsequent purification and refolding steps were performed exactly as described previously (21).
Generation of Antisera
A polyclonal antiserum (#657) was raised in sheep at the Scottish Antibody Production Unit (SAPU, Lanarkshire, UK) using recombinant human DUSP9/MKP-4 protein as antigen. Phospho-specific antibodies recognizing either human or murine DUSP9/MKP-4 phosphorylated on Ser-58 were also raised in sheep (SAPU) using either peptide RLRRGSLSVRA or RLRRGSMSVRS corresponding to residues 53–63 of human or murine DUSP9/MKP-4, respectively. The phosphoserine residue is in bold and underlined. The antisera were affinity-purified as described previously (22).
Protein Phosphatase Assays
Phosphatase activities and catalytic activation of wild-type and mutant forms of DUSP9/MKP-4 were assayed using p-NPP hydrolysis exactly as described previously (23).
In Vitro Phosphorylation and GST Pulldown Assays
Overnight cultures of E. coli transformed with pGEX, pGEX.MKP-4-1–267wt, pGEX.MKP-4-1–267 S58A, or pGEX.PTP-SL-(147–288) were diluted 1:10 in LB plus ampicillin and incubated for a further 2 h at 37 °C before induction with isopropyl 1-thio-β-d-galactopyranoside at either 0.1 m (pGEX and pGEX.PTP-SL-(147–288)) or 0.3 m (pGEX.MKP-4 wt and pGEX.MKP-4 S58A) for 2 h. For in vitro phosphorylation assays, 3-ml (pGEX) or 9-ml (pGEX.MKP-4-1–267wt and pGEX.MKP-4-1–267 S58A) volumes were then centrifuged, and the pellets were resuspended in 300 μl of PBS, 0.1% Triton X-100. Cell suspensions were then sonicated and centrifuged. Supernatants were separated, 50 μl of glutathione-Sepharose (GE Healthcare) was then added, and reactions were incubated with shaking for 15 min at 4 °C. Proteins were then washed 4 times with PBS and once with 1 ml of PKA reaction buffer (40 mm Tris-HCl, pH 7.5, 20 mm MgCl2 10 μm ATP) at 4 °C. Phosphorylation was carried out in a volume of 30 μl of PKA reaction buffer containing 0.65 units/μl PKAc (Promega) and 2.5 μCi of [γ-32P]ATP incubated for 45 min at 30 °C with shaking. Reactions were stopped by the addition of SDS sample buffer and analyzed by SDS-PAGE. Gels were stained with Coomassie Blue before drying and autoradiography.
For GST pulldown assays, E. coli were grown and induced as before. 3-ml (pGEX.PTP-SL-(147–288)) or 9 ml (pGEX.MKP-4-1–267wt and pGEX.MKP-4-1-267 S58A) culture volumes were centrifuged, and the pellets were resuspended in 600 μl of PBS, 0.1% Triton X-100. Cell suspensions were then sonicated and centrifuged, and the supernatants were divided into two aliquots. 30 μl of glutathione-Sepharose was then added to each aliquot, and proteins were incubated for 15 min at 4 °C with shaking. Proteins were washed 4 times with PBS and once with PKA reaction buffer containing 0.2 mm ATP before incubation in 45 μl of PKA reaction buffer containing 0.2 nm ATP either with or without 0.5 units/μl PKAc (Promega) for 3 h at 25 °C with shaking. Proteins were then washed once with PBS at 4 °C. HEK293 cells were lysed in a buffer containing 100 mm Tris-HCl, pH 7.5, 300 mm NaCl, 2% octylphenoxypoly-ethoxyethanol (Igepal), 100 mm PMSF, 0.2 mg/ml aprotinin 0.5 m NaF, 200 mm Na4P2O7. 1.5 ml of this lysate was then added to each reaction and incubated for 2 h at 4 °C with shaking. After incubation, proteins were washed 4 times with HNTG buffer (20 mm Hepes, pH 7.5, 150 mm NaCl, 0.1% Triton X-100, 10% glycerol, 10 mm Na4P2O7) and once with PBS at 4 °C before the addition of SDS sample buffer and analysis by SDS page and Western blotting.
Yeast Two-hybrid Assays
Assays were performed according to the manufacturer's instructions (Matchmaker 3 kit, Clontech) exactly as described previously (18).
Kinase Assays
The activity of wild-type and mutant forms of DUSP9/MKP-4 toward either ERK2 or p38α was determined by co-transfection of COS-1 cells with expression vectors encoding either HA-tagged ERK2 or p38α together with increasing amounts of an expression vector encoding either wild-type or mutant forms of DUSP9/MKP-4. After transfection, cells were exposed to an appropriate activating stimulus (15% fetal calf serum for ERK2 or 10 μg/ml anisomycin for p38α) for 30 min followed by cell lysis. Proteins were then analyzed by SDS-PAGE and Western blotting. HA-tagged MAP kinases were immunoprecipitated before assay of kinase activity toward myelin basic protein exactly as described previously (6).
Cell Culture and Treatments
Immortalized mouse embryo fibroblasts (MEFs) and COS-1 cells were cultured and transfected exactly as described previously (6, 24). To stimulate PKA activity, cells were treated with 10 μm forskolin for 30 min either with or without a 30-min preincubation with 1 μm okadaic acid to inhibit protein phosphatase activity. Cells were then lysed, and proteins were either immunoprecipitated or analyzed directly by SDS-PAGE and Western blotting.
Immunoprecipitation and Analysis of Phospho-DUSP9/MKP-4
Freshly dissected murine placental tissue or cultured MEFs were lysed in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.2 150 mm NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100). Lysates were then sonicated and cleared by centrifugation, and the protein concentration was determined using Bradford reagent (Bio-Rad). Sheep polyclonal antiserum against murine DUSP9/MKP-4 (#302, 3 μg/immunoprecipitation) was incubated with protein G-Sepharose beads (Sigma, 10 μl/immunoprecipitation) overnight at 4 °C in an excess of phosphate-buffered saline (PBS). Beads were then washed with 10 volumes of sodium borate, pH 9.0, at room temperature, and coupling was performed by incubation with dimethyl pimelimidate (20 mm) for 30 min. Reactions were terminated by washing the beads in 0.2 m ethanolamine for 2 h, and any loosely bound IgG was removed by washing briefly in 50 mm glycine, pH 2.5. The pH was corrected with 0.2 m Tris, pH 8.0, and the beads were washed extensively in PBS.
Coupled beads were incubated overnight with 500 μg of placental or MEF lysate at 4 °C with inversion. Beads were then washed four times in radioimmune precipitation assay buffer containing protease and PhosStop phosphatase inhibitors (Roche Applied Science) before the addition of LDS sample buffer (Invitrogen) supplemented with 0.1% β-mercaptoethanol and heating to 95 °C for 15 min. Proteins were analyzed by SDS-PAGE and Western blotting. Murine DUSP9/MKP-4 protein was detected using the sheep polyclonal antiserum (#657) raised against human DUSP9/MKP-4 protein. DUSP9/MKP-4 phosphorylation was monitored using the antiserum raised against the murine DUSP9/MKP-4 Ser-58 phosphopeptide. For phosphatase treatments the final two washes of the immunoprecipitation protocol were not carried out. Instead, all samples were washed twice in 1× PMP metallophosphatase buffer (New England Biolabs) supplemented with 1 mm MnCl2. λ-Protein phosphatase (2000 units) was then added. and the reactions were incubated at 30 °C for 2 h with occasional agitation. After the addition of LDS sample buffer (Invitrogen) supplemented with 0.1% β-mercaptoethanol and heating to 95 °C for 15 min, proteins were analyzed by SDS-PAGE and Western blotting as described above.
RESULTS
A Conserved KIM Mediates the Inactivation of Both ERK1/2 and p38α by DUSP9/MKP-4
Amino acid sequence alignment of DUSP9/MKP-4, DUSP7/MKP-X, and DUSP6/MKP-3 reveals that DUSP9/MKP-4 contains a potential KIM in which two arginine residues (Arg-52 and Arg-53 in DUSP9/MKP-4) essential for the interaction of DUSP6/MKP-3 with ERK1 and -2 are conserved (Fig. 1A). Although mutation of Arg-52, -53, and -55 to alanine does not affect the intrinsic catalytic activity of DUSP9/MKP-4 toward p-NPP in vitro, it does prevent the catalytic activation of DUSP9/MKP-4 by recombinant ERK2 and p38α MAPKs (Fig. 1, B and C), indicating that the KIM mediates a functional interaction with these kinases. To confirm this, we first used a yeast two-hybrid assay to examine the interaction of DUSP9/MKP-4 with ERK2, JNK, and p38α MAPKs. Wild-type DUSP9/MKP-4 clearly interacts with ERK2 and p38α and only weakly with JNK. In contrast, the R52A/R53A/R55A mutant (DUSP9-KIM) shows no significant level of interaction with all three MAPKs (Fig. 1, D and E). These results are consistent with our previous data, which demonstrated that DUSP9/MKP-4 could dephosphorylate both ERK1/2 and p38α but not JNK when expressed in mammalian cells (6). Finally, we transfected increasing amounts of wild type and the KIM mutant of DUSP9/MKP-4 into COS-1 cells and assayed the phosphorylation of both ERK1/2 and p38α by Western blotting. Expression of wild-type DUSP9/MKP-4 significantly reduced the phosphorylation of both ERK1/2 and p38α, whereas the KIM mutant is unable to dephosphorylate either kinase (Fig. 1, F and G). We conclude that the ability of DUSP9/MKP-4 to interact with and dephosphorylate both ERK1/2 and p38α MAPKs in vivo is mediated by a conserved KIM.
FIGURE 1.
A conserved KIM mediates the recognition and inactivation of ERK1/2 and p38α by DUSP9/MKP-4. A, shown is amino acid sequence alignment of the region containing the kinase interaction motif of human (Hs), bovine (Bt), murine (Mm), and rat (Rn) DUSP9/MKP-4 with human (Hs) DUSP6/MKP-3 and DUSP7/MKP-X sequences. Amino acid residues are numbered. Basic residues essential for substrate interaction (KIM) are shaded gray, and the putative PKA consensus motif (RRXS) in DUSP9/MKP-4 is highlighted in bold type. The Ser-58 phospho-acceptor is marked with an asterisk. B, mutation of the KIM does not affect the intrinsic phosphatase activity of DUSP9/MKP-4. Time-dependent hydrolysis of p-NPP by either wild-type (■) or mutant (□) DUSP9/MKP-4 was monitored by measuring the change in optical density (OD) at 405 nm. Assays were performed in triplicate, and mean values with associated errors are shown. C, catalytic activation of DUSP9/MKP-4 is KIM-dependent. Catalytic activation is expressed as -fold increase in the initial rate of p-NPP hydrolysis by 5 μg of recombinant phosphatase toward p-NPP assayed in the presence of 5 μg of the indicated MAP kinase. Assays were performed in triplicate, and mean values with associated errors are shown. D and E, analysis of yeast two-hybrid interactions between DUSP9/MKP-4 or DUSP9/MKP-4 KIM and a panel of mitogen- and stress-activated MAPK isoforms is shown. D, pGBKT7, pGBKT7.DUSP9/MKP-4, and pGBKT7.DUSP9/MKP-4 KIM were transformed into PJ69-4A yeast cells and mated with PJ69-4α yeast cells containing empty pGADT7 or the GAL4 activation domain fusions pGADT7.ERK2, pGADT7.JNK1, or pGADT7.p38α. Yeast diploids expressing both binding domain and activation domain fusions were selected on synthetic dropout media deficient in leucine and tryptophan (−Leu/−Trp). Diploid colonies were resuspended and replated for analysis onto −Leu/−Trp or −Leu/−Trp/−His/−Ade-selective plates. Protein-protein interactions were assessed by growth on this selective medium. E, semiquantitative analysis of the two-hybrid interactions based on the level of induction of the β-galactosidase gene is shown. Assays were performed in triplicate, and mean values with associated errors are shown. F and G, the dephosphorylation of ERK2 and p38α MAPKs by DUSP9/MKP-4 is KIM-dependent. COS-1 cells were transfected with either HA-tagged ERK2 (F) or HA-tagged p38α (G) expression constructs (1 μg of plasmid) together with increasing (0, 100, or 250 ng) amounts of plasmid encoding either Myc-tagged wild-type DUSP9/MKP-4 or DUSP9/MKP-4 KIM mutant. Cells were then either left untreated (−) or stimulated (+) with either serum (F) or anisomycin (G) before lysis and analysis of proteins by SDS-PAGE and Western blotting using the indicated antibodies.
The KIM within DUSP9/MKP-4 Contains a PKA Phosphorylation Site
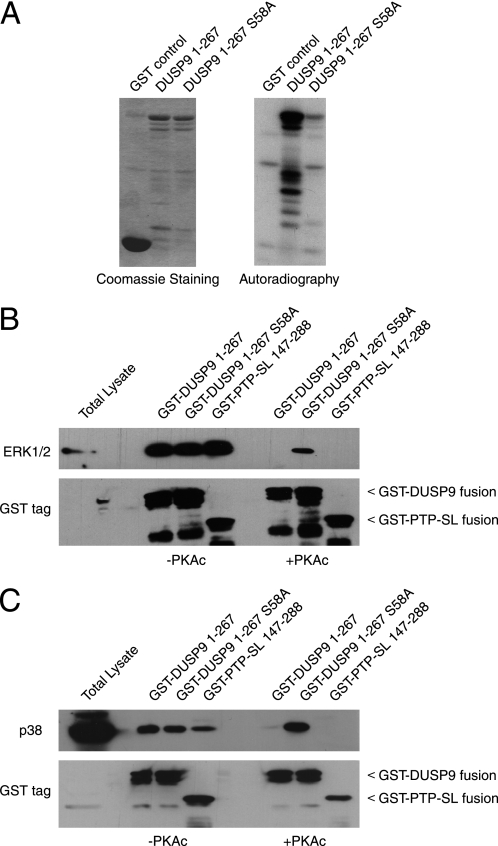
Examination of our amino acid sequence alignment reveals that the KIM within DUSP9/MKP-4 is unique among the three cytoplasmic MKPs in containing a second di-arginine motif. Furthermore, this latter motif forms part of a PKA consensus sequence (55RRXS58) that is conserved in mammals (Fig. 1A). The proximity of this phospho-acceptor to the KIM suggests that modification of Ser-58 might modulate the ability of DUSP9/MKP-4 to interact with MAPK substrates. Interestingly, a subset of protein-tyrosine phosphatases (PTPs), which includes the striatal-enriched phosphatase (STEP/PTPN5), STEP-related phosphatase (PTP-SL/PTPBR7/PTPRR), and the lymphoid-specific phosphatase hematopoietic PTP (He-PTP/LC-PTP/PTPN7) also recognize ERK1/2 and p38α via a conserved KIM (25). Furthermore, in the case of striatal-enriched phosphatase, PTP-SL, and He-PTP, this motif also overlaps with a PKA consensus site, and phosphorylation of this site attenuates the binding of these enzymes to their MAPK substrates (26–28). To explore the possibility that DUSP9/MKP-4 might also be regulated by PKA, we expressed the MAPK binding domain of DUSP9/MKP-4 (residues 1–267) as a GST fusion protein in which the Ser-58 phospho-acceptor was either intact or mutated to alanine. These proteins were then incubated in vitro with PKAc in the presence of [γ-32P]ATP and analyzed by SDS-PAGE and autoradiography. Wild-type GST-DUSP9/MKP-4 1–267 clearly shows high levels of 32P incorporation after incubation with PKAc (Fig. 2A). In contrast, much lower levels of incorporation were seen with either GST alone or the S58A mutant of GST-DUSP9/MKP-4 1–267, demonstrating that this residue is the PKA target. To determine whether this modification might affect the ability of DUSP9/MKP-4 to bind to either ERK1/2 or p38α, GST-DUSP9/MKP-4 fusion proteins were phosphorylated by PKAc in vitro as above in the presence of cold ATP, incubated with Rat-1 cell lysates, and precipitated with glutathione-Sepharose. As a positive control, a GST fusion of the wild-type MAPK binding domain of PTP-SL (residues 147–288) was used. Samples were analyzed by SDS-PAGE, and the presence of the MAP kinases ERK1/2 or p38α was detected by Western blotting. Clearly, all three GST fusion proteins can interact with both ERK and p38α in the absence of PKA. In contrast, incubation of these fusion proteins with PKA in the presence of ATP and Mg2+ abolished the interaction of both wild-type GST-DUSP9/MKP-4 1–267 and GST-PTP-SL-(147–288) with both MAPKs. In contrast, GST-DUSP9/MKP-4 1–267, in which the Ser-58 phospho-acceptor site was mutated to alanine, retained its ability to bind both ERK1/2 and p38α even in the presence of PKA (Fig. 2B). We conclude that Ser-58 can be phosphorylated by PKA in vitro and that this modification can reduce the affinity of the MAPK binding domain of DUSP9/MKP-4 for both ERK1/2 and p38α.
FIGURE 2.
Ser-58 within the amino-terminal domain of DUSP9/MKP-4 is phosphorylated by PKA in vitro, and this prevents binding to both ERK2 and p38α. A, GST alone or GST fusions of either DUSP9/MKP-4 1–267 or DUSP9/MKP-4 1–267 S58A were expressed in bacterial cells, purified from cell lysates, and incubated with recombinant PKA and [γ-32P]ATP in vitro. Proteins were then analyzed by SDS-PAGE and Coomassie Blue staining (left panel) or by autoradiography to show 32P incorporation (right panel). B and C, Ser-58 phosphorylation abolishes interaction with MAP kinases in vitro. GST-DUSP9/MKP-4 1–267, GST-DUSP9/MKP-4 1–267 S58A, or GST-PTP-SL-(147–288) fusion proteins were incubated either in the absence (−PKAc) or presence (+PKAc) of PKA. HEK293 cell lysates were then added and incubated for 2 h, and GST fusion proteins were then pulled down using glutathione-Sepharose before analysis by SDS-PAGE and Western blotting. MAP kinases were detected using antibodies against either ERK1/2 (B) or p38α (C). The presence of GST fusion proteins was verified using an antibody against GST (B and C, lower panels). In the first of each panel, total lysate (20 μg) was loaded.
Modification of Ser-58 Reduces the Affinity of DUSP9/MKP-4 for Both ERK2 and p38α and Leads to Impaired Inactivation of These MAPKs
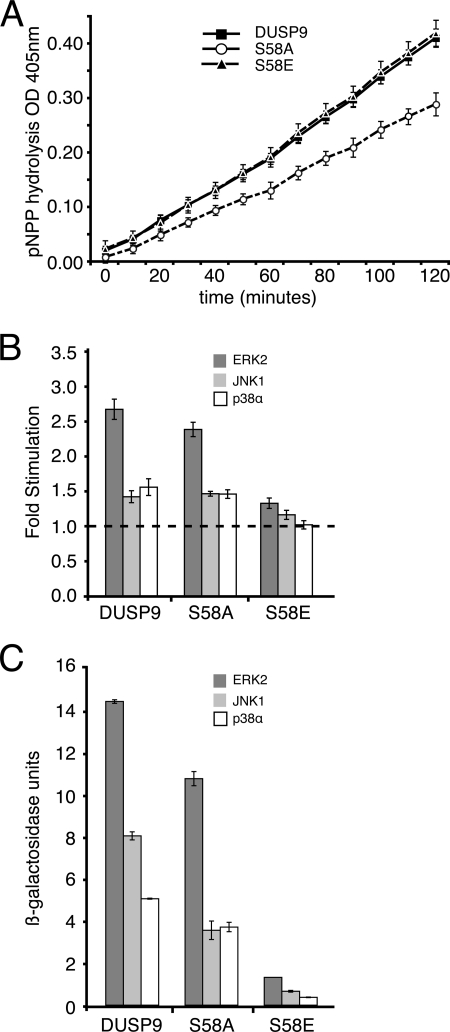
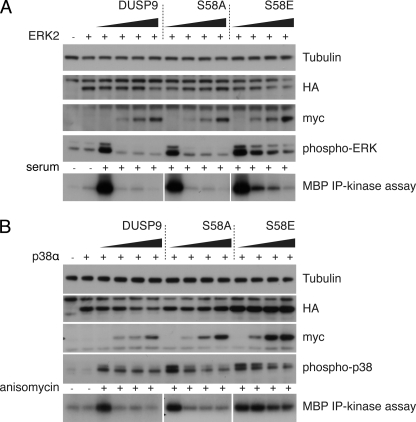
To explore the effects of PKA-mediated phosphorylation of DUSP9/MKP-4 in the context of the full-length protein, we first determined the effects of mutating the Ser-58 phospho-acceptor site to either alanine or glutamic acid on the catalytic activation of DUSP9/MKP-4 by recombinant MAPKs in vitro. Although neither mutation significantly affected the intrinsic catalytic activity of DUSP9/MKP-4 toward p-NPP in vitro, the addition of recombinant MAP kinases to these reactions clearly shows that although the activity of both wild-type and the S58A mutant of DUSP9/MKP-4 is significantly increased on the addition of either ERK2 or p38α, the protein containing the phospho-mimetic S58E mutation shows a much reduced level of catalytic activation in response to these kinases (Fig. 3, A and B). Thus, in agreement with our GST pulldown assays (Fig. 2B), this result indicates that modification of Ser-58 impairs the ability of DUSP9/MKP-4 to interact with both ERK2 and p38α. To explore this in a more physiological context, we first made use of the yeast two-hybrid assay. This clearly shows that although both wild-type DUSP9/MKP-4 and DUSP9/MKP-4 S58A interact with ERK2, JNK and p38α the phospho-mimetic S58E mutant shows a greatly reduced ability to bind to these MAPKs (Fig. 3B). Finally, we compared the ability of wild-type DUSP9/MKP-4 and the Ser-58 mutants to dephosphorylate and inactivate endogenous ERK1/2 and p38α MAPKs when expressed in COS-1 cells. Both wild-type DUSP9/MKP-4 and the S58A mutant efficiently dephosphorylated and inactivated both MAPKs. In contrast, the S58E mutant was significantly less active toward both ERK1/2 and p38α (Fig. 4, A and B). We conclude that phosphorylation of Ser-58 in the context of full-length DUSP9/MKP-4 leads to a reduced binding affinity toward both ERK1/2 and p38α MAPKs, resulting in decreased attenuation of MAPK signaling by this phosphatase.
FIGURE 3.
Phospho-mimetic substitution of Ser-58 blocks the catalytic activation and binding of full-length DUSP9/MKP-4 to ERK2 and p38α. A, mutation of Ser-58 does not affect the intrinsic phosphatase activity of DUSP9/MKP-4. Time-dependent hydrolysis of p-NPP by either wild-type (■), mutant S58A (○), or mutant S58E (▴) DUSP9/MKP-4 was monitored by measuring the change in optical density (OD) at 405 nm. Assays were performed in triplicate, and mean values with the associated errors are shown. B, catalytic activation of DUSP9/MKP-4 is substantially reduced by substitution of Ser-58 for glutamic acid. Catalytic activation is expressed as the -fold increase in the initial rate of p-NPP hydrolysis by 5 μg of recombinant phosphatase toward p-NPP assayed in the presence of 5 μg of the indicated MAP kinase. Assays were performed in triplicate, and mean values with associated errors are shown. C, the binding of DUSP9/MKP-4 to MAPKs is substantially reduced by substitution of Ser-58 for glutamic acid. pGBKT7, pGBKT7.DUSP9/MKP-4, pGBKT7.DUSP9/MKP-4 S58A, or pGBKT7.DUSP9/MKP-4 S58E were transformed into PJ69-4A yeast cells and mated with PJ69-4α yeast cells containing empty pGADT7 or the GAL4 activation domain fusions pGADT7.ERK2, pGADT7.JNK1, or pGADT7.p38α. Semiquantitative analysis of the two-hybrid interactions was based on the level of induction of the β-galactosidase gene. Assays were performed in triplicate, and mean values with associated errors are shown.
FIGURE 4.
Phospho-mimetic substitution of Ser-58 greatly reduces the activity of DUSP9/MKP-4 toward MAPK substrates in COS-1 cells. COS-1 cells were transfected with expression constructs (1 μg of plasmid) encoding either HA-tagged ERK2 (A) or HA-tagged p38α (B) together with increasing (0, 50, 100, or 250 ng) amounts of plasmid encoding either Myc-tagged DUSP9/MKP-4, DUSP9/MKP-4 S58A, or DUSP9/MKP-4 S58E. Cells were then either left untreated (−) or stimulated (+) with either serum (A) or anisomycin (B) before lysis. Proteins were then either analyzed by SDS-PAGE and Western blotting using the indicated antibodies (upper panels), or HA-tagged MAPKs were immunoprecipitated (IP), and kinase activities assayed in the presence of [γ-32P]ATP using myelin basic protein (MBP) as the substrate followed by SDS-PAGE and autoradiography (bottom panels).
Phosphorylation of DUSP9/MKP-4 Is PKA-dependent
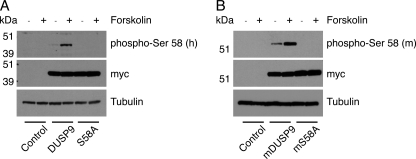
To monitor the phosphorylation of DUSP9/MKP-4 in intact cells, we first raised antibodies against a human DUSP9/MKP-4 KIM phospho-Ser-58 peptide. After affinity purification, this antiserum was able to recognize recombinant wild-type human DUSP9/MKP-4 protein but not mutant DUSP9/MKP-4 S58A protein after incubation with PKAc and ATP in vitro (data not shown). However, this antiserum did not recognize murine DUSP9/MKP-4 under similar conditions. Examination of the amino acid sequence of the KIM peptide in murine DUSP9/MKP-4 revealed four conservative amino acid changes including the substitution of methionine by leucine at the residue immediately carboxyl-terminal to the serine phospho-acceptor. A second antiserum raised against the murine phosphopeptide recognized recombinant murine DUSP9/MKP-4 only after incubation with PKAc and ATP in vitro (data not shown). To verify that DUSP9/MKP-4 could be phosphorylated in vivo, expression vectors encoding either myc-tagged wild type or the S58A mutant of human DUSP9/MKP-4 were transfected into COS-1 cells. After 24 h cells were either left untreated or exposed to the PKA agonist forskolin before analysis by SDS-PAGE and Western blotting. Activation of the PKA pathway clearly results in Ser-58 phosphorylation of wild type but not the S58A mutant of DUSP9/MKP-4 (Fig. 5A). Similar results were obtained on expression of either wild type or the S58A mutant of murine DUSP9/MKP-4 in COS-1 cells using the antibody raised against the murine DUSP9 phosphopeptide (Fig. 5B).
FIGURE 5.
Both human and murine DUSP9/MKP-4 are phosphorylated on Ser-58 in response to forskolin in COS-1 cells. COS-1 cells were transfected with expression constructs (500 ng of plasmid) encoding either myc-tagged wild-type human DUSP9/MKP-4 or DUSP9/MKP-4 S58A (A) or wild-type murine DUSP9/MKP-4 or DUSP9/MKP-4 S58A (B). Cells were either left untreated (−) or stimulated with forskolin (+) before lysis and analysis of proteins by SDS-PAGE and Western blotting using antisera raised against either human (A) or murine-specific Ser-58 (B) phosphopeptides. An antibody against tubulin was used as a loading control, whereas ectopic expression of wild-type and mutant DUSP9/MKP-4 was detected using an anti-myc monoclonal antibody.
Endogenous DUSP9/MKP-4 Is Phosphorylated in Response to PKA Activation
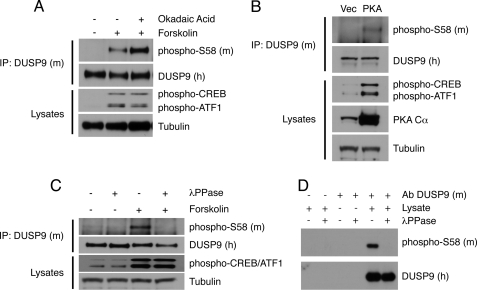
DUSP9/MKP-4 mRNA and protein expression is tissue-specific. The major sites of expression during early development are in the liver and extraembryonic tissues. In contrast, DUSP9/MKP-4 is absent in the adult liver but is found in the testis and kidney (8). A survey of commonly used laboratory cell lines revealed that DUSP9/MKP-4 is expressed in MEFs where both mRNA and protein can be detected by RT-PCR and Western blotting, respectively (data not shown). To establish that endogenous DUSP9/MKP-4 can be phosphorylated on Ser-58, MEFs were either left untreated or exposed to forskolin both in the absence and presence of the serine/threonine phosphatase inhibitor okadaic acid. Cells were lysed, and DUSP9/MKP-4 protein was then immunoprecipitated before analysis by SDS-PAGE and Western blotting using our anti-murine phospho-Ser-58 antiserum. We clearly detected phospho-Ser-58 DUSP9/MKP-4 in cells exposed to forskolin, and this occurred concomitantly with activation of PKA as evidenced by increased phosphorylation of the PKA targets CREB and ATF1. Furthermore, levels of Ser-58 phosphorylation are significantly increased in the presence of okadaic acid, indicating that phosphorylation of this site is subject to regulation by type 1/2 serine/threonine phosphatases (Fig. 6A). Modification of endogenous DUSP9/MKP-4 was also observed when a constitutively active form of the catalytic subunit of PKA (PKA Cα) was expressed in these cells (Fig. 6B), and the incubation of DUSP9/MKP-4 immunoprecipitated from forskolin-treated cells with λ-phosphatase reversed the phosphorylation of Ser-58 (Fig. 6C). Finally, we obtained placentas from mouse embryos at day 13.5 and subjected tissue lysates to immunoprecipitation and Western blotting. We readily detected phospho-Ser-58 DUSP9/MKP-4 in these lysates, and again this signal was lost on incubation of the immunoprecipitates with λ-phosphatase (Fig. 6D). We conclude that endogenous DUSP9/MKP-4 can be modified in response to activation of PKA in cultured cells. Furthermore, this modification is present at significant levels in normal murine extraembryonic tissues, where this phosphatase performs an essential function in regulating MAPK signaling during early development.
FIGURE 6.
Endogenous DUSP9/MKP-4 is phosphorylated on Ser-58 in both MEFs and murine placental tissue. A, immortalized MEFs were either left untreated or exposed to forskolin (10 μm) either in the absence or presence of the phosphatase inhibitor okadaic acid (1 μm). Cells were then lysed, and endogenous DUSP9/MKP-4 was immunoprecipitated (IP) using a polyclonal antibody (#302) against murine DUSP9/MKP-4 followed by SDS-PAGE and Western blotting using a polyclonal antibody (#657) raised against human DUSP9/MKP-4 and an antibody raised against a murine-specific Ser-58 phosphopeptide (upper two panels). Input lysates were also analyzed by SDS-PAGE and Western blotting using antibodies against phospho-CREB/ATF1, or as a loading control, tubulin (lower two panels). B, Ser-58 phosphorylation is enhanced by expression of a constitutively active mutant of PKA. Immortalized MEFs were transfected with either empty vector (Vec) or an expression vector encoding a constitutively active PKA Cα subunit. Cells were then lysed, and endogenous DUSP9/MKP-4 was immunoprecipitated using a polyclonal antibody (#302) against murine DUSP9/MKP-4 followed by SDS-PAGE and Western blotting using a polyclonal antibody (#657) raised against human DUSP9/MKP-4 and an antibody raised against a murine-specific Ser-58 phosphopeptide (upper two panels). Input lysates were also analyzed by SDS-PAGE and Western blotting using antibodies against phospo-CREB/ATF1, PKA Cα, or as a loading control, tubulin (lower three panels). C, immortalized MEFs were either left untreated or exposed to forskolin (10 μm). Cells were then lysed, and endogenous DUSP9/MKP-4 was immunoprecipitated using a polyclonal antibody (#302) against murine DUSP9/MKP-4. Immunoprecipitates were then incubated either in the absence (−) or presence (+) of λ-phosphatase (λPPase) before analysis by SDS-PAGE and Western blotting using a polyclonal antibody (#657) raised against human DUSP9/MKP-4 and an antibody raised against the murine Ser-58 phosphopeptide (upper two panels). Input lysates were also analyzed by SDS-PAGE and Western blotting using antibodies against phospo-CREB/ATF1 and, as a loading control, tubulin (lower two panels). D, endogenous murine DUSP9/MKP-4 was immunoprecipitated from placental lysates using a polyclonal antibody (Ab, #302) against murine DUSP9/MKP-4 with appropriate controls (no antibody and antibody but no lysate), and the samples were then divided into two aliquots and incubated either in the absence (−) or presence (+) of λ-phosphatase (λPPase). Proteins were then analyzed by SDS-PAGE and Western blotting using a polyclonal antibody (#657) raised against recombinant human DUSP9/MKP-4 and an antibody raised against the murine Ser-58 phosphopeptide as indicated.
DISCUSSION
Here we demonstrate that DUSP9/MKP-4 recognizes its cognate MAPK substrates ERK1/2 and p38α via a conserved KIM. However, this docking sequence is unique among this family of dual-specificity MAPK phosphatases in containing a second di-arginine motif that forms part of a conserved consensus site for PKA phosphorylation. We demonstrate that the Ser-58 phospho-acceptor within this motif is efficiently phosphorylated by PKA both in vitro and in vivo and that modification of Ser-58 has the potential to attenuate the interaction of DUSP9/MKP-4 with both ERK1/2 and p38α MAP kinases resulting in a reduced ability of DUSP9/MKP-4 to negatively regulate these signaling pathways. These results strongly suggest that DUSP9/MKP-4 represents a point of cross-talk between the cAMP and MAPK signaling pathways at which PKA activity would positively reinforce signaling through both the classical Ras/ERK1/2 and p38 MAPKs.
The cAMP and MAPK signaling pathways have long been known to interact. Initial observations that cAMP can inhibit proliferation in a variety of cell types can be explained by the ability of PKA to inhibit C-Raf activity (29). Subsequent work has uncovered additional mechanisms of cross-talk between cAMP signaling and the Ras/MAPK pathway by which ERK1/2 activity and cell proliferation are stimulated in response to cAMP. The latter effects appear to be cell type-specific and may also involve modulation of Raf isoform activity but can also be mediated by the interaction of cAMP signaling at multiple levels in MAPK pathways (29). These include the switching of G-protein-coupled receptors from Gs-mediated stimulation of adenyl cyclase to Gi -coupled activation of ERK1/2 (30). Thus the ability of PKA to phosphorylate DUSP9/MKP-4 would represent an additional level of positive interaction between cAMP and MAPK signaling pathways in DUSP9/MKP-4-expressing tissues such as placenta, kidney, and testis.
Although our work identifies DUSP9/MKP-4 as the first member of the dual-specificity MKPs for which substrate binding may be regulated in response to PKA signaling, this mode of regulation has been described in a small subset of PTPs, which selectively target ERK1/2 and p38α in neuronal and hematopoietic tissues (26, 27). To date, the physiological significance of this modification is unclear. The neurotransmitter dopamine leads to the phosphorylation of striatal-enriched phosphatase via activation of D1 receptors and PKA in brain striatal tissue slices (28), and transient phosphorylation of the KIM within He-PTP was observed after T-cell antigen receptor ligation (31). Presumably KIM phosphorylation will affect the ability of these enzymes to inhibit the ERK and p38 MAP kinases, and our results suggest that this mechanism of pathway cross-talk is a conserved, but not universal, feature of protein phosphatases, which are dedicated to the regulation of MAPK signaling pathways in mammalian cells.
Unlike many dual-specificity MKPs, which are mitogen- or stress-inducible genes, DUSP9/MKP-4 mRNA and protein are constitutively expressed in murine fibroblasts. Furthermore, we show that Ser-58 is rapidly modified in the presence of forskolin. Post-translational modification of the KIM would allow the activity of DUSP9/MKP-4 toward its MAPK substrates to be regulated very rapidly without the need for new mRNA and protein synthesis. Furthermore, our observation that PKA-inducible phosphorylation of endogenous DUSP9/MKP-4 on Ser-58 is significantly enhanced in the presence of okadaic acid suggests that this modification might be reversed by a type 2 serine/threonine phosphatase such as PP2A, thus allowing dynamic regulation of DUSP9/MKP-4 activity.
Finally, we have demonstrated that endogenous DUSP9/MKP-4 is constitutively phosphorylated in murine placental tissue where this phosphatase plays an essential role in the regulation of MAPK signaling during early development (8). This suggests that the regulation of DUSP9/MKP-4 substrate binding by KIM phosphorylation represents a physiologically relevant mechanism of regulation. Our phospho-DUSP9/MKP-4-specific antiserum should allow a more extensive analysis of the tissue and cell distribution of phospho-DUSP9/MKP-4 both during development and under physiological conditions where cAMP signaling many influence cell and tissue fate.
Acknowledgment
We thank Dr. Anthony Zeleznik (University of Pittsburgh) for the kind gift of the mammalian expression vector encoding the constitutively active mutant of PKA.
This work was supported in part by Ministerio de Ciencia e Innovación Grant SAF2009-10226, Generalitat Valenciana Grants AP-040/10 and ACOMP/2010/222, Instituto de Salud Carlos III (Spain and Fondo Europeo de Desarrollo Regional ISCIII-RETICS RD06/0020/0049), and EU Research Training Network MRTN-CT-2006-035830 (to R. C.-M.). This work was also supported by Cancer Research UK CR-UK Stress Response Laboratory Programme Grant C8227/A12053 (to S. M. K.).
- DUSP9/MKP-4
- dual specificity phosphatase-9/MAP kinase phosphatase 4
- ATF1
- activating transcription factor 1
- CREB
- cAMP-response element-binding protein
- KIM
- kinase interaction motif
- MEF
- mouse embryo fibroblast; p-NPP, para-nitrophenyl phosphate
- PTP
- protein-tyrosine phosphatase.
REFERENCES
- 1. Owens D. M., Keyse S. M. (2007) Oncogene 26, 3203–3213 [DOI] [PubMed] [Google Scholar]
- 2. Camps M., Nichols A., Arkinstall S. (2000) FASEB J. 14, 6–16 [PubMed] [Google Scholar]
- 3. Dickinson R. J., Keyse S. M. (2006) J. Cell Sci. 119, 4607–4615 [DOI] [PubMed] [Google Scholar]
- 4. Groom L. A., Sneddon A. A., Alessi D. R., Dowd S., Keyse S. M. (1996) EMBO J. 15, 3621–3632 [PMC free article] [PubMed] [Google Scholar]
- 5. Muda M., Theodosiou A., Rodrigues N., Boschert U., Camps M., Gillieron C., Davies K., Ashworth A., Arkinstall S. (1996) J. Biol. Chem. 271, 27205–27208 [DOI] [PubMed] [Google Scholar]
- 6. Dickinson R. J., Williams D. J., Slack D. N., Williamson J., Seternes O. M., Keyse S. M. (2002) Biochem. J. 364, 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muda M., Boschert U., Smith A., Antonsson B., Gillieron C., Chabert C., Camps M., Martinou I., Ashworth A., Arkinstall S. (1997) J. Biol. Chem. 272, 5141–5151 [DOI] [PubMed] [Google Scholar]
- 8. Christie G. R., Williams D. J., Macisaac F., Dickinson R. J., Rosewell I., Keyse S. M. (2005) Mol. Cell. Biol. 25, 8323–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adams R. H., Porras A., Alonso G., Jones M., Vintersten K., Panelli S., Valladares A., Perez L., Klein R., Nebreda A. R. (2000) Mol. Cell 6, 109–116 [PubMed] [Google Scholar]
- 10. Giroux S., Tremblay M., Bernard D., Cardin-Girard J. F., Aubry S., Larouche L., Rousseau S., Huot J., Landry J., Jeannotte L., Charron J. (1999) Curr. Biol. 9, 369–372 [DOI] [PubMed] [Google Scholar]
- 11. Emanuelli B., Eberlé D., Suzuki R., Kahn C. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3545–3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu H., Dembski M., Yang Q., Yang D., Moriarty A., Tayber O., Chen H., Kapeller R., Tartaglia L. A. (2003) J. Biol. Chem. 278, 30187–30192 [DOI] [PubMed] [Google Scholar]
- 13. Voight B. F., Scott L. J., Steinthorsdottir V., Morris A. P., Dina C., Welch R. P., Zeggini E., Huth C., Aulchenko Y. S., Thorleifsson G., McCulloch L. J., Ferreira T., Grallert H., Amin N., Wu G., Willer C. J., Raychaudhuri S., McCarroll S. A., Langenberg C., Hofmann O. M., Dupuis J., Qi L., Segrè A. V., van Hoek M., Navarro P., Ardlie K., Balkau B., Benediktsson R., Bennett A. J., Blagieva R., Boerwinkle E., Bonnycastle L. L., Bengtsson, Boström K., Bravenboer B., Bumpstead S., Burtt N. P., Charpentier G., Chines P. S., Cornelis M., Couper D. J., Crawford G., Doney A. S., Elliott K. S., Elliott A. L., Erdos M. R., Fox C. S., Franklin C. S., Ganser M., Gieger C., Grarup N., Green T., Griffin S., Groves C. J., Guiducci C., Hadjadj S., Hassanali N., Herder C., Isomaa B., Jackson A. U., Johnson P. R., Jørgensen T., Kao W. H., Klopp N., Kong A., Kraft P., Kuusisto J., Lauritzen T., Li M., Lieverse A., Lindgren C. M., Lyssenko V., Marre M., Meitinger T., Midthjell K., Morken M. A., Narisu N., Nilsson P., Owen K. R., Payne F., Perry J. R., Petersen A. K., Platou C., Proença C., Prokopenko I., Rathmann W., Rayner N. W., Robertson N. R., Rocheleau G., Roden M., Sampson M. J., Saxena R., Shields B. M., Shrader P., Sigurdsson G., Sparsø T., Strassburger K., Stringham H. M., Sun Q., Swift A. J., Thorand B., Tichet J., Tuomi T., van Dam R. M., van Haeften T. W., van Herpt T., van Vliet-Ostaptchouk J. V., Walters G. B., Weedon M. N., Wijmenga C., Witteman J., Bergman R. N., Cauchi S., Collins F. S., Gloyn A. L., Gyllensten U., Hansen T., Hide W. A., Hitman G. A., Hofman A., Hunter D. J., Hveem K., Laakso M., Mohlke K. L., Morris A. D., Palmer C. N., Pramstaller P. P., Rudan I., Sijbrands E., Stein L. D., Tuomilehto J., Uitterlinden A., Walker M., Wareham N. J., Watanabe R. M., Abecasis G. R., Boehm B. O., Campbell H., Daly M. J., Hattersley A. T., Hu F. B., Meigs J. B., Pankow J. S., Pedersen O., Wichmann H. E., Barroso I., Florez J. C., Frayling T. M., Groop L., Sladek R., Thorsteinsdottir U., Wilson J. F., Illig T., Froguel P., van Duijn C. M., Stefansson K., Altshuler D., Boehnke M., McCarthy M. I. (2010) Nat. Genet. 42, 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Camps M., Nichols A., Gillieron C., Antonsson B., Muda M., Chabert C., Boschert U., Arkinstall S. (1998) Science 280, 1262–1265 [DOI] [PubMed] [Google Scholar]
- 15. Tárrega C., Ríos P., Cejudo-Marín R., Blanco-Aparicio C., van den Berk L., Schepens J., Hendriks W., Tabernero L., Pulido R. (2005) J. Biol. Chem. 280, 37885–37894 [DOI] [PubMed] [Google Scholar]
- 16. James P., Halladay J., Craig E. A. (1996) Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 18. Slack D. N., Seternes O. M., Gabrielsen M., Keyse S. M. (2001) J. Biol. Chem. 276, 16491–16500 [DOI] [PubMed] [Google Scholar]
- 19. Pulido R., Zúñiga A., Ullrich A. (1998) EMBO J. 17, 7337–7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Escamilla-Hernandez R., Little-Ihrig L., Orwig K. E., Yue J., Chandran U., Zeleznik A. J. (2008) Mol. Endocrinol 22, 1842–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Keyse S. M., Emslie E. A. (1992) Nature 359, 644–647 [DOI] [PubMed] [Google Scholar]
- 22. Perander M., Aberg E., Johansen B., Dreyer B., Guldvik I. J., Outzen H., Keyse S. M., Seternes O. M. (2008) Biochem. J. 411, 613–622 [DOI] [PubMed] [Google Scholar]
- 23. Dowd S., Sneddon A. A., Keyse S. M. (1998) J. Cell Sci. 111, 3389–3399 [DOI] [PubMed] [Google Scholar]
- 24. Staples C. J., Owens D. M., Maier J. V., Cato A. C., Keyse S. M. (2010) J. Biol. Chem. 285, 25928–25940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keyse S. M. (2000) Curr. Opin. Cell Biol. 12, 186–192 [DOI] [PubMed] [Google Scholar]
- 26. Blanco-Aparicio C., Torres J., Pulido R. (1999) J. Cell Biol. 147, 1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saxena M., Williams S., Taskén K., Mustelin T. (1999) Nat. Cell Biol. 1, 305–311 [DOI] [PubMed] [Google Scholar]
- 28. Paul S., Snyder G. L., Yokakura H., Picciotto M. R., Nairn A. C., Lombroso P. J. (2000) J. Neurosci. 20, 5630–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dumaz N., Marais R. (2005) FEBS J. 272, 3491–3504 [DOI] [PubMed] [Google Scholar]
- 30. Baillie G. S., Sood A., McPhee I., Gall I., Perry S. J., Lefkowitz R. J., Houslay M. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 940–945 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31. Nika K., Hyunh H., Williams S., Paul S., Bottini N., Taskén K., Lombroso P. J., Mustelin T. (2004) Biochem. J. 378, 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]