Background: Resistance to β-lactam antibiotics is regulated by the bla operon.
Results: The fate of BlaR1, BlaI, and β-lactamase in the course of induction of resistance is evaluated.
Conclusion: BlaR1 fragments in a specific way to allow recovery from induction of resistance.
Significance: The processes of induction of resistance and recovery from it have been explained.
Keywords: Antibiotic Resistance, Infectious Diseases, Proteolytic Enzymes, Signal Transduction, Staphylococcus aureus
Abstract
The fates of BlaI, the gene repressor protein for the bla operon, BlaR1, the β-lactam sensor/signal transducer, and PC1 β-lactamase in four strains of Staphylococcus aureus upon exposure to four different β-lactam antibiotics were investigated as a function of time. The genes for the three proteins are encoded by the bla operon, the functions of which afford inducible resistance to β-lactam antibiotics in S. aureus. BlaR1 protein is expressed at low copy number. Acylation of the sensor domain of BlaR1 by β-lactam antibiotics initiates signal transduction to the cytoplasmic domain, a zinc protease, which is activated and degrades BlaI. This proteolytic degradation derepresses transcription of all three genes, resulting in inducible resistance. These processes take place within minutes of exposure to the antibiotics. The BlaR1 protein was shown to undergo fragmentation in three S. aureus strains within the time frame relevant for manifestation of resistance and was below the detection threshold in the fourth. Two specific sites of fragmentation were identified, one cytoplasmic and the other in the sensor domain. This is proposed as a means for turnover, a process required for recovery from induction of resistance in S. aureus in the absence of the antibiotic challenge. In S. aureus not exposed to β-lactam antibiotics (i.e. not acylated by antibiotic) the same fragmentation of BlaR1 is still observed, including the shedding of the sensor domain, an observation that leads to the conclusion that the sites of proteolysis might have evolved to predispose the protein to degradation within a set period of time.
Introduction
The BlaR1 protein of methicillin-resistant Staphylococcus aureus (MRSA)3 is an integral membrane protein involved in sensing of the presence of β-lactam antibiotics and transduction of the information to the cytoplasm (Fig. 1). Antibiotic acylates the C-terminal sensor domain located on the surface of the plasma membrane by a mechanism involving a surface loop of the protein (1) and an elaborate process that leads to N-decarboxylation of a lysine within the antibiotic-binding site (2–4). The acylated species enjoys a longevity that often exceeds the doubling time of most S. aureus strains; hence, a single BlaR1 modification by the antibiotic is sufficient for the entire bacterial generation (5). It has been proposed that the cytoplasmic domain of BlaR1 is a zinc protease (6). Hence, the surface sensing of the antibiotic (the signal) is amplified by the protease activity of the cytoplasmic domain. As we will disclose in this report, the copy numbers of the BlaR1 protein are very small, so this catalytic activity is critical for signal amplification.
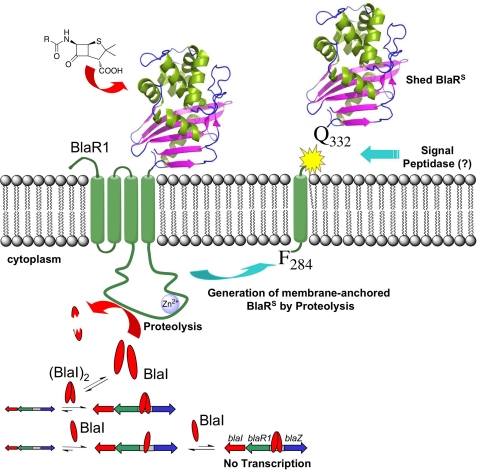
FIGURE 1.
Schematic of the proposed mechanism of activation and turnover of BlaR1. The process of antibiotic recognition on the cell surface leads to activation of the zinc protease in the cytoplasmic domain. The protease activity degrades BlaI, resulting in derepression of the antibiotic resistance genes, including that for BlaR1 itself. BlaR1 experiences fragmentation at two discrete sites, including one that sheds the sensor domain (BlaRS) from the membrane.
It is also argued that once the signal is transduced to the cytoplasmic side, the zinc protease domain experiences autolytic fragmentation between amino acids Arg-293 and Arg-294, an event that was proposed to activate the protease domain from latency (6). The now active protease domain would proteolyze the gene repressor BlaI, inducing expression of BlaI, β-lactamase, and BlaR1 itself as a mechanism against β-lactam antibiotics.
Here, we examine the time dependence of the events involving BlaI, β-lactamase, and BlaR1 within four strains of S. aureus in the presence of four β-lactam antibiotics. We disclose herein that BlaR1 in some strains of S. aureus experiences the aforementioned fragmentation within a time frame relevant to induction of resistance. Furthermore, we document and characterize three BlaR1 species, two of which are fragmentation products. One fragmentation takes place in the cytoplasmic domain, and another takes place in the sensor domain. The proteolytic event in the sensor domain leads to shedding of the domain into the medium from the surface of the membrane. These fragmentation events are proposed herein as regulatory events necessary for the recovery from the induction of resistance in the absence of antibiotic.
EXPERIMENTAL PROCEDURES
Strains
S. aureus strains NRS128 (also designated NCTC8325 (RN0031)), NRS123 (also designated MW2, C1999000459, USA400, and 99065), NRS70 (also designated N315), and NRS144 (also designated RN4220) were obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) program. S. aureus strain MRSA252 was purchased from the American Type Culture Collection (ATCC® number BAA-1720).
Minimal Inhibitory Concentration Determinations
The minimal inhibitory concentrations (MICs) of β-lactam compounds were determined by the broth microdilution method, as recommended by the Clinical and Laboratory Standards Institute (7). The MICs were determined in BBLTM Mueller Hinton II broth using a bacterial inoculum of 5 × 105 cfu/ml. All plates were incubated at 37 °C for 20 h before the results were interpreted. The procedure for MIC determination in the presence of recombinant BlaRS (rBlaRS)is described in the supplemental material.
S. aureus Cultures Grown in the Presence of Sub-MIC Concentrations of β-Lactam Antibiotics
We compared the levels of BlaI and BlaR1 proteins in S. aureus NRS128, NRS123, NRS70, and MRSA252. The different cultures were first grown to exponential phase, at which point different β-lactam antibiotics were added at sub-MIC concentrations, and the cultures were allowed to grow for different durations to a maximum of 3 h. The concentrations of antibiotics were as described in supplemental Table S1: 3.2-fold below the MIC for penicillin G (PEN), ampicillin (AMP), and oxacillin (OXA); and 6.4-fold below MIC for 2-(2′-carboxyphenyl)-benzoyl-6-aminopenicillanic acid (CBAP) (in order to make it comparable with the concentrations used in a key previous publication) (6). For each strain, 250 ml of Luria-Bertani (LB) medium in a 3-liter Erlenmeyer flask was inoculated with 250 μl of an overnight culture (stationary phase). The cultures were grown at 37 °C, 220 rpm, until A625 reached 0.8 (∼4 h after inoculation). At this point, the cultures were divided in 30-ml aliquots in sterile 125-ml Erlenmeyer flasks. One 30-ml culture was grown in the absence of β-lactam compounds as the non-induced control; the other aliquots were supplemented with the corresponding concentration of each one of the β-lactam antibiotics assayed. Cultures were then grown at 37 °C, 220 rpm. Five-ml aliquots were taken from each culture at 15 min, 30 min, 1 h, and 3 h. After 3 h of growth, all non-induced cultures reached an A625 of 1.9–2.2. After 3 h, the β-lactam-induced NRS128 cultures reached an A625 of 1.8–1.9; the β-lactam-induced NRS123 cultures reached an A625 of 1.2–1.3 upon induction with OXA and CBAP and of 1.8–2 upon induction with PEN and AMP; the β-lactam-induced NRS70 cultures reached an A625 of ∼1.2 upon induction with OXA and CBAP; and the β-lactam-induced MRSA252 cultures reached an A625 of 2.1–2.2. Each 5-ml aliquot was centrifuged for 30 min at 3200 × g and 4 °C, and the supernatant and cell pellets were separated. The supernatants (culture media) were kept at 4 °C, and β-lactamase activity was assayed immediately; the cell pellets were frozen at −20 °C. For BlaR1 and BlaI protein detection in whole-cell extracts, the cell pellets were thawed and resuspended in Lysis Buffer 1 (100 mm sodium phosphate, pH 7.5, 50 mm NaHCO3, 1× Complete EDTA-free protease inhibitor mixture, 1 mm EDTA, 20 mm MgCl2, 15 μg/ml DNase I, 15 μg/ml RNase A, 200 μg/ml lysostaphin) and incubated for 30 min at 37 °C. Total protein was quantified using the BCA protein assay kit (Pierce, Thermo Scientific); for each sample, a 60- or 80-μg portion of the total protein was loaded in Tris-glycine SDS-polyacrylamide gels. The protocol for membrane protein preparation is described in the supplemental material.
Recombinant BlaR1 Expression in Escherichia coli
The blaR1 gene was amplified from plasmid pI258 of S. aureus NRS128 using the polymerase chain reaction (PCR) and cloned in pET-24a(+) vector between NdeI and XhoI sites (supplemental material). This construct, designated pET-24a(+)_BlaRHis6, allowed inducible expression of BlaR1 from the T7 promoter, with eight additional amino acids on the C terminus: LEHHHHHH (the first two amino acids introduced by the XhoI cloning site, and the His6 tag). E. coli BL21 StarTM(DE3) competent cells were transformed with plasmid pET-24a(+)_BlaRHis6, and transformants were selected on kanamycin plates. Recombinant BlaR1 (rBlaR1) was expressed in E. coli BL21 StarTM(DE3) harboring plasmid pET-24a(+)_BlaRHis6, upon induction with isopropyl β-d-1-thiogalactopyranoside at 28 °C. Membrane proteins were prepared by ultracentrifugation from a 500-ml culture. A detailed protocol for the induction and membrane protein preparation is described in the supplemental material.
BlaRS and BlaI Recombinant Proteins and Specific Antibodies
rBlaRS and rBlaI expressed in E. coli were purified as described previously (5, 8). Rabbit polyclonal antibody against BlaRS (anti-BlaRS, 1.93 mg/ml) and rabbit polyclonal antibody against BlaI (anti-BlaI, 1.86 mg/ml) were generated by immunization of a rabbit with either purified rBlaRS or rBlaI protein, respectively (EZBiolab). The antibodies were (NH4)2SO4-precipitated from the sera or purified by Protein A chromatography by EZBiolab. Stabilized goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (anti-rabbit-HRP) was purchased from Pierce, Thermo Scientific.
Western Blot
For the detection of BlaR1, the samples were resolved in 10, 11, or 12% Tris-glycine SDS-PAGE. For the detection of BlaI, the samples were separated in 15% Tris-glycine SDS-PAGE. After electrophoresis, the proteins were transferred to 0.45-μm nitrocellulose membranes (Trans-Blot® transfer medium, Bio-Rad), using Tris-glycine-methanol buffer (25 mm Tris, 192 mm glycine, pH 8.3, 20% methanol) at 4 °C, 2 h, 200 mA, or to 0.45 μm polyvinylidene fluoride (PVDF) membranes, using CAPS buffer (10 mm CAPS, pH 11.0, 10% methanol), at 4 °C, 1.5 h, 140 mA, when indicated. The membranes were blocked at 4 °C overnight in 10 ml of Blotto (100 mm Tris-HCl, pH 7.5, 150 mm NaCl, 3% bovine serum albumin, 3% nonfat milk, 0.02% sodium azide). In the case of Western blot analysis on S. aureus extracts, the membranes were blocked with Blotto supplemented with 100 μg/ml human IgG (Sigma), and 1× Protein A/G blocker (GenScript), in order to block the IgG-binding proteins present in the cell extracts (including Protein A). BlaI and BlaR1 were detected by immunoblot analysis using the rabbit anti-BlaI and anti-BlaRS primary antibodies, respectively (1:1000 dilution in both cases), and a 1:15,000 dilution of anti-rabbit-HRP secondary antibody. The antibodies were diluted in Tween/Tris-buffered saline (100 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Tween 20) with 1.1% nonfat dry milk. In the case of Western blot analyses on S. aureus extracts, the antibody dilutions were also supplemented with 10 μg/ml human IgG and 1× Protein A/G blocker. The membranes were washed with Tween/Tris-buffered saline. The immune complexes were detected using an enhanced chemiluminiscence system (SuperSignal® West Dura Extended Duration substrate) according to the manufacturer's instructions and CL-XPosureTM films (Thermo Scientific). Alternatively, the ONE-HOUR IP-WesternTM kit (rabbit; GenScript) was used to detect BlaR1, according to the manufacturer's instructions. For each membrane, 40 μg of anti-BlaRS antibody were incubated with 100 μl of IP-WB1 solution for 2 h at room temperature.
β-Lactamase Activity Assays
The presence of PC1 β-lactamase in the media of S. aureus cultures was evaluated using the chromogenic β-lactam nitrocefin as substrate. The medium was separated from the cells by centrifugation for 30 min at 3200 × g at 4 °C. A 1-ml portion of culture was first equilibrated to room temperature, and the reaction was started by the addition of 10 μl of a 10 mm nitrocefin stock solution in dimethyl sulfoxide (DMSO) (final nitrocefin concentration was 99 μm). Nitrocefin hydrolysis was monitored at 500 nm, for 2 min at room temperature. The initial rate was obtained from the slope of the time course, which was linear in the first 2 min of the reaction. At a saturating concentration of nitrocefin, the initial rate measured is a good estimation of the Vmax, which is directly proportional to the concentration of PC1 β-lactamase in the medium sample.
Immunoaffinity Purification of Full-length BlaR1 and Its C-terminal Fragments
Immobilized anti-BlaRS antibody (anti-BlaRS/Protein A resin) was prepared as described earlier (9). A detailed protocol is provided in the supplemental material. The anti-BlaRS/Protein A resin were stored at 4 °C.
Prior to specific immunoprecipitation of BlaR1 and its proteolytic fragments from either S. aureus whole-cell extracts or culture media, the S. aureus IgG-binding proteins were removed by incubation with IgG-SepharoseTM 6 Fast Flow (GE Healthcare). Because the anti-BlaRS antibody can efficiently immunoprecipitate the denatured target (as determined in preliminary experiments with purified rBlaRS; data not shown), the samples were supplemented with SDS and boiled for 5 min. For the immunoprecipitation of the BlaR1 C-terminal fragment from culture medium, the medium was first incubated with IgG-SepharoseTM 6 Fast Flow, and the supernatant was then supplemented with SDS and boiled. For immunoprecipitation of BlaR1 from cell extracts, the cells were first resuspended in buffer with SDS and boiled, and after centrifugation, the supernatant was incubated with IgG-SepharoseTM 6 Fast Flow. In either case, the sample was then supplemented with Triton X-100 (2.5% final concentration), mixed with anti-BlaRS/Protein A resin, and incubated at 4 °C overnight. After a brief centrifugation, the supernatant (unbound proteins) was collected, and the resin (bound protein) was washed. The bound proteins were eluted with 2× Laemmli sample buffer (non-reducing), and a small aliquot was resolved by SDS-PAGE and transferred to PVDF membrane. BlaR1 protein and fragments were identified by immunoblot analysis, as described above. A detailed protocol is provided in the supplemental material.
For in-gel digestion and mass spectrometry analysis, the eluted proteins were separated on SDS-PAGE and stained with Coomassie Brilliant Blue, and the protein band of interest was cut from the gel and processed, as described below. For Edman degradation, the eluted proteins were separated on SDS-PAGE, transferred to 0.2-μm PVDF membrane (Bio-Rad), and stained with Coomassie Brilliant Blue; the protein band of interest was cut from the membrane and sent for analysis (Proseq Inc., Boxford, MA).
Preparative Purification of Recombinant BlaR1 Expressed in E. coli
E. coli BL21 StarTM(DE3) cells harboring pET-24a(+)_BlaRHis6 plasmid were induced with isopropyl β-d-1-thiogalactopyranoside to express rBlaR1 protein, as described above. Cells were centrifuged (30 min at 3000 × g), and 200 mg of wet pellet was resuspended in 3 ml of 25 mm Hepes buffer, pH 7.5, supplemented with 1 mm EDTA and 1× Complete EDTA-free protease inhibitor mixture (Roche Applied Science). Cells were lysed by sonication at 4 °C, supplemented with 2% final concentration of SDS, and boiled for 5 min. After centrifugation (15 min at 8000 × g) the soluble fraction was collected and supplemented with 10% glycerol and 62.5 mm Tris-HCl buffer, pH 6.8. The total soluble proteins were separated on 10% Tris-glycine SDS-PAGE using a PrepCell (Bio-Rad, model 491, 28-mm gel tube) continuous electrophoresis unit. Two-ml fractions were collected (at 1 ml/min). The presence of rBlaR1 in each fraction was evaluated in 20-μl aliquots by immunoblot analysis using anti-BlaRS-specific antibody, as described above. The fractions containing rBlaR1 were pooled, and Triton X-100 was added to a final concentration of 1% before immunoaffinity purification using the anti-BlaRS/Protein A resin as described above. Proteins bound to the resin were eluted with non-reducing Laemmli sample buffer and separated again on 10% SDS-PAGE using a PrepCell continuous electrophoresis unit. This purification step allowed separation of rBlaR1 from the immunoglobulins. The presence of rBlaR1 in the fractions was followed by immunoblot analysis. Fractions containing the pure rBlaR1 protein were pooled and concentrated using a 10-kDa Amicon Ultra device (Millipore). Non-reducing Laemmli sample buffer was added to this concentrated protein fraction, and the sample was run on 10% SDS-PAGE and transferred to a 0.2-μm PVDF membrane. rBlaR1 was visualized by Coomassie staining. The protein band was cut and sent for N terminus determination by Edman degradation (Proseq Inc.).
In-gel Trypsin and Pepsin Digestions and LC/MS/MS Analysis
Individual bands were excised with a clean razor blade, placed into individual tubes, and digested in gel essentially according to previously published work (10). Briefly, gels were dehydrated and rehydrated with alternating addition of 2:1 acetonitrile (ACN), 50 mm ammonium bicarbonate. Samples were reduced with 20 mm dithiothreitol for 30 min at 60 °C and alkylated with 10 mm iodoacetamide for 20 min at room temperature. Bands intended for pepsin digestion were then exchanged with alternating washes of ACN and 10 mm HCl. The samples were then dried in a SpeedVac. Gel bands were swollen in ∼20 μl of sequencing grade trypsin (20 ng/μl) or pepsin (75 ng/μl) and allowed to absorb for 30 min at 4 °C. A 50–100-μl portion of 50 mm ammonium bicarbonate (trypsin digestions) or 10 mm HCl (pepsin digestions) was added to cover the band and then incubated overnight at 37 °C for complete proteolysis or 6 h for the pepsin digestion. The liquid was removed and pooled with an additional portion of 30% ACN, 2% formic acid and then dried in a SpeedVac. In-gel pepsin digests were quenched first by the addition of 1 m NH4OH and then reacidified.
Samples were resuspended in 15 μl of 0.5% formic acid and then Zip-tip-cleaned (Millipore). A 2-μl aliquot of the sample (10 μl total) was injected directly onto a 75 μm × 15-cm column (in-house packed with Jupiter 90 Å Proteo C12), running at 300 nl/min. Sample was washed for 12 min with 2% ACN, 0.1% formic acid and then separated over a 60-min gradient from 2 to 35% B (B = 98% ACN, 0.1% formic acid). Analysis by MS/MS was performed on a QTrap 5500 mass spectrometry system (ABSciex) running in ion trap IDA mode coupled to a two-dimensional Eksignet Ultra Nano ultrahigh performance liquid chromatograph. Acquisition parameters were essentially identical to those reported earlier (11). Two summed survey scans (EMS) were followed by six data-dependent full-scan MS/MS scans (enhanced product ion) by the TOP6 method. Dynamic fill time was used with a total ion current target of 1 × 107 ions for EMS and 1 × 108 ions for enhanced product ion scans. Data files were searched using the Mascot (Matrix Science) and Paragon (ABSciex) search engines. Briefly, mass tolerance was set to 2.0, and MS/MS was set to 0.8 Da. Two missed cleavages were considered to decrease protease specificity. For Mascot, carbamidomethylation of Cys was defined as a permanent modification, and variable modifications of carbamidomethylation were considered on Lys residues and peptide N termini. Deamidation on Asn and Gln and oxidation of Met were also considered as variable modifications. Paragon utilizes a custom set of modifications, which are detailed in the literature (12). As one example of the instances of full-length BlaR1 identification, 15 peptides with confidence >95% were detected and sequenced. Sequence coverage was 30% with respect to the full-length protein entry and 62% based on the new N terminus reported here. False positive assessment was performed in accordance with the Molecular and Cellular Proteomics guidelines (13) and are included as fully annotated spectra and sequences within (supplemental Figs. S6, S7, S9–S13, S15, and S16). In order to address the question of which peptide(s) are at the N terminus of the protein, the data were searched using semitryptic tolerances. This enables the search engine to identify peptides that are non-tryptic for either the N or C terminus but not both. This allows for the processed N terminus of a protein to be detected and determined. Non-canonical search tolerances are automatically considered in the Paragon search algorithm and are included in these data. All BlaR1 spectra were manually annotated for confirmation.
Multiple-reaction monitoring (MRM LC/MS/MS)
Targeted detection and sequencing of specific peptides from BlaR1 for enhanced sensitivity and quantification were performed on samples prepared identically. Generation of the MRM transitions was determined using MRM-initiated detection and sequencing as well as the empirical MS/MS data from the LC/MS/MS of the gel bands (11, 14, 15). In-gel-digested E. coli-expressed rBlaR1 was used to determine appropriate peptide transitions for analysis. Briefly, three or four transitions were generated for each peptide from the empirical MS/MS spectra obtained from data-dependent analysis and search results. MRM acquisition was performed on a QTrap 5500 running in triple-quadrupole mode. Quantification was performed with peak areas using Multi-Quant (AB Sciex), and relative peak ratios for identical peptides are illustrated between samples (supplemental Figs. S9 and S13).
RESULTS
Time Dependence of Activation of the bla System; Monitoring of BlaR1, BlaI, and β-Lactamase Levels
In order to evaluate the time course of activation of the bla system, we monitored the levels of the three proteins encoded in the operon (BlaR1, BlaI, and β-lactamase) in S. aureus strains NRS70, NRS123, NRS128, and MRSA252. The process of induction was with concentrations of antibiotic below the MICs of CBAP, PEN, AMP, and OXA (supplemental Table S1). CBAP is an excellent inducer of the system, but it is not a clinical drug and is no longer available commercially. Ampicillin is an extended spectrum penicillin, penicillin G is a β-lactamase-sensitive narrow spectrum penicillin, and oxacillin is a β-lactamase-resistant narrow spectrum penicillin (in the same class as methicillin).
All four strains used in this study contain the complete bla system. The genes of the bla operon are localized in the chromosome of strain MRSA252 and in plasmids in the other three strains (plasmid pI258 in NRS128, plasmid pMW2 in strain NRS123, and plasmid pN315 in NRS70). Strains NRS70 and MRSA252 also contain the complete mec operon (localized in plasmid pN315 and in the chromosome, respectively). Strain NRS123 also presents the mec operon (localized in plasmid pMW2), but the mecI gene is missing, and the mecR1 gene has a deletion and codes only for amino acids 1–325. This strain is believed to rely on BlaR1 and BlaI for the regulation of induction of β-lactamase and PBP2a.
The BlaI proteins from the different strains used here have 98–100% identity (100% identity between BlaI from NRS128 and MRSA252) and only 59–60% identity with MecI of strains NRS70 and MRSA252 (16, 17). The BlaR1 proteins from the different strains that we used exhibit 93–99% identity (99% identity between BlaR1 from NRS128 and MRSA252) and only 30% identity with MecR1 from strain MRSA252 (with 40% homology in the sensor domain region). The β-lactamases from the different strains exhibit 98–100% identity (100% identity for the enzymes from NRS128 and MRSA252).
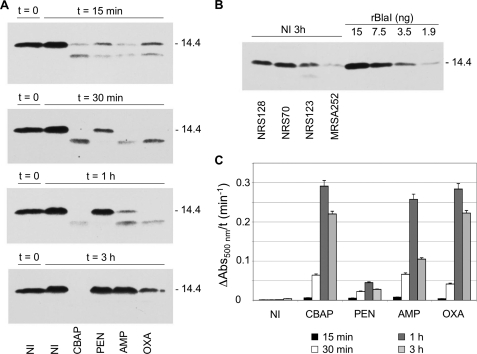
We will discuss first the changes observed with BlaI. In the absence of antibiotic, BlaI, a protein of 14.4 kDa, remains stable and intact (Fig. 2A). As we determined earlier, concentrations of BlaI in NRS128 and NRS70 are 2.3–6.4 μm (NRS128) and 1.3–3.6 μm (NRS70) in the cells at the exponential phase of growth and 0.98–2.7 μm (NRS128) and 0.75–2.1 μm (NRS70) in the stationary phase (8). All four antibiotics initiate the process of degradation of BlaI within minutes. As depicted in Fig. 2 for strain NRS128, induction with CBAP was rapid (turnover of BlaI) and was sustained for the 3-h monitoring period. With the other three antibiotics as well, induction was rapid, but we observed gradual expression of BlaI, which was no longer degraded toward the end of the time course (Fig. 2A). This pattern was the same for strains NRS70, NRS123, and MRSA252 (supplemental Fig. S1). In general, CBAP was the best inducer, and oxacillin was the second best, but with penicillin G, ampicillin, and oxacillin, induction was gradually reversed in the course of 3 h. A comparison among the different strains showed that the concentration of BlaI in strain NRS123 was somewhat lower than those in NRS128 and NRS70 but that it was significantly lower in strain MRSA252 (Fig. 2B).
FIGURE 2.
Time course of BlaI turnover and resynthesis in whole-cell extracts, and of PC1 β-lactamase activity in the media, after induction of the bla system by β-lactam antibiotics in S. aureus NRS128 (the results for the three other strains are given in the supplemental material). A, the BlaI protein was detected in whole-cell extracts (60-μg portion of total protein) of non-induced (NI) and induced (15 min, 30 min, 1 h, and 3 h) cultures by Western blot analysis, using anti-BlaI antibody. The proteins were separated by SDS-PAGE (15% gels) and transferred to nitrocellulose membrane, and the IgG-binding proteins were blocked with human IgG and Protein A/G blocker (GenScript) prior to the immunoblot analysis. Numbers to the right indicate the position of migration of the molecular mass markers (kDa). B, comparison of the amount of BlaI in whole-cell extracts of the strains NRS128, NRS123, NRS70, and MRSA252 (60-μg portion of total protein), using purified rBlaI as a standard. The cultures were grown in the absence of antibiotics for an additional 3 h after A625 of 0.8 was reached (non-induced condition at 3 h). C, the chromogenic β-lactam antibiotic nitrocefin was used as a reporter to monitor the β-lactamase levels in the culture media of non-induced cultures and after induction with CBAP, penicillin G, ampicillin, and oxacillin for different times (15 min (black bars), 30 min (white bars), 1 h (dark gray bars), and 3 h (light gray bars). All cultures were exposed to different antibiotics at the same A value (A625 = 0.8). Error bars, S.E.
With the antibiotic-induced degradation of BlaI, derepression of the resistance genes ensues. We monitored activity of β-lactamase in the media by the chromogenic nitrocefin assay in the same time course for degradation of BlaI. Within the first 15 min of monitoring, we already noted the enzyme activity, which increased in time in the presence of all four antibiotics and reached a maximum within 1 h (see Fig. 2C for NRS128). The levels of β-lactamase activity dropped subsequently to varying degrees in strain NRS128, depending on the antibiotic, but significant activity persisted for the duration of the experiment. In other strains, the β-lactamase activity showed a similar trend with some antibiotics, although with others, it kept on increasing (Figs. S2 and S3). As an example, with strain NRS70, the β-lactamase activity kept on increasing during the 3-h monitoring. There is correlation between the ability of the S. aureus PC1 β-lactamase to turn over these penicillins and the above observations (18, 19). MRSA emerged clinically in the face of the challenge by the then-new β-lactamase-resistant penicillins, which include oxacillin and methicillin. Whereas CBAP never became clinical, it is a very poor substrate for the PC1 β-lactamase (18); hence, it persists in the medium and is indeed the best inducer. Oxacillin is also a poor substrate for the PC1 β-lactamase (18), and it is a somewhat poorer inducer than CBAP. Ampicillin and penicillin G are good substrates and in general worse inducers (18, 19). Hence, qualitatively, if the antibiotic survives in the medium longer, it has the ability to maintain induction for a longer duration. The process is actually quite complex. We do not discount that the longevity of the acylated surface domain of BlaR1 with different antibiotics (5) and the ability of those species individually to manifest signal transduction to varying degrees should also play a role in these processes. They do, and they contribute to differences seen with various antibiotics in general.
Derepression of the transcription of the genes of the bla operon upon β-lactam induction also should result in increased expression of the β-lactam sensor/signal transducer membrane protein BlaR1 because the regulatory mechanism is the same. Here the situation becomes complex. Our discussion below will center primarily on the better studied strain NRS128, but we will also highlight the main differences observed with the other strains.
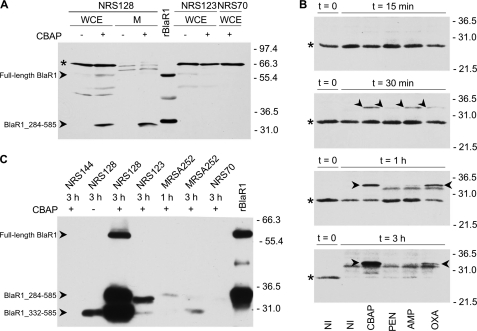
Visualization of full-length BlaR1 from NRS128 in Western blot assays using rabbit antibodies was hindered by binding of the primary antibody to the various IgG-binding proteins (including Protein A) in the 45–70 kDa region (supplemental Fig. S4). These proteins could not be blocked completely with human IgG and hence prevented visualization of full-length BlaR1. We pursued an alternative strategy using the ONE-HOUR IP-WesternTM kit (GenScript) to improve the blocking of the nonspecific interactions with IgG-binding proteins (Fig. 3A). Unfortunately, this method suffered from lower sensitivity, which also precluded clear detection of full-length BlaR1 (supplemental Fig. S5). Previous reports have identified a protein band of ∼69 kDa as full-length BlaR1 in gels (6, 20, 21), based on its theoretical mass of 69.3 kDa (amino acids 1–585). In the study of BlaR1 from S. aureus (6), the complement of the genes for the bla system was introduced into the strain S. aureus COL; hence, it is not exactly what we have done in the present study, where we have used S. aureus NRS128 directly. As described below, full-length BlaR1 is present at very low copies in S. aureus, even after induction by antibiotics. The band at slightly over 66 kDa that one sees in Fig. 3A is not full-length BlaR1 because it is a soluble protein (not membrane-bound). This assertion is also supported by other lines of evidence; the 66-kDa band was also detected in protein extracts of strains that lack the bla operon, as in strain S. aureus COL (supplemental Fig. S5); it is likely to be one of the S. aureus IgG-binding proteins. The full-length BlaR1 that we cloned in E. coli expressed well, as seen in whole-cell extract (Fig. 3A) and in membranes (data not shown). We know unequivocally from mass spectrometry sequence analyses of the protein bands at roughly 60 kDa and 33–35 kDa in E. coli (Fig. 3A, arrowheads) that they are BlaR1 species (supplemental Figs. S6 and S7). Hence, rBlaR1 expressed in E. coli runs as a 60-kDa protein although its theoretical mass is 70.3 kDa (amino acids 1–593; where the difference with BlaR1 from NRS128 is the C-terminal His6 tag). BlaR1 is an integral membrane protein with four predicted membrane-spanning regions, and as such it might not migrate according to the migration patterns of soluble proteins used as size standards in SDS-PAGE (22). However, it is also possible that BlaR1 would possess an N-terminal signal peptide sequence that could be cleaved after translocation in the membrane, giving rise to a smaller protein. The 33–35-kDa BlaR1 C-terminal fragment in E. coli was slightly bigger than the fragment seen in CBAP-induced NRS128, consistent with the mass difference due to the C-terminal His6 tag present in the E. coli expressed protein. Careful inspection of this gel showed vanishingly small quantities (Fig. 3A, asterisk) of a protein species corresponding to the 60-kDa BlaR1 band that is seen in E. coli in the case of the lanes for the samples from S. aureus. However, the quantities were so low that they could as well be nonspecific signals. We saw a couple of other similarly faint bands on the same gel, at lower molecular weights in the whole-cell extract of the strain NRS128, which might also be nonspecific. We estimate from synthetic peptides and serial dilutions from in-gel digests of BlaR1 expressed in E. coli that our targeted LC/MS/MS methods are sensitive down to a few dozen amol (10−18 mol) of extracted peptide digest on column (data not shown). This corresponds to a detection level of protein in gel of ∼50–500 pg in total. However, even with this extremely high level of sensitivity, we were unable to reliably detect peptide sequences in the 60-kDa CBAP-induced NRS128 bands corresponding to BlaR1. However, we saw a more abundant C-terminal 33–35-kDa BlaR1 fragment in the case of CBAP-induced strain NRS128 in both whole cells and the membrane fraction (Fig. 3A). This species accumulated in NRS128 with time (from 30 min to 3 h), and it was clearly detected upon induction with CBAP and oxacillin, the better inducers, and in small amounts with ampicillin at 30 min of induction (Fig. 3B). This fragment was not detected in whole-cell extracts of strains NRS123 and NRS70, but it was present in MRSA252 after 1 h of induction with all four antibiotics, although at lower quantity (Fig. 3A and supplemental Fig. S8). Thus, with this analysis in hand, we have detected a fragmentation product of the full-length BlaR1 and might have detected small traces of the full-length BlaR1 as well. However, to verify that indeed the trace that we detect in S. aureus is full-length BlaR1, the sample had to be enriched.
FIGURE 3.
Time dependence of accumulation of BlaR1 species in S. aureus strain NRS128 after activation of the bla system by β-lactams. A, BlaR1 detection in whole-cell extracts (WCE) and in membrane fractions (M) of NRS128, NRS123, and NRS70 cells, compared with rBlaR1. The S. aureus samples correspond to non-induced cultures (−) or cultures induced for 3 h with CBAP (+). The proteins were separated by SDS-PAGE (12% gel) and transferred to nitrocellulose membrane, and BlaR1 was detected by Western blot using anti-BlaRS antibodies and the ONE-HOUR IP-WesternTM kit (GenScript). The arrowheads indicate bands corresponding to the BlaR1 species, labeled accordingly. The faint band seen in S. aureus whole-cell extracts could correspond to full-length BlaR1 but could also be a nonspecific signal. Recombinant BlaR1 has an additional C-terminal tag, which includes the His6 tag. The asterisk indicates nonspecific bands (also see supplemental Fig. S5). B, BlaR1 levels in whole-cell extracts (80-μg portion of the total protein) of non-induced S. aureus NRS128 cultures (NI) and in S. aureus NRS128 cultures induced for different times (15 min, 30 min, 1 h, and 3 h) with the indicated antibiotic (the results for the three other strains are given in the supplemental material). The proteins were separated by SDS-PAGE (12% gels) and transferred to nitrocellulose membrane, and BlaR1 was detected by Western blot using anti-BlaRS antibodies and blocking with human IgG, Protein A/G blocker (GenScript). The arrowheads indicate bands corresponding to the 33–35-kDa membrane-anchored C-terminal BlaR1 fragment (BlaR1_284–585). The asterisks indicate nonspecific bands. C, BlaR1 immunoprecipitated from non-induced and β-lactam-induced S. aureus and from E. coli. BlaR1 was immunoaffinity purified with anti-BlaRS/Protein A resin from whole-cell extracts of NRS128, NRS123, NRS70, and MRSA252 non-induced (−) and 1- and/or 3-h CBAP-induced bacteria (+). Immunoprecipitated rBlaR1 from E. coli is also shown. The eluted proteins were separated by SDS-PAGE (11% gel) and transferred to PVDF membrane, and BlaR1 was visualized by Western blot using anti-BlaRS antibodies and the ONE-HOUR IP-WesternTM kit (GenScript). Numbers on the right indicate the position of migration of the molecular mass markers (kDa).
In-gel trypsin digestion of the 60-kDa and 33–35-kDa protein bands from whole-cell E. coli extracts allowed identification of several peptides from both the sensor and N-terminal domains of BlaR1 in both bands (supplemental Figs. S6 and S7). This confirms the presence of full-length BlaR1 in the 60-kDa protein and indicates that after fragmentation, both the N and C terminal halves of BlaR1 migrate in the 33–35 kDa region, of which only the C-terminal half is detected by the Western blot analysis, but both samples are present under the band. This last observation would appear to indicate that full-length BlaR1 in E. coli (which seemingly migrates as a 60-kDa protein but indeed has a theoretical mass of 70.3 kDa) is not running according to its true mass in SDS-PAGE, an observation that is made also for other membrane proteins (22).
BlaR1 Immunoprecipitation and N-terminal Sequencing
We enriched BlaR1 by immunoprecipitation with a specific antibody. We first treated the S. aureus extracts (NRS128, NRS123, NRS70, and MRSA252, both non-induced and CBAP-induced) with a human IgG-Sepharose resin, followed by incubation with BlaR1-specific antibody immobilized onto a Protein A resin. This method enriched samples such that by Western blotting we clearly detected the 60-kDa protein in CBAP-induced strain NRS128, together with two smaller fragments (the aforementioned 33–35-kDa fragment and a 30-kDa fragment; Fig. 3C). We will come back to the third species, the 30-kDa fragment, later. Mass spectrometry sequencing unequivocally identified all three as BlaR1 species. Recombinant BlaR1 expressed in E. coli was also immunoprecipitated and produced two of the protein bands seen in CBAP-induced NRS128 (Fig. 3C): the 60-kDa BlaR1 and a 33–35-kDa proteolysis product. In CBAP-induced NRS123-, NRS70-, and MRSA252-enriched samples, we still could not detect full-length BlaR1, which indicated that the levels of BlaR1 expression after induction in these strains are much lower than in NRS128 (Fig. 3C). The detection limit of purified recombinant BlaR1 sensor domain by Western blot was estimated at 6–13 ng, using the ONE-HOUR IP-WesternTM kit. In addition, we hasten to add that the yield of immunoprecipitation of full-length BlaR1 and of its C-terminal fragments could be different, given the high hydrophobic nature of the former segment. Its decreased solubility could result in higher losses during the immunoprecipitation procedure. In strain MRSA252, a 33–35-kDa fragment was observed after 1-h induction (Fig. 3C), and the smaller 30-kDa fragment was observed after 3-h induction (Fig. 3C); the quantities of both fragments were significantly lower than that seen in NRS128. In NRS123, we could also see the 33–35-kDa fragment seen in NRS128 (Fig. 3C). However, even after enrichment by immunoprecipitation, we still could not detect the 33–35-kDa BlaR1 proteolysis product in CBAP-induced NRS70 (Fig. 3C). It is important to point out that in samples from non-induced NRS128, we were also able to detect the 30-kDa BlaR1 fragment seen in CBAP-induced NRS128, although we still could not detect full-length BlaR1. The presence of this 30-kDa fragment in the absence of antibiotic indicates that BlaR1 fragmentation takes place even when the sensor domain of BlaR1 is not acylated by β-lactams (i.e. a non-induced system). This is an important observation for reversal of induction that we will return to subsequently.
The BlaR1-enriched samples were used for determination of the N-terminal sequences of the fragments. The N-terminal sequence of the 33–35-kDa fragment in recombinant BlaR1 from E. coli was determined to be F284NGKKSLLKR … by Edman degradation (Proseq Inc.). This produces a protein of 36.6-kDa theoretical mass, including the eight additional C-terminal amino acids introduced with the His6 tag (LEHHHHHH). We also tried to identify the N-terminal peptide using in-gel digestion LC/MS/MS analysis. Because the N terminus of the 33–35-kDa membrane-associated C-terminal BlaR1 fragment was previously reported as amino acid residue Arg-294 (6), we could not use trypsin digestion for this experiment because this protease would cleave after Arg-293. Hence, we chose pepsin instead, which would not cut at position Arg-293 and would generate peptides of appropriate size and coverage of the sequence for analysis. In-gel pepsin digestion of immunoprecipitated rBlaR1 from E. coli and MRM LC/MS/MS analysis allowed identification of the N-terminal peptide F284NGKKSL (supplemental Figs. S9 and S10), a determination that turned out to be identical to the N-terminal sequence by Edman degradation. We were also able to identify peptide KR293RLINIKEANL within the protein coverage, which indicates that cleavage in this strain did not occur between residues Arg-293 and Arg-294. The N-terminal sequence of the 33–35-kDa fragment from CBAP-induced NRS128 could not be determined by Edman degradation due to its significantly low quantity. Alternatively, we identified the F284NGKKSLL peptide via in-gel pepsin digestion and MRM LC/MS/MS analysis (supplemental Fig. S9), which is identical to the N-terminal peptide observed for the 36.6-kDa fragment of BlaR1 expressed in E. coli. This important finding reveals that in S. aureus strain NRS128, BlaR1 undergoes fragmentation upon exposure of the organism to β-lactam antibiotics at the very same sequence position where BlaR1 cloned in E. coli does. The importance of this observation is 2-fold. First, BlaR1 expressed in E. coli sequesters into the membrane correctly as a functional protein. Second, the fragmentation is now likely to be autolytic because a potential other protease effector that could possibly exist in S. aureus for the proteolytic processing of BlaR1 is not necessarily present in E. coli.
In immunoaffinity-purified samples, full-length BlaR1 from S. aureus strain NRS128 and that expressed in E. coli run in SDS-polyacrylamide gels as a protein of approximate apparent mass of 60 kDa (Figs. 3C and 4A). Again, the reader is reminded that the recombinant sample from E. coli is a bit larger, because of the His6 tag. Given the previously mentioned discrepancy with the theoretical mass of full-length BlaR1 as assessed by SDS-PAGE, we attempted to determine the N terminus of full-length BlaR1 in immunoprecipitated samples of CBAP-induced NRS128 and of BlaR1 expressed in E. coli. The protein quantity from the immunoaffinity-purified sample from E. coli, with the highest expression, was still insufficient for N-terminal sequencing by Edman degradation.
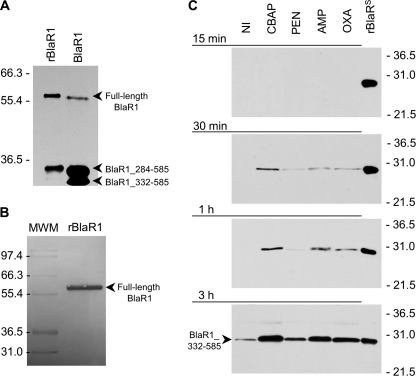
FIGURE 4.
Fragmentation of BlaR1 in CBAP-induced NRS128 and in E. coli and the shedding of its sensor domain in antibiotic-induced S. aureus NRS128. A, full-length BlaR1 in CBAP-induced NRS128 (right lane) and in E. coli (left lane) have comparable sizes, with that in E. coli larger by the size of the His6 tag moiety. BlaR1 immunoprecipitated from CBAP-induced NRS128 and from E. coli was separated by SDS-PAGE (10% gel), transferred to PVDF membrane, and detected by Western blot using the anti-BlaRS antibodies and the ONE-HOUR IP-WesternTM kit (GenScript). B, recombinant BlaR1 (∼60-kDa) obtained from E. coli whole-cell extracts by means of the preparative purification for Edman degradation, visualized in the Coomassie-stained PVDF membrane. Numbers on the left of A and B indicate the position of migration of the molecular mass markers (kDa). C, detection of the C-terminal domain of BlaR1 (BlaRS or BlaR1_332–585) in media of the S. aureus NRS128 culture after induction with different β-lactam antibiotics (CBAP, PEN, AMP, and OXA) for different times (15 min, 30 min, 1 h, and 3 h). The results for the three other strains are given in the supplemental material. The proteins were separated by SDS-PAGE (12% gels) and transferred to nitrocellulose membrane, and BlaR1 was detected by Western blot using antibodies specific to recombinant BlaRS and the ONE-HOUR IP-WesternTM kit. Numbers on the right indicate the position of migration of the molecular mass markers (kDa).
In order to address the identification of the N terminus of the 60-kDa BlaR1 band, we next purified large quantities of the 60-kDa BlaR1 protein expressed in E. coli for N-terminal sequencing by Edman degradation. We used a combination of immunoprecipitation and size exclusion chromatography using a PrepCell continuous electrophoresis unit. This protocol produced a sufficient quantity of purified full-length BlaR1 (Fig. 4B). N-terminal sequencing by Edman degradation was subsequently performed, but the N terminus of the 60-kDa BlaR1 could not be determined because the protein appears to be N-terminally blocked (Proseq Inc.). N-terminal blocking of bacterial proteins can be due to N-acetylation or N-formylation of methionine. N-Acetylation is an N-terminal modification in prokaryotes, which has so far been reported for proteins associated with translation (23) and secreted proteins (11). N-Formylmethionine is used to initiate protein synthesis in eubacteria (24), and although deformylation is generally highly efficient, high levels of expression in E. coli can result in partial retention of the formyl group (25, 26). However, it is also possible that the N-terminal modification is an artifact of the protein preparation.
Because Edman degradation was not successful, we attempted to determine the N terminus by partial proteolysis (independent in-gel digestions by trypsin and pepsin) and LC/MS/MS analysis. Notwithstanding considerable time and resource that was dedicated to this effort, a definitive N terminus could not be identified by any of these attempts. However, the peptides that were isolated and sequenced indicated that the protein was full-length BlaR1. In-gel trypsin digestion and LC/MS/MS analysis identified the peptide F60NNVNNQAPTVESK (supplemental Fig. S11). In-gel pepsin digestion of the same BlaR1 sample allowed identification of the peptides I48PFIPIKF and F58KFNNVNNQAPTVE (supplemental Fig. S12). The tryptic peptide F60NNVNNQAPTVESK was also identified in the 60-kDa BlaR1 band immunoprecipitated from CBAP-induced NRS128 (supplemental Fig. S13). In light of the fact that pepsin is predicted to cleave at many sites throughout the sequence of amino acids 1–48, we could not definitively discount the possibility that the N terminus could reside anywhere between amino acids 1 and 48. Unfortunately, technical challenges with analysis with all other proteases compatible with in-gel digestion (Lys-C and CNBr) did not shed additional light on the position of the actual N terminus within the amino acid sequence of residues 1–48 (data not shown). Chemical labeling of the N terminus is also unlikely to be successful because the Edman analysis revealed it to be blocked. Therefore, we are able to disclose that the 60-kDa BlaR1 from S. aureus strain NRS128 or that expressed in E. coli exhibits an N terminus that would start either at Ile-48 or at a point before this residue. Furthermore, our experiments reveal that the N terminus is blocked. This might be a natural process for membrane sequestration of full-length BlaR1.
Shedding of the BlaR1 Sensor Domain
As indicated earlier, we had identified three BlaR1 species from strain NRS128 (Fig. 3C). A striking and totally unexpected observation that we made was that the 30-kDa fragment containing the sensor domain accumulated in the culture media as a shed protein (Fig. 4C). The shedding of the domain took place even in the absence of induction by antibiotic. However, in the presence of antibiotics, we clearly saw more of the shed domain, which might be due to the up-regulation of expression of BlaR1. The process of shedding was more vigorous in the presence of the best inducer, CBAP, but it was seen for all four antibiotics that we used. Hence, as expression of BlaR1 is induced, one detects more of the shed domain in the medium. This shedding of the sensor domain of BlaR1 was not observed in the strains NRS123 and NRS70 within the time frame of our monitoring but was also observed in strain MRSA252 (supplemental Fig. S14). NRS123 and NRS70 seem to express BlaR1 at significantly lower levels upon induction, so we cannot rule out the possibility that the sensor domain is being shed but is below detection thresholds. The 30-kDa BlaR1 fragment was isolated by immunoprecipitation from the growth medium of strain NRS128 induced for 3 h with CBAP, and the N terminus was identified to be Q332SITD … by mass spectrometry (supplemental Figs. S15 and S16). This result indicated that the cleavage that releases the sensor domain occurs between amino acids Gly-331 and Glu-332. This type of processing was recently shown to be due to the action of type I signal peptidase in Staphylococcus epidermidis (27). BlaR1 from S. epidermidis has a high probability type I signal peptidase recognition sequence just to the C-terminal side of the last predicted transmembrane helix, and cleavage after Gly-331 was predicted for BlaR1 from S. epidermidis. Our experiments revealed that in strains of S. aureus that harbor BlaR1 sequences predicted to be substrates for type I signal peptidase, one sees shedding of the surface domain. We propose that the 35.5-kDa membrane-anchored BlaR1 fragment (amino acids 284–585) observed in these strains might be the substrate of the S. aureus type I signal peptidase (Fig. 1).
Mechanistic Implications of BlaR1 Fragmentation
Does the shedding event have a mechanistic implication? The function for the surface domain of BlaR1 that is accepted by the dogma is one of sensing the presence of β-lactam antibiotics. This function necessitates transduction of information into the cytoplasm for derepression of the antibiotic resistance genes, which could be envisioned for BlaR1 as an integral membrane protein. It is less obvious if the fragmentation that leads to the shedding of the sensor domain has any advantage for S. aureus in countering the effects of antibiotic.
We wondered whether the release of the sensor domain would serve the function of a vanguard by sequestering the incoming antibiotic, buying time for the organism to recruit its resources for induction of full-blown resistance mechanisms. To test for this, we determined the concentration of the shed domain within the growth medium by Western blotting. After 3 h of induction in the presence of antibiotic CBAP, we detected a mere 0.11 μg/ml (3.7 nm) of the BlaRS in the medium, a low concentration that no doubt is intimately linked to the low copy numbers of BlaR1 in the membrane. Regardless, the addition of recombinant purified BlaRS to the growth medium of strain NRS144, an S. aureus strain devoid of the bla system, increased the MIC by 2–4-fold for the antibiotic only after the concentration of the shed domain reached >11 μg/ml (supplemental Table S2). This argues against a beneficial contribution to the manifestation of resistance by the shedding event because the concentration of BlaRS in solution does not easily reach these high values. Furthermore, we know that actual manifestation of resistance in S. aureus strains takes minutes and not hours. Under these conditions, the increase in MIC in the presence of the purified sensor domain is due to the uncommon covalent antibiotic sequestration mechanism, documented earlier for unrelated systems (28). The sensor domain of BlaR1 is acylated by β-lactam antibiotics with long half-lives (3, 5). The absence of multiple turnover of antibiotic explains why high concentrations of BlaRS are required for an effect on the MIC to be observed.
In light of the above experiments, the alternative explanation for why shedding takes place is that it is a process in turnover of BlaR1, one step in multiple proteolytic fragmentation of the protein that turns it over. All proteins experience turnover, but the case for BlaR1 is special because it provides the mechanism for the recovery of the organism from the cost of recruitment of resistance mechanisms in the absence of antibiotics. As the exposure to antibiotics is removed, either by lack of availability or by the action of β-lactamase, the organism's mobilization for induction of resistance mechanisms should be reversed. Fragmentation of BlaR1 would afford this. This turnover of BlaR1 must also be present in all other strains, except the quantities of BlaR1 and its fragments in various strains (e.g. NRS123 and NRS70) are low to the degree that we were not able to visualize them. That BlaR1 has to experience turnover is a certainty in general, as is the case for any protein, but it becomes a requirement for recovery from mobilization against antibiotics in the absence of the antibiotic pressure.
DISCUSSION
The BlaR1 protein of S. aureus is a central player in resistance by this organism to β-lactam antibiotics. Its role as a β-lactam sensor/signal transducer is fully appreciated, but the details of its molecular events are less understood. The process of β-lactam sensing is elaborate, and it involves an uncommon N-carboxylated lysine that experiences N-decarboxylation on acylation of the protein with the antibiotic (2, 4). This N-decarboxylation of the lysine within the antibiotic-binding site affords longevity to the acyl-protein species, which in turn initiates signaling events from the surface of the membrane to the cytoplasm. The events of the transmembrane signaling are not known but must involve conformational changes in the protein that propagate from the membrane surface to the cytoplasm.
The events at the cytoplasmic side are not understood either, but these processes lead to the activation of the zinc protease domain of the protein that initiates proteolysis that derepresses the antibiotic resistance genes. An earlier study suggested that an autolytic proteolysis at the cytoplasmic domain, triggered by acylation of the surface domain by the antibiotic, was the activation step (6). Our results do not rule this out, although we see a lag in BlaR1 fragmentation compared with BlaI proteolysis. Nonetheless, a very significant observation is that in strains NRS128 and MRSA252, BlaR1 fragments are observed even in the absence of induction by antibiotics. Upon induction, although the manifestation of resistance commenced within minutes, BlaR1 fragmentation is observed only at longer times. Hence, the full picture is not in hand. As in any other signal transduction event, conformational changes are key. Our recent report that a conformational change is involved in activation of the cytoplasmic zinc protease AmpD suggests that a similar process for activation of the cytoplasmic domain of BlaR1 might be operative (29).
We documented that the onset of proteolytic processing of BlaI, leading to gene derepression and manifestation of the β-lactamase activity, commences within minutes of incubation of S. aureus with antibiotics and lasts for as long as the antibiotic persists in the medium. We saw a bit of variation in the time course of these events with different β-lactam antibiotics, but the general trend holds. These observations are consistent with earlier microbiological and clinical observations (30, 31).
We see fragmentation of BlaR1 in S. aureus, even in the absence of induction by β-lactams, and within the time frame relevant for manifestation of antibiotic resistance. However, the accumulation of BlaR1 fragments takes place much later than BlaI degradation because the levels of BlaR1 also start to increase as a consequence of the induction process. The intuitive conclusion from this observation is that BlaR1 fragmentation itself might not be a requirement for the onset of resistance, but we cannot be certain. So, what is the ramification of fragmentation of BlaR1 in S. aureus? We reported recently that under conditions that simulated the in vivo situation, 10% of the bla operon is derepressed at any given time (8). This leads to the leakiness of the system, which allows for some minimal background expression of BlaR1, BlaI, and β-lactamase. This leakiness actually might have evolved and might be critical for the survival of S. aureus to respond to the presence of β-lactams because it produces sufficient BlaR1 in the absence of the antibiotic to serve as a vanguard for its detection. This minimal background formation of BlaR1 obviously leads to its ultimate insertion into the membrane, an environment that is strictly regulated.
It has been suggested that ∼30% of prokaryotic gene products are sequestered into the membrane (32–34). There should be a limit of how much protein can be populated within this environment. We noted in the present study that in the absence of antibiotic, one still sees proteolysis of BlaR1. Hence, as the quantity of BlaR1 goes up (due to the leakiness of the gene, prior to induction), it experiences proteolytic turnover, perhaps by the regulatory processes involved in membrane homeostasis. As derepression takes place upon exposure of the organism to the antibiotic, the copy number of BlaR1 goes up; so does its proteolytic turnover, as documented by our study. Hence, proteolysis of BlaR1 is probably a protein turnover event within the membrane environment, yet our results indicate that the proteolytic fragmentation is highly specific and takes place at particular places in the sequence of the protein. The cytoplasmic sites are probably locations of autolytic cleavage, as discussed earlier, and the one on the surface that leads to shedding of the sensor domain has been shown to be mediated by signal peptidase in S. epidermidis.
In the foregoing discussion, we have argued that BlaR1 is inherently prone to proteolysis. Its implication is that BlaR1 proteolysis is probably a means for reversal of induction. We have shown that when antibiotic acylates the sensor domain, the complex often is stable for the duration of the bacterial generation, at times much longer (5). When the challenge of antibiotic is withdrawn, by the action of β-lactamase or by lack of sufficient exposure, the organism has to reverse the mobilization of resistance. This process begins with degradation of BlaR1 in a stepwise process, as reported in this paper.
Supplementary Material
Acknowledgments
S. aureus isolates NRS128, NRS123, NRS70, and NRS144 were obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) program, supported under NIAID, National Institutes of Health, Contract HHSN272200700055C. We acknowledge Dr. Maxim Suvorov for construction of the vector pET-24a(+)_BlaRHis6.
This work was supported, in whole or in part, by National Institutes of Health Grant AI 33170.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S16.
- MRSA
- methicillin-resistant S. aureus
- MIC
- minimal inhibitory concentrations
- CBAP
- 2-(2′-carboxyphenyl)-benzoyl-6-aminopenicillanic acid
- PEN
- penicillin G
- AMP
- ampicillin
- OXA
- oxacillin
- BlaRS
- C-terminal sensor domain of BlaR1
- rBlaRS
- recombinant C-terminal sensor domain of BlaR1 expressed in E. coli
- rBlaR1
- recombinant BlaR1 expressed in E. coli
- rBlaI
- recombinant BlaI expressed in E. coli
- ACN
- acetonitrile
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid
- MRM
- multiple-reaction monitoring
- EMS
- enhanced mass spectrometry.
REFERENCES
- 1. Hanique S., Colombo M. L., Goormaghtigh E., Soumillion P., Frère J. M., Joris B. (2004) J. Biol. Chem. 279, 14264–14272 [DOI] [PubMed] [Google Scholar]
- 2. Thumanu K., Cha J., Fisher J. F., Perrins R., Mobashery S., Wharton C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10630–10635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cha J., Mobashery S. (2007) J. Am. Chem. Soc. 129, 3834–3835 [DOI] [PubMed] [Google Scholar]
- 4. Borbulevych O., Kumarasiri M., Wilson B., Llarrull L. I., Lee M., Hesek D., Shi Q., Peng J., Baker B. M., Mobashery S. (2011) J. Biol. Chem. 286, 31466–31472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Golemi-Kotra D., Cha J. Y., Meroueh S. O., Vakulenko S. B., Mobashery S. (2003) J. Biol. Chem. 278, 18419–18425 [DOI] [PubMed] [Google Scholar]
- 6. Zhang H. Z., Hackbarth C. J., Chansky K. M., Chambers H. F. (2001) Science 291, 1962–1965 [DOI] [PubMed] [Google Scholar]
- 7. Wikler M. A., Cockerill F. R., 3rd, Bush K., Dudley M. N., Eliopoulos G. M., Hardy D. J., Hecht D. W., Hindler J. F., Patel J. B., Powell M., Turnidge J. D., Weinstein M. P., Zimmer B. L., Ferraro M. J., Swenson J. M. (2006) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard, 7th Ed., Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Llarrull L. I., Prorok M., Mobashery S. (2010) Biochemistry 49, 7975–7977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hernandez-Barrantes S., Toth M., Bernardo M. M., Yurkova M., Gervasi D. C., Raz Y., Sang Q. A., Fridman R. (2000) J. Biol. Chem. 275, 12080–12089 [DOI] [PubMed] [Google Scholar]
- 10. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 11. DiGiuseppe Champion P. A., Champion M. M., Manzanillo P., Cox J. S. (2009) Mol. Microbiol. 73, 950–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) Mol. Cell Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 13. Jones A. R., Orchard S. (2008) Mol. Cell Proteomics 7, 2067–2068 [DOI] [PubMed] [Google Scholar]
- 14. Unwin R. D., Griffiths J. R., Leverentz M. K., Grallert A., Hagan I. M., Whetton A. D. (2005) Mol. Cell Proteomics 4, 1134–1144 [DOI] [PubMed] [Google Scholar]
- 15. Anderson L., Hunter C. L. (2006) Mol. Cell Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 16. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 17. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. (2010) Nucleic Acids Res. 38, W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo F., Huynh J., Dmitrienko G. I., Viswanatha T., Clarke A. J. (1999) Biochim. Biophys. Acta 1431, 132–147 [DOI] [PubMed] [Google Scholar]
- 19. Christensen H., Martin M. T., Waley S. G. (1990) Biochem. J. 266, 853–861 [PMC free article] [PubMed] [Google Scholar]
- 20. Hardt K., Joris B., Lepage S., Brasseur R., Lampen J. O., Frère J. M., Fink A. L., Ghuysen J. M. (1997) Mol. Microbiol. 23, 935–944 [DOI] [PubMed] [Google Scholar]
- 21. Zhu Y., Englebert S., Joris B., Ghuysen J. M., Kobayashi T., Lampen J. O. (1992) J. Bacteriol. 174, 6171–6178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rath A., Glibowicka M., Nadeau V. G., Chen G., Deber C. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones J. D., O'Connor C. D. (2011) Proteomics 11, 3012–3022 [DOI] [PubMed] [Google Scholar]
- 24. Kozak M. (1983) Microbiol. Rev. 47, 1–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tang J., Hernández G., LeMaster D. M. (2004) Protein Expr. Purif. 36, 100–105 [DOI] [PubMed] [Google Scholar]
- 26. Sugino Y., Tsunasawa S., Yutani K., Ogasahara K., Suzuki M. (1980) J. Biochem. 87, 351–354 [DOI] [PubMed] [Google Scholar]
- 27. Powers M. E., Smith P. A., Roberts T. C., Fowler B. J., King C. C., Trauger S. A., Siuzdak G., Romesberg F. E. (2011) J. Bacteriol. 193, 340–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Antunes N. T., Frase H., Toth M., Mobashery S., Vakulenko S. B. (2011) Biochemistry 50, 6387–6395 [DOI] [PubMed] [Google Scholar]
- 29. Carrasco-López C., Rojas-Altuve A., Zhang W., Hesek D., Lee M., Barbe S., André I., Ferrer P., Silva-Martin N., Castro G. R., Martínez-Ripoll M., Mobashery S., Hermoso J. A. (2011) J. Biol. Chem. 286, 31714–31722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Collins J. F. (1979) in β-Lactamases (Hamilton-Miller J. M. T., Smith J. T. eds) pp. 351–368, Academic Press, London [Google Scholar]
- 31. Clarke S. R., Dyke K. G. (2001) J. Antimicrob. Chemother. 47, 377–389 [DOI] [PubMed] [Google Scholar]
- 32. Wallin E., von Heijne G. (1998) Protein Sci. 7, 1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weiner J. H., Li L. (2008) Biochim. Biophys. Acta 1778, 1698–1713 [DOI] [PubMed] [Google Scholar]
- 34. Bernsel A., Daley D. O. (2009) Trends Microbiol. 17, 444–449 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.