Background: The pathology of Marfan syndrome is caused by insufficient fibrillin-1 microfibril formation in connective tissues.
Results: Successful improvement of Marfan syndrome manifestations are induced by the direct administration of recombinant ADAMTSL6β.
Conclusion: This study demonstrated critical importance of microfibril regeneration in preventing Marfan syndrome.
Significance: Our current data support a new concept that the regeneration of microfibrils using ADAMTSL6β is essential for improving Marfan syndrome.
Keywords: Adenovirus, Connective Tissue, Development, Extracellular Matrix Proteins, Fibrillin, Genetic Diseases, ADAMTSL
Abstract
Marfan syndrome (MFS) is a systemic disorder of the connective tissues caused by insufficient fibrillin-1 microfibril formation and can cause cardiac complications, emphysema, ocular lens dislocation, and severe periodontal disease. ADAMTSL6β (A disintegrin-like metalloprotease domain with thrombospondin type I motifs-like 6β) is a microfibril-associated extracellular matrix protein expressed in various connective tissues that has been implicated in fibrillin-1 microfibril assembly. We here report that ADAMTSL6β plays an essential role in the development and regeneration of connective tissues. ADAMTSL6β expression rescues microfibril disorder after periodontal ligament injury in an MFS mouse model through the promotion of fibrillin-1 microfibril assembly. In addition, improved fibrillin-1 assembly in MFS mice following the administration of ADAMTSL6β attenuates the overactivation of TGF-β signals associated with the increased release of active TGF-β from disrupted fibrillin-1 microfibrils within periodontal ligaments. Our current data thus demonstrate the essential contribution of ADAMTSL6β to fibrillin-1 microfibril formation. These findings also suggest a new therapeutic strategy for the treatment of MFS through ADAMTSL6β-mediated fibrillin-1 microfibril assembly.
Introduction
Marfan syndrome (MFS)2 is a severe, systemic disorder of connective tissue formation and can lead to aortic aneurysms, ocular lens dislocation, emphysema, bone overgrowth, and severe periodontal disease (1–3). MFS has an estimated prevalence of 1 in 5,000–10,000 individuals (3, 4). Fibrillin-1 comprises one of the major insoluble extracellular matrix components in connective tissue microfibrils and provides limited elasticity to tissues through fibrillin-1 microfibril formation (5, 6). Various mouse models of MFS have been established via gene targeting or missense mutations, with germ line mutations in fibrillin-1 leading to progressive connective tissue destruction due to fibrillin-1 fragmentation in association with an insufficiency of fibrillin-1 microfibril formation (7–10). Hence, it is largely accepted that MFS is caused by insufficient fibrillin-1 microfibril formation in various connective tissues (11, 12).
A variety of MFS therapies have been developed, including surgical therapy for aortic root aneurysms that are life-threatening (12), traditional medical therapies, such as β-adrenergic receptor blockade, for slow aortic growth and to decrease the risk of aortic dissection, and novel approaches based on new insights, such as the pathogenesis of insufficient fibrillin-1 microfibril formation and deregulation of TGF-β activation (2). It has been demonstrated also that deregulation of TGF-β activation contributes to MFS pathogenesis and that matrix sequestration of TGF-β is critical for the regulated activation and signaling of the extracellular fibrillin-1 microfibrils of connective tissues (8). These observations predict that the clinical features of MFS-like manifestations are caused by alterations in TGF-β signaling networks (13). Loeys-Dietz syndrome, which is caused by heterozygous mutations in the genes encoding TGF-β receptors 1 and 2, is another autosomal dominant disorder with MFS-like manifestations, such as aortic root aneurysms, aneurysms and dissections throughout the arterial tree, and generalized arterial tortuosity (4). Importantly, systemic antagonism of TGF-β signaling through the administration of a TGF-β-neutralizing antibody or losartan, an angiotensin II type 1 receptor blocker, has been shown to have a beneficial effect on alveolar septation and muscle hypoplasia (8, 10). These observations provide a proof of principle for the use of TGF-β antagonism is in a general therapeutic strategy for MFS and other disorders of the TGF-β signaling network. However, another potential therapeutic strategy that remains to be investigated is the reconstruction of the microfibril in connective tissues through the expression or administration of a microfibril-associated molecule that regulates or stabilizes fibrillin-1 microfibril formation. To investigate this concept, it will be necessary to identify molecular mechanisms of microfibril formation and an appropriate fibrillin-1 microfibril-associated molecule.
ADAMTSL (A disintegrin-like metalloprotease domain with thrombospondin type I motifs-like) is a subgroup of the ADAMTS superfamily that shares particular protein domains with the ADAMTS protease, including thrombospondin type I repeats, a cysteine-rich domain, and an ADAMTS spacer, but lacks the catalytic and disintegrin-like domains (14). A recent study has demonstrated that ADAMTSL2 mutations cause geleophysic dysplasia, an autosomal recessive disorder similar to MFS, through the dysregulation of TGF-β signaling (15). A homozygous mutation in ADAMTSL4 also causes autosomal recessive isolated ectopia lentis, another disease similar to MFS that is characterized by the subluxation of the lens as a result of disruption of the zonular fibers (16). The novel ADAMTSL family molecules ADAMTSL6α and -6β were recently identified by in silico screening for novel ECM proteins produced from a mouse full-length cDNA data base (FANTOM). These proteins are localized in connective tissues, including the skin, aorta, and perichondrocytes. Among the ADAMTSL6 family, ADAMTSL6β has been shown to associate with fibrillin-1 microfibrils through its direct interaction with the N-terminal region of fibrillin-1 and thereby promotes fibrillin-1 matrix assembly in vitro and in vivo (17). These findings suggest a potential clinical application of ADAMTSL6β as a novel MFS therapy by promoting fibrillin-1 microfibril assembly and regulating TGF-β activation.
In our current study, we report that ADAMSL6β is essential for the development and regeneration of the connective tissue periodontal ligament (PDL), a tooth-supporting tissue located between the root and alveolar bone that is morphologically similar to the ligament tissue that is capable of withstanding mechanical force. Using mgR/mgR mice as an animal model of MFS microfibril disorder, we demonstrate that ADAMSL6β expression can rescue fibrillin-1 microfibril formation through the promotion of fibrillin-1 microfibril assembly. PDL provides a useful experimental model not only for investigating the molecular pathogenesis of MFS but also for evaluating novel therapeutic strategies for the improvement of microfibril disorders. This is because the principal elastic fiber system of PDL is composed of fibrillin-1 microfibrils and does not contain significant amounts of elastin (18–20). This composition also suggests that PDL will have an increased susceptibility to breakdown in MFS compared with other elastic tissues composed of both elastin and fibrillin-1. Furthermore, the restoration of fibrillin-1 assembly following administration of recombinant ADAMTSL6β regulates the overactivation of TGF-β signaling, which is associated with an increased release of active TGF-β from disrupted fibrillin-1 microfibrils. The results of our present study demonstrate for the first time that ADAMTSL6β is essential for fibrillin-1 microfibril formation and suggest a novel therapeutic approach to the treatment of MFS through the promotion of ADAMTSL6β-mediated fibrillin-1 microfibril assembly.
EXPERIMENTAL PROCEDURES
Animals
C57BL/6 mice were purchased from CLEA Japan, Inc. (Tokyo, Japan). mgR/mgR mice were generously provided by Dr. Francesco Ramirez (Mount Sinai Medical Center, New York). All mouse care and handling conformed to the National Institutes of Health guidelines for animal research. All experimental protocols were approved by the Tokyo University of Science Animal Care and Use Committee.
Histochemical Analysis
Frontal sections of C57BL mouse heads at embryonic day 13 (E13), E15, E17, and postnatal day 1 (P1) were prepared as described above. Fresh frozen sections of P7 and P35 mice were prepared using the Kawamoto tape method, according to the manufacturer's instructions (Leica Microsystems, Tokyo, Japan) (21), and 10-μm sagittal sections were generated. Cells were fixed with 4% paraformaldehyde and blocked with 1% BSA. The primary antibody used was an anti-Adamtsl6 polyclonal antibody (R1–1) (17), anti-fibrillin-1 polyclonal antibody (pAB9543), anti-FIBRILLIN-1 monoclonal antibody (clone 69, Chemicon, Temecula, CA), and anti-FLAG M2 monoclonal antibody (Sigma-Aldrich). The secondary antibodies used were Alexa 488 or Alexa 555 anti-rabbit or anti-mouse IgG (Invitrogen), followed by nuclear staining with DAPI. An anti-Adamtsl6 polyclonal antibody was labeled with Alexa 488 by using the Zenon antibody labeling kit according to the manufacturer's instructions (Invitrogen) for double immunostaining with an anti-fibrillin-1 polyclonal antibody. For visualization of oxytalan fibers, sections were oxidized for 15 min in 10% Oxone (Merck) and subsequently stained with aldehyde fuchsin as described previously (20). Fluorescence images were sequentially collected using a confocal microscope featuring 403-, 488-, and 543-nm laser lines (LSM510; Carl Zeiss MicroImaging, Jena, Germany). The in situ hybridization methodology and probe design are described in the supporting information.
ADAMTSL6β cDNA
As described previously (17), ADAMTSL6β or Adamtsl6β was cloned into p3XFLAG-CMV-14 to generate p3XFLAG-CMV-ADAMTSL6β. The coding sequence of the cDNA was confirmed to be identical to the published sequence. The ADAMTSL6β coding sequence containing the Kozak consensus sequence and tagged with the FLAG epitope at its C terminus end was then subcloned into the pcDNA4 expression vector (Invitrogen) or into the pDONR221 vector via a BP reaction (Invitrogen) to generate adenovirus or lentivirus, respectively.
Generation of Adenovirus
Recombinant adenovirus was constructed by homologous recombination between the expression cosmid cassette (pAxCAwt) and the parental virus genome in 293 cells (Riken, Tsukuba, Japan) as described previously (22) using an adenovirus construction kit (Takara, Ohtsu, Japan).
Generation of Lentivirus
Recombinant lentivirus carrying ADAMTSL6β was constructed via the recombination of pDONR221-containing ADAMTSL6β segments into CSII-CMV-RfA using a LR reaction to generate CSII-CMV- ADAMTSL6β. CSII-CMV-RfA was kindly provided by Dr. Hiroyuki Miyoshi (Riken, Tsukuba, Japan). Lentiviruses were produced essentially as described previously (23). Next, a 500-μl aliquot of producer cell culture fluid was added to human periodontal ligament (HPDL) (passage 7) or MFS periodontal ligament (MHPDL) (passage 7) cells in the presence of Polybrene (7.5 μg/ml). Stably transduced cells were maintained in the medium described above.
For knockdown experiments, miRNA expression vectors were constructed according to the manufacturer's protocol (Invitrogen). Two sets of Adamtsl6β miRNAs to target sense (5′-TGCTGAATAACAGGTAGCTGACAAACGTTTTGGCCACTGACTGACGTTTGTCATACCTGTTATT-3′) and antisense (5′-CCTGAATAACAGGTATGACAAACGTCAGTCAGTGGCCAAAACGTTTGTCAGCTACCTGTTATTC-3′) transcripts were used to generate lentiviruses for the knockdown of Adamtsl6β. A control miRNA was purchased from Invitrogen.
Infection of Developing Tooth Germ with Adenovirus
To investigate the effects of Adamtsl6β on PDL formation in mgR/mgR mice, developing tooth germs were dissected from E14.5 mgR/mgR mouse embryos as described above and then infected with adenovirus that had been concentrated using the Adeno-X Maxi purification kit (Clontech) at 4 °C for 48 h in accordance with the manufacturer's recommendations. The adenovirus-infected tooth germs were then further incubated at 37 °C for 6 days in an in vitro organ culture as described previously (24).
Expression and Purification of Recombinant Adamtsl6β
The expression and purification of recombinant Adamtsl6β was performed using 293F cells (Invitrogen) and nickel-agarose (Qiagen, Hilden, Germany) as described previously (17). Briefly, pSecTag2A containing an Adamtsl6β segment fused with Myc and His tags at its C terminus was transfected into 293F cells, which were cultured for 3 days. Conditioned medium was applied to a nickel-agarose column for the purification of recombinant Adamtsl6β. The purified protein was dialyzed against PBS and stored at −80 °C.
Tissue Culture and in Vitro Microfibril Assembly Assay
The establishment of immortalized human periodontal cells and MHPDL cells has been described previously (25). Cells were incubated with α-minimum essential medium (Sigma) containing 10% fetal bovine serum (FBS; BioWhittaker, Walkersville, MD), 50 μg/ml ascorbic acid, and 100 units/ml streptomycin and penicillin in a humidified atmosphere of 5% CO2 at 37 °C. HPDL or MHPDL cells were plated onto 12-mm-thick coverglass coated with poly-l-Lys (Iwaki, Tokyo, Japan) placed in 24-well plates at 6 × 104 cells/well and incubated for 14 days. For the addition of purified recombinant mouse Adamtsl6β, C-terminal histidine-tagged mouse Adamtsl6β was prepared as described previously (17). Adamtsl6β protein (10, 5, 2.5, 1.25, or 0.625 μg) and the cells were incubated for 3 days. The cells were fixed with 4% paraformaldehyde and immunostained as described above.
RNA Preparation and Real-time RT-PCR
Total RNA was isolated from cells using Isogen (Nippon Gene Co., Ltd., Tokyo, Japan) as described previously (26). cDNAs were synthesized from 1-μg aliquots of total RNA in a 20-μl reaction containing 10× reaction buffer, 1 mm dNTP mixture, 1 unit/μl RNase inhibitor, 0.25 unit/μl reverse transcriptase (M-MLV reverse transcriptase; Invitrogen), and 0.125 μm random 9-mers (Takara, Tokyo, Japan). The mRNA expression levels were determined using Power SYBR® Green PCR Master Mix (Applied Biosystems), and products were analyzed with an AB 7300 real-time PCR system (Applied Biosystems). Specific primers for human fibrillin-1 (forward, 5′-AATGAGCTGAATGGCTGTTACAA-3′; reverse, 5′-ACCATATGCTATATATTCTTCGATAACAAT-3′), mouse fibrillin-1 (forward, 5′-AAGGGGTTAATGTCATGATGTCAC-3′; reverse, 5′-CCACACAAGAACATAAAACCAAGG-3′), and mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward, 5′-ACTGAGCAAGAGAGGCCCTATCC-3′; reverse, 5′-CCTAGGCCCCTCCTGTTATTATGG-3′) were used for real-time PCR. The primers for human GAPDH have been described previously (27).
Pull-down Assay
Pull-down assays to demonstrate direct interactions between Adamtsl6β and TGF-β1 proteins were performed as described previously by Nakajima et al. (28). Briefly, purified recombinant mouse Adamtsl6β (5 μg) was incubated with 0.1 μg of recombinant TGF-β1 proteins (Wako, Osaka, Japan) for 1 h at 4 °C in 0.3 ml of binding buffer (20 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 1% Triton X-100). We next added 12.5 μl of nickel-magnet (Promega, Madison, WI) to the reactions and incubated them for 30 min at 25 °C. The precipitates were washed three times with binding buffer, eluted by 250 mm imidazole, and subjected to SDS-PAGE. The proteins were blotted and visualized with the corresponding antibody.
Cell Culture
The method used to culture the mouse dental follicle cells has been described previously (29). To examine the effects of Adamtsl6β upon the TGF-β1-induced expression of periostin, mouse dental follicle cells were cultured in a 12-well plate at a density of 5 × 104 cells/well in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS until they reached confluence. At this point, the medium was replaced with DMEM containing 0.2% FBS. After 12 h, the cells were treated with recombinant mouse Adamtsl6β. After 12 h, the cells were treated with TGF-β1 (10 ng/ml) for 3 days and subjected to real-time PCR analysis.
Tooth Replantation Model
The tooth replantation experiments were performed as described previously (30). Briefly, the upper first molar from 4-week-old C57BL/6(SLC) mice was extracted under deep anesthesia. Extracted teeth were then replanted into the original cavity to allow the natural repair of the PDL. The replanted teeth were collected at 3, 7, and 14 days after transplantation and subjected to immunohistochemical analysis using the Kawamoto tape method or in situ hybridization as described in the supplemental Methods.
Generation of Transgenic Bioengineered Tooth Germ
Molar tooth germs were dissected from the mandibles of E14.5 mice. The isolation of mesenchyme and epithelium and the dissociation of mesenchymal cells have been described previously (24). Dissociated cells were cultured on tissue culture plates in DMEM containing 10% fetal calf serum and then infected with Adamtsl6β adenovirus for 8 h. After incubation for 24 h, mesenchymal cells overexpressing Adamtsl6β were collected via trypsin digestion and precipitated by centrifugation in a siliconized tube, and the supernatant was completely removed. The cell density of the precipitated, adenovirus-infected mesenchymal cells after removal of supernatant reached a concentration of 5 × 108 cells/ml, as described previously (24). Transgenic bioengineered tooth germ was reconstituted with dissociated adenovirus-infected mesenchymal cells and epithelial tissue using our previously described three-dimensional cell manipulation system, the organ germ method (24). The transgenic bioengineered tooth germs were incubated for 10 min at 37 °C, placed on cell culture inserts (0.4-μm pore diameter; BD Biosciences), and then further incubated at 37 °C for 6 days in an in vitro organ culture as described previously (24). Mesenchymal cells infected with lentiviruses carrying Adamtsl6β miRNAi were used to generate Adamtsl6β miRNAi-transgenic bioengineered tooth germ by incubation for 10 min at 37 °C, placement on cell culture inserts (0.4-μm pore diameter; BD Biosciences), and then a further incubation at 37 °C for 12 days in an in vitro organ culture as described previously (24).
Local Administration of ADAMTSL6β Using a PDL Injury Model
To create gels for injection, 2 μl of recombinant Adamtsl6β (10 μg/μl) (see supplemental Methods) and 1.5 μl of PKH67 green fluorescent cell linker (Sigma-Aldrich) were suspended in a 9-μl gel drop of collagen liquid Cellmatrix type I-A (Nitta Gelatin) composed of acid-soluble collagen isolated from pig tendon. The lower first molars of 4-week-old C57BL/6(SLC) mice were extracted under deep anesthesia. Following blood coagulation, at 3 days after tooth extraction, the alveolar bony wall of the proximal site of the lower second molar tooth was surgically removed to expose the PDL. The PDL was then disrupted by dislocation of the second molar tooth with lingual to buccal side movement. The collagen drop containing recombinant Adamtsl6β was then inserted into the damaged PDL. Mouse mandibles were collected 17 days after insertion, and immunohistochemical analysis was performed using the Kawamoto tape method as described in the supplemental Methods.
RESULTS
ADAMTSL6β Regulates Microfibril Assembly in Various Connective Tissues
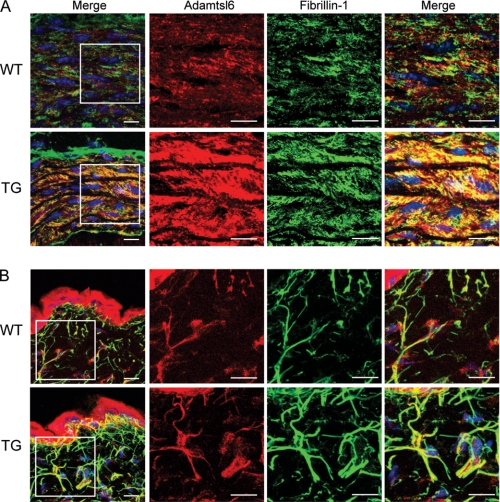
To investigate whether Adamtsl6β plays a critical role in microfibril assembly in connective tissues, we generated Adamtsl6β-transgenic mice (Tsl6β-TG mice) in which the transgene is expressed in the whole body. Because Adamtsl6 has been shown to be expressed in the aorta and skin, we investigated microfibril assembly of these tissues in the Tsl6β-TG mice. Immunohistochemical analysis revealed that Adamtsl6-positive microfibril assembly was barely detectable in WT mice but strongly induced in the aorta of Tsl6β-TG mice (Fig. 1A). Confocal microscopy analysis further revealed that Adamtsl6- and fibrillin-1-positive microfibrils are clearly increased in the aorta and that microfibril assembly is also induced in the skin of Tsl6β-TG mice. This confirmed that Adamtsl6 induces fibrillin-1 microfibril assembly in connective tissue, such as the aorta and skin (Fig. 1B).
FIGURE 1.
Immunohistochemical analysis of Tsl6β-TG mice. Immunofluorescence detection of ADAMTSL-6 proteins and fibrillin-1. Frontal cryosections were prepared from the skin (A) or aortas (B) of wild type (top) or Tsl6β-TG (bottom) littermates and subjected to double immunostaining with antibodies against Adamtsl6 (red) and fibrillin-1 (green). The boxed areas in the leftmost panels are shown at higher magnification in the rightmost panels. Immunohistochemical analysis indicated that the expression of Adamtsl6- and fibrillin-1-positive microfibrils was markedly increased in the aorta and skin of Tsl6β TG mice compared with WT mice. The merged images illustrate that these fibrils are colocalized in the skin and aorta.
ADAMTSL6β Is Involved in Microfibril Formation during PDL Development and Wound Healing
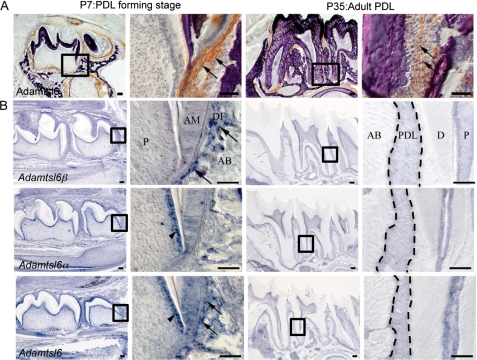
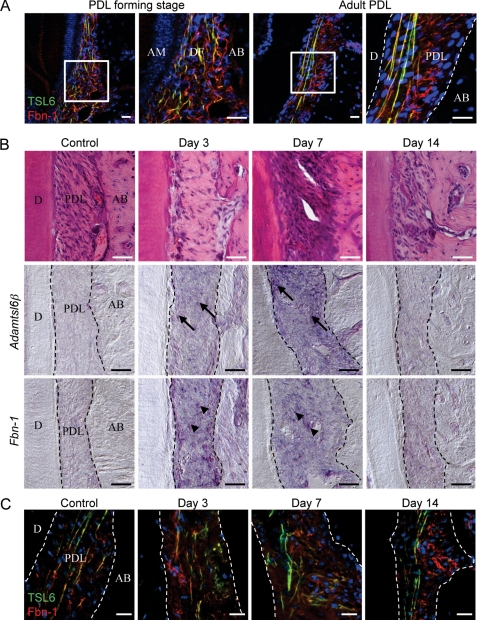
To investigate whether Adamtsl6β contributes to connective tissue formation, we first examined its expression patterns during embryonic tooth germ development and in the PDL formation stage after birth as a model of connective tissue formation. Development of the PDL proceeds as follows: 1) the dental follicle (DF), the origin of the PDL, is formed at the CAP stage of tooth germ formation; 2) the DF differentiates during the tooth root-forming stage; and 3) the DF differentiates into the PDL to be inserted into collagen fibers known as Sharpey's fibers in the cementum matrix and alveolar bone, which resembles tendinous tissue (26, 31). In situ hybridization analysis revealed that Adamtsl6β is barely expressed in the DF, the origin of PDL formation in the surrounding tooth germ (supplemental Fig. S1B), but was very strongly expressed in the PDL-forming stage of the DF at P7 (Fig. 2B). However, Adamtsl6β expression was significantly down-regulated in the adult PDL at P35 (Fig. 2B). In contrast to Adamtsl6β, Adamtsl6α was found to be expressed in odontoblasts but not to be expressed in either the DF during the PDL-forming stage or in the adult PDL (supplemental Fig. S1B and Fig. 2B). Immunohistochemical analysis further revealed that Adamtsl6 is only weakly expressed in the early stage (bell stage) DF (supplemental Fig. S1A) but became detectable in assembled microfibril-like structures during the PDL-forming stage of the DF and in organized microfibrils in the adult PDL (Fig. 2A). Using confocal microscopy analysis, Adamtsl6 was observed to colocalize with fibrillin-1 to form immature microfibrillar-like structures at the PDL-forming stage of the DF, which were then observed as fully assembled mature microfibril structures in the adult PDL (Fig. 3A).
FIGURE 2.
Expression patterning of Adamtsl6α and -β during the PDL-forming stage. A, sagittal sections of lower molar at P7 and P35 were immunostained with anti-Adamtsl6 antibody. The image in the box on the left is shown at higher magnification on the right. Note that Adamtsl6 protein was deposited as microfibril aggregates in DF at the PDL-forming stage (arrows) and formed a mature microfibrillar assembly in adult PDL (arrows). Bar, 100 μm. B, in situ hybridization analysis using a specific probe for Adamtsl6α and -β and control probe are shown. Adamtsl6β mRNA at the P1-late bell stage of dental follicle formation in the tooth germ is indicated by arrows. Expression of Adamtsl6α was detected in the odontoblast (arrowheads) by specific probes. In contrast, control probes that detected the conserved region of Adamtsl6 recognized both odontoblast (arrowhead) and DF (arrows). Bar, 100 μm. AB, alveolar bone; AM, ameloblast; D, dentin; P, pulp; DF, dental follicle; PDL, periodontal ligament.
FIGURE 3.
Adamtsl6β is involved in fibrillin-1 microfibril formation during PDL development and wound healing. A, localization of fibrillin-1- and Adamtsl6-positive microfibrils were analyzed during the PDL formation stage (left) and in adult PDL (right) using antibodies against Adamtsl6 (TSL6; green) and fibrillin-1 (Fbn-1; red). Formation of microfibrils positive for anti-Adamtsl6β and anti-fibrillin-1 was detectable during the process of PDL formation. B, frontal section of control side PDL and injured PDL 3, 7, and 14 days after replantation of the tooth were analyzed by hematoxylin and eosin staining (top) and in situ hybridization analysis of Adamtsl6β (middle) or fibrillin-1 (Fbn-1; bottom) expression in PDL. Cells positive for Adamtsl6β and fibrillin-1 mRNA expression are indicated by arrows or arrowheads, respectively. C, immunohistochemical analysis using anti-Adamtsl6 (TSL6; green) and anti-fibrillin-1 (Fbn-1; red) antibodies indicated that expression of Adamtsl6- and fibrillin-1-positive microfibrils was markedly increased 3 and 7 days after injury. A merged image illustrates that these fibrils were colocalized during the wound healing processes.
Using an Adamtsl6 antibody, positively stained fibers were observed in the adult PDL that were almost identical to those marked by aldehyde fuchsin staining and are indicative of microfibrils (supplemental Fig. S2). This suggested that Adamtsl6 was a component of microfibrils. Because developmental processes involve similar mechanisms to wound healing, we next determined whether Adamtsl6β is involved in PDL microfibril assembly during wound healing using a tooth replantation model (supplemental Fig. S3A) (30). Histochemical analysis revealed an injured PDL with an irregular architecture at 3 days after replantation, although gradual healing then occurred at between 7 and 14 days after replantation (Fig. 3B and supplemental Fig. S3B). During these processes, Adamtsl6β and fibrillin-1 mRNA expression were found to be clearly induced in the PDL at 3–7 days after replantation but to decrease again by 14 days after replantation (Fig. 3B).
Similar to these gene expression patterns, Adamtsl6- and fibrillin-1-positive microfibrillar-like structures resembling those seen in the DF during the PDL-forming stage were markedly increased in the damaged PDL at 3–7 days after replantation. These structures had evolved into mature microfibrils by 14 days after replantation (Fig. 3C and supplemental Fig. S3B). In contrast to these gene expression patterns, the expression of periostin, a PDL differentiation marker, was detected at 7 days after replantation (supplemental Fig. S3C). These data indicate that fibrillin-1 microfibril formation is induced in the early stages of both PDL development and wound healing and that Adamtsl6β is involved in these processes.
ADAMTSL6β Regulates PDL Formation through Fibrillin-1 Microfibril Assembly
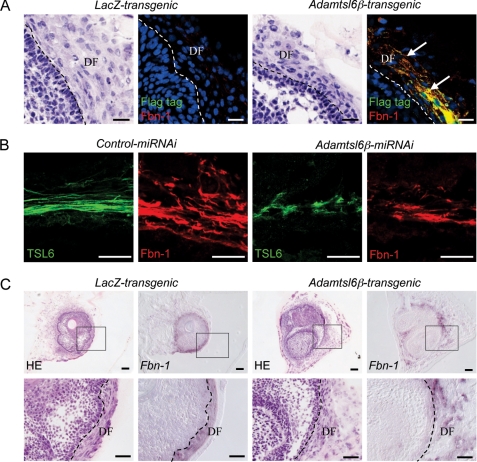
We have recently developed a new three-dimensional single cell processing technique, the organ germ method, which can be used to generate bioengineered tooth germ reconstituted from E14.5 molar tooth germ-derived epithelial and mesenchymal cells (24). Utilizing this system, we developed a transgenic bioengineered tooth germ by overexpressing exogenous genes in mesenchymal cells derived from tooth germ using adenovirus (supplemental Fig. S4, A and B). Because transgenic bioengineered tooth germ was found to accurately reproduce PDL development (supplemental Fig. S4, C and D), we generated Adamtsl6β-transgenic bioengineered tooth germ to examine the contributions of Adamtsl6β to PDL formation. Following immunohistochemical staining, Adamtsl6β-transgenic bioengineered tooth germs showed clear colocalization between fibrillin-1 microfibrils and Adamtsl6β (Fig. 4A) after 6 days of culture. Conversely, fibrillin-1 microfibrils were barely detectable in control LacZ-transgenic bioengineered tooth germ (Fig. 4A).
FIGURE 4.
Adamtsl6β contributes to PDL formation through regulation of fibrillin-1 microfibril assembly. A, immunohistochemical analysis of LacZ-transgenic tooth germ (LacZ-transgenic) or Adamtsl6β-transgenic bioengineered tooth germ (Adamtsl6β-transgenic) using double immunostaining with anti-FLAG (Flag tag; green) and anti-fibrillin-1 (Fbn-1; red). Microfibrils positive for Adamtsl6β and fibrillin-1 expression are indicated by arrows. B, immunohistochemical analysis of control miRNAi-transgenic bioengineered tooth germ (Control miRNAi) or Adamtsl6β miRNAi-transgenic tooth germ (Adamtsl6β-miRNAi) using double immunostaining with anti-Adamtsl6 (TSL6; green) and anti-fibrillin-1 (Fbn-1; red). C, hematoxylin and eosin staining (HE) and in situ hybridization analysis of fibrillin-1 mRNA expression (Fbn-1) in LacZ transgenic tooth germ (LacZ-transgenic) or Adamtsl6β transgenic bioengineered tooth germ (Adamtsl6β-transgenic). The image in the box at the top is shown at higher magnification at the bottom (C).
To confirm the role of Adamtsl6β in regulating microfibril formation in the DF from bioengineered tooth germ, we generated Adamtsl6β miRNAi-transgenic bioengineered tooth germ to suppress Adamtsl6β expression. Immunohistochemical analysis subsequently revealed that the Adamtsl6β miRNAi-transgenic germ exhibited poor Adamtsl6- and fibrillin-1-positive microfibril formation after 12 days of culture. However, no changes were observed in control miRNAi-transgenic bioengineered tooth germ, further indicating that Adamtsl6β regulates microfibril assembly during PDL formation (Fig. 4B). We next evaluated whether the promotion of fibrillin-1 microfibril assembly was the result of increased mRNA expression. In situ hybridization analysis revealed that fibrillin-1 mRNA expression was similar in LacZ- and Adamtsl6β-transgenic bioengineered tooth germ (Fig. 4C). These data indicate that Adamtsl6β is capable of recruiting fibrillin-1 to assembling microfibrils without increasing the fibrillin-1 transcript levels.
ADAMTSL6β Negatively Regulates TGF-β-induced Periostin Gene Expression during PDL Formation
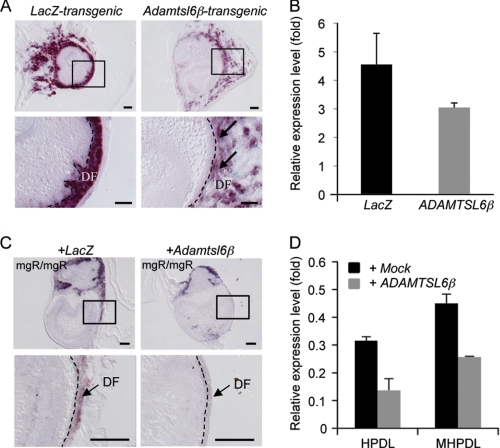
To investigate whether Adamtsl6β regulates PDL formation, we analyzed the expression of genes that function in PDL formation, including type I collagen, type XII collagen, periostin, and f-spondin (27). Among these genes, periostin, the protein product of which is known to be induced by TGF-β (32, 33), was markedly down-regulated in Adamtsl6β-transgenic bioengineered tooth germ (Fig. 5B and supplemental Fig. S5). In situ hybridization analysis further revealed the strong expression of periostin in the DF from LacZ-transgenic tooth germ when compared with the DF from Adamtsl6β-transgenic tooth germ (Fig. 5A). Real-time PCR analysis confirmed the suppression of periostin expression in Adamtsl6β-transgenic tooth germ (Fig. 5B).
FIGURE 5.
ADAMTSL6β negatively regulates periostin expression. A, in situ hybridization for periostin mRNA expression in LacZ transgenic tooth germ (LacZ-transgenic) or Adamtsl6β transgenic bioengineered tooth germ (Adamtsl6β-transgenic). The image in the top box is shown at higher magnification in the bottom box. Down-regulation of periostin mRNA expression in Adamtsl6β transgenic tooth germ is indicated by the arrows. B, total RNA extracted from LacZ (LacZ)- or Adamtsl6β (Adamtsl6β)-transgenic bioengineered tooth germ. cDNA was synthesized and subjected to quantitative real-time PCR for the expression of periostin and GAPDH transcripts. Levels of GAPDH transcript were used to normalize cDNA levels. Levels of GAPDH were set at 1, and relative expression levels are shown. Data are presented as triplicates, and the means ± S.D. are shown. C, in situ hybridization analysis of Adamtsl6β-adenovirus-infected mgR/mgR mouse tooth germ showed reduced periostin expression when compared with LacZ-adenovirus-infected mgR/mgR mouse tooth germ. Bar, 100 μm. D, real-time PCR analysis of periostin mRNA in HDPL and MHPDL cells transduced with mock or Adamtsl6β. Periostin mRNA expression was down-regulated in HPDL and MHPDL cells overexpressing Adamtsl6β as compared with expression in mock-overexpressed HPDL and MHPDL cells.
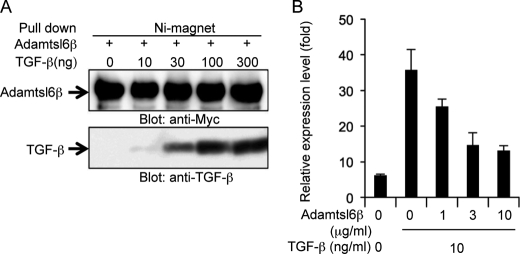
To evaluate whether Adamtsl6β negatively regulates periostin gene expression in our MFS model system, we analyzed Adamtsl6β adenovirus-infected tooth germ obtained in MFS mice that were homozygous for a targeted hypomorphic allele (mgR/mgR) of fibrillin-1 (7). In situ hybridization analysis showed that periostin expression was remarkably reduced in developing tooth germ from mgR/mgR mice after infection with Adamtsl6β-adenovirus compared with LacZ-adenovirus-infected tooth germ (Fig. 5C). We next investigated the effects of ADAMTSL6β on human PDL cells obtained from an MFS patient with severe periodontitis (MHPDL) (25). As expected, ADAMTSL6β overexpression in these MHPDL cells clearly reduced periostin expression when compared with mock-infected cells. Interestingly, the level of periostin expression in MHPDL cells with ADAMTSL6β overexpression was comparable with that of normal HPDL cells (Fig. 5D), raising the possibility that ADAMTSL6β negatively regulates TGF-β and thereby reduces periostin expression. We evaluated this possibility by testing the ability of His-tagged recombinant Adamtsl6β to bind to TGF-β1. Interactions between Adamtsl6β and TGFβ1 were barely detectable at the 10 ng/ml concentrations, but strong associations between these proteins could be detected in a dose-dependent manner at 30, 100, or 300 ng/ml by pull-down analysis using nickel-magnetic beads (Fig. 6A). To then test the effects of recombinant Adamtsl6β on TGF-β activity, we measured the periostin expression levels in mouse dental follicle cells treated with Adamtsl6β in the presence or absence of TGF-β1. Recombinant Adamtsl6β inhibited TGF-β-induced periostin expression in a dose-dependent manner (Fig. 6B). These data suggest that Adamtsl6β directly binds to TGF-β to reduce periostin expression during the PDL-forming stage in both normal and MFS model settings.
FIGURE 6.
Adamtsl6β binds TGF-β to negatively regulate periotsin gene expression. A, His tag recombinant Adamtsl6β was incubated with TGF-β1 at the indicated concentrations followed by treatment with nickel-magnetic beads. Co-precipitates were detected using the corresponding antibodies, and Adamtsl6β was found to bind to TGF-β1 directly. B, mouse dental follicle cells were cultured with TGF-β1 for 3 days in the presence of recombinant mouse Adamtsl6β. Periostin mRNA levels were quantified by real-time PCR analysis. Adamtsl6β inhibited expression of periostin in a dose-dependent manner. Error bars, S.D.
The Local Administration of ADAMTSL6β Improves Wound Healing Ability in an MFS Model
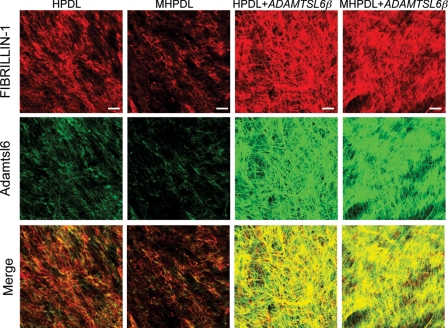
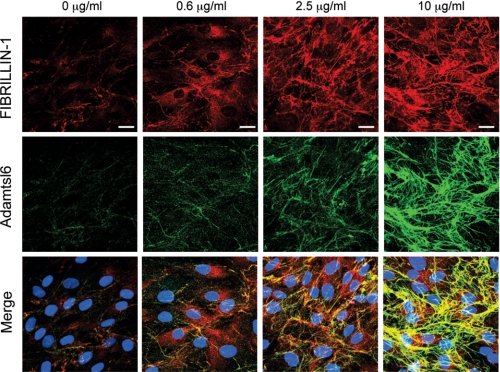
We next investigated whether ADAMTSL6β alleviates fibrillin-1 microfibril disorder in MHPDL cells, which exhibit reduction in fibrillin-1 microfibril assembly (Fig. 7). The overexpression of ADAMTSL6β strongly induced fibrillin-1 microfibril assembly in MHPDL cells compared with the mock-infected controls (Fig. 7, top and middle). Merged images revealed that ADAMTSL6β colocalizes with fibrillin-1 in MHPDL cells that overexpress ADAMTSL6β (Fig. 7, bottom). We have previously shown that recombinant Adamtsl6β induces fibrillin-1 microfibril assembly in MG63 cells (17). Thus, we next investigated whether recombinant Adamtsl6β improves the symptoms of MHPDL microfibril disorder. We found that recombinant Adamtsl6β induces fibrillin-1 microfibril assembly in a dose-dependent manner in MHPDL cells during a 3-day incubation in culture (Fig. 8, top and middle). Staining with an anti-Adamtsl6 polyclonal antibody indicated that exogenous Adamtsl6 colocalizes with fibrillin-1 (Fig. 8, bottom). Endogenous fibrillin-1 was only marginally detectable in MHPDL cells. However, an abundant fibrillin-1 network formation was evident in the presence of high concentrations (10 μg/ml) of recombinant Adamtsl6β (Fig. 8, top). These results indicate that Adamtsl6β improved fibrillin-1 MHPDL microfibril assembly.
FIGURE 7.
Overexpression of ADAMTSL6β improves microfibril disorder in PDL from an MFS patient. Immunohistochemical analysis of HPDL or MHPDL cells transduced with mock or ADAMTSL6β using anti-fibrillin-1 (top) and anti-ADAMTSL6 (middle) antibodies. The data show that ADAMTSL6β induces fibrillin-1 microfibril assembly in MHPDL cells. The bottom images were produced by superimposition of the upper and middle images, together with DAPI nuclear staining.
FIGURE 8.
Recombinant Adamtsl6β improves microfibril disorder in PDL from MFS patients. Immunohistochemical analysis with anti-fibrillin-1 (top) and anti-Adamtsl6 (middle) antibodies reveals a marked improvement in fibrillin-1 microfibril assembly (arrows) in MHPDL cells incubated with purified recombinant Adamtsl6β at 0.6, 2.5, and 10 μg/ml for 3 days. The bottom images were produced by superimposition of the upper and middle images, together with DAPI nuclear staining.
To investigate whether Adamtsl6β was capable of improving microfibril assembly in vivo, we investigated the PDL from mgR/mgR mice and by histochemical analysis observed a disorganized structure with a disrupted cell alignment, both of which are characteristic MFS morphologies (Fig. 9A). Immunohistochemical analysis clearly revealed fragmented Adamtsl6β- and fibrillin-1-positive microfibrils when compared with wild type mice (Fig. 9A, arrows). We next infected Adamtsl6β adenovirus into the DF of developing tooth germ isolated from mgR/mgR mouse embryos at E14.5 to evaluate the improvements in fibrillin-1 microfibril disorder during PDL formation. By histochemical analysis, we found that the overexpression of Adamtsl6β resulted in an improved DF morphology with compact and aligned cells (Fig. 9B). DF tooth germ infected with LacZ showed low cell numbers and an irregular architecture. Immunohistochemical analysis subsequently revealed that Adamtsl6β overexpression strongly induces fibrillin-1 microfibril assembly in the tooth germ from mgR/mgR mice, whereas no assembly was observed in LacZ-infected tooth germ (Fig. 9B, arrows). These data indicate that Adamtsl6β can indeed restore the impaired microfibrils in mgR/mgR mice.
FIGURE 9.
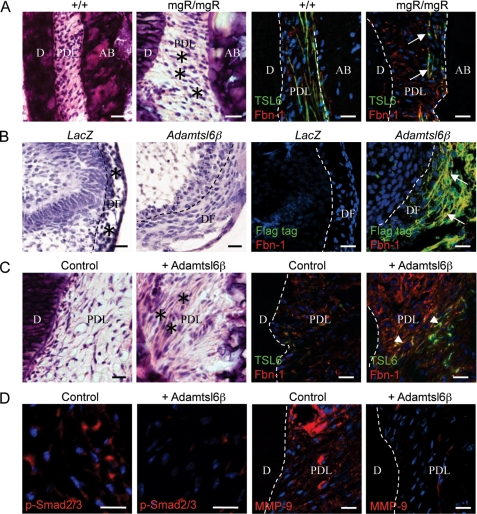
Local administration of Adamtsl6β improves microfibril disorder and attenuates TGF-β signaling in PDL from an MFS model. A, hematoxylin and eosin staining of a PDL revealing a markedly abnormal architecture in mgR/mgR mice when compared with wild type. Notable is the loosening of the PDL with an irregular cell alignment and an expanded cell-cell distance (asterisks). Immunohistochemical analysis using Adamtsl6 (TSL6; green) and fibrillin-1 (Fbn-1; red) antibodies revealed a clear disruption of fibrillin-1- and Adamtsl6-positive microfibrils in the PDL from mgR/mgR mice (arrows). B, hematoxylin and eosin staining of LacZ-infected mgR/mgR mouse tooth germ (LacZ) revealing an abnormal architecture with an irregular cell alignment (asterisks) when compared with Adamtsl6β-infected mgR/mgR mouse tooth germ (Adamtsl6β). Immunohistochemical analysis using FLAG (Flag-tag; green) and fibrillin-1 (Fbn-1; red) antibodies showed an improvement in fibrillin-1 microfibril assembly in Adamtsl6β-infected mgR/mgR mouse tooth germ (arrows). C, histological analysis of the injured PDL in mgR/mgR mice after the local administration of control gel or gel containing recombinant Adamtsl6β for 17 days. Hematoxylin and eosin staining revealed PDL healing after the injection of gel containing recombinant Adamtsl6β (asterisks) compared with the control. Immunohistochemical analysis further showed an improvement in fibrillin-1 microfibril assembly (arrowheads) induced by the injection of recombinant Adamtsl6β. D, immunohistochemical analysis of pSmad2/3 and MMP-9 expression in the injured PDL of mgR/mgR mice after the local administration of control gel or gel containing recombinant Adamtsl6β for 17 days. The suppression of nuclear accumulation of pSmad2/3 and MMP-9 expression is evident after injection of recombinant Adamtsl6β compared with the control gel.
We next investigated whether Adamtsl6β might be developed as a novel therapeutic for MFS microfibril disorder. Collagen gel containing recombinant Adamtsl6β was locally administrated into an experimentally damaged PDL in mgR/mgR mice (supplemental Fig. S6, A and B). Fluorescence microscopic analysis revealed that the collagen gel implanted in damaged PDL was still present at 17 days after injection (supplemental Fig. S6C). Histochemical analysis showed that a damaged PDL could still be observed at 7 days after injection of the collagen gel containing recombinant Adamtsl6β (supplemental Fig. S6D). However, healing and improved cell alignment were apparent in the PDL of wild type mice at 17 days after injection (Fig. 9C, asterisk). Immunohistochemical analysis further revealed that the reorganization of fibrillin-1- and Adamtsl6-positive microfibril assembly could be observed after 17 days of incubation (Fig. 9C (arrowheads) and supplemental Fig. S6E). In contrast, the administration of control collagen gel failed to induce PDL healing, and an irregular cell morphology and poor fibrillin-1 microfibril formation could still be observed (Fig. 9C and supplemental Fig. S6E).
The enhanced activation of TGF-β has been suggested to directly contribute to tissue destruction in MFS (12). Ligand-activated TGF-β receptors induce the phosphorylation of Smad2 and Smad3 (pSmad2/3), which form a heteromeric complex with Smad4 that translocates to the nucleus and mediates the expression of target genes (34). The nuclear accumulation of pSmad2/3 has been detected in affected tissues in an MFS mouse model, including the aorta and skeletal muscle (10, 35). Consistent with these results, we observed the nuclear accumulation of pSmad2/3 in PDL from mgR/mgR mice 17 days after injection of control collagen gel in our current experiments (Fig. 9D). However, the local administration of Adamtsl6β markedly suppressed the nuclear localization of pSmad2/3. Further evidence for the Adamtsl6β suppression of TGF-β signaling is derived from the previous analysis of matrix metalloprotease (MMP)-9, which is known to be induced by TGF-β and is expressed in abnormal smooth muscle cells in the early vascular lesions that contribute to elastolysis (36, 37). In contrast to control collagen gel administration, the expression of MMP-9 was markedly suppressed by the administration of collagen gel containing recombinant Adamtsl6β (Fig. 9D). These results illustrate that reorganization of microfibrils by recombinant Adamtsl6β prevents the pathological activation of TGF-β by structurally damaged fibrillin-1 in MFS.
DISCUSSION
Our current experiments successfully demonstrate that ADAMTSL6β has an essential role in PDL development and regeneration through the promotion of fibrillin-1 assembly and the negative regulation of TGF-β signaling. We also demonstrate in our present analyses that the local administration of ADAMTSL6β can rescue the disease manifestations of MFS in a mouse model, raising the possibility that this extracellular matrix protein could be used as a novel therapeutic agent for the treatment of MFS. Hence, our data show for the first time that the restoration of properly formed microfibrils by ADAMTSL6β is essential not only for improvement of the predominant symptoms of MFS but also for the suppression of excessive TGF-β signaling induced by microfibril disassembly.
To clarify the role of ADAMTSL6β in PDL formation, experiments were performed to determine whether ADAMTSL6β-mediated fibrillin-1 microfibril assembly is critical for PDL development and regeneration. The formation of fibrillin-1 microfibril networks has been shown to be essential for the development and growth of individual organ systems (38). Vascular smooth muscle cells are gradually organized via the formation of elastic fibers and interconnecting fibrillin-1 microfibrils during aortic media generation, resulting in the organization of elastic lamellae as the main determinant of arterial function (39). In addition to providing mechanical stability, previous studies have demonstrated that the organization of fibrillin-1 microfibril assemblies contributes to the regulation of the activities of signaling molecules, such as TGF-β and BMP-7 (40, 41). During digit formation, fibrillin-1 may be a positive regulator that dictates the functional sites for cytokine concentration. In other tissues, fibrillin-1 acts as a negative regulator of signaling through cytokine sequestration (2, 7). Thus, the importance of microfibril network formation has been demonstrated in several disparate settings. However, the importance of the molecular mechanisms governing fibrillin-1 assembly during organogenesis has been hampered by the unanswered issue of the actual factor that drives microfibril assembly. Our present study demonstrates that ADAMTSL6β, an inducing factor for microfibril assembly (18), regulates development and regeneration of PDL. Furthermore, ADAMTSL6β-mediated fibrillin-1 microfibril assembly may accelerate the sequestration of large latent complexes of TGF-β or active TGF-β, thereby negatively regulating the expression of TGF-β regulatory targets, such as periostin (35, 42). Hence, our data provide significant insight into the molecular mechanisms by which ADAMTSL6β controls fibrillin-1 microfibril assembly and TGF-β signaling during organogenesis.
The establishment of fibrillin-1 microfibril assembly mechanisms is ultimately critical for the development of new MFS therapeutic approaches (12). The pathogenetic relevance of MFS is highlighted by the fact that microfibril assembly is frequently disrupted in patients with various fibrillinopathies (43). MFS fibrillinopathies have been explained by the structural insufficiency of fibrillin-1 microfibrils, leading to activation of TGF-β and its regulatory targets (7, 44). Although many recent publications have addressed the organization of fibrillins in microfibrils (45–48), relatively little information has been available regarding the mechanisms and components involved in microfibril formation. A previous study reported that the transgenic expression of wild-type fibrillin-1 alleles in a missense mutation (C1039G) heterozygous mouse model of MFS effectively rescues the aortic phenotype (11). From these data, essential improvements in fibrillin-1 microfibril formation represent a productive therapeutic strategy for the reduction of MFS disease severity (3).
In contrast to our present findings, another recent study has indicated that fibronectin is an essential component in the assembly of fibrillin-1 through its interaction with the C-terminal region of fibrillin-1, thus suggesting the possibility of improved microfibril assembly through regulation of fibrillin-1-associated proteins (49, 50). Our present data demonstrate, however, that the exogenous application of recombinant ADAMTSL6β improves fibrillin-1 microfibril assembly in an MFS mouse model. Hence, ADAMTSL6β reinforcement of fibrillin-1 microfibrils may represent a new, viable treatment for MFS. Although the mechanisms by which ADAMTSL6β accelerated fibrillin-1 microfibril assembly remain to be determined, in another study using the MFS mouse model and MHPDL cells, which are PDL cells obtained from an MFS patient, ADAMTSL6β seems to recruit available normal fibrillin-1 molecules and induce microfibril assembly with a resulting improvement in microfibril mechanical stability. These findings indicate that ADAMTSL6β is capable of enhancing microfibrils even in animals with a fibrillin-1 haploinsufficiency. Thus, ADAMTSL6β is potentially a novel therapeutic target for the treatment of MFS.
Recent evidence has suggested that restoration of microfibril assembly plays an important role in the prevention of pathological activation of TGF-β signaling in MFS (12). TGF-β is secreted from cells as a large latent complex consisting of TGF-β, latency-associated peptide, and LTBP-1 to be sequestered by fibrillin-1 (51). The promotion of fibrillin-1 microfibril assembly is therefore critical for the prevention of tissue destruction mediated by abnormal TGF-β signaling in MFS. In the present study, we have demonstrated that the reinforcement of fibrillin-1 microfibril assembly and the inhibition of TGFβ1 function by ADAMTSL6β facilitate wound healing in the PDL of mgR/mgR mice.
In conclusion, we provide evidence for the contributions of ADAMTSL6β-mediated fibrillin-1 microfibril assembly to PDL development, regeneration, and alleviation of MFS manifestations. We thereby introduce the concept that a fibrillin-1-associated protein, such as ADAMTSL6β, which induces microfibril assembly, should be considered in the development of future mechanism-based therapeutics for the improvement of connective tissue disorders, such as MFS. Our data suggest that the reinforcement of fibrillin-1 assembly by ADAMTSL6β accelerates the sequestration of newly synthesized large latent complexes into fibrillin-1. Further studies will help to clarify the nature of the interactions between ADAMTSL6β, fibrillin-1, TGF-β, and LTBP-1 and reveal how ADAMTSL6β expression suppresses TGF-β signaling. It will also be necessary to develop methodologies for the systemic administration of ADAMTSL6β to induce fibrillin-1 microfibril assembly in connective tissue for the treatment of life-threatening conditions, such as aortic aneurysm. Because elastolysis occurs continuously in aortic aneurysms in MFS, chronic administration of ADAMTSL6β may be required for the stabilization of microfibrils to prevent progressive tissue destruction. This approach will facilitate drug discovery for treating MFS and related connective tissue disorders.
Supplementary Material
Acknowledgments
We thank Dr. Lynn Sakai for providing the anti-fibrillin-1 polyclonal antibody (pAb9543) and Dr. Francesco Ramirez for providing the mgR/mgR mice. We also thank Drs. Zenzo Isogai, Takamasa Yokoi, Momotoshi Shiga, Tomoko Wada, and Hayato Ohshima for valuable advice and discussions during the course of this work.
This work was supported by Health and Labour Sciences Research Grants from the Ministry of Health, Labour, and Welfare (No. 23164001) (to M. S.), a Grant-in-aid for Scientific Research (B) (No. 23659980) (to M. S.), and Program for Development of Strategic Research Center in Private Universities supported by MEXT(2010) (to M. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods and Figs. S1–S6.
- MFS
- Marfan syndrome
- PDL
- periodontal ligament
- En
- embryonic day n
- Pn
- postnatal day n
- MHPDL
- MFS periodontal ligament
- HPDL
- human periodontal ligament
- DF
- dental follicle.
REFERENCES
- 1. Straub A. M., Grahame R., Scully C., Tonetti M. S. (2002) J. Periodontol. 73, 823–826 [DOI] [PubMed] [Google Scholar]
- 2. Ramirez F., Dietz H. C. (2007) J. Cell Physiol. 213, 326–330 [DOI] [PubMed] [Google Scholar]
- 3. Judge D. P., Dietz H. C. (2005) Lancet 366, 1965–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dietz H. C., Loeys B., Carta L., Ramirez F. (2005) Am. J. Med. Genet. C Semin. Med. Genet. 139, 4–9 [DOI] [PubMed] [Google Scholar]
- 5. Sakai L. Y., Keene D. R., Engvall E. (1986) J. Cell Biol. 103, 2499–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramirez F., Dietz H. C. (2007) Curr. Opin. Genet. Dev. 17, 252–258 [DOI] [PubMed] [Google Scholar]
- 7. Pereira L., Lee S. Y., Gayraud B., Andrikopoulos K., Shapiro S. D., Bunton T., Biery N. J., Dietz H. C., Sakai L. Y., Ramirez F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3819–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neptune E. R., Frischmeyer P. A., Arking D. E., Myers L., Bunton T. E., Gayraud B., Ramirez F., Sakai L. Y., Dietz H. C. (2003) Nat. Genet. 33, 407–411 [DOI] [PubMed] [Google Scholar]
- 9. Carta L., Pereira L., Arteaga-Solis E., Lee-Arteaga S. Y., Lenart B., Starcher B., Merkel C. A., Sukoyan M., Kerkis A., Hazeki N., Keene D. R., Sakai L. Y., Ramirez F. (2006) J. Biol. Chem. 281, 8016–8023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Habashi J. P., Judge D. P., Holm T. M., Cohn R. D., Loeys B. L., Cooper T. K., Myers L., Klein E. C., Liu G., Calvi C., Podowski M., Neptune E. R., Halushka M. K., Bedja D., Gabrielson K., Rifkin D. B., Carta L., Ramirez F., Huso D. L., Dietz H. C. (2006) Science 312, 117–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Judge D. P., Biery N. J., Keene D. R., Geubtner J., Myers L., Huso D. L., Sakai L. Y., Dietz H. C. (2004) J. Clin. Invest. 114, 172–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Judge D. P., Dietz H. C. (2008) Annu. Rev. Med. 59, 43–59 [DOI] [PubMed] [Google Scholar]
- 13. Ng C. M., Cheng A., Myers L. A., Martinez-Murillo F., Jie C., Bedja D., Gabrielson K. L., Hausladen J. M., Mecham R. P., Judge D. P., Dietz H. C. (2004) J. Clin. Invest. 114, 1586–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hirohata S., Wang L. W., Miyagi M., Yan L., Seldin M. F., Keene D. R., Crabb J. W., Apte S. S. (2002) J. Biol. Chem. 277, 12182–12189 [DOI] [PubMed] [Google Scholar]
- 15. Le Goff C., Morice-Picard F., Dagoneau N., Wang L. W., Perrot C., Crow Y. J., Bauer F., Flori E., Prost-Squarcioni C., Krakow D., Ge G., Greenspan D. S., Bonnet D., Le Merrer M., Munnich A., Apte S. S., Cormier-Daire V. (2008) Nat. Genet. 40, 1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ahram D., Sato T. S., Kohilan A., Tayeh M., Chen S., Leal S., Al-Salem M., El-Shanti H. (2009) Am. J. Hum. Genet. 84, 274–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsutsui K., Manabe R., Yamada T., Nakano I., Oguri Y., Keene D. R., Sengle G., Sakai L. Y., Sekiguchi K. (2010) J. Biol. Chem. 285, 4870–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sawada T., Sugawara Y., Asai T., Aida N., Yanagisawa T., Ohta K., Inoue S. (2006) J. Histochem. Cytochem. 54, 1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tsuruga E., Irie K., Yajima T. (2002) J. Dent. Res. 81, 771–775 [DOI] [PubMed] [Google Scholar]
- 20. Staszyk C., Gasse H. (2004) Anat. Histol. Embryol. 33, 17–22 [DOI] [PubMed] [Google Scholar]
- 21. Kawamoto T. (1990) J. Histochem. Cytochem. 38, 1805–1814 [DOI] [PubMed] [Google Scholar]
- 22. Nishimura R., Hata K., Harris S. E., Ikeda F., Yoneda T. (2002) Bone 31, 303–312 [DOI] [PubMed] [Google Scholar]
- 23. Yamamoto T., Miyoshi H., Yamamoto N., Yamamoto N., Inoue J., Tsunetsugu-Yokota Y. (2006) Blood 108, 3305–3312 [DOI] [PubMed] [Google Scholar]
- 24. Nakao K., Morita R., Saji Y., Ishida K., Tomita Y., Ogawa M., Saitoh M., Tomooka Y., Tsuji T. (2007) Nat. Methods 4, 227–230 [DOI] [PubMed] [Google Scholar]
- 25. Shiga M., Saito M., Hattori M., Torii C., Kosaki K., Kiyono T., Suda N. (2008) Cell Tissue Res. 331, 461–472 [DOI] [PubMed] [Google Scholar]
- 26. Handa K., Saito M., Yamauchi M., Kiyono T., Sato S., Teranaka T., Sampath Narayanan A. (2002) Bone 31, 606–611 [DOI] [PubMed] [Google Scholar]
- 27. Nishida E., Sasaki T., Ishikawa S. K., Kosaka K., Aino M., Noguchi T., Teranaka T., Shimizu N., Saito M. (2007) Gene 404, 70–79 [DOI] [PubMed] [Google Scholar]
- 28. Nakajima M., Kizawa H., Saitoh M., Kou I., Miyazono K., Ikegawa S. (2007) J. Biol. Chem. 282, 32185–32192 [DOI] [PubMed] [Google Scholar]
- 29. Yokoi T., Saito M., Kiyono T., Iseki S., Kosaka K., Nishida E., Tsubakimoto T., Harada H., Eto K., Noguchi T., Teranaka T. (2007) Cell Tissue Res. 327, 301–311 [DOI] [PubMed] [Google Scholar]
- 30. Hasegawa T., Suzuki H., Yoshie H., Ohshima H. (2007) Cell Tissue Res. 329, 259–272 [DOI] [PubMed] [Google Scholar]
- 31. Nanci A., Bosshardt D. D. (2006) Periodontol 2000 40, 11–28 [DOI] [PubMed] [Google Scholar]
- 32. Goetsch S. C., Hawke T. J., Gallardo T. D., Richardson J. A., Garry D. J. (2003) Physiol. Genomics 14, 261–271 [DOI] [PubMed] [Google Scholar]
- 33. Cheng J., Grande J. P. (2002) Exp. Biol. Med. 227, 943–956 [DOI] [PubMed] [Google Scholar]
- 34. Heldin C. H., Miyazono K., ten Dijke P. (1997) Nature 390, 465–471 [DOI] [PubMed] [Google Scholar]
- 35. Cohn R. D., van Erp C., Habashi J. P., Soleimani A. A., Klein E. C., Lisi M. T., Gamradt M., ap Rhys C. M., Holm T. M., Loeys B. L., Ramirez F., Judge D. P., Ward C. W., Dietz H. C. (2007) Nat. Med. 13, 204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chou Y. T., Wang H., Chen Y., Danielpour D., Yang Y. C. (2006) Oncogene 25, 5547–5560 [DOI] [PubMed] [Google Scholar]
- 37. Bunton T. E., Biery N. J., Myers L., Gayraud B., Ramirez F., Dietz H. C. (2001) Circ. Res. 88, 37–43 [DOI] [PubMed] [Google Scholar]
- 38. Charbonneau N. L., Ono R. N., Corson G. M., Keene D. R., Sakai L. Y. (2004) Birth Defects Res. C Embryo Today 72, 37–50 [DOI] [PubMed] [Google Scholar]
- 39. Brooke B. S., Karnik S. K., Li D. Y. (2003) Trends Cell Biol. 13, 51–56 [DOI] [PubMed] [Google Scholar]
- 40. Gregory K. E., Ono R. N., Charbonneau N. L., Kuo C. L., Keene D. R., Bächinger H. P., Sakai L. Y. (2005) J. Biol. Chem. 280, 27970–27980 [DOI] [PubMed] [Google Scholar]
- 41. Ramirez F., Rifkin D. B. (2009) Curr. Opin. Cell Biol. 21, 616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Massagué J. (2008) Mol. Cell 29, 149–150 [DOI] [PubMed] [Google Scholar]
- 43. Vollbrandt T., Tiedemann K., El-Hallous E., Lin G., Brinckmann J., John H., Bätge B., Notbohm H., Reinhardt D. P. (2004) J. Biol. Chem. 279, 32924–32931 [DOI] [PubMed] [Google Scholar]
- 44. Hutchinson S., Furger A., Halliday D., Judge D. P., Jefferson A., Dietz H. C., Firth H., Handford P. A. (2003) Hum. Mol. Genet. 12, 2269–2276 [DOI] [PubMed] [Google Scholar]
- 45. Baldock C., Siegler V., Bax D. V., Cain S. A., Mellody K. T., Marson A., Haston J. L., Berry R., Wang M. C., Grossmann J. G., Roessle M., Kielty C. M., Wess T. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11922–11927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee S. S., Knott V., Jovanovi J., Harlos K., Grimes J. M., Choulier L., Mardon H. J., Stuart D. I., Handford P. A. (2004) Structure 12, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hubmacher D., El-Hallous E. I., Nelea V., Kaartinen M. T., Lee E. R., Reinhardt D. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6548–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kuo C. L., Isogai Z., Keene D. R., Hazeki N., Ono R. N., Sengle G., Bächinger H. P., Sakai L. Y. (2007) J. Biol. Chem. 282, 4007–4020 [DOI] [PubMed] [Google Scholar]
- 49. Sabatier L., Chen D., Fagotto-Kaufmann C., Hubmacher D., McKee M. D., Annis D. S., Mosher D. F., Reinhardt D. P. (2009) Mol. Biol. Cell 20, 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kinsey R., Williamson M. R., Chaudhry S., Mellody K. T., McGovern A., Takahashi S., Shuttleworth C. A., Kielty C. M. (2008) J. Cell Sci. 121, 2696–2704 [DOI] [PubMed] [Google Scholar]
- 51. Isogai Z., Ono R. N., Ushiro S., Keene D. R., Chen Y., Mazzieri R., Charbonneau N. L., Reinhardt D. P., Rifkin D. B., Sakai L. Y. (2003) J. Biol. Chem. 278, 2750–2757 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.