Abstract
Bacterial fibronectin-binding proteins (FnBPs) contain a large intrinsically disordered region (IDR) that mediates adhesion of bacteria to host tissues, and invasion of host cells, through binding to fibronectin (Fn). These FnBP IDRs consist of Fn-binding repeats (FnBRs) that form a highly extended tandem β-zipper interaction on binding to the N-terminal domain of Fn. Several FnBR residues are highly conserved across bacterial species, and here we investigate their contribution to the interaction. Mutation of these residues to alanine in SfbI-5 (a disordered FnBR from the human pathogen Streptococcus pyogenes) reduced binding, but for each residue the change in free energy of binding was <2 kcal/mol. The structure of an SfbI-5 peptide in complex with the second and third F1 modules from Fn confirms that the conserved FnBR residues play equivalent functional roles across bacterial species. Thus, in SfbI-5, the binding energy for the tandem β-zipper interaction with Fn is distributed across the interface rather than concentrated in a small number of “hot spot” residues that are frequently observed in the interactions of folded proteins. We propose that this might be a common feature of the interactions of IDRs and is likely to pose a challenge for the development of small molecule inhibitors of FnBP-mediated adhesion to and invasion of host cells.
Keywords: Fibronectin, Host-Pathogen Interactions, Isothermal Titration Calorimetry, Protein Complexes, Protein Structure, Protein-Protein Interactions
Introduction
In the last 20 years there has been increasing recognition of the functional importance of regions of protein sequence that lack a stable tertiary fold under physiological conditions (1, 2). Previously, the “structure-function paradigm” asserted that a protein must be stably folded to be functional. However, it has since been shown that intrinsically disordered regions (IDRs)4 are involved in key molecular recognition events in both physiological (3) and pathological processes (4) and that IDRs often become structured on binding (5).
Our previous work has shown that cell wall-attached proteins expressed by some staphylococci and streptococci species and also the Lyme disease-causing spirochete, Borrelia burgdorferi, contain IDRs that bind the protein fibronectin (Fn) through a highly unusual mechanism of protein-protein recognition that we call a tandem β-zipper (6–11). This work investigates the thermodynamics of this interaction by studying the binding of Fn to SfbI, an Fn-binding protein (FnBP) from Streptococcus pyogenes.
Fn is a chordate glycoprotein (12). It is found as a disulfide-linked dimer in human plasma and in an insoluble fibrillar form as a component of extracellular matrices. Fn plays an essential role in development (13, 14) and in other processes requiring cell migration, such as wound healing and tissue remodeling (15). Fn is a modular protein, and the N-terminal domain (NTD) contains a string of five Fn type I (F1) modules (1–5F1) whose sequences are highly conserved across vertebrates (16). IDRs within bacterial FnBPs interact with these F1 modules.
S. pyogenes is an important human pathogen that causes common infections of the throat and skin such as pharyngitis and impetigo, and it can also cause more severe invasive infections such as streptococcal toxic shock syndrome and necrotizing fasciitis (reviewed in Ref. 17). SfbI mediates bacterial invasion of epithelial cells and endothelial cells (18–20) by binding to Fn, which acts as a bridge between SfbI and host cell integrins. Internalization might allow S. pyogenes to avoid anti-microbial drugs and host-defense mechanisms (reviewed in Refs. 21, 22) and thus is a potential target for new therapeutics.
Biophysical studies have shown that an FnBP from Staphylococcus aureus contains a series of intrinsically disordered Fn-binding repeats (FnBRs) that undergo a disorder-to-order transition on binding either 2–4F1 or 2–5F1 from the NTD of Fn (10, 24, 25). In this tandem β-zipper interaction, the FnBR extends the triple-stranded anti-parallel β-sheet of sequential F1 modules by forming an additional strand anti-parallel to the C-terminal strand (strand E) of each F1 module (7, 25). SfbI from S. pyogenes contains five FnBRs that each bind the NTD of Fn (7, 8, 22). SfbI-5, the most C-terminal FnBR, also has a 1F1-binding motif and can therefore bind all five F1 modules (1–5F1). To date, 1F1-binding motifs have only been identified in the C-terminal streptococcal FnBRs (7, 22).
The thermodynamics of protein-protein interactions involving two folded proteins have been well studied. Mutational analyses show that the free energy of binding is often concentrated within a few hot spot residues (26) (change in Gibbs free energy of binding, ΔΔG0, on mutation of ≥2 kcal/mol) rather than being distributed evenly across the interface (27). The ability to identify hot spot residues is particularly important for the discovery of drugs that target protein-protein interactions (28), and there are now a number of web servers that will perform such predictions (29–31). Much less is known about the thermodynamics of the interactions of IDRs.
In this study we demonstrate that SfbI-5 is an IDR. We use a Hidden Markov Model (HMM) to identify the most conserved FnBR residues across bacterial species. We show that for the NTD/SfbI-5 tandem β-zipper interaction, residues that are highly conserved across FnBRs, and that form very similar intermolecular interactions in NTD·FnBR complexes across bacterial species, are not hot spot residues. Therefore, unlike many interfaces formed between folded proteins, for SfbI-5 the binding energy is distributed across the interface. We suggest this might be a feature of the rather extended interactions that many IDRs form relative to folded proteins (32). The distributed energy of binding will pose a challenge to the development of small molecules that inhibit FnBP-mediated adhesion to and invasion of host cells.
EXPERIMENTAL PROCEDURES
Proteins, Proteolytic Fragments, Synthetic Peptides, Protein Expression, and Purification
The synthetic peptide PyTT5, corresponding to residues 560–577 of SfbI (UniProt accession Q01924), was purchased from Alta Bioscience (Birmingham, UK). The N and C termini were capped by acetylation and amidation, respectively. Human plasma Fn was purchased from Sigma (product no. F0895). The N-terminal domain from human plasma Fn, containing 1–5F1, was purchased as a 30-kDa proteolytic fragment from Sigma (product no. F9911) and is referred to as pNTD herein. The N and C termini were determined by mass spectrometry (MS) using an apex ultra FTMS (Bruker Daltonics).
2F13F1 (residues 62–151 of mature human Fn) was expressed in Pichia pastoris and purified using procedures similar to those described previously (7, 33). SfbI-5 (corresponding to residues 541–591 of SfbI) was expressed as a recombinant glutathione S-transferase (GST) fusion using the pGEX-5X-SfbI-5 construct described previously (7). Escherichia coli BL21 Gold competent cells (Stratagene) were transformed with pGEX-5X-SfbI-5 using a rapid protocol (34). A culture of a single transformed colony inoculated into LB-Amp broth (1% (w/v) tryptone, 0.5% (w/v) yeast extract, 1% NaCl (w/v), 100 μg/ml ampicillin) was grown for ∼18 h at 37 °C and then diluted into LB-Amp broth and incubated with shaking at either 30 or 37 °C. Cultures were induced to overexpress the GST fusion with isopropyl β-d-thiogalactopyranoside when the OD at 600 nm was between 0.6 and 1.2 and were then grown for a further 17 h (30 °C) or 3 h (37 °C). The GST fusion protein was purified from cleared cell lysates using glutathione-Sepharose (GE Healthcare).
For isothermal titration calorimetry (ITC) and NMR experiments, SfbI-5 was cleaved from GST with Factor Xa (New England Biolabs), purified by reversed-phase high performance liquid chromatography (8), and lyophilized. The mass of the purified product was confirmed by MS. 15N,13C-labeled SfbI-5 was produced using similar methods but using isotope-labeled minimal media.
Site-directed Mutagenesis
All SfbI-5 mutants were created through site-directed mutagenesis of pGEX-5X-SfbI-5 using the QuikChange II mutagenesis kit (Stratagene) and the primers in supplemental Table 1. In-house DNA sequencing confirmed the presence of mutations. SfbI-5 mutants were expressed, purified, and lyophilized as for wild-type SfbI-5, and their masses were confirmed by MS.
ITC
The binding of pNTD to SfbI-5 and SfbI-5 mutants was measured in PBS (140 mm NaCl, 2.7 mm KCl, 10 mm Na2PO4, 1.8 mm KH2PO4) at 37 °C using a VP-ITC microcalorimeter (MicroCal) and a procedure very similar to that reported previously (8). For each SfbI-5 peptide, an ITC experiment consisted of a binding titration and a control titration; the SfbI-5 peptide titrated into PBS with or without pNTD, respectively. The experiments were performed with pNTD (1.4–4 μm) in the cell and the peptide (20–50 μm) in the syringe. The experiments with the following SfbI-5 peptides were performed twice: SfbI-5, F554A, K556A, K556E, D574A, and T575A; and those with T558A were performed three times. Data were analyzed and fitted to a single site binding model using nonlinear regression analysis in Origin 7.0 software (MicroCal).
The concentration of pNTD was determined by absorbance at 280 nm using a theoretical extinction coefficient, based on the amino acid sequence, of 63,130 m−1 cm−1. The concentration of the SfbI-5 peptide was initially estimated from the weight of lyophilized material and then determined for one syringe sample for the following SfbI-5 peptides by quantitative amino acid analysis (Alta Bioscience, Birmingham): SfbI-5, F554A, K556A, K556E, T558A, and D574A. Use of these peptide concentrations resulted in molar ratio of binding (n) values within 12% of 1.0. Thus, for all other titrations, the peptide concentrations were corrected during analysis to give an n value of 1. Heats of dilution were either the injection of the appropriate peptide into buffer or the average peak area of the last 5 or 10 injections from the binding titration. For those SfbI-5 mutants whose ITC experiments were repeated, the mean value of each parameter was used for subsequent analysis.
NMR Spectroscopy
A standard suite of triple resonance experiments was performed for sequential assignment of 15N,13C-labeled SfbI-5 (1 mm, pH 5.5, 10% (v/v) D2O, 0.02% (w/v) NaN3). All were acquired using standard Bruker pulse sequences on a Bruker AVII 700 MHz spectrometer equipped with a triple resonance probe and z axis gradients at 25 °C. Data were processed using NMRPipe (35) and analyzed using CCPN Analysis (36).
Secondary structure propensity (SSP) was calculated from the assigned chemical shifts of the SfbI-5 1HN, 15NH, 13Cα, 13Cβ, and 13C′ nuclei (supplemental Table 2) using the SSP PerlScript kindly made available for download by Marsh et al. (37). The input used for the script was the experimentally determined SfbI-5 chemical shifts and the statistically derived random coil dataset from Wang and Jardetzky (38). Appropriate sequence correction values for the random coil dataset (38) were added to the corresponding observed SfbI-5 chemical shifts before SSP score calculation.
X-ray Crystallography
Crystals of 2F13F1·PyTT5 (0.3 and ∼3.5 mm, pH 7.7) were grown using sitting drop vapor diffusion from a 1:1 dilution with the well solution. The well solution was either 20 mm MgCl2, 0.1 m HEPES, pH 7.5, 22% w/v polyacrylic acid, or 0.1 m Tris, pH 7.0, 1.5 m (NH4)2SO4 for the lower and higher resolution data, respectively. Crystals were flash-cooled in liquid nitrogen using 20 or 30% (v/v) glycerol as the cryoprotectant.
Data Collection and Refinement
The lower resolution data were collected in-house at 120 K and the higher resolution data at 100 K at the European Synchrotron Radiation Facility on the ID23-1 beamline. Statistics are provided in supplemental Table 3. Data were processed and scaled using MOSFLM/SCALA (39–41). Successful solution of the lower resolution data were obtained by molecular replacement using models of 2F1 and 3F1 (PDB code 2RKZ) (25), searched for separately within Phaser (42). This structure was refined as described previously (25) and was used (without PyTT5) as the starting point for refinement of the higher resolution data using REFMAC (43).
Creation of a Hidden Markov Model
A profile HMM (44) was assembled using the sequences of known FnBRs (22) and using structural data (25) to inform where gaps in the alignment would be more likely. The HMM was used to search for other FnBRs in the UniRef90 sequence database (45). A set of nonredundant FnBRs was created from those identified in the search, in which each FnBR had no more than 70% pairwise identity to any other FnBR in the set. An HMM representing the nonredundant FnBRs was used in a second search of the database. This iterative process was repeated three times, until no further FnBRs were identified. An HMM logo was produced (46) using a final HMM of all nonidentical FnBRs.
Model of 1–5F1·SfbI-5
The model of 1–5F1·SfbI-5 was built from the structure of 2F13F1·PyTT5, an alignment between 4F15F1 and S. aureus FnBPA peptide structures (PDB codes 2RKY and 2RL0) (25), and 1F1 and the 1F1-binding region from the structure of 1F12F1 in complex with B3T (PDB code 1O9A) (7) using the program MODELLER (47). The 1F12F1 interface is not well defined even in the presence of peptide (7).
Surface Plasmon Resonance (SPR)
Interactions of SfbI-5 with human Fn and pNTD were studied using SPR. Binding studies were performed using a BIAcore T100 instrument (GE Healthcare) at 25 °C. GST-tagged wild-type or mutant (T558A) SfbI-5 was immobilized onto the experimental flow cell of a certified C1 sensor chip (GE Healthcare) by amine coupling using N-hydroxysuccinimide and ethyl-(dimethylaminopropyl)carbodiimide. Ethanolamine was subsequently used to block the surface. The reference flow cell underwent an identical but blank immobilization and blocking protocol. The SPR running buffer was HBSP (10 mm HEPES, pH 7.4, 150 mm NaCl, 0.05% (v/v) polysorbate 20). Flow rates of 20 and 30 μl/min were used for data acquisition and regeneration, respectively. The analyte contact and dissociation times were 180 and 600 or 1200 s, respectively. The longer dissociation time was required for the interaction of wild-type SfbI-5 with Fn, to ensure dissociation of a significant amount of analyte from the ligand. Regeneration contact and stabilization times were 540 and 3000 s, respectively. 0.1 m NaOH was used for regeneration. A kinetic data series contained sensorgrams measured for seven or eight sequential 2-fold analyte dilutions from 13.9 nm for both Fn and pNTD. Kinetic data series were fitted to a Langmuir 1:1 binding model by Biacore T100 evaluation software (GE Healthcare) using a constant zero bulk refractive index parameter. An upper limit for analyte concentration for sensorgram analysis was determined to ensure reliable curve-fitting indicated by low χ2 values. For the interaction between pNTD and SfbI-5 T558A, steady state affinity analysis was used to determine the binding affinity by Biacore T100 evaluation software (GE Healthcare) using a constant zero offset.
RESULTS
SfbI-5 Is an IDR
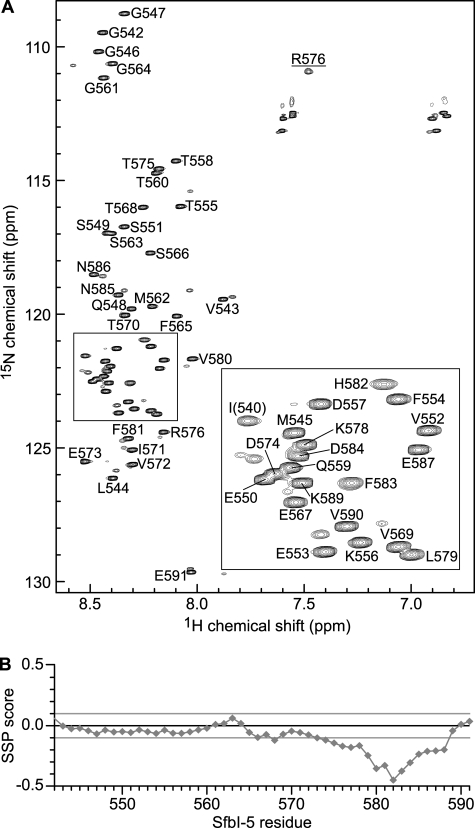
SfbI-5 binds to pNTD with the highest affinity (Kd ∼3 nm at 37 °C) (8) of the FnBRs studied to date. Although SfbI-5 is predicted to be an IDR based on its sequence, this has not been confirmed experimentally. In the absence of pNTD, the 15N-1H HSQC NMR spectrum of 15N,13C-labeled SfbI-5 has the narrow line widths and low chemical shift dispersion in the 1H dimension typical of an IDR (Fig. 1A). To confirm this observation, standard NMR methods were used to assign chemical shifts to the 1HN, 15NH, 13Cα, 13Cβ, and 13C′ nuclei (supplemental Table 2). The secondary structure propensity for each residue was determined by calculation of its SSP score, using the assigned chemical shifts and those predicted for a sequence-corrected random coil (Fig. 1B) (37, 38).
FIGURE 1.
SfbI-5 is intrinsically disordered. A, 1H,15N HSQC spectrum of 15N,13C-labeled SfbI-5. The peaks in the spectra were assigned to the 1HN and 15NH resonances of SfbI-5 residues using standard procedures. Each peak is labeled according to its position in the full-length SfbI-5 sequence. The peak annotated I(540) is not native to full-length SfbI but is directly N-terminal to Pro-541 from native SfbI, and the peak annotated R576 is from the side chain of Arg-576. The other smaller unassigned peaks in close proximity to assigned resonances are likely to arise from residues near a proline as a result of the proline being in a cis-conformation. Inset is an enlarged area of the boxed region of the spectrum. B, sequence-corrected SSP score was calculated for SfbI-5 using all available chemical shift information. SSP scores of +1 and −1 represent a stable α-helix and β-strand, respectively. A score of 0 represents random coil.
No evidence of stable secondary structure or significant SSP was detected in the 2–5F1-binding region of SfbI-5. This is consistent with previous circular dichroism results that streptococcal and staphylococcal FnBRs are IDRs (48), and also with a previous NMR analysis of an FnBR-containing region in S. aureus FnBPA (6). However, unlike other FnBRs in SfbI, SfbI-5 contains a 1F1-binding region (residues 577–588) (7), and this region has SSP for β-strand conformations (Fig. 1B). In particular, residues 580–584 have an SSP score of less than −0.3, indicating that within this region more than a third of the SfbI-5 molecules exhibit ϕ and ψ angles typical of β-strand at any given time. This provides a plausible explanation for our previous observation that the 1F12F1-binding motif from SfbI-5 bound with higher affinity than SfbI peptides that bind 2F13F1 or 4F15F1 (8).
Conservation of FnBR Residues
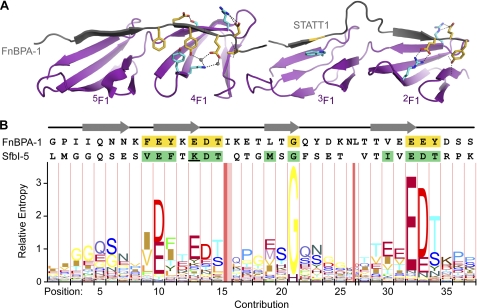
Fig. 2 shows an HMM logo (46) assembled from an alignment of all nonredundant staphylococcal, streptococcal, and spirochetal FnBRs identified in the sequence databases. Fig. 2 shows that, particularly in the FnBR region that binds 2F13F1, there are residues that are highly conserved across all FnBRs.
FIGURE 2.
Conserved FnBR residues identified with an iterative HMM search of the UniRef90 database. A, shown above the HMM logo are the crystal structures of 2F13F1 and 4F15F1 (purple) bound to FnBPA-1 peptides (gray) shown in ribbon representation (25). Conserved FnBPA-1 and F1 module side chains important in the interaction are shown in stick representation and are colored yellow or cyan, respectively. Hydrogen bonds are shown as black dashed lines and two bridging water molecules as gray spheres. B, sequences of FnBPA-1 (residues 476–514) and the SfbI-5 2–5F1-binding region (residues 544–578) are aligned to the columns in the HMM logo. Conserved FnBPA-1 side chains are also highlighted yellow in the FnBPA-1 sequence, and SfbI-5 residues chosen for mutation to alanine are highlighted green. The SfbI-5 residue Lys-556 is underlined; this residue was also mutated separately to glutamate. Regions of FnBPA-1 that form a β-strand when bound to F1 modules are indicated above the sequences by gray arrows. The logo was drawn with the LogoMat-M server (46).
Effect of Mutation of Conserved FnBR Residues on Affinity of the Interaction of SfbI-5 with 1–5F1
The effect of mutating conserved SfbI-5 residues on the binding of SfbI-5 to 1–5F1 was investigated using ITC. The 2F1- and 4F1-binding regions contain the most conserved FnBR residues (Fig. 2). Additionally, we have previously shown that, of the 2–5F1-binding regions, only the 2F1- and 4F1-binding regions show significant binding to F1 module pairs that contain their target F1 module (8). Thus, we focused the mutation analysis on these regions and on the highly conserved Gly in the 3F1-binding region. Also, for comparison, we mutated two residues with lower levels of conservation, one in the 2F1- and one in the 3F1-binding region.
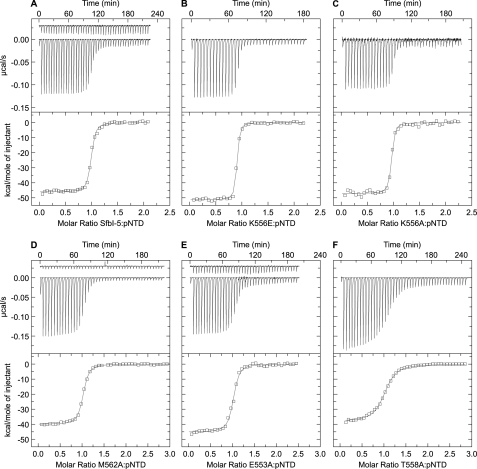
As can be observed from Table 1 and Fig. 3, when conserved residues in SfbI-5 were mutated to alanine, the binding became weaker relative to wild-type SfbI-5. In the one case where a residue in SfbI-5 that was not the consensus FnBR residue for that position (Fig. 2) was mutated to the consensus residue (K556E), the binding was significantly tighter relative to wild-type SfbI-5. Thus, overall, these results are consistent with the HMM logo in Fig. 2. However, whereas mutation of all the conserved residues weakened the interaction, for even the most conserved FnBR residues, the effect of mutation was relatively modest when compared with the effect of mutating hot spot residues on the interactions of folded proteins (defined here as those that contribute ≥2 kcal/mol to the interaction (27)).
TABLE 1.
Thermodynamic parameters for the interaction between pNTD and SfbI-5 (wild-type and mutants) determined by ITC at 37 °C, pH 7.4
| SfbI-5 or SfbI-5 mutant | Kd | ΔH0 | ΔS0 | ΔG0 | ΔΔG0 |
|---|---|---|---|---|---|
| nm | kcal/mol | cal mol−1K−1 | kcal/mol | kcal/mol | |
| SfbI-5 | 3.5 ± 0.2 | −45.4 ± 0.7 | −108 | −11.99 ± 0.04 | 0.00 ± 0.06 |
| V552A | 10.6 ± 0.8 | −44.3 ± 0.2 | −106 | −11.30 ± 0.05 | 0.69 ± 0.06 |
| E553A | 8.4 ± 0.8 | −45.2 ± 0.3 | −109 | −11.45 ± 0.06 | 0.54 ± 0.08 |
| F554A | 31.5 ± 4.9 | −42.9 ± 0.7 | −104 | −10.63 ± 0.10 | 1.36 ± 0.10 |
| K556A | 1.6 ± 0.2 | −45.7 ± 1.4 | −107 | −12.46 ± 0.08 | −0.47 ± 0.09 |
| K556E | 0.8 ± 0.2 | −49.0 ± 2.4 | −116 | −12.90 ± 0.18 | −0.90 ± 0.18 |
| D557A | 13.8 ± 1.0 | −43.2 ± 0.2 | −103 | −11.14 ± 0.05 | 0.85 ± 0.06 |
| T558A | 55.4 ± 2.7 | −37.9 ± 1.6 | −89.0 | −10.29 ± 0.03 | 1.70 ± 0.05 |
| M562A | 8.2 ± 0.4 | −40.0 ± 0.1 | −92.2 | −11.46 ± 0.03 | 0.53 ± 0.05 |
| G564A | 28.7 ± 2.1 | −38.5 ± 0.2 | −89.6 | −10.69 ± 0.04 | 1.30 ± 0.06 |
| I571A | 7.7 ± 0.3 | −44.5 ± 0.1 | −106 | −11.50 ± 0.03 | 0.49 ± 0.05 |
| E573A | 13.9 ± 0.8 | −43.6 ± 0.2 | −105 | −11.14 ± 0.03 | 0.85 ± 0.06 |
| D574A | 41.0 ± 5.9 | −46.3 ± 1.2 | −116 | −10.47 ± 0.09 | 1.52 ± 0.10 |
| T575A | 11.1 ± 0.5 | −44.9 ± 0.4 | −108 | −11.27 ± 0.03 | 0.72 ± 0.05 |
FIGURE 3.
ITC studies of SfbI-5 (wild-type and mutants) binding to pNTD. A, wild-type; B, K556E; C, K556A; D, M562A; E, E553A; and F, T558A. In each sub-figure, the top panel shows the experimental trace as a solution of SfbI-5 (or SfbI-5 mutant) was titrated into a pNTD solution. When a control titration was used to account for heats of dilution it is also shown but is displaced by −0.03 μcal/s. The bottom panel shows the processed molar heats of binding normalized to the ratio between SfbI-5 (or SfbI-5 mutant) to pNTD. For each titration, the curve from nonlinear least squares regression analysis, using a single-site binding model, is shown in gray. Thermodynamic parameters derived from these experiments are in Table 1.
Key Interactions Are Conserved between S. aureus FnBPA and S. pyogenes SfbI in Binding to 2F13F1
To test whether residues conserved across FnBRs play the same functional role in the binding of different organisms to Fn, we determined the structure of 2F13F1 in complex with PyTT5 (the 2F13F1-binding peptide from SfbI-5) and compared it with our previously determined structures of homologous peptides from S. aureus FnBPA in complex with 2F13F1 (2F13F1·STATT1 and 2F13F1·STATT5; PDB codes 2RKZ and 3CAL, respectively (25)).
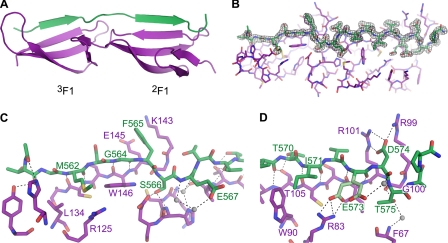
Fig. 4 shows the 1.7 Å resolution structure of PyTT5 in complex with 2F13F1 (crystallographic parameters are provided in supplemental Table 3). It is clear that PyTT5 binds the E strands of 2F1 and 3F1 through a tandem β-zipper mechanism (Fig. 4, A and B), with PyTT5 residues 562–565 and 570–574 adopting ϕ and ψ angles typical of the β-strand. As expected, there is little difference in the F1 module structures; comparison of the structure with that of 2F13F1·STATT1 shows a root mean square deviation of 2.11 Å between 2F13F1 in the two structures.
FIGURE 4.
Structure of 2F13F1·PyTT5. PyTT5 (green) binds along the E strands of both modules in 2F13F1 (purple). A, one of the two 2F13F1·PyTT5 complexes in the asymmetric unit. B, 2F13F1·PyTT5 structure showing the difference density at a level of 1σ (gray) when the PyTT5 molecule is deleted. C, interaction between 2F13F1 and the 3F1-binding and linker regions of PyTT5 shown as a stick representation. D, interaction between 2F1 and the 2F1-binding region of PyTT5 shown as a stick representation. In both C and D, hydrogen bonds and salt bridges are black dashes, and invariant bridging water molecules are shown as gray spheres. Residues mentioned in the text are labeled.
The side chains of the PyTT5 3F1-binding region (residues 560–565) interact with 3F1 primarily via van der Waals contacts, with Met-562 contacting Leu-134 and Arg-125 in 3F1, and Phe-565 stacking against aliphatic side-chain atoms of Lys-143 and Glu-145 (Fig. 4C). Furthermore, a glycine at position 564 allows PyTT5 to adopt a β-strand conformation without a steric clash with Trp-146 in 3F1 (Fig. 4C). Thus, the highly conserved glycine identified in Fig. 2 performs the same function in this structure as in the two structures of 2F13F1 with S. aureus peptides (25).
The PyTT5 linker region (residues 566–569) does not loop away from 2F13F1 as in the 2F13F1·STATT1 and 2F13F1·STATT5 structures (25), although it is the same length as the linker region in STATT5 (25). Instead, the PyTT5 linker binds to the interface between 2F1 and 3F1 primarily via hydrogen bonds; the side chains of bacterial residues Ser-566 and Glu-567 participate in hydrogen bond networks involving residues in both 2F1 and 3F1 (Fig. 4C).
The PyTT5 2F1-binding region (residues 570–577) binds to 2F1 via both van der Waals and hydrogen bond interactions. The two key van der Waals contacts are between the side chains of the bacterial residues Ile-571 and Asp-574 and the side chains of Trp-90 and Arg-99 of 2F1, respectively. Thr-570 forms a hydrogen bond with the side chain of Thr-105 in 2F1, adopting a staggered conformation favored in β-sheets (49). The side chain of Glu-573 exists in two conformations in both 2F13F1·PyTT5 complexes in the asymmetric unit with ∼50% occupancy in each conformation. One of the conformations results in a salt bridge to the side chain of Arg-83 in 2F1 (Fig. 4D) as found for the corresponding side chain in both complexes of S. aureus peptides with 2F13F1 (25). The second Glu-573 side-chain conformation forms a hydrogen bond to the carbonyl group of Asp-574 via a bridging water molecule (Fig. 4D). The side chain of Asp-574 forms a salt bridge with both Arg-101 and Arg-99, and Thr-575 forms hydrogen bonds to the backbone atoms of Gly-100 and Phe-67, although the latter is via a bridging water molecule (Fig. 4D). Therefore, the PyTT5 residues corresponding to the conserved “E(D/E)(T/S)” motif identified in Fig. 2 form interactions in the complex that are very similar to those of the corresponding residues in the 2F13F1·STATT1 and 2F13F1·STATT5 complexes (25).
PyTT5 Efficiently Forms an Extended Interface with 2F13F1
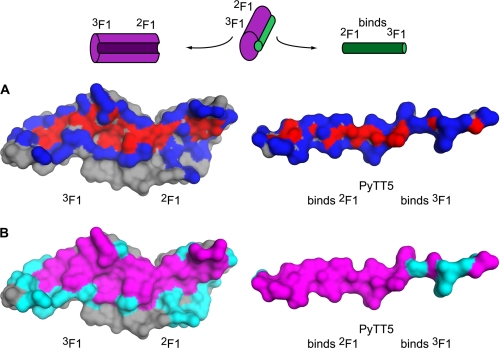
The extent of the interface between PyTT5 and 2F13F1 was analyzed using ProFace (50). A protein interface can be divided into two categories at both the atom and residue level (51, 52). At the atom level, the interface can be divided into buried and accessible interface atoms, where buried atoms lose all accessible surface area (ASA) on complex formation, and accessible atoms lose some ASA. The ProFace analysis showed that every residue of PyTT5 is involved in the interface with 2F13F1, with at least one atom of each residue in PyTT5 having some buried surface area at the interface (Fig. 5A; Table 2).
FIGURE 5.
Interface between 2F13F1 and PyTT5. Surface representation of 2F13F1 and PyTT5 (left and right, respectively) showing the interface between 2F13F1 and PyTT5. Indicated are the 2F1 and 3F1 modules, as are the regions of PyTT5 that bind 2F1 and 3F1. The schematic above the interfaces shows how the complex was opened like a book to reveal the interface. A, interface is shown at the atom level and is represented as buried and accessible atoms, which are colored red and blue, respectively. B, interface is shown at the residue level and is represented as core (magenta) and rim (cyan) residues. Residue and atom accessibility was calculated from the output of ProFace (50).
TABLE 2.
Analysis of the interfaces in the 2F13F1-PyTT5 complex by change in ASA on complex formation
Statistics for the interface between 2F13F1 chain B and PyTT5 chain D were calculated with the ProFace server (50). Local density is a measure of the atomic packing at the interface (64).
| Complex |
2F13F1-PyTT5 (molecule, chain) |
||
|---|---|---|---|
| 2F13F1, B | PyTT5, D | Complex | |
| Interface area | |||
| Total | 851.4 Å2 | 1095.6 Å2 | 1947.0 Å2 |
| Core | 653.1 Å2 | 853.0 Å2 | 1506.1 Å2 |
| Rim | 241.5 Å2 | 242.6 Å2 | 484.1 Å2 |
| Interface area/surface area | 0.17 | 0.41 | 0.24 |
| No. of interface atoms | |||
| Total | 122 | 89 | 211 |
| Buried | 44 | 22 | 66 |
| Accessible | 78 | 67 | 145 |
| No. of interface residues | |||
| Total | 38 | 18 | 56 |
| Core | 20 | 14 | 34 |
| Rim | 18 | 4 | 22 |
| Fraction of nonpolar atoms | 0.57 | 0.60 | 0.58 |
| Nonpolar interface area | 443.5 Å2 | 592.3 Å2 | 1035.8 Å2 |
| Fraction of fully buried atoms | 0.36 | 0.25 | 0.31 |
| Local density | 36.56 | 32.70 | |
At the residue level, the interface can be divided into core and rim residues, with core residues containing at least one buried interface atom and rim residues containing at least one accessible interface atom and no buried interface atoms. By this definition, 14 of the 18 residues (78%) in PyTT5 were core residues, and the remaining residues were rim residues (32%). In comparison, 2F13F1 had 20 core residues (22%) and 18 rim residues (20%) out of a total of 89 residues (Table 2). This indicates that each residue of PyTT5 contacts, on average, more residues of 2F13F1 than vice versa. It also indicates that a much greater percentage of PyTT5 is involved in the interface compared with 2F13F1. This is further shown by the fact that 40% of the ASA of PyTT5 is buried at the interface, compared with 17% of the ASA of 2F13F1. Thus, PyTT5 is a very efficient ligand. These properties have been noted in other complexes involving IDRs (32, 53, 54).
SfbI-5 Is Predicted to Form a Large, Extended Interface with 1–5F1
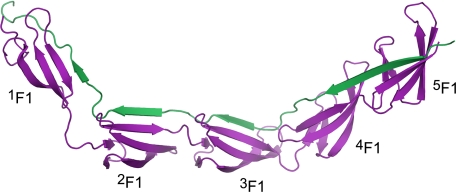
Structural data are now available for a single 1F12F1·FnBR peptide complex (7), three 2F13F1·FnBR (25) peptide complexes, and two 4F15F1·FnBR peptide complexes (25), so it is intriguing to consider the extent of the interface between SfbI-5 and 1–5F1. ProFace analysis of a model of 1–5F1 bound to SfbI-5 (Fig. 6) predicts that the total buried surface area of the interface would be ∼5150 Å2. This is a very large interface for a heterocomplex. In fact, comparison between the predicted interface area of 1–5F1·SfbI-5 and the observed interface areas from a set of heterodimer complex structures from the Protein Data Bank (32) reveals that the predicted interface is larger than any of the interfaces in the dataset and more than 1500 Å2 larger than the largest complex involving an IDR.
FIGURE 6.
Model of the 1–5F1-SfbI-5 complex. Ribbon representation of a model of SfbI-5 (green) in complex with 1–5F1 (purple).
Furthermore, 40% of the ASA of SfbI-5 is buried in the interface, compared with 18% of the ASA of 1–5F1. Thus, the reasons PyTT5 is an efficient ligand for binding to 2F13F1 are also predicted to make SfbI-5 an efficient ligand when in complex with 1–5F1. For a globular protein to bury a similar amount of surface area, it would have to have two to three times more residues. Therefore, a probable biological advantage of the intrinsic disorder of SfbI-5 is large interface formation (and tight binding) with fewer resources committed on the part of S. pyogenes (53).
Effect of Mutation of Conserved FnBR Residues on Affinity of the Interaction of SfbI-5 with Fn
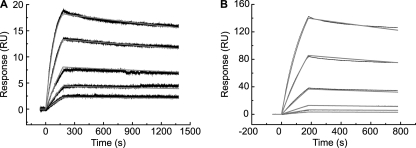
In vivo, SfbI-5 interacts with 1–5F1 in the context of intact Fn. Thus, the effect of the mutation of conserved FnBR residues on binding to plasma Fn was also determined (Fig. 7). SPR was first used to determine the dissociation constants for binding to pNTD (to compare with values obtained using ITC). Using the Van't Hoff isochore to adjust for the temperature difference, we estimate that the ITC experiments would yield an affinity of 0.18 nm if performed at 25 °C compared with the SPR-measured value of 0.14 nm. The effects of mutation were also similar, with the T558A mutation causing a 19-fold reduction in the affinity when measured using SPR (Table 3) compared with a 16-fold reduction in affinity when measured using ITC (Table 1). When binding of Fn to wild-type SfbI-5 and the T558A mutant was compared, the mutation resulted in an ∼5-fold reduction in the affinity. Thus, the T558A mutation has an even smaller effect on binding to Fn.
FIGURE 7.
SPR studies of GST-SfbI-5 (wild-type and T558A) binding to Fn. Either GST-SfbI-5 (A) or SfbI-5 T558A (B) was immobilized on the chip. Dimeric plasma Fn was used as the analyte. SPR response curves, base-line corrected, are shown in black; and the best fits to the data of Langmuir 1:1 models are shown in gray. Parameters derived from these experiments are shown in Table 3.
TABLE 3.
Parameters for the interaction of pNTD and Fn with SfbI-5 and the T558A mutant determined using SPR at 25 °C, pH 7.4
| Construct | k1a | k−1a | Kd | ΔG0 | ΔΔG0 |
|---|---|---|---|---|---|
| × 107m−1s−1 | × 10−4s−1 | nm | kcal/mol | kcal/mol | |
| pNTD | |||||
| SfbI-5 | 2.89 | 39.7 | 0.137 ± 0.002 | −13.44 ± .01 | 0.00 |
| T558A | NDb | NDb | 2.6 ± 0.3 | −11.71 ± .07 | 1.74 ± 0.07 |
| Fn | |||||
| SfbI-5 | 0.462 | 1.405 | 0.030 ± 0.001 | −14.35 ± .00 | 0.00 |
| T558A | 0.170 | 2.33 | 0.137 ± 0.004 | −13.44 ± .02 | 0.91 ± 0.02 |
a For each experiment, the errors for k1, k−1, and Kd were calculated from the errors in fitting the data from a single kinetic series to a Langmuir 1:1 binding model. The fitting model minimizes the errors associated with each variable. For all experiments, the errors for k1 and k−1 came out to zero when expressed to a suitable number of significant figures.
b ND means not determined. The data obtained for T558A·pNTD did not fit a kinetic model and was therefore fitted to a steady state affinity model. Thus, there are no kinetic data available for this experiment.
DISCUSSION
SfbI-5 Is an IDR
Comparison of the chemical shifts of the SfbI-5 1HN, 15NH, 13Cα, 13Cβ, and 13C′ nuclei with sequence-corrected random coil values revealed that SfbI-5 is an IDR, with no evidence of any stable secondary structure. However, the C-terminal 1F1-binding region of SfbI-5 showed significant propensity for β-strand formation (Fig. 1B). Based on a sequence alignment between SfbI-5 and the Streptococcus dysgalactiae FnBR peptide B3T, whose structure has been solved in complex with 1F12F1 (7), the residues with high β-strand propensity are predicted to form a β-strand when bound to 1F1. Therefore, this region might be a preformed structural element primed for 1F1 recognition (55).
Conserved Residues in Streptococcal and Staphylococcal FnBRs Perform Similar Functions
The crystal structure of PyTT5 in complex with 2F13F1 shows that the peptide binds to 2F13F1 via a tandem β-zipper (Fig. 4A). Comparison of this structure with those of FnBPA peptides in complex with 2F13F1 (25) revealed that residues that were conserved between these peptides formed similar interactions with 2F13F1. Given the sequence similarity between the other F1-binding regions of SfbI with FnBR peptides that have been solved in complex with 4F15F1 and 1F12F1 (7, 25), it is highly likely that SfbI-5 will bind to 1F1 and 4F15F1 via a tandem β-zipper. Thus, we prepared a model of the intact complex that highlights the extraordinary efficiency of the interaction (Fig. 6).
High efficiency of extended interface formation has been proposed as a potential advantage of intrinsic disorder (53), and overall, the interface area per IDR residue is higher in protein-protein heterocomplexes containing an IDR than for complexes containing two folded proteins (32). The tandem β-zipper interaction of SfbI-5 with 1–5F1 provides a clear demonstration of the efficiency of extended interface formation and the high affinity that can be achieved by an IDR. As seen previously for FnBR peptide interactions with F1 modules, there is little change in the 2F13F1 structure on PyTT5 binding.
Analysis of ΔΔG0 Values
The effect of mutating conserved SfbI-5 residues on the binding of SfbI-5 to pNTD was determined by calculating the ΔΔG0 for binding of the SfbI-5 mutants relative to wild-type SfbI-5 (Table 1). Most of the effects of the mutations can be rationalized from the structures based on the loss of buried atoms or intermolecular hydrogen bonds involving the mutated residue.
Some of the effects of mutations could also have been predicted based on residue conservation (Fig. 2). For example, K556A had a small but favorable ΔΔG0 of −0.47 kcal/mol and K556E had a more favorable ΔΔG0 of −0.90 kcal/mol. This result would be predicted from Fig. 2 as glutamate is the most conserved residue at this position. Also, for G564A, the unfavorable change in free energy is likely caused by changes in surrounding interactions that are required to avoid a steric clash between the alanine side chain and Trp-146 of 3F1 (Fig. 4C).
However, a comparison of Table 1 and Fig. 2 shows that overall there is not a strong correlation between the level of conservation of a residue and the ΔΔG0 value for mutation of that residue. The largest effect is observed for T558A, yet Thr-558 is not, by some way, the most highly conserved residue. Ile-571 and Met-562 were mutated because they are less well conserved (Fig. 2) and were predicted to form β-strand interactions with an F1 module primarily via backbone-backbone hydrogen bonds, a prediction that was confirmed by the 2F13F1·PyTT5 structure (Fig. 4, C and D). Yet the small ΔΔG0 values (0.49 and 0.53 kcal/mol, respectively) for mutation of these residues are similar to that observed for the mutation of a highly conserved acidic residue (Glu-553) in the conserved E(D/E)(T/S) motif within the 2F1-binding region.
Energy of the NTD/SfbI-5 Interaction Is Distributed across the Interface
All mutations of conserved residues reduced the affinity of binding, but ΔΔG0 values were relatively small (≤2.0 kcal/mol) compared with similar analyses on mutating interfaces in globular complexes (56, 57). Some analyses use a different threshold (e.g. ΔΔG0 >1 kcal/mol (31)) for defining hot spot residues. The latter threshold would mean that four SfbI-5 residues were hot spots (Phe-554, Thr-558, Gly-564, and Asp-574). Interestingly, only one of these residues is from the 2F1-binding region, yet it is this region that binds more tightly to 2F13F1 (8). However, regardless of the threshold chosen, it is clear from Table 1 that in the NTD/SfbI-5 interaction, the binding energy is distributed across the large interface rather than being concentrated in a few residues.
It has been observed that hot spots tend to be core residues, and tend not to be rim residues. This is because core residues form side-chain interactions with the protein partner, and rim residues act like an “o-ring” and shield the core residues from water (58). However, the extended nature of PyTT5, when in complex with 2F13F1, means that PyTT5 does not contain an o-ring of rim residues (Fig. 5B). Based on our model (Fig. 6), intact SfbI-5, when in complex with 1–5F1, would also lack o-ring residues. The reason this might lead to the absence of hot spots can be better understood by considering the interface at the level of atoms.
At the atom level of protein/protein interactions, the main chain is not directly affected by site-directed mutagenesis to alanine (51). Also, when accessible interface atoms are removed by mutation, they can be replaced by water molecules at a lower energetic cost than buried atoms (51). By this rationale, hot spot residues are likely to contain a buried interface atom that is deleted when the residue is mutated to alanine. Only 2 of 22 (9.1%) buried interface atoms in PyTT5 but 14 of 44 (32%) in 2F13F1 fulfill these criteria. Therefore, it appears the extended nature of the interface between SfbI-5 and 1–5F1, which is facilitated by the intrinsic disorder of free SfbI-5, is likely to preclude the presence of hot spot residues in SfbI-5. A more even distribution of the free energy of binding might be a more general property of complexes containing IDRs that have extended interfaces (32) that are unlikely to shield important interacting residues from water.
We used the HotPOINT server (29, 59) to predict hot spots in 2F13F1·PyTT5 and S. aureus FnBR peptides in complex with 2F13F1 (2RKZ and 3CAL) and 4F15F1 (2RKY and 2RL0) (25). Consistent with our ITC results, this analysis also suggests an overall lack of hot spots in the FnBR peptides with only a single conserved residue in the FnBR peptides (and only in 2RL0) predicted to be a hot spot.
A Paradox and a Challenge
We have shown that conserved residues in FnBRs of S. pyogenes and S. aureus play equivalent functional roles in binding to Fn and that single interactions can be sacrificed (in vitro) while maintaining a high affinity interaction. But why then are the FnBR residues so conserved? Are the conserved residues important for an aspect not probed by our pNTD interaction assay or are the small changes in affinity observed functionally significant?
Other potential functional roles for the conserved residues that are not probed by the pNTD binding assay are the disruption of intramolecular Fn interactions and/or maintenance of disorder in the FnBR. Long range intramolecular interactions between 2–5F1 and Fn type 3 modules (60) have recently been shown to mask motogenic sites within the collagen-binding domain of Fn. Changes in chemical shifts in the N-terminal strand (A strand) and E strands of F1 modules on F3 module binding (60) suggests that the F3- and SfbI-5-binding sites on 2–5F1 partially overlap. The faster association rate observed for binding of wild-type SfbI-5 to pNTD compared with Fn (Table 3) is consistent with the FnBR-binding site being somewhat cryptic in Fn. Therefore, we reasoned that conserved residues might play a role in unmasking the binding site, and thus, the effect of their mutation would only be observed when binding to Fn (rather than pNTD) was measured. However, the mutation that had the largest effect on SfbI-5 binding to pNTD (T558A) had an even smaller effect on the Kd for binding to intact Fn, suggesting that the interaction with Fn also lacks hot spots in the IDR.
Conserved residues might also play a role in maintaining the disordered state of FnBRs, similar to conservation of structurally important residues in a folded protein. By definition, disordered proteins do not make enough energetically favorable intramolecular contacts to overcome the unfavorable entropy of folding to fold into a stable tertiary structure. This is achieved by a characteristic amino acid composition, with IDRs having on average more charged and polar residues and fewer hydrophobic residues (61). It also requires each residue to limit the number of favorable interactions it makes with its neighboring residues (62). Therefore, there might be selective pressure for a disorder-promoting residue at a given position in an IDR depending on its neighboring sequence. Whether this selective pressure exists in FnBRs is hard to identify given current knowledge of IDR residue conservation.
Only streptococcal FnBPs and, in each case, only the C-terminal FnBR in the protein contain a 1F1-binding motif. These 1–5F1-binding FnBRs bind NTD with higher affinity than 2–5F1-binding FnBRs in SfbI (8). For example, in our previous study SfbI-5 bound pNTD with a Kd of ∼2 nm at 37 °C, whereas SfbI-4 bound with a Kd of 60 nm at 25 °C (8). Although S. aureus FnBPs only contain 2–5F1-binding FnBRs, the C-terminal FnBR (FnBPA-11) also binds NTD with the highest affinity, i.e. Kd <1 nm at 25 °C compared with 5 nm for FnBPA-1 (10). Thus, selective pressure for the maintenance of high affinity binding at the most C-terminal FnBR might operate in vivo, for example, at the level of integrin-mediated bacterial invasion.
The extended, multidomain nature of the tandem β-zipper interface is a plausible (and intuitive) explanation for the apparent absence of hot spots in SfbI-5. The mutation analysis of the NTD/SfbI-5 interaction presented here provides experimental support for this intuition. This interaction is, as yet, unusual with the only other reported example in LIM/Ldb interactions (54). However, extended interactions appear to be favored by IDRs (32), and thus, we suggest that IDRs in general might be more likely to form interactions that lack well defined hot spots.
Rational design strategies for drugs that target protein/protein interactions are still in the relatively early stages of development (28). The additional challenges involved in the design of drugs that target interactions involving IDRs have recently been highlighted (63). The more even distribution of binding energy over the interface, as observed here, than is commonly observed in the binding of folded proteins will add a further challenge, specifically for development of inhibitors of FnBP-mediated invasion of host cells and perhaps more generally for interactions involving IDRs.
Supplementary Material
Acknowledgments
We thank Adam Dowle for the mass spectrometry of 1–5F1, Sam Hart for x-ray data collection, Peter Ashton for assistance with bioinformatics, and David Williamson and Andrew Brentnall for assistance with the acquisition and processing of the NMR spectra. We thank the ESRF, Grenoble, for excellent data collection facilities and the Wellcome Trust for funding for the Biacore T100.
This work was supported by a British Heart Foundation Senior Basic Science Fellowship (to J. R. P.), an Overseas Research Students Awards Scheme Scholarship, scholarships from the Universities of Sydney and York (to N. N.), and by Biotechnology and Biological Sciences Research Council Grant D010608/1.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3.
The atomic coordinates and structure factors (code 3zrz) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- IDR
- intrinsically disordered region
- FnBP
- fibronectin-binding protein
- Fn
- fibronectin
- FnBR
- Fn-binding repeat
- NTD
- N-terminal domain
- F1
- Fn type 1
- ITC
- isothermal titration calorimetry
- PDB
- Protein Data Bank
- HMM
- Hidden Markov Model
- SSP
- secondary structure propensity
- SPR
- surface plasmon resonance
- ASA
- accessible surface area.
REFERENCES
- 1. Dunker A. K., Brown C. J., Lawson J. D., Iakoucheva L. M., Obradović Z. (2002) Biochemistry 41, 6573–6582 [DOI] [PubMed] [Google Scholar]
- 2. Tompa P. (2005) FEBS Lett. 579, 3346–3354 [DOI] [PubMed] [Google Scholar]
- 3. Liu J., Perumal N. B., Oldfield C. J., Su E. W., Uversky V. N., Dunker A. K. (2006) Biochemistry 45, 6873–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Uversky V. N., Oldfield C. J., Dunker A. K. (2008) Annu. Rev. Biophys. 37, 215–246 [DOI] [PubMed] [Google Scholar]
- 5. Dyson H. J., Wright P. E. (2002) Curr. Opin. Struct. Biol. 12, 54–60 [DOI] [PubMed] [Google Scholar]
- 6. Penkett C. J., Redfield C., Jones J. A., Dodd I., Hubbard J., Smith R. A., Smith L. J., Dobson C. M. (1998) Biochemistry 37, 17054–17067 [DOI] [PubMed] [Google Scholar]
- 7. Schwarz-Linek U., Werner J. M., Pickford A. R., Gurusiddappa S., Kim J. H., Pilka E. S., Briggs J. A., Gough T. S., Höök M., Campbell I. D., Potts J. R. (2003) Nature 423, 177–181 [DOI] [PubMed] [Google Scholar]
- 8. Schwarz-Linek U., Pilka E. S., Pickford A. R., Kim J. H., Höök M., Campbell I. D., Potts J. R. (2004) J. Biol. Chem. 279, 39017–39025 [DOI] [PubMed] [Google Scholar]
- 9. Raibaud S., Schwarz-Linek U., Kim J. H., Jenkins H. T., Baines E. R., Gurusiddappa S., Höök M., Potts J. R. (2005) J. Biol. Chem. 280, 18803–18809 [DOI] [PubMed] [Google Scholar]
- 10. Meenan N. A., Visai L., Valtulina V., Schwarz-Linek U., Norris N. C., Gurusiddappa S., Höök M., Speziale P., Potts J. R. (2007) J. Biol. Chem. 282, 25893–25902 [DOI] [PubMed] [Google Scholar]
- 11. Kim J. H., Singvall J., Schwarz-Linek U., Johnson B. J., Potts J. R., Höök M. (2004) J. Biol. Chem. 279, 41706–41714 [DOI] [PubMed] [Google Scholar]
- 12. Tucker R. P., Chiquet-Ehrismann R. (2009) Int. J. Biochem. Cell Biol. 41, 424–434 [DOI] [PubMed] [Google Scholar]
- 13. George E. L., Georges-Labouesse E. N., Patel-King R. S., Rayburn H., Hynes R. O. (1993) Development 119, 1079–1091 [DOI] [PubMed] [Google Scholar]
- 14. Watt F. M., Hodivala K. J. (1994) Curr. Biol. 4, 270–272 [DOI] [PubMed] [Google Scholar]
- 15. Magnusson M. K., Mosher D. F. (1998) Arterioscler. Thromb. Vasc. Biol. 18, 1363–1370 [DOI] [PubMed] [Google Scholar]
- 16. DeSimone D. W., Norton P. A., Hynes R. O. (1992) Dev. Biol. 149, 357–369 [DOI] [PubMed] [Google Scholar]
- 17. Cunningham M. W. (2000) Clin. Microbiol. Rev. 13, 470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rohde M., Müller E., Chhatwal G. S., Talay S. R. (2003) Cell. Microbiol. 5, 323–342 [DOI] [PubMed] [Google Scholar]
- 19. Molinari G., Talay S. R., Valentin-Weigand P., Rohde M., Chhatwal G. S. (1997) Infect. Immun. 65, 1357–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ozeri V., Rosenshine I., Mosher D. F., Fässler R., Hanski E. (1998) Mol. Microbiol. 30, 625–637 [DOI] [PubMed] [Google Scholar]
- 21. Nitsche-Schmitz D. P., Rohde M., Chhatwal G. S. (2007) Thromb. Haemost. 98, 488–496 [PubMed] [Google Scholar]
- 22. Schwarz-Linek U., Höök M., Potts J. R. (2006) Microbes Infect. 8, 2291–2298 [DOI] [PubMed] [Google Scholar]
- 23. Mosher D. F. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1193–1195 [DOI] [PubMed] [Google Scholar]
- 24. Penkett C. J., Dobson C. M., Smith L. J., Bright J. R., Pickford A. R., Campbell I. D., Potts J. R. (2000) Biochemistry 39, 2887–2893 [DOI] [PubMed] [Google Scholar]
- 25. Bingham R. J., Rudiño-Piñera E., Meenan N. A., Schwarz-Linek U., Turkenburg J. P., Höök M., Garman E. F., Potts J. R. (2008) Proc. Natl Acad. Sci. U.S.A. 105, 12254–12258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clackson T., Wells J. A. (1995) Science 267, 383–386 [DOI] [PubMed] [Google Scholar]
- 27. Bogan A. A., Thorn K. S. (1998) J. Mol. Biol. 280, 1–9 [DOI] [PubMed] [Google Scholar]
- 28. Arkin M. R., Wells J. A. (2004) Nat. Rev. Drug Discov. 3, 301–317 [DOI] [PubMed] [Google Scholar]
- 29. Tuncbag N., Keskin O., Gursoy A. (2010) Nucleic Acids Res. 38, W402–W406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Darnell S. J., Page D., Mitchell J. C. (2007) Proteins 68, 813–823 [DOI] [PubMed] [Google Scholar]
- 31. Kortemme T., Kim D. E., Baker D. (2004) Sci. STKE 2004 pl2. [DOI] [PubMed] [Google Scholar]
- 32. Mészáros B., Tompa P., Simon I., Dosztányi Z. (2007) J. Mol. Biol. 372, 549–561 [DOI] [PubMed] [Google Scholar]
- 33. Bright J. R., Pickford A. R., Potts J. R., Campbell I. D. (2000) Methods Mol. Biol. 139, 59–69 [DOI] [PubMed] [Google Scholar]
- 34. Pope B., Kent H. M. (1996) Nucleic Acids Res. 24, 536–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 36. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 37. Marsh J. A., Singh V. K., Jia Z., Forman-Kay J. D. (2006) Protein Sci. 15, 2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Y., Jardetzky O. (2002) J. Am. Chem. Soc. 124, 14075–14084 [DOI] [PubMed] [Google Scholar]
- 39. Leslie A. G. (1992) Joint CCP4 + ESF-EACBM Newsletter on Protein Crystallography, No. 26, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 40. Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 41. Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 15299374 [Google Scholar]
- 42. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallog. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 44. Eddy S. R. (1998) Bioinformatics 14, 755–763 [DOI] [PubMed] [Google Scholar]
- 45. Suzek B. E., Huang H., McGarvey P., Mazumder R., Wu C. H. (2007) Bioinformatics 23, 1282–1288 [DOI] [PubMed] [Google Scholar]
- 46. Schuster-Böckler B., Schultz J., Rahmann S. (2004) BMC Bioinformatics 5, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 48. House-Pompeo K., Xu Y., Joh D., Speziale P., Höök M. (1996) J. Biol. Chem. 271, 1379–1384 [DOI] [PubMed] [Google Scholar]
- 49. Hutchinson E. G., Sessions R. B., Thornton J. M., Woolfson D. N. (1998) Protein Sci. 7, 2287–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saha R. P., Bahadur R. P., Pal A., Mandal S., Chakrabarti P. (2006) BMC Struct. Biol. 6, 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lo Conte L., Chothia C., Janin J. (1999) J. Mol. Biol. 285, 2177–2198 [DOI] [PubMed] [Google Scholar]
- 52. Chakrabarti P., Janin J. (2002) Proteins Struct. Funct. Genet. 47, 334–343 [DOI] [PubMed] [Google Scholar]
- 53. Gunasekaran K., Tsai C. J., Kumar S., Zanuy D., Nussinov R. (2003) Trends Biochem. Sci. 28, 81–85 [DOI] [PubMed] [Google Scholar]
- 54. Deane J. E., Ryan D. P., Sunde M., Maher M. J., Guss J. M., Visvader J. E., Matthews J. M. (2004) EMBO J. 23, 3589–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fuxreiter M., Simon I., Friedrich P., Tompa P. (2004) J. Mol. Biol. 338, 1015–1026 [DOI] [PubMed] [Google Scholar]
- 56. Schreiber G., Fersht A. R. (1995) J. Mol. Biol. 248, 478–486 [DOI] [PubMed] [Google Scholar]
- 57. Wells J. A., de Vos A. M. (1993) Annu. Rev. Biophys. Biomol. Struct. 22, 329–351 [DOI] [PubMed] [Google Scholar]
- 58. Janin J., Bahadur R. P., Chakrabarti P. (2008) Q. Rev. Biophys. 41, 133–180 [DOI] [PubMed] [Google Scholar]
- 59. Tuncbag N., Gursoy A., Keskin O. (2009) Bioinformatics 25, 1513–1520 [DOI] [PubMed] [Google Scholar]
- 60. Vakonakis I., Staunton D., Ellis I. R., Sarkies P., Flanagan A., Schor A. M., Schor S. L., Campbell I. D. (2009) J. Biol. Chem. 284, 15668–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Radivojac P., Iakoucheva L. M., Oldfield C. J., Obradovic Z., Uversky V. N., Dunker A. K. (2007) Biophys. J. 92, 1439–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dosztányi Z., Csizmók V., Tompa P., Simon I. (2005) J. Mol. Biol. 347, 827–839 [DOI] [PubMed] [Google Scholar]
- 63. Dunker A. K., Uversky V. N. (2010) Curr. Opin. Pharmacol. 10, 782–788 [DOI] [PubMed] [Google Scholar]
- 64. Bahadur R. P., Chakrabarti P., Rodier F., Janin J. (2004) J. Mol. Biol. 336, 943–955 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.