Background: The mechanisms account for the miR-125b dysregulation in cancer cells.
Results: miR-125b, activating by CDX2, a homeobox transcription factor, regulates cell differentiation through repression of the core binding factor.
Conclusion: This study revealed a novel regulatory pathway including transcription factors and miRNA in the pathogenesis of hematopoietic malignancies.
Significance: The results potentially provide the knowledge necessary to design novel targeted therapies against the disease.
Keywords: Cell Differentiation, Leukemia, MicroRNA, Myeloid Cell, Transcription Factors
Abstract
MicroRNA-125b (miR-125b), a small noncoding RNA molecule, has been found to be deregulated and functions as an oncogene in many cancers including hematopoietic malignancies. However, the mechanisms accounting for miR-125b dysregulation remain to be elucidated. The present study aims to identify the factors that might contribute to up-regulation of miR-125b in human hematopoietic malignancies and its downstream targets for lineage-specific differentiation. We at first reported that CDX2, a homeobox transcription factor, binds to promoter regions of the miR-125b gene and activates transcriptional regulation of miR-125b in malignant myeloid cells. We further revealed that increasing levels of CDX2 in malignant myeloid cells activate miR-125b expression, which in turn inhibits core binding factor β (CBFβ) translation, thereby counteracting myeloid cell differentiation, at least for granulocytic lineage, and promoting leukemogenesis. Interestingly, we found that this novel pathway including CDX2, miR-125b, and CBFβ was mediated by undergoing all-trans-retinoic acid induction. Once differentiation ensues with all-trans-retinoic acid treatment, CDX2 activity decreases, leading to a reduction in miR-125b transcription and up-regulation of CBFβ in myeloid cells and in patients. The study provides a new mechanism that contributes to hematopoietic malignancies, which could involve deregulation of miR-125b and its up- and downstream factors. As altered expression of miRNAs has been reported in a wide range of malignancies, delineating the underlying molecular mechanisms of aberrant miRNA expression and characterizing the upstream and downstream factors will help to understand important steps in the pathogenesis of these afflictions.
Introduction
MicroRNAs (miRNAs),4 a class of small noncoding RNAs that range from 19 to 25 nucleotides in length, can silence specific target genes through translational repression or direct mRNA degradation (1, 2). They have been found to control a number of fundamental biological processes such as development, differentiation, proliferation, apoptosis, and stress responses etc. The deregulated expression of specific miRNAs that modulate the expression of oncogenes and tumor suppressors is associated with the development of malignancies (3, 4). MicroRNA-125b (miR-125b), one of the earliest discovered miRNAs, has been found deregulated in many cancers including hematopoietic malignancies. Notably, its expression in vivo was related to the stage of the maturation block underlying the subtypes of myeloid leukemia, and has been found to arrest myeloid cell differentiation in the pathogenesis of the disease (5, 6). For example, Bousquet et al. (5) demonstrated that miR-125b was required to promote the cells proliferation of promyelocytic blasts and arrest them differentiated to granulocyte/monocyte lineages. Mice in which miR-125b is overexpressed exhibit myeloid-dominated bone marrow and spleens as well as myeloid cell infiltration into the liver (7). Although miR-125b is important for modulating these physiological events, the mechanisms accounting for the aberrant expression of miR-125b in cancer cells, especially in hematopoietic malignancies, have not yet been elucidated.
The expression of tissue-specific genes is usually controlled by tissue-specific or tissue-enriched transcription factors. Recently, several miRNAs have been identified as direct transcriptional targets of transcription factors. For example, PU.1 activates transcription of miR-424, and this up-regulation stimulates monocyte differentiation through miR-424-dependent translational repression of NFI-A (8). PU.1, together with interferon regulatory factors IRF-1 and IRF-9, also binds to the promoter region of miR-342. IRF-1 maintains miR-342 at low levels, whereas the binding of PU.1 and IRF-9 on the promoter region upon all-trans-retinoic acid (ATRA)-mediated differentiation up-regulates miR-342 expression (9). In some cases, specific miRNAs and their transcriptional activators happen to be in regulatory feedback loops in which they control each other (10, 11). These loops may represent regulatory mechanisms allowing relatively small variations in miRNA concentration to induce drastic changes in cellular transcriptional patterns. In hematopoietic malignancies, several critical hematopoietic transcription factors, including C/EBPα, PU.1, CDX2, and GATA1, have been reported to play important roles in myeloid cell differentiation. Among them, homeobox transcription factors, which are classically known as regulators of axial elongation and anterior-posterior patterning during early embryogenesis, and CDX2, play a pivotal role during hematopoietic development (7, 8).
A few studies have identified miR-125b as targets and most are involved in cancer cell proliferation or apoptosis. Highly expressed miR-125b down-regulated BAK1, thereby inducing androgen-independent growth in prostate cancer cells (9). miR-125b could inhibit cell proliferation, cell cycle progression, and metastasis of hepatocellular carcinoma cells through suppression of the oncogene LIN28B (10). Functional analysis in breast cancer cells revealed overexpression of the miR-125b suppression proliferation by down-regulation of ERBB2 and ERBB3 (11). Recently, miR-125b was reported to be functional as a tumor suppressor by regulating the ETS1 proto-oncogene in invasive breast cancer (12). miR-125b targets multiple signaling pathways that manifest its important role in malignancies. However, targets for lineage-specific differentiation are not well elucidated.
In this study, we link expression of the miR-125b to transcription factors in myeloid cells. We revealed that up-regulation of miR-125b is due to activation by the homeobox transcription factor CDX2 and in turn inhibits myeloid cell differentiation. We further demonstrate that miR-125b directly targets core binding factor β (CBFβ) in myeloid cells and mediated myeloid cell differentiation, at least for granulocytic lineage. This pathway is activated by ATRA induction. These results shed light on the ATRA-dependent therapy mechanism and potentially provides the knowledge necessary to design novel targeted therapies for hematopoietic malignancy.
EXPERIMENTAL PROCEDURES
Cell Cultures and Clinical Samples
The human myeloid leukemia cell lines NB4, HL60, and K562 were maintained in RPMI 1640 medium containing 10% fetal bovine serum (Invitrogen). The 293T and HCT15 cell lines were maintained in DMEM containing 10% fetal bovine serum (Invitrogen).
A total of 80 acute myeloblastic leukemia (AML) samples including five subtypes (from AML-M1 to AML-M5) and 10 normal samples from the First and Second Affiliated Hospital of Sun Yat-sen University were enrolled in this study. Patient characteristics were available for all patients (supplemental Table S1). Bone marrow was collected from patients by bone marrow puncture at diagnosis or at follow-up after therapy. Written informed consent for the biological studies was obtained from the parents/guardians. The study was approved by the Ethics Committee of the affiliated hospitals of Sun Yat-sen University.
Constructs
The pcDNA6.2-125b plasmid was generated by cloning a fragment containing pre-miR-125b (from −198 to −160 bp relative to the 5′-end of mature miR-125b) into the pcDNA6.2 vector (Invitrogen). As a negative control, the pcDNA6.2-GW/EmGFP-miR-neg control plasmid (Invitrogen), which contains an insert that can form a hairpin structure that is processed into mature miRNA, but is predicted not to target any known vertebrate gene, was used. Synthesized 52-nucleotide DNA oligos containing the miR-125b response element were annealed and ligated into XhoI/NotI-digested psiCheck-2 vector (Promega) immediately downstream of the Renilla luciferase gene to obtain the wild type. For mutation analysis, the 6-nucleotide core seed-matched site on the CBFβ 3′-UTR (TCAGGG) was replaced by its complementary bases (AGTCCC). To express CDX2, C/EBPα, GATA1, and PU.1, the full-length coding sequence of each gene was amplified from NB4 RNA by RT-PCR and subsequently cloned into the pcDNA3 expression vector (Invitrogen).
For promoter analysis, a DNA fragment extending from position −1512 to +41 bp relative to pri-miR-125b was cloned into the promoterless pGL3 basic vector (Promega) for the pro-miR125b vector. Site-directed mutagenesis in pro-miR125b was performed with the MutanBEST Kit (TaKaRa).
All cloned products were verified by sequencing. Primers and other oligonucleotides are shown under supplemental Table S2.
Cell Electroporation
HL60 and NB4 cell (2 × 105 cells/sample) transfections were nucleofected using the Neon® Transfection System (Invitrogene) with 1 μg of plasmid vectors or 100 pmol of oligonucleotides in a 10-μl reaction. The miR-125b mimics and mimics-scrambled (mimics negative control, mimics-NC) were purchased from GenePharm (Shanghai, China), the anti-miR-125b, anti-scrambled (anti-negative control, anti-NC), si-h-CBFβ, and siRNA scrambled (si-NC) were purchased from RiboBio (Guangzhou, China). The transfected cells were collected for RNA or protein analysis at 48 h after transfection.
RNA Analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Total RNA was quantified by real-time PCR in the ABI Step One Detection System (Applied Biosystems) using SYBR® Premix Ex TaqTM II (TaKaRa). Mature miR-125b was detected in total RNA using the Hairpin-itTM miRNAs Real-time PCR Quantitation Kit containing a stem-loop like RT primer and miRNA-specific PCR primers (GenePharma, Shanghai, China). U6 small RNA was also amplified as an internal control using the same system and specific primer sets. mRNA quantitation was performed in the same system using endogenous β-actin mRNA as an internal control. The primers for real time PCR are shown under supplemental Table S2. Δ-Δ Ct values were normalized with those obtained from the amplification of the internal control. All reactions were performed in triplicate.
Immunoblot Analysis
Cells were lysed in RIPA buffer (Pierce) or 1% SDS. Proteins were fractionated by electrophoresis on a 10% SDS-polyacrylamide gel, electroblotted onto a PVDF membrane (Millipore), and reacted with anti-CBFβ (Santa Cruz), anti-CDX2 (BioGenex), anti-GAPDH (Protein Tech Group), or anti-β-tubulin (Cell Signaling Technology). Immunoreactivity was determined using the ECL method (Millipore) according to the manufacturer's instructions.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP was performed using the ChIP assay kit (Upstate) according to the manufacturer's instructions. DNA/protein cross-linking was achieved by incubating the cells for 20 min at 37 °C in 1% formaldehyde. After sonication, chromatin was immunoprecipitated overnight with 6 μl of anti-CDX2 antibody (BioGenex) or 2 μg of normal IgG. Genomic regions of about 130 bp containing the putative CDX2 binding site and an approximately 170-bp fragment 5 kbp upstream of the miR-125b promoter as negative control were amplified by PCR using the specific primers as shown under supplemental Table S2.
EMSA for in Vitro DNA Binding
The nuclear extract from HCT15 cells transfected with the CDX2 expression vector was prepared using the Nuclear-Cytosol Extraction Kit (KeyGEN) following the manufacturer's instructions. Probe oligonucleotides were annealed and labeled with [γ-32P]ATP using Klenow enzyme. DNA binding reactions included labeled probe (0.07 pmol) and nuclear extract (6 μg) and were incubated with or without a competitor probe at 25 °C for 30 min following the Promega Gel Shift Assay System instructions (Promega). To perform supershifts, anti-CDX2 antibodies (2 μl) were added to the reaction and incubated for another 20 min. The sequences of the oligonucleotides used for the assay are shown under supplemental Table S2.
Luciferase Assays
For promoter analysis, 293T cells at ∼80% confluence were transfected with wild-type miR-125b or mutant promoter luciferase constructs (500 ng) and the internal control Renilla luciferase plasmid pRL-TK (10 ng, Promega) using Lipofectamine 2000 (Invitrogen). In selected experiments (assays to examine the effects of transcription factors on miR-125b promoter activity), various combinations of pcDNA control vector (50 ng) and vectors encoding CDX2, C/EBPα, GATA1, or PU.1 (50 ng) were cotransfected. Cell extracts were prepared 24 h after transfection, and the luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega).
In the 3′-UTR assay, 293T and K562 cells at ∼80% confluence were co-transfected with 200 ng of the psiCHECK-2-derived reporter vectors and 50 ng of miR-125b or miR-neg expression vector. Firefly and Renilla luciferase activities were measured immediately using dual luciferase assays (Promega) at 24 h after transfection according to the manufacturer's instructions.
Statistical Analysis
Quantitative RT-PCR and luciferase data are presented as the mean ± S.D. from at least three independent experiments. Except where specified, the differences between groups were analyzed using two-tailed, one-way analysis of variance Student's t test. p < 0.05 was considered statistically significant.
RESULTS
Dysregulation of miR-125b in AML Correlates with Activation of the Homeobox Gene CDX2
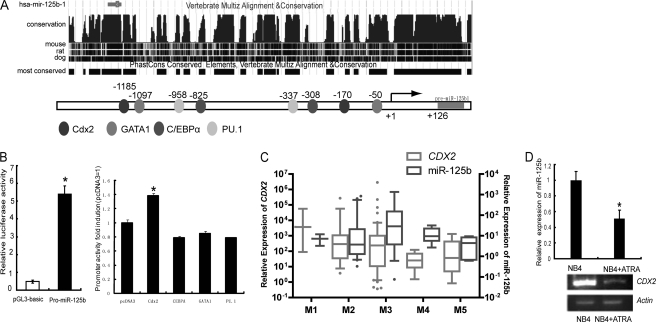
Considerable evidence has suggested that transcription factors control the expression of miRNAs (13–16). To identify factors that influence miR-125b expression, we first analyzed the promoter of miR-125b. The transcriptional start site for pri-miR-125b was identified 126 nucleotides upstream of the 5′ end of the pre-miRNA (17). We used the TESS program, a positional weight matrices from the TRANSFAC, JASPAR, IMD, and transcription factor consensus DNA-binding motifs (18) to search for transcription factor binding sites on the promoter of miR-125b. We found that hematopoietic transcription factor binding sites are enriched on the promoter of miR-125b, including sites for CDX2, PU.1 C/EBPα, and GATA1 (Fig. 1A). To examine whether there is any contribution from the 5′-untranslated region of miR-125b to promoter expression, we next cloned the putative miR-125b promoter into the basic pGL3-luc reporter (pro-miR-125b) to study the function. As shown in Fig. 1B, the miR-125b promoter activated the expression of the downstream reporter, indicating that the predicted miR-125b promoter is a true promoter (Fig. 1B, left). We then coexpressed hematopoietic transcription factors CDX2, PU.1 C/EBPα, and GATA1, respectively, with the pro-miR-125b to test the influence of these factors on the activity of the promoter. The results showed that only CDX2 could stimulate the miR-125b promoter, whereas PU.1, C/EBPα, and GATA1 did not show contributions on transcription activity (Fig. 1B, right), suggesting that CDX2 might be the activator of miR-125b.
FIGURE 1.
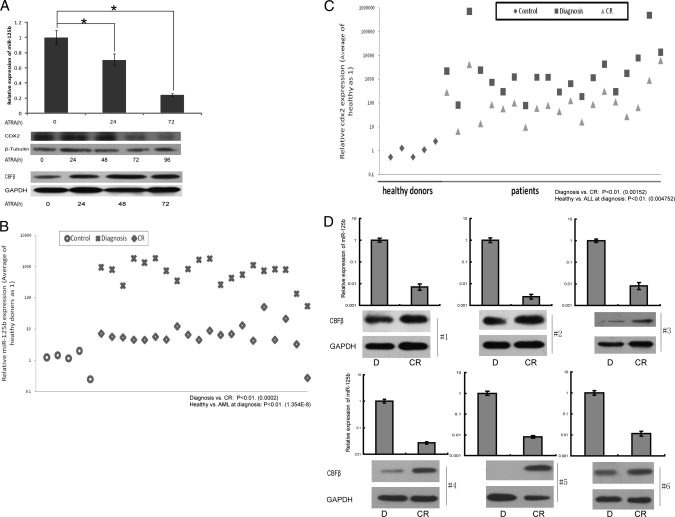
Transcription factor binding sites in the miR-125b promoter. A, upper panel, conservation of the miR-125b 5′ proximal genomic region. Lower panel, the 5′ proximal promoter region of miR-125b with putative binding sites for several hematopoietic transcription factors including CDX2, C/EBPα, GATA1, and PU.1. B, luciferase reporter assay performed in 293T cells. Left, miR-125b promoter construct (Pro-miR-125b) activity. Right, miR-125b promoter construct co-transfected with empty vector (pcDNA3), Cdx2, C/EBPα, GATA1, or PU.1. The data shown are relative to the control promoter activity (co-transfected with empty vector). At least three independent experiments were performed. *, p < 0.05 versus control. C, expression levels of miR-125b and CDX2 in 60 myeloid leukemia patients of different subtypes (AML-M1 to M5) were analyzed by qRT-PCR. Data are presented as the fold-change of miR-125b or CDX2 expression in patient samples with respect to their expression in bone marrow mononuclear cells from 10 healthy donors. The average expression of miR-125b or CDX2 in each subtype was statistically compared with the average normal value. Bottom and top of the boxes are the 25th and 75th percentiles, respectively; the bar in the box is the median; the ends represent the 90 or 10%, respectively. D, the expression of miR-125b and CDX2 was positively correlated under 1 μm ATRA treatment for the indicated time in NB4 cells. Upper, miR-125b expression detected by qRT-PCR. Lower, CDX2 expression was detected by semi-qPCR. Three independent experiments were performed.
CDX2, a homeobox transcription factor, has recently been described as aberrantly expressed in myeloid and lymphoblastic leukemia and as promoting leukemogenesis. To further evaluate the consistent expression profiles of CDX2 and miR-125b, we used qRT-PCR to measure the co-expression level of CDX2 and miR-125b in clinical samples of myeloblastic leukemia and 60 AML patients were applied, including five subtypes of AML (from AML-M1 to AML-M5). Five bone marrow samples from healthy donors were analyzed as controls. AML arises from the differentiation arrest of the myeloid precursor and malignant proliferation in the bone marrow and blood. It can be divided into different subtypes based on cell type and the degree of maturity and is an excellent model for studying the genetic regulation of differentiation and cancer progression (19). To confirm that the primers for miR-125b cannot amplify miR-125a, which belongs to the miR-125 family, we used the primers to amplify miR-125b in the miR-125a overexpressed cells, and the results showed no increases for miR-125b expression (supplemental Fig. S1A), demonstrating that the designed primers are specifically miR-125b. We also tested the efficiency of real time PCR amplification by a dilution curve, the results showed that the efficiencies of both miR-125b and the internal control are higher than 95% (supplemental Fig. S1B). As shown in Fig. 1C, the expression of miR-125b in AML is positively correlated with CDX2 expression. Up-regulated expression of both miR-125b and CDX2 was detected in almost all of the subtypes of AML, strongly implying the relationship of up-regulation of both miR-125b and CDX2 and their contribution to the differentiation block of the myeloid cell. We also monitored the expression levels of CDX2 and miR-125b in vitro in NB4 cells under ATRA induction for 72 h (Fig. 1D). We found that the expression level of the CDX2 mRNA was decreased and positively correlated with miR-125b after the ATRA treatment, which also suggested their potential function to inhibit cell differentiation. Taken together, these findings suggest that in human AML, high expression of miR-125b is due to the activation of CDX2, and the positive correlation between CDX2 and miR-125b was found in both myeloid leukemia samples and myeloid leukemia cell line.
CDX2 Is Involved in the Transcriptional Regulation of miR-125b
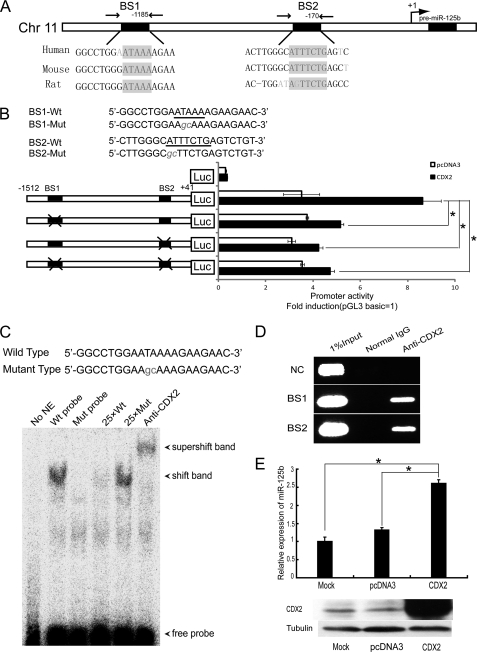
The close correlation between the high expression levels of CDX2 and miR-125b perturbation in human myeloid leukemia let us to speculate that the initiation of high CDX2 expression levels might be a key step in the development of myeloid precursor and malignant proliferation with aberrant miR-125b expression. Two binding sites for the CDX2 transcription factor located upstream of the transcription start site were identified. These binding sites are highly conserved in mammals (Fig. 2A). To test whether the putative binding sites for CDX2 on the miR-125b promoter are responsible for its transcriptional activity, promoter-luciferase fusion constructs containing site-specific mutations were produced (Fig. 2B). The results of the luciferase reporter assay in the 293T cell line showed that these two binding sites are essential for CDX2 to activate miR-125b transcription (Fig. 2B). To establish direct binding of CDX2 to the miR-125b promoter, we performed an electrophoretic mobility shift assay using a miR-125b DNA fragment containing the consensus site for CDX2. We found that incubating the labeled wild-type DNA fragment with nuclear extract from HCT15 cells transiently transfected with CDX2 resulted in the formation of a discrete protein-DNA complex, but no shift band when incubated with the mutation probe. These complexes could be effectively supershifted by inclusion of CDX2 antibodies and their formation could be blocked by the unlabeled DNA fragment (Fig. 2C). These data suggest that CDX2 can directly bind to the miR-125b promoter.
FIGURE 2.
CDX2 is responsible for miR-125b activation. A, schematic representation of the miR-125b genomic region. The sequence and location of the Cdx2-binding sites are indicated by black boxes (BS1 and BS2), whereas the arrows point to the regions amplified by PCR in the ChIP experiments. B, upper panel, mutant sequences of the miR-125b promoter reporter vector are shown. Lower panel, 293T cells were co-transfected with a CDX2 expression vector or empty vector and various luciferase reporter vectors containing the miR-125b promoter for 24 h, followed by measurement of luciferase activity. C, electrophoretic mobility shift assay showing direct binding of CDX2 to the miR-125b promoter on BS1. Nuclear extracts with wild-type (lanes 2 and 4–6) or mutant probes (lane 3) were analyzed for their ability to bind to the promoter. D, interaction of CDX2 with the putative CDX2 binding sites on the promoter region of miR-125b were examined in NB4 cells using a ChIP assay. One percent of input DNA was used as a positive control for PCR. The IgG-immunoprecipitated chromatin was used as a control for CDX2. A irrelevant sequence 5 kbp upstream of miR-125b promoter was used as a negative control (NC). E, upper panel, the level of miR-125b in NB4 cells transfected with water, pcDNA3, or CDX2 expression vector was examined by qPCR. Lower panel, the level of CDX2 in NB4 cells transfected with water, pcDNA3, or CDX2 expression vector was examined by Western blot. *, p < 0.05.
To further examine whether CDX2 physically interacts with the miR-125b promoter region in vivo, we performed ChIP. DNA fragments containing the two potential CDX2 binding sites were immunoprecipitated from NB4 cells by CDX2 antibodies, suggesting that CDX2 is actually interacting with the corresponding promoter sites (Fig. 2D). No enrichment was seen in the negative control region and when mock immunoprecipitations were carried out with normal mouse IgG antibody.
Next, we performed overexpression studies to test whether CDX2 could up-regulate miR-125b expression in myeloid cells. A construct expressing CDX2 and a negative pcDNA3 vector were transfected into two myeloid cell lines, NB4 and HL60, and the expression of miR-125b was analyzed by qRT-PCR. As shown in Fig. 2E and supplemental Fig. S2, cells transfected with the CDX2-expressing vector displayed obvious up-regulation of miR-125b. These results show the involvement of CDX2 in the transcriptional regulation of miR-125b, and they also suggest that miR-125b functions as an effector of CDX2 during myeloid differentiation. Collectively, these findings strongly indicate that CDX2 directly interacts with the miR-125b promoter and positively regulates miR-125b expression in the development of myeloid precursor and malignant proliferation.
miR-125b Counteracts Myeloid Cell Differentiation through Repression of the CBFβ
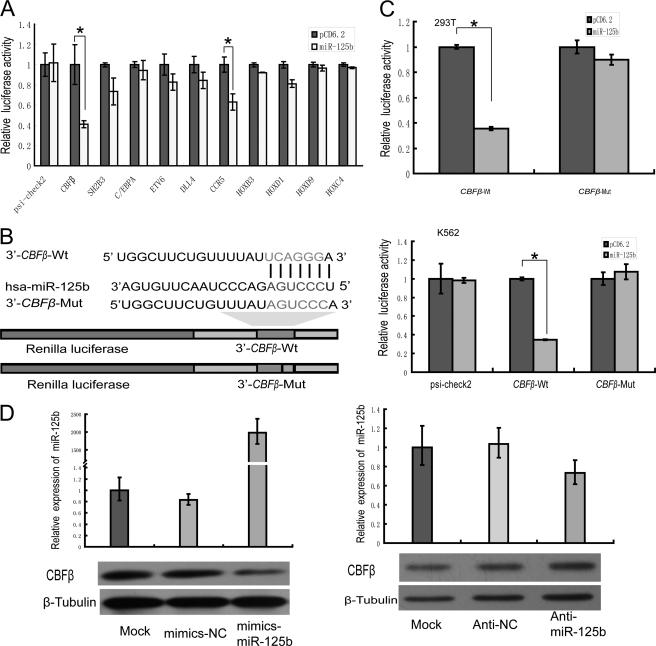
Given the importance of miR-125b in the pathogenesis of hematopoietic malignancies and/or to myeloid cell differentiation, the downstream targets in this regulatory pathway are of interest. Among the hundreds of predicted regulatory targets of miR-125b, most targets are oncogenes or tumor suppressor genes involved in cancer cell proliferation or apoptosis (9, 20, 21). Targets for miR-125b associated with hematopoietic-specific differentiation have not yet been reported. We noticed that the core binding factor CBFβ, the chemokine receptor CCR5, and several members of the HOX gene family (which are known to be important genes involved in myeloid and lymphoid differentiation) are also predicted targets. To confirm whether the CBFβ, CCR5, and HOX genes are bona fide targets of miR-125b, the 3′-UTRs of these genes were inserted downstream of a luciferase ORF. Among them, we confirmed that CBFβ and CCR5 were repressed by miR-125b in 293T cells, with a repression rate of more than 25% as measured by the luciferase assay after transfection (Fig. 3A). Remarkably, we found that the core binding factor CBFβ had the most reduced activity, and the predicted binding sites are conserved among species (supplemental Fig. S3), suggesting that CBFβ is likely to be the most important target regulated by miR-125b. A construct containing a mutated sequence of the miRNA-binding site, 3′ CBFβ-mut, was also produced as a control (Fig. 3B). Luciferase activity measurements indicated specific repression of the wild-type substrate by miR-125b and no effect when the target site was mutated (Fig. 3, B and C). Altogether, these data demonstrate that CBFβ may be the target of miR-125b and that the putative binding sites are critical for miR-125b-mediated regulation of CBFβ expression.
FIGURE 3.
CBFβ is a direct target of miR-125b. A, 293T cells were co-transfected putative targets of miR-125b, including the CBFβ, SH2B3, C/EBPα, ETV6, DLL4, CCR5, and HOX genes, with miR-125b expression vector (miR-125b) or control vector (pCD6.2) using Lipofectamine 2000 reagent. Renilla luciferase activity was normalized to firefly luciferase activity, and the results were expressed relative to the pCD6.2 group. The 3′-UTRs of these genes were predicted to bind to miR-125b. Only CBFβ and CCR5 showed a reduction in activity, and CBFβ showed the greatest reduction in activity (50 plus 5%). B, schematic representation of the constructs used in the luciferase assay. The sequences shown below indicate the putative miR-125b target site on the wild-type 3′-UTR (construct CBFβ-wt), its mutated derivative (construct CBFβ-mut), and the pairing regions of miR-125b. C, 293T cells (upper panel) or K562 cells (lower panel) were co-transfected psi-check2 with either CBFβ-wt or CBFβ-mut, plus either pre-miR-125b (miR-125b) or empty vector (pCD6.2). Renilla luciferase activity was expressed relative to the pCD6.2 group. D, NB4 cells were electroporated with water or 100 pmol of mimics NC (mimics-scramble), mimics-miR-125b (left panel), anti-NC (anti-scramble), or anti-miR-125b (right panel). The levels of miR-125b were examined by qPCR. Cells lysates were prepared for Western blotting with an antibody against CBFβ, and the expression of β-tubulin served as a loading control. *, p < 0.05.
To further confirm that the CBFβ protein is suppressed by miR-125b, we performed both overexpression and knock-down of miR-125b in NB4 cells. As shown in Fig. 3D, when NB4 cells were transfected with the miR-125b mimics, the CBFβ protein was significantly reduced. Alternatively, when NB4 cells were transfected with the miR-125b inhibitor, CBFβ protein was expression increased (Fig. 3D). These data indicate that CBFβ is an important functional target of miR-125b in malignant myeloid cells.
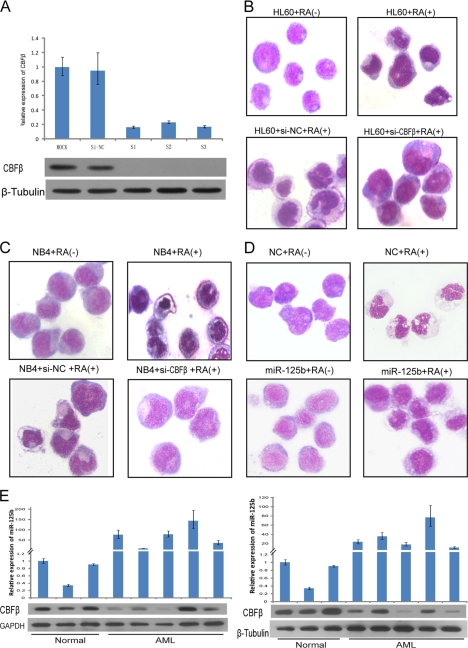
CBFβ is a subunit of the CBF. Recent studies have revealed that CBF is essential for normal hematopoiesis and that its aberrant expression might induce leukemia (22–24). To analyze the physiological relevance of CBFβ in counteracting myeloid cell differentiation, RNAi against CBFβ was performed. Transfection of NB4 cells with different siRNAs against the CBFβ mRNA resulted in a strong decrease in the levels of endogenous CBFβ mRNA and protein (Fig. 4A). To test if down-regulation of CBFβ is able to block the differentiation of HL60 and NB4 leukemic cells upon chemical treatment, we performed RNAi experiments with the addition of ATRA. These experimental conditions allowed us to get predominant maturation of HL60 cells, and NB4 cells treated with ATRA underwent characteristic granulocytic differentiation. We observed that in both HL60 and NB4 cell lines, down-regulation of CBFβ significantly prevented differentiation toward the granulocytic lineage. The maturation arrest was shown by morphology (Fig. 4, B and C). The manipulation by overexpression of miR-125b has a similar result with that using RNAi against CBFβ (Fig. 4D). These results show that CBFβ down-regulation is important for myeloid cell differentiation, at least for granulocytic lineage, therefore indicating that one of the pathways by which miR-125b prevents granulocytopoiesis is through CBFβ repression. This hypothesis was further confirmed in vivo with clinical samples (Fig. 4E). The expression levels of CBFβ and miR-125b were measured in a panel of AML-M3 primary samples, a subtype of AML, characterized by the accumulation of abnormal promyelocytes, which represents a unique model for differentiation therapy of human neoplasia (25). We found that the levels of CBFβ and miR-125b were inversely correlated in 80% of cases. Taken together, these results demonstrated that down-regulated CBFβ by miR-125b is associated with the hematopoietic malignancies.
FIGURE 4.
CBFβ plays an important role in myeloid cell differentiation, especially to the granulocytic lineage. A, qRT-PCR and Western blot analyses of the effects of different siRNAs against CBFβ. NB4 cells were transfected with water, siRNA-NC (siRNA-scramble), or three siRNA strands targeting CBFβ. B and C, morphological analysis of NB4 and HL60 cells at day 3 after electroporation with H2O, si-NC (siRNA negative control), or si-CBFβ following treatment with 1 μm ATRA. Transient transfection of the cells with si-CBFβ blocked the granulocytic differentiation induced by ATRA. A representative experiment is shown. D, morphological analysis of miR-125b overexpressed NB4 cells at day 3 of treatment with 1 μm ATRA. E, aberrant expression of CBFβ and miR-125b in 10 AML-M3 primary samples, 3 healthy donors were used as controls.
A CDX2/miR-125b/CBFβ Pathway during ATRA-mediated Myelocytic Differentiation
Based on the above results, we hypothesized that a dynamic relationship exists between ATRA, CDX2, miR-125b, and CBFβ. To investigate this hypothesis, we analyzed the levels of CDX2, miR-125b, and CBFβ in the in vitro system with NB4 and HL60 cells under ATRA induction. NB4 and HL60, two leukemia cell lines, are useful models for hematopoietic cell lineage differentiation as they respond to ATRA treatment thus allowing cells to reach a granulocyte terminal differentiation (16, 19, 26). The expression profile of miR-125b was detected using qRT-PCR. We observed that miR-125b progressively decreased down to 30% at 72 h in NB4 cells during differentiation induction with ATRA (Fig. 5A, upper panel). As expected, the Western blot analysis showed that the CDX2 protein behaves very similar to miR-125b, which is highly expressed at the earlier stage of induction and persistently decreases (Fig. 5A, middle panel). Conversely, our results showed that the CBFβ expression profile was increased after ATRA treatment (Fig. 5A, lower panel). Similar results were obtained in HL60 cells, which confirmed our hypothesis in vitro (supplemental Fig. S4).
FIGURE 5.
An ATRA-mediated CDX2/miR-125b/CBFβ pathway in myeloid cell lines and clinical samples. A, expression levels of CDX2, miR-125b, and CBFβ in NB4 cells using 1 μm ATRA induction for the indicated times. CDX2 was positively correlated with miR-125b expression, whereas CBFβ showed negative co-expression. *, p < 0.05. B and C, expression levels of CDX2 and miR-125b, in 20 pairs of AML-M3 patients before and after therapy. Average expression levels were statistically paired and compared with the average value of patient samples at primary diagnosis without treatment. Data were analyzed using analysis of variance followed by t test for pairwise comparisons to compare the values of means from two related samples in a “diagnosis and complete remission” scenario, Tukey-Kramer's test for multiple comparisons was used to compare the values of means between healthy and AML at diagnosis. D, miR-125b and CBFβ protein levels in 6 pairs of AML-M3 patients at diagnosis (D) and complete remission (CR).
To further validate the association of CDX2, miR-125b, and CBFβ in vivo, we measured the expression levels of three molecules in a panel of clinical samples, including 5 normal controls and 20 pairs of AML-M3 patients before and after therapy. AML-M3 paired patients at diagnosis/complete remission were chosen because ATRA is widely used in the treatment of AML-M3 in conventional therapy (27, 28). We found that the expression levels of miR-125b and CDX2 were highly expressed in all samples at diagnosis and markedly reduced in all of the diagnosis/complete remission samples compared (Fig. 5, B and C), indicating that the expression level of miR-125b mRNA positively correlates with CDX2 mRNA, and their expressions respond with ATRA treatment.
We next analyzed the levels of CBFβ protein in the same samples used above. The results showed that the CBFβ protein was elevated in ∼55% of diagnosis/complete remission patients (11/20) receiving therapy (the Spearman correlation coefficient was −0.95). The results from six pairs of samples are shown in Fig. 5D. Thus, there exists a negative correlation between the level of miR-125b and that of the CBFβ protein with respect to disease treatment.
DISCUSSION
In this study, we link expression of the miR-125b to homobox transcription factor CDX2 in myeloid cells and myeloid neoplasia, CDX2 leads to overexpression of miR-125b, which in turn regulates the repression of CBFβ, an important factor required for lineage differentiation, thereby counteracting myeloid cell differentiation, at least for granulocytic lineage. This is the first study on the mechanisms accounting for miR-125b dysregulation in hematopoietic malignancies. miR-125b is critical in multiple cellular processes, especially in controlling proliferation and differentiation (20, 21, 29). For instance, a recent report has also shown that miR-125b causes a dose-dependent myeloproliferative disorder that progress into myeloid leukemia. Overexpressing miR-125b in mice leads to myeloid-dominated bone marrow and spleen as well as myeloid cell infiltration into the liver (30), and myeloid cell differentiation arrest by miR-125b was found in acute myeloid leukemia (5). All these reports implied that miR-125b was significantly related to leukemia cell differentiation in vivo and might be an important factor in clinical therapeutics. Clarification of its regulatory mechanism is necessary to better understand the role of miR-125b in myeloid differentiation and provide a basis for the potential application of miR-125b in targeted therapeutics against hematopoietic malignancy.
Three observations indicate that there is a causal relationship between aberrant CDX2 and miR-125b expression and myeloid leukemogenesis. First, CDX2 displayed a similar expression pattern as miR-125b in both hematopoietic malignant cells and clinical samples. Second, two CDX2 consensus binding sites were mapped on the promoter region of the miR-125b gene and deletion of either site caused a dramatic decrease in the miR-125b promoter activity. EMSA and ChIP assays showed that CDX2 directly interacts with the miR-125b promoter in vitro and in vivo. Third, the direct interaction of CDX2 with the miR-125b promoter was further confirmed upon ATRA treatment. Our results demonstrate that down-regulation of CDX2 due to ATRA treatment suppresses the expression of miR-125b, which in turn leads to ATRA-mediated myeloid cell differentiation.
CDX2 has been demonstrated to play an essential role in the self-renewal of trophoblast stem cells (31) and has been linked to increased self-renewal in myeloid leukemias (32). Recent studies have shown that aberrant expression of CDX2 in the hematopoietic compartment is a frequent event in the pathogenesis of myeloid and lymphoblastic leukemias and promotes leukemogenesis (32–35). These data suggest a role for CDX2 as part of a common effector pathway that promotes the proliferative capacity and self-renewal potential of myeloid progenitor cells. Together with our observations, we have linked both important factors, CDX2 and miR-125b, during transformation converges to enhance self-renewal in myeloid cells. Another interesting finding is that in human ataxia telangiectasia, CDX2 is a transcriptional repressor of miR-125b (36). However, we found that CDX2 is a transcriptional activator of miR-125b in hematopoietic malignancies, mainly in myeloid leukemia. These data further support the notion that the same gene may play different roles in different situations or diseases. Furthermore, we tried to detect a new pathway of hematopoietic lineage differentiation of which both CDX2 and miR-125b may be components. Therapeutic targeting of these pathway subsets may be effective at combating hematopoietic cancers.
Given the specific expression of miR-125b during development of hematopoietic malignancies, and that miR-125b is an important effector for the commitment to differentiation, miR-125b appears to have a high hierarchical position among the factors regulating this hematopoietic lineage. Among the numerous putative targets of miR-125b, CBFβ mRNA was validated as a true functional target. Our results showed that RNAi against CBFβ in ATRA-treated promyelocytic cells arrests granulopoietic differentiation, suggesting that CBFβ down-regulation is indeed required for progression to granulopoietic differentiation. Notably, in vitro and in vivo experiments showed that miR-125b represses CBFβ. Previous studies have shown that CBFβ and its interacting component Runx1 are both necessary for HSC formation at the initial stage of definitive hematopoiesis and that only CBFβ is required for subsequent differentiation of the lymphoid and myeloid lineages (22, 37). CBFβ has also been reported to contribute to the inhibition of granulocyte differentiation for AML1-ETO (38) and is essential for maintaining the proliferation capacity of myeloid progenitors (39). Therefore, CBFβ appears to counteract differentiation in myeloid lineages. These results indicate that miR-125b may contribute to leukemogenesis partly through differentiation arrest by down-regulating CBFβ, at least in granulopoietic differentiation.
A recent study has shown that NFI-A and C/EBPα control granulocytic differentiation by regulating miR-223 (13). More recently, another study has proven that PU.1, together with interferon regulatory factors IRF-1 and IRF-9, also controls granulocytic differentiation by regulating miR-342 (16). Here, we provide another example that a CDX2/miR-125b/CBFβ pathway acts in ATRA-mediated myeloid cell differentiation, at least for a granulocytic lineage. We confirmed the existence of the CDX2/miR-125b/CBFβ pathway in a well established system for the differentiation/maturation of myeloid NB4 or HL60 cell lines and in clinical AML-M3 samples. Because AML-M3 (a acute promyelocytic leukemia) represents a unique model for differentiation therapy of human neoplasia (25), thus the discoveries in this study represent a new mechanism in hematopoietic cell differentiation block and indicated more therapeutic molecular targets.
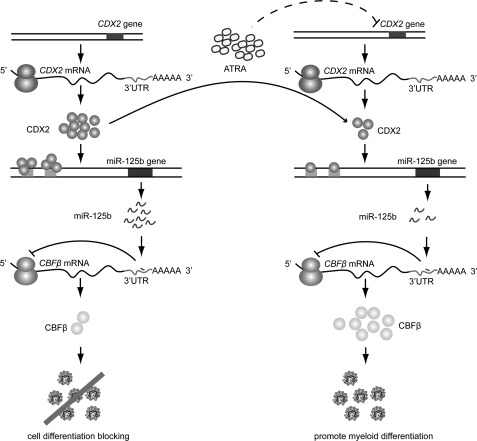
Taken together, as modeled in Fig. 6, we propose a CDX2/miR-125b/CBFβ pathway in human myeloid leukemia pathogenesis and therapy. In the development of hematopoietic malignancies, increasing levels of CDX2 activate miR-125b, which in turn inhibits CBFβ translation, thereby counteracting myeloid cell differentiation and promoting leukemogenesis. Once differentiation ensues, for example, upon induction of chemotherapeutics such as ATRA, CDX2 activity decreases, leading to reduced transcription of miR-125b. This reduction in CDX2 further causes expression changes of target genes, mediated at least partly by CBFβ; however, the mechanisms underlying the aberrant expression of CDX2 are not yet fully understood. In conclusion, our study provides a new mechanism that contributes to hematopoietic malignancies, which could involve the deregulation of miR-125b and its up- and downstream factors. These findings not only provide insights into the interplay between transcription factors and miR-125b during myeloid cell differentiation, but also potentially provide the knowledge necessary to design novel targeted therapies against the disease.
FIGURE 6.
Schematic representation of the CDX2/miR-125b/CBFβ pathway in the pathogenesis of hematopoietic malignancies and ATRA-dependent therapy. Left row, in the AML pathogenesis, increasing CDX2 levels activate miR-125b, which in turn inhibits CBFβ translation, thereby counteracting myeloid cell differentiation and promoting leukemogenesis. Right row, once differentiation ensues, for example, under ATRA induction, CDX2 activity decreases, reducing the transcription of miR-125b. This mechanism further contributes to ATRA-dependent therapy through high expression of target genes, mediated at least in part by CBFβ.
Acknowledgments
We thank the hospitals that provided samples for the analysis, Xue-Qun Luo at the First Affiliated Hospital of Sun Yat-sen University, and Dr. Hai-Xia Guo at the Second Affiliated Hospital of Sun Yat-sen University.
This work was supported by National Science and Technology Department 973 Program Grants 2011CBA01105, 2011CB811301, and 2009ZX09103-641, National Natural Science Foundation of China Grants 30672254 and 30872784, and the Fundamental Research Funds for the Central Universities.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S4.
- miRNA
- microRNA
- ATRA
- all-trans-retinoic acid
- CBFβ
- core binding factor β
- qPCR
- quantitative PCR
- AML
- acute myeloblastic leukemia.
REFERENCES
- 1. Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 2. Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. (2005) Nature 433, 769–773 [DOI] [PubMed] [Google Scholar]
- 3. Chen C. Z. (2005) N. Engl. J. Med. 353, 1768–1771 [DOI] [PubMed] [Google Scholar]
- 4. Esquela-Kerscher A., Slack F. J. (2006) Nat. Rev. Cancer 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 5. Bousquet M., Quelen C., Rosati R., Mansat-De Mas V., La Starza R., Bastard C., Lippert E., Talmant P., Lafage-Pochitaloff M., Leroux D., Gervais C., Viguié F., Lai J. L., Terre C., Beverlo B., Sambani C., Hagemeijer A., Marynen P., Delsol G., Dastugue N., Mecucci C., Brousset P. (2008) J. Exp. Med. 205, 2499–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang H., Luo X. Q., Zhang P., Huang L. B., Zheng Y. S., Wu J., Zhou H., Qu L. H., Xu L., Chen Y. Q. (2009) PLoS One 4, e7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chawengsaksophak K., James R., Hammond V. E., Köntgen F., Beck F. (1997) Nature 386, 84–87 [DOI] [PubMed] [Google Scholar]
- 8. Chawengsaksophak K., de Graaff W., Rossant J., Deschamps J., Beck F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7641–7645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shi X. B., Xue L., Yang J., Ma A. H., Zhao J., Xu M., Tepper C. G., Evans C. P., Kung H. J., deVere White R. W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19983–19988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liang L., Wong C. M., Ying Q., Fan D. N., Huang S., Ding J., Yao J., Yan M., Li J., Yao M., Ng I. O., He X. (2010) Hepatology 52, 1731–1740 [DOI] [PubMed] [Google Scholar]
- 11. Scott G. K., Goga A., Bhaumik D., Berger C. E., Sullivan C. S., Benz C. C. (2007) J. Biol. Chem. 282, 1479–1486 [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y., Yan L. X., Wu Q. N., Du Z. M., Chen J., Liao D. Z., Huang M. Y., Hou J. H., Wu Q. L., Zeng M. S., Huang W. L., Zeng Y. X., Shao J. Y. (2011) Cancer Res. 71, 3552–3562 [DOI] [PubMed] [Google Scholar]
- 13. Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M. L., Nervi C., Bozzoni I. (2005) Cell 123, 819–831 [DOI] [PubMed] [Google Scholar]
- 14. Fontana L., Pelosi E., Greco P., Racanicchi S., Testa U., Liuzzi F., Croce C. M., Brunetti E., Grignani F., Peschle C. (2007) Nat. Cell Biol. 9, 775–787 [DOI] [PubMed] [Google Scholar]
- 15. Rosa A., Ballarino M., Sorrentino A., Sthandier O., De Angelis F. G., Marchioni M., Masella B., Guarini A., Fatica A., Peschle C., Bozzoni I. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19849–19854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Marchis M. L., Ballarino M., Salvatori B., Puzzolo M. C., Bozzoni I., Fatica A. (2009) Leukemia 23, 856–862 [DOI] [PubMed] [Google Scholar]
- 17. Zhou R., Hu G., Liu J., Gong A. Y., Drescher K. M., Chen X. M. (2009) PLoS Pathog. 5, e1000681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suh E., Chen L., Taylor J., Traber P. G. (1994) Mol. Cell. Biol. 14, 7340–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tenen D. G. (2003) Nat. Rev. Cancer 3, 89–101 [DOI] [PubMed] [Google Scholar]
- 20. Le M. T., Teh C., Shyh-Chang N., Xie H., Zhou B., Korzh V., Lodish H. F., Lim B. (2009) Genes Dev. 23, 862–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le M. T., Xie H., Zhou B., Chia P. H., Rizk P., Um M., Udolph G., Yang H., Lim B., Lodish H. F. (2009) Mol. Cell. Biol. 29, 5290–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kundu M., Chen A., Anderson S., Kirby M., Xu L., Castilla L. H., Bodine D., Liu P. P. (2002) Blood 100, 2449–2456 [DOI] [PubMed] [Google Scholar]
- 23. Hart S. M., Foroni L. (2002) Haematologica 87, 1307–1323 [PubMed] [Google Scholar]
- 24. Talebian L., Li Z., Guo Y., Gaudet J., Speck M. E., Sugiyama D., Kaur P., Pear W. S., Maillard I., Speck N. A. (2007) Blood 109, 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tallman M. S., Nabhan C., Feusner J. H., Rowe J. M. (2002) Blood 99, 759–767 [DOI] [PubMed] [Google Scholar]
- 26. Roussel M. J., Lanotte M. (2001) Oncogene 20, 7287–7291 [DOI] [PubMed] [Google Scholar]
- 27. Melnick A., Licht J. D. (1999) Blood 93, 3167–3215 [PubMed] [Google Scholar]
- 28. Lo Coco F., Zelent A., Kimchi A., Carducci M., Gore S. D., Waxman S. (2002) Cancer Res. 62, 5618–5621 [PubMed] [Google Scholar]
- 29. Lee Y. S., Kim H. K., Chung S., Kim K. S., Dutta A. (2005) J. Biol. Chem. 280, 16635–16641 [DOI] [PubMed] [Google Scholar]
- 30. O'Connell R. M., Chaudhuri A. A., Rao D. S., Gibson W. S., Balazs A. B., Baltimore D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14235–14240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuckenberg P., Buhl S., Woynecki T., van Fürden B., Tolkunova E., Seiffe F., Moser M., Tomilin A., Winterhager E., Schorle H. (2010) Mol. Cell. Biol. 30, 3310–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scholl C., Bansal D., Döhner K., Eiwen K., Huntly B. J., Lee B. H., Rücker F. G., Schlenk R. F., Bullinger L., Döhner H., Gilliland D. G., Fröhling S. (2007) J. Clin. Invest. 117, 1037–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rawat V. P., Thoene S., Naidu V. M., Arseni N., Heilmeier B., Metzeler K., Petropoulos K., Deshpande A., Quintanilla-Martinez L., Bohlander S. K., Spiekermann K., Hiddemann W., Feuring-Buske M., Buske C. (2008) Blood 111, 309–319 [DOI] [PubMed] [Google Scholar]
- 34. Thoene S., Rawat V. P., Heilmeier B., Hoster E., Metzeler K. H., Herold T., Hiddemann W., Gökbuget N., Hoelzer D., Bohlander S. K., Feuring-Buske M., Buske C. (2009) Leukemia 23, 649–655 [DOI] [PubMed] [Google Scholar]
- 35. Riedt T., Ebinger M., Salih H. R., Tomiuk J., Handgretinger R., Kanz L., Grünebach F., Lengerke C. (2009) Blood 113, 4049–4051 [DOI] [PubMed] [Google Scholar]
- 36. Smirnov D. A., Cheung V. G. (2008) Am. J. Hum. Genet. 83, 243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Link K. A., Chou F. S., Mulloy J. C. (2010) J. Cell. Physiol. 222, 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roudaia L., Cheney M. D., Manuylova E., Chen W., Morrow M., Park S., Lee C. T., Kaur P., Williams O., Bushweller J. H., Speck N. A. (2009) Blood 113, 3070–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuo Y. H., Landrette S. F., Heilman S. A., Perrat P. N., Garrett L., Liu P. P., Le Beau M. M., Kogan S. C., Castilla L. H. (2006) Cancer Cell 9, 57–68 [DOI] [PubMed] [Google Scholar]