Background: Okenone is a unique ketocarotenoid found in purple sulfur bacteria; its diagenesis product, okenane, is an important geochemical biomarker.
Results: Novel C-4/4′ and C-2 carotene ketolases were identified in Thiodictyon sp. CAD16.
Conclusion: The complete biosynthetic pathway for okenone was elucidated.
Significance: Okenone biosynthesis is oxygen-independent and occurs under anoxic and microoxic conditions, so okenane is not a biomarker specific for anoxic conditions.
Keywords: Carotene, Carotenoid, Enzymes, Photosynthesis, Photosynthetic Pigments, Biomarkers, Carotenogenesis, Ketolase, Okenone, Purple Sulfur Bacteria
Abstract
Okenone is a unique ketocarotenoid found in many purple sulfur bacteria; it is important because of its unique light absorption and photoprotection properties. Okenane, a compound formed by diagenetic reduction of okenone, is an important biomarker in geochemical analyses of sedimentary rocks. Despite its ecological and biogeochemical importance, the biochemical pathway for okenone synthesis has not yet been fully described. The genome sequence of an okenone-producing organism, Thiodictyon sp. strain CAD16, revealed four genes whose predicted proteins had strong sequence similarity to enzymes known to produce ψ-end group modifications of carotenoids in proteobacteria. These four genes encoded homologs of a 1,2-carotenoid hydratase (CrtC), an O-methyltransferase (CrtF), and two paralogs of carotenoid 3,4-desaturases (CrtD). Expression studies in lycopene- or neurosporene-producing strains of Escherichia coli confirmed the functions of crtC and crtF, but the crtD paralogs encoded enzymes with previously undescribed functions. One enzyme, CruS, was only distantly related to CrtD desaturases, was bifunctional, and performed a 3,4-desaturation and introduced a C-2 keto group into neurosporene derivatives in the presence of dioxygen. The enzyme encoded by the other crtD paralog also represents a new enzyme in carotenogenesis and was named cruO. CruO encodes the C-4/4′ ketolase uniquely required for okenone biosynthesis. The identification of CruO and the demonstration of its biochemical activity complete the elucidation of the biosynthetic pathway for okenone and other related ketocarotenoids.
Introduction
Purple bacteria are a major group of anoxygenic, chlorophototrophic proteobacteria. They mainly occur in anoxic or microoxic aquatic environments in which they usually grow either photoautotrophically or chemolithoautotrophically by oxidizing sulfide and other reduced sulfur compounds (1–5). Because oxygen represses pigment synthesis and expression of genes for components of the photosynthetic apparatus, phototrophic growth of most of these organisms only occurs under anoxic conditions (aerobic anoxygenic phototrophs are an exception (6)). Two physiologically and phylogenetically separate groups are distinguished: the purple non-sulfur bacteria (Alphaproteobacteria and Betaproteobacteria) and purple sulfur bacteria (PSB2; Gammaproteobacteria) (5, 7, 8). The PSB comprise two lineages, Chromatiaceae and Ectothiorhodospiraceae, and more than 25 genera are recognized within these two groups (1, 4, 5).
Like purple non-sulfur bacteria (9, 10), PSB possess type-2 photochemical reaction centers composed of subunits PufL, PufM, and PufH; a membrane-anchored, tetraheme cytochrome, cytochrome c554, is often associated with the reaction centers as well (11). PSB characteristically synthesize either bacteriochlorophyll a or b (12), and these organisms generally synthesize carotenoids that belong to one of three main families: spirilloxanthin, rhodopinal (uncommon), or okenone, which are named after commonly encountered compounds of each family (13). Okenone is of special interest because its distinctive absorption properties confer a strong advantage to PSB that colonize deeper water layers (4). Consistent with this function, energy transfer studies have shown that excitonic energy is efficiently transferred from okenone to bacteriochlorophyll a in purified light-harvesting complexes in vitro (14). Okenone is also an important geochemical biomarker molecule. Okenane, the diagenesis product of okenone, has been detected in rock formations up to 1.64 billion years old (15). Okenane has been suggested to be an “unambiguous indicator for purple sulfur bacteria” (15), and the detection of this compound has been used as strong evidence to support suggestions that oceans were stratified and sulfidic up into the photic zone between ∼1,800 and 800 million years ago (e.g. see Refs. 15–17).
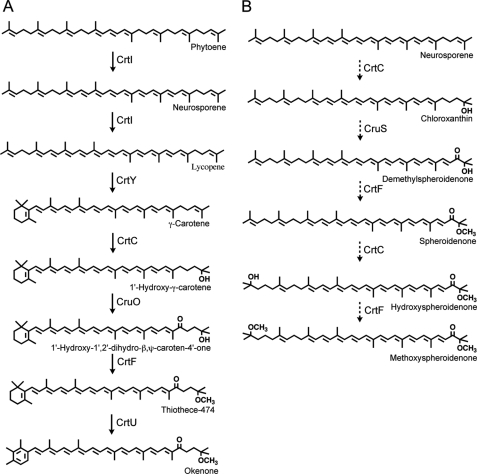
Okenone (1′-methoxy-1′,2′-dihydro-χ,ψ-caroten-4′-one) (see Fig. 9) was first described in 1963 (18), and the proposed structure was verified by total synthesis in 1967 (19). Okenone is a monocyclic, aromatic carotenoid that has a χ-ring as one end group. The open-chain ψ-end is methoxylated at the C-1′ atom and also has a keto group at C-4′ (13, 19). Thiodictyon sp. CAD16, which is a PSB isolated from Lake Cadagno, Switzerland (20), synthesizes okenone as its only carotenoid under anoxic, chlorophototrophic growth conditions (21). The two gene products, CrtY and CrtU, required for the synthesis of the χ-ring of okenone in this PSB were recently identified (21). The CrtY-type, lycopene monocyclase cyclizes one ψ-end group of the precursor, lycopene, and produces a β-ring. The β-ring of γ-carotene is subsequently converted into a χ-ring by CrtU, a γ-carotene desaturase/methyltransferase that is phylogenetically more closely related to CrtU proteins of green sulfur bacteria than to the mechanistically similar but phylogenetically distant CruE proteins of cyanobacteria (21). Based upon knowledge of carotenoid biosynthesis in other organisms, the following reactions are predicted to modify the remaining ψ-end group. The 1′,2′-double bond must be hydrated so that the hydroxyl group resides at the C-1′ position. Using S-adenosyl-l-methionine as the methyl donor, an O-methyltransferase would subsequently methylate the hydroxyl group (13, 22). The introduction of a keto group at the C-4′ would complete this predicted pathway.
FIGURE 9.
Proposed pathways for biosynthesis of okenone and methoxyspheroidenone. A, pathway leading to okenone. This pathway is not oxygen-dependent, and the enzymatic reactions were confirmed in E. coli. The reaction order shown is the most likely reaction order that has been deduced from information available in the literature and from the experiments described herein, but the timing of the ring desaturation/methyl transfer reaction is still uncertain. B, putative pathway for oxygen-dependent synthesis of methoxyspheroidenone. The dashed arrows indicate that the enzymatic functions have been confirmed in E. coli, but none of the carotenoids of this pathway were detected in Thiodictyon sp. CAD16 and T. marina cells grown under dark, microoxic conditions.
Most purple bacteria produce carotenoids lacking cyclic end groups, which often have methoxy groups at one or both ends of the molecule (13). Both required reactions, the hydration and the subsequent methylation of the introduced hydroxyl group, have been previously characterized in purple bacteria. The hydroxylation of the ψ-end group is carried out by 1,2-hydratases (CrtC) (23–25), and the methylation is performed by an O-methyltransferase (CrtF) (23, 26, 27). An alternative enzyme, CruF, can perform the 1,2-hydration reaction in Cyanobacteria, Chloroflexi, and some other bacteria (28). Three carotenoid ketolases, CrtO, CrtW, and CrtA, have been identified in chlorophototrophic bacteria. CrtO and CrtW introduce keto groups at the 4- and/or 4′-positions of β-rings in Cyanobacteria, Chloroflexi, and Acidobacteria (29–32). CrtA occurs in purple bacteria, and this enzyme introduces a keto group at the C-2 and C-2′-position of ψ-end groups (23, 33). A recent study reported that CrtA is a non-P450 heme protein and that the enzyme in Escherichia coli can introduce a hydroxyl group at C-1 and C-1′ in addition to the keto group at the C-2 and C-2′ positions in various substrates (34). CrtA and CrtO require oxygen to perform their ketolation reactions. CrtW is apparently a member of the di-iron oxygenase/membrane fatty acid desaturase family of proteins that may (30) or may not (35) require oxygen as a substrate.
In this study, we completed the elucidation of the biosynthetic pathway for okenone biosynthesis in Thiodictyon sp. CAD16. The enzymes responsible for the modifications of the ψ-end group were identified, specifically that for the introduction of the C-4′ keto group. This oxygen-independent enzyme, denoted CruO, defines a fourth family of bacterial carotenoid ketolases. Serendipitously, we also discovered a gene encoding an enzyme that has the combined activities of CrtD (3,4-desaturase) and CrtA (2-ketolase) when the protein is produced in E. coli. This oxygen-dependent enzyme defines a fifth family of bacterial carotenoid ketolases.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Thiodictyon sp. CAD16 is a purple sulfur bacterium (Chromatiaceae) isolated from Lake Cadagno, Switzerland (20). The draft genome of this organism was determined by pyrosequencing in the laboratory of Dr. Stephan C. Schuster3 and in the Genomics Core Facility of The Pennsylvania State University. Data from GX-20 FLX, titanium, and paired end chemistries were combined to produce the draft genome sequence. Three genes, crtC, crtF, and crtD, for carotenoid biosynthesis were identified by blast searches using sequences derived from other carotenoid-synthesizing organisms as queries. The regions encoding the genes for okenone biosynthesis in Thiodictyon sp. CAD16 have been deposited in GenBankTM under the accession numbers JN206680 (cluster shown in Fig. 1) and JN206681 (crtF). Draft genome sequences for Thiocapsa marina DSM 5653T and Marichromatium purpuratum strain DSM 1591T were determined as part of a community sequencing project in collaboration with the Department of Energy-Joint Genome Institute and will soon be available through GenBank.
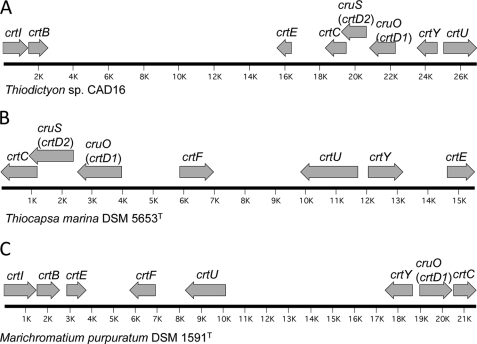
FIGURE 1.
Organization of carotenoid genes of PSB. A, Thiodictyon sp. CAD16; B, T. marina DSM 5653T; and C, M. purpuratum DSM 1591T.
Recombinant DNA procedures were performed by standard methods using chemically competent E. coli strain TOP10F′. The expression of carotenoid genes was performed in E. coli BL21(DE3) cells. E. coli cells were grown in LB medium supplemented with 34 μg of chloramphenicol ml−1, 50 μg of ampicillin ml−1, 50 μg of streptomycin ml−1, and 30 μg of kanamycin ml−1 as required. For expression of genes for carotenoid biosynthesis, cells were grown at 37 °C to stationary phase.
Cloning of Carotenoid Genes and Construction of E. coli Strains Expressing Genes of Carotenogenesis
PCR amplification of genes encoding enzymes for carotenogenesis was performed using genomic template DNA from Thiodictyon sp. CAD16, the Phusion proofreading polymerase (Finnzymes, Vantaa, Finland), and oligonucleotide primers (see supplemental Table S1). PCR products were digested with restriction enzymes as required for the sites introduced by the oligonucleotide primers and PCR amplifications and were cloned into pCOLA Duet vector or pCDF Duet vector (Novagen-EMD, La Jolla, CA). DNA sequencing was performed to verify all recombinant gene constructions. Plasmids pAC::LYC and pAC::NEUR (36) and combinations of pCOLA::crtD1(cruO), pCOLA::crtD2(cruS), pCOLA::crtD1(cruO)::crtD2(cruS), pCDF::crtC, pCDF::crtF, and pCDF::crtC::crtF were transformed into chemically competent BL21(DE3) cells, and cells were plated on Luria-Bertani medium amended with the respective antibiotics. Single transformant colonies were picked and grown to stationary phase at 37 °C without induction with isopropyl β-d-thiogalactoside in LB medium supplemented with the same antibiotics. In vitro tests of the functions of carotenoid genes were performed with E. coli BL21(DE3) cells harboring plasmid pCOLA::crtD1(cruO) or plasmid pCDF::crtC. Cells were grown to stationary phase at 37 °C without induction with isopropyl β-d-thiogalactoside in LB medium supplemented with the appropriate antibiotics. 5 ml of cells were pelleted by centrifugation in a tabletop centrifuge and washed in 25 mm 3-(N-morpholino)propanesulfonate buffer, pH 7.2. Cells were resuspended in 450 μl of the same buffer and broken by sonication. 2–6 μg of carotenoids dissolved in 50 μl of methanol were added to the crude cell extract, and the solution was incubated overnight in the dark at 37 °C. Carotenoids were extracted from the reaction mixture with diethyl ether and dried under a stream of nitrogen gas.
Pigment Analysis
Pigments were extracted from cell pellets of E. coli by sonication in acetone/methanol (7:2, v/v), and the resulting extracts were analyzed by high performance liquid chromatography (HPLC). The HPLC system included a binary pump (model G1312A), vacuum degasser (model G1379A), manual injector (model G1328A), and a diode array detector (model G1315B) (1100 series, Agilent Technologies, Palo Alto, CA); Agilent ChemStation software was used to control the system. The HPLC column was an analytical 5-μm Discovery C18 column (25 cm × 4.6 mm; Supelco, Bellefonte, PA). Solvent A was methanol/acetonitrile/water (42:33:25 by volume), and solvent B was methanol/ethyl acetate/acetonitrile (50:30:20 by volume). The solvent B concentration was increased linearly from 70 to 90% over 5 min and then increased to 100% over 10 min. Solvent B was held at 100% for 10 min and returned within 1.0 min to 70%. The flow rate was held constant at 1.00 ml min−1. The same HPLC system and program as described above were used to purify selected carotenoids. Carotenoid-containing fractions were collected and dried under a stream of nitrogen gas. For better resolution, carotenoid fractions were chromatographed on an analytical 5-μm Prontosil C30 column (4.6 × 250 mm; MAC-MOD Analytical, Chadds Ford, PA). Solvent A was methanol/methyl tert-butyl ether/water (66:30:4 by volume), and solvent B was methyl tert-butyl ether/methanol/acetonitrile (50:30:20 by volume). The solvent B concentration was increased linearly from 20 to 60% over 15 min and then held at 60% constant for 15 min followed by a linear increase to 100% over 5 min. Solvent B was held at 100% for 10 min and returned within 0.1 min to 20%. The flow rate was held constant at 1.00 ml min−1. The HPLC system was the same as described above. Sodium borohydride reduction was performed as described by Britton (37, 38) to test for the presence of aldehyde or keto groups in purified carotenoids. Reduced carotenoids were extracted with diethyl ether and reanalyzed on the HPLC system using the protocol described in Frigaard et al. (39). Carotenoids were identified by comparison of elution times of unknowns with those for known standards, absorption spectral properties, behavior after chemical modification, and mass spectrometry.
Mass Spectrometry
Pigment-containing fractions were collected as described above and dried under a stream of nitrogen gas. Mass spectrometric analyses were performed on a Waters LCT Premier time-of-flight (TOF) mass spectrometer (Waters Corp. (Micromass Ltd.), Manchester, UK). The mass spectrometer was operated with MassLynxTM software Version 4.0. Dried carotenoid samples were dissolved in acetonitrile, and samples were introduced into the mass spectrometer using a Waters 2695 HPLC system. Samples were analyzed using flow injection analysis in which the sample is injected into the mobile phase and passes directly into the mass spectrometer where the analytes are ionized and detected. The mobile phase (flow rate was 0.15 ml min−1) used was 75% acetonitrile (LC-MS grade) and 25% aqueous ammonium acetate (10 mm). The nitrogen drying gas temperature was set to 200 °C at a flow of 6 liters min−1, and the capillary voltage was 2,200 V. The mass spectrometer was set to scan from 100 to 650 m/z in positive ion mode and used electrospray ionization.
Phylogenetic Analyses
For phylogenetic analyses, protein sequences with similarity to CrtD1 (CruO) and CrtD2 (CruS) were identified and recovered from public databases using the blastP algorithm (40) and were aligned with ClustalX Version 2.0 (41). Phylogenetic trees were calculated by using PROTDIST with the Jones-Taylor-Thornton matrix of the Phylogeny Inference Package (PHYLIP Version 3.6.8) (42). Phylogenetic trees were inferred from the distances by using the NEIGHBOR program of the Phylogeny Inference Package. Bootstrap analyses (100 resamplings) were performed with SEQBOOT of the Phylogeny Inference Package.
RESULTS
Organization of Carotenogenesis Genes in Three Okenone-producing Purple Sulfur Bacteria
Homologs of several known carotenogenesis genes were identified in the draft genome sequence of the okenone-producing bacterium Thiodictyon sp. CAD16 (21) (Fig. 1A). All of these genes except crtF occurred in a 26-kb region of the genome (Fig. 1A). The clustered genes included crtE (geranylgeranyl pyrophosphate synthase), crtB (phytoene synthase), crtI (phytoene desaturase), crtC (hydroxyneurosporene synthase), two paralogous crtD genes encoding proteins with distant sequence similarity to C-3,4 desaturase, crtU (γ-carotene desaturase), and crtY (lycopene cyclase). Importantly, no close homologs of the ketolases CrtO, CrtW, and CrtA were identified in the genome. For reasons that will be described below, the crtD1 gene will be redesignated cruO, and the crtD2 gene will be redesignated cruS. The crtY and crtU genes as well as the two crtD paralogs (i.e. cruO and cruS) and crtC are clustered together; crtE was found downstream from crtC (see Fig. 1A). The crtI and crtB genes were located about 12 kb away and apparently form a dicistronic operon. The crtF gene, encoding a protein with similarity to hydroxyneurosporene-O-methyltransferase, was also identified, but this gene was encoded on a separate contig that apparently is not linked to this region of the genome. An unusual aspect of the gene organization is that the crtD2/cruS and crtC sequences overlap over an extended region of 303 bp; the overlapping genes are in different reading frames.
T. marina DSM 5653T is a marine, okenone-producing PSB (43), and the organization of its genes for carotenogenesis is shown in Fig. 1B. The crtI and crtB genes were closely linked and probably form a dicistronic operon, but these two genes are encoded on a separate contig in the draft genome sequence that is unlinked to the larger gene cluster and are separated from it by at least 268 kb. The organization of the remaining genes is similar to that in Thiodictyon sp. CAD16 except that crtF occurs between crtU and cruO. Interestingly, the organization of the two crtD paralogs and crtC as well as the organization of the overlapping crtD2/cruS and crtC sequences (297-bp overlap, different reading frames) are similar to the arrangement in Thiodictyon sp. CAD16 (Fig. 1B).
M. purpuratum DSM 1591T is also a marine, okenone-producing PSB (44). All of the genes encoding enzymes of carotenogenesis in M. purpuratum occur within a 21-kb region of its genome (Fig. 1C). In contrast to the situation in T. marina and Thiodictyon sp. CAD16, only a single crtD homolog, which is most similar to crtD1/cruO of Thiodictyon sp. CAD16, was found upstream from crtC, and the sequence of this gene did not overlap with that of crtC (Fig. 1C). No ortholog of crtD2/cruS was identified in the draft genome of M. purpuratum, which obviously suggested that the product of this gene is not likely to be required for the synthesis of okenone. The crtF gene was also identified within this region, but its orientation relative to crtU differs from that in T. marina. The observed differences in the organization of these carotenogenesis genes suggests that multiple recombination events have led to substantial rearrangements of these genes in the three strains examined. In all three organisms, okenone was the only carotenoid produced under photoautotrophic growth conditions (21). Okenone was also the only carotenoid produced by T. marina when cells were grown under dark microoxic, chemolithoautotrophic conditions. Thiodictyon sp. CAD16 also produced exclusively okenone under various growth conditions, including photoautotrophic, photomixotrophic, and dark microoxic, chemoautotrophic conditions.4
Modification of ψ-End Group by CrtC and CrtF
To understand the functions of some of the carotenogenesis genes of Thiodictyon sp. CAD16 better, the crtC, crtF, crtD1/cruO, and crtD2/cruS genes were expressed in E. coli BL21(DE3) strains that produced either lycopene or neurosporene (36). An E. coli strain harboring pAC::LYC and thus producing lycopene was transformed with the expression plasmid pCDFDuet::crtC encoding the putative 1,2-hydratase of Thiodictyon sp. CAD16. The resulting transformed cells produced lycopene, 1-hydroxylycopene, and 1,1′-dihydroxylycopene, products that have the same absorption spectra (and therefore chromophore) but different retention times on the HPLC (supplemental Fig. S1A) (45, 46). Cells harboring plasmids pAC::LYC, pCPL1 (encoding the CruA-type lycopene monocyclase of Chlorobaculum tepidum (47), and pCDFDuet::crtC only produced small amounts of γ-carotene and 1′-hydroxy-γ-carotene (data not shown). Similar results were obtained when the crtY gene from Thiodictyon sp. CAD16 was used (data not shown). These results suggested that hydroxylation occurred more rapidly than ring cyclization in E. coli and that hydroxylation prevented cyclization of lycopene to form γ-carotene.
To determine whether this interpretation of the results was correct, γ-carotene was synthesized in E. coli and used as the substrate in an in vitro assay with an extract containing CrtC. When purified γ-carotene was incubated with a crude extract of E. coli cells harboring plasmid pCDFDuet::crtC, 1′-hydroxy-γ-carotene was efficiently produced (Fig. 2, peak 1). CrtC from Thiodictyon sp. CAD16 could also use neurosporene as a substrate and produced 1- and 1′-hydroxyneurosporene and 1,1′-dihydroxyneurosporene as the products (supplemental Fig. S1B). These results and those described above indicated that Thiodictyon sp. CAD16 CrtC hydrates the double bonds of the ψ-end groups of lycopene, 1-hydroxylycopene, γ-carotene, neurosporene, and 1- and 1′-hydroxy-neurosporene.
FIGURE 2.
HPLC elution profile of pigments produced in in vitro assay with γ-carotene and crude extract of E. coli BL21(DE3) harboring plasmid pCDFDuet::crtC. The in-line absorption spectra of peaks 1 and 2 are shown in the numbered boxes below the elution profile. Peak 1, 1′-hydroxy-γ-carotene; peak 2, γ-carotene.
To test the function of the O-methyltransferase (CrtF), E. coli was transformed with plasmids pAC::LYC and pCDFDuet::crtC::crtF. Six carotenoids, all of which had the same absorption spectrum as lycopene, could be resolved by HPLC from this strain (Fig. 3). The addition of a single hydroxyl group by hydration of the 1,2-double bond caused the resulting 1-hydroxylycopene to elute ∼4.6 min earlier than lycopene, and the addition of a second hydroxyl group by hydration of the 1′,2′-double bond caused 1,1′-dihydroxylycopene to elute ∼8.8 min earlier than lycopene (supplemental Fig. S1A and Fig. 3). O-Methylation of a hydroxyl group caused the retention time to increase by ∼2 min. Therefore, the six carotenoids produced were 1,1′-dihydroxylycopene (Fig. 3, peak 1), 1-methoxy-1′-hydroxylycopene (Fig. 3, peak 2), 1-hydroxylycopene (Fig. 3, peak 3), 1,1′-dimethoxylycopene (Fig. 3, peak 4), 1-methoxylycopene (Fig. 3, peak 5), and lycopene (Fig. 3, peak 6). E. coli cells transformed with pAC::NEUR, pCDFDuet::crtC::crtF, and pCOLADuet::crtD2(cruS) showed that 2-ketocarotenoids (see below) could also be methylated at their 1- and/or 1′-hydroxy groups (data not shown).
FIGURE 3.
HPLC elution profile of pigments produced in E. coli BL21(DE3) harboring plasmids pAC::LYC and pCDFDuet::crtC::crtF. The in-line absorption spectra of peaks 1 to 6 were identical; the spectrum of the compound numbered 1 is shown in the numbered box below the elution profile. Peak 1, 1,1′-dihydroxylycopene; peak 2, 1-methoxy-1′-hydroxylycopene; peak 3, 1-hydroxylycopene; peak 4, 1,1′-dimethoxylycopene; peak 5, 1-methoxylycopene; peak 6, lycopene. The insertion of a hydroxyl group into lycopene or 1-hydroxylycopene causes a compound to elute ∼4.4 min earlier. The methylation of a hydroxyl group increases the retention time by ∼2 min.
Modification of ψ-End Groups by CrtD1 (CruO)
Although the CrtD1/CruO and CrtD2/CruS gene products have some sequence similarity to C-3,4 carotenoid desaturases (∼50% identity and 55% similarity (48)), these enzymes had no activity with the substrates neurosporene, lycopene, and γ-carotene (data not shown). When E. coli was transformed with pAC::LYC, pCDFDuet::crtC, and pCOLADuet::crtD1(cruO), four carotenoids were produced (Fig. 4), and none had the absorption or HPLC elution properties of lycopene or its mono- or dihydroxy derivatives (see Fig. 4 and supplemental Fig. S1A). The compound in peak 1 eluted earlier (i.e. was more hydrophilic) than 1,1′-dihydroxylycopene, and its absorption spectrum showed a bathochromic shift of ∼30 nm compared with lycopene and its derivatives. The compounds in peaks 2, 3, and 4 eluted earlier than 1-hydroxylycopene, and each had bathochromic shifts of ∼15 nm compared with lycopene and its derivatives (Fig. 4). Furthermore, the fine structure of the absorption spectra decreased in each of the four new carotenoids (Fig. 4). The increased hydrophilicity, bathochromic shift, and reduced fine structure in the absorption spectrum are consistent with the presence of one keto group in compounds 2, 3, and 4 and two keto groups in compound 1.
FIGURE 4.
HPLC elution profile of pigments produced in E. coli BL21(DE3) harboring plasmids pAC::LYC, pCDFDuet::crtC, and pCOLADuet::crtD1(cruO). The in-line absorption spectra of the compounds numbered 1 to 4 are shown in the numbered boxes below the elution profile. Peak 1, carotenoid with reduced fine structure and a bathochromic shift of ∼30 nm compared with lycopene; peaks 2, 3, and 4, carotenoids with reduced fine structure and bathochromic shifts of ∼15 nm compared with lycopene. For additional details, see the text.
The carotenoids in peaks 1 and 2 were purified and subjected to reduction with sodium borohydride. The long reaction time required (30 min); the hypsochromic shifts of 29 and 19 nm, respectively; and the increased fine structure of the resulting products (Fig. 5) confirmed that keto groups were present in both compounds (37, 38). The absorption spectra of the resulting products were very similar to those of lycopene and its 1-hydroxy and 1,1′-dihydroxy derivatives (Fig. 5). HPLC analyses of the reduced carotenoids implied the presence of four hydroxyl groups in the product from the carotenoid of peak 1 and three hydroxyl groups in the product from the carotenoid of peak 2 (supplemental Fig. S2). Furthermore, the carotenoid in peak 1 of Fig. 4 had an m/z ratio of 601.4, and the carotenoid in peak 2 had an m/z ratio of 587.4 (supplemental Fig. S3). These data collectively indicated that the carotenoid in peak 1 of Fig. 4 contained two hydroxyl groups and two keto groups, whereas the carotenoid in peak 2 of Fig. 4 contained two hydroxyl groups and a single keto group. Therefore, compounds 1 and 2 are 4,4′-diketo-1,1′-dihydroxylycopene and 4-keto-1,1′-dihydroxylycopene, respectively (Fig. 4). Compounds 3 and 4 were not fully identified, but the absorption spectra indicated that each also carried a single keto group and probably a single hydroxyl group. The absorption spectrum of compound 3 further suggested that it is likely to be a cis-isomer of 4-keto-1-hydroxylycopene; the compound in peak 4 is probably 4-keto-1-hydroxylycopene.
FIGURE 5.
Test for keto groups in carotenoids by reduction with sodium borohydride. A, carotenoid corresponding to the peak of the elution profile shown in Fig. 4. Panel 1, spectrum of carotenoid dissolved in isopropanol; panel 2, spectrum 2 min after the addition of sodium borohydride; panel 3, spectrum 30 min after the addition of sodium borohydride. Complete reduction of the carotenoid occurred after 30 min, resulting in a hypsochromic shift of 29 nm and an increase in the fine structure. The resulting carotenoid had a spectrum identical to that of lycopene. B, carotenoid corresponding to peak 2 of the elution profile shown in Fig. 4. Panel 1, spectrum of carotenoid dissolved in isopropanol; panel 2, spectrum 2 min after the addition of sodium borohydride; panel 3, spectrum 30 min after the addition of sodium borohydride. Complete reduction of the carotenoid occurred after 30 min, resulting in a hypsochromic shift of 19 nm and an increase of the fine structure. The resulting carotenoid had a spectrum identical to that of lycopene.
1-Hydroxylycopene and 1,1′-dihydroxylycopene were extracted from E. coli cells harboring pAC::LYC and pCDFDuet::crtC, and these substrates were incubated with a whole-cell extract from E. coli cells harboring pCOLADuet::crtD1(cruO). The results showed that the CrtD1/CruO protein alone was capable of introducing the 4-keto group into these substrates (Fig. 6). Growth of E. coli harboring plasmids pAC::LYC, pCDFDuet::crtC, and pCOLADuet::crtD1(cruO) under anoxic conditions showed that oxygen is not required for the formation of the keto group by CrtD1/CruO (supplemental Fig. S4). Analyses of carotenoids produced in E. coli harboring plasmids pAC::NEUR, pCDFDuet::crtC, and pCOLADuet::crtD1(cruO) showed that 1-hydroxyneurosporene, 1′-hydroxyneurosporene, and 1,1′-dihydroxyneurosporene were also substrates for CrtD1/CruO (supplemental Fig. S5). γ-Carotene was extracted from E. coli cells harboring pAC::LYC and pCPL1, and this substrate was incubated with whole-cell extracts from E. coli cells harboring pCDFDuet::crtC and E. coli cells harboring pCOLADuet::crtD1(cruO). Traces of a carotenoid with the spectral features of thiothece-474 were observed (49) (data not shown). Small amounts of the same carotenoid were detected in E. coli cells transformed with pAC::LYC, pCPL1, pCDFDuet::crtC, and pCOLADuet::crtD1(cruO). This carotenoid was absent in E. coli cells transformed with pAC::LYC, pCDFDuet::crtC, and pCOLADuet::crtD1(cruO) and E. coli cells transformed with pAC::LYC, pCPL1, and pCDFDuet::crtC (data not shown). These observations showed that 1′-hydroxy-γ-carotene was a substrate for CrtD1/CruO. The data obtained here demonstrated that CrtD1/CruO introduces a keto group at the 4- and/or 4′-positions of various carotenoid substrates, including acyclic and monocyclic compounds, and at both the 4- and 4′-positions in some acyclic substrates. Because this gene product performs a unique reaction that has not previously been characterized in carotenoid biosynthesis, i.e. the introduction of the 4-keto groups of ψ-ends with hydrated 1,2-double bonds, this gene has been redesignated cruO.
FIGURE 6.
HPLC elution profile of pigments produced in in vitro assay with hydroxylated lycopene derivatives and crude extract of E. coli BL21(DE3) harboring plasmid pCOLADuet::crtD1(cruO). The in-line absorption spectra of the compounds numbered 1 to 5 in the elution profile are shown in the numbered boxes. Peak 1, carotenoid with reduced fine structure and a bathochromic shift of ∼15 nm compared with lycopene; peak 2, 1,1′-dihydroxylycopene; peak 3, carotenoid with reduced fine structure and a bathochromic shift of ∼15 nm compared with lycopene; peak 4, 1-hydroxylycopene; peak 5, lycopene. For additional details, see the text.
Modification of ψ-End Groups by CrtD2/CruS
E. coli strains transformed with plasmids pAC::LYC, pCDFDuet::crtC, and pCOLADuet::crtD1(cruO)::crtD2(cruS) (instead of pCOLADuet::crtD1(cruO)) produced the same carotenoids as cells producing CrtC and CruO (see Fig. 4 and above). However, when E. coli was transformed with pAC::NEUR, pCDFDuet::crtC, and pCOLADuet::crtD2(cruS), new carotenoids were produced (Fig. 7). Besides 1-hydroxyneurosporene/1′-hydroxyneurosporene and 1,1′-dihydroxyneurosporene, two carotenoids with reduced fine structure and a bathochromic shift of ∼40 nm compared with neurosporene were produced (Fig. 7, peaks 1 and 3). The compounds in peaks 1 and 3 were purified by HPLC and subsequently reduced with sodium borohydride. The long reaction time of 30 min, the hypsochromic shift of 29 nm, and the increased fine structure in the absorption spectra of the products (supplemental Fig. S6) confirmed the presence of a keto group in these two compounds. Moreover, the absorption spectra of the reduced compounds indicated the presence of 10 conjugated double bonds rather than the nine found in neurosporene. The compounds purified from peaks 1 and 3 had m/z ratios of 587.4 and 569.4, respectively (supplemental Fig. S7). These data indicated that the compound in peak 1 contained two hydroxyl groups, one keto group, and one double bond more than neurosporene, whereas the compound in peak 3 contained one hydroxyl group, one keto group, and one double bond more than neurosporene. It was inferred that the compound in peak 1 was 1′-hydroxydemethylspheroidenone and that the compound in peak 3 was demethylspheroidenone. To verify the formation of a keto group at C-2 position, authentic spheroidenone produced by Rhodobacter sphaeroides cells grown under microoxic conditions was used as a chromatographic standard. E. coli cells transformed with pAC::NEUR, pCDFDuet::crtC::crtF, and pCOLADuet::crtD2(cruS) produced spheroidenone as well as other related carotenoids (supplemental Fig. S8). The spheroidenone produced coeluted with authentic spheroidenone when the two samples were coinjected in the HPLC system, and their absorption spectra were identical (supplement Fig. S8). When E. coli cells harboring plasmids pAC::NEUR, pCDFDuet::crtC, and pCOLADuet::crtD2(cruS) were grown under anoxic conditions, no keto groups were introduced, and the elution profile was identical to that shown in supplemental Fig. S1B. E. coli strains transformed with plasmids pAC::LYC, pCDFDuet::crtC, and pCOLADuet::crtD2(cruS) showed that 1,1′-dihydroxylycopene and 1-hydroxylycopene were not substrates for CrtD2/CruS (data not shown). An in vitro assay with γ-carotene and whole-cell extracts from E. coli cells harboring pCDFDuet::crtC and E. coli cells harboring pCOLADuet::crtD2(cruS) showed that 1′-hydroxy-γ-carotene was also not a substrate (data not shown). These results showed that CrtD2/CruS introduces a 2-keto group and a 3,4-double bond into neurosporene, 1-hydroxy-neurosporene, or 1,1′-dihydroxyneurosporene. These oxygen-dependent activities are equivalent to the combined activities of CrtA (33) and CrtD (48), respectively. To reflect the fact that this is a previously undescribed enzymatic activity in carotenoid biosynthesis, the crtD2 gene has been redesignated cruS.
FIGURE 7.
HPLC elution profile of pigments produced in E. coli BL21(DE3) harboring plasmids pAC::NEUR, pCDFDuet::crtC, and pCOLADuet::crtD2(cruS). The in-line absorption spectra of the compounds in peaks 1 and 2 are shown in the numbered boxes below the elution profile. Peaks 1 and 3, carotenoids with reduced fine structure and a bathochromic shift of ∼40 nm compared with neurosporene; peak 2, 1,1′-dihydroxyneurosporene; peak 4, 1-hydroxyneurosporene and/or 1′-hydroxyneurosporene; peak 5, neurosporene.
Phylogenetic Analysis of Carotenoid Genes
Fig. 8 shows a phylogenetic analysis of CrtD, CruO, and CruS proteins from various purple bacteria. The CruO and CruS ketolases are only very distantly related to other members (22, 29, 32) of the pyridine nucleotide-disulfide oxidoreductase superfamily, including the β-carotene ketolase CrtO found in cyanobacteria and other organisms (22, 29, 32). Thus, CrtO proteins were not included, and only CrtD-like proteins of purple bacteria were included in this analysis. As can be seen from the branch lengths, the CrtD proteins of purple bacteria are highly divergent. However, the CruO and CruS clades appear to be distinctive and are well separated from other CrtD proteins. Aside from the proteins and organisms discussed here, proteins related to CruO and CruS were only detected in a contig derived from a marine metagenome.
FIGURE 8.
Neighbor-joining phylogenetic tree of CrtD desaturases and their homologs. Bootstrap numbers from 100 resamplings are indicated at the nodes.
DISCUSSION
In this study, the enzymes that modify the ψ-end of okenone were identified in Thiodictyon sp. CAD16, a PSB that produces okenone as the only carotenoid when grown under dark microoxic or anoxic conditions in the light. Because Thiodictyon sp. CAD16 and other okenone-producing bacteria are not genetically manipulatable, the genes encoding these enzymes were heterologously expressed in E. coli. These three enzymes introduce the methoxyl group at C-1′ (CrtC and CrtF) and the keto group at C-4′ (CruO) of the ψ-end of okenone. Together with the results of a previous study concerning CrtY and CrtU (21), the key enzymes of okenone biosynthesis have now been characterized. Although heterologous expression of the carotenoid genes cannot prove the order in which the reactions occur in the native organism, a pathway for the biosynthesis of okenone in Thiodictyon sp. CAD16 can be proposed, and similarly, a putative pathway for the synthesis of methoxyspheroidenone can also be proposed (Fig. 9). As is the case in other bacteria, lycopene is the substrate for the cyclization reaction to produce γ-carotene (21, 50, 51). CrtU probably acts on γ-carotene as occurs in GSB (45), but because thiothece-474 is found in small amounts in some okenone-producing bacterium (52), CrtU could also catalyze the last step in okenone biosynthesis. However, although the yield of χ,ψ-carotene was low, heterologous expression of CrtU in C. tepidum showed that γ-carotene can be a substrate for CrtU (21). CruO and CrtF only modified hydroxylated carotenoids, and similarly, CruO was active with 1-hydroxy-γ-carotene and 1-hydroxylycopene but was not active with lycopene. Because CrtF has a very broad substrate spectrum (26) and because Rubrivivax gelatinosus CrtD cannot desaturate 1-methoxy carotenoids (48), we propose that the ψ-end modifications are catalyzed in sequence by CrtC, CruO, and finally CrtF. Based on the findings described here and elsewhere (21), the most likely sequence of reactions for the conversion of phytoene into okenone are summarized in Fig. 9A. The putative pathway for the synthesis of methoxyspheroidenone includes the hydroxylation of neurosporene. 1- or 1′-Hydroxylated neurosporene are then substrates for CruS introducing a desaturation and a keto group at C-2 position. Consecutive methylation and hydroxylation reactions then lead to methoxyspheroidenone (Fig. 9B).
The 1,2-hydratases of several purple bacteria, including Rhodobacter capsulatus and R. gelatinosus, have been characterized previously, and similar enzymes from other chlorophototrophic and heterotrophic bacteria have also been identified (23–25, 45, 53–55). CrtC from R. capsulatus modifies neurosporene or lycopene, but this enzyme is unable to introduce a second hydroxyl group, and thus, monohydroxylated carotenoids are produced in this organism. On the other hand, CrtC from R. gelatinosus hydroxylates various acyclic carotenes and their 1-hydroxy derivatives (24). CrtC from Thiodictyon sp. CAD16 resembled CrtC of R. gelatinosus with respect to its broad substrate specificity and its ability to synthesize 1,1′-dihydroxy carotenoids. The 1,2-hydratase of Thiodictyon sp. CAD16 could hydroxylate a monocyclic carotene, γ-carotene, a reaction that is also catalyzed by CrtC from C. tepidum (45). In the experiments conducted here, hydroxylation of lycopene prevented cyclization. This observation implies that Thiodictyon sp. cells must have a mechanism to ensure that cyclization occurs before hydroxylation, but the nature of this process is currently unknown.
Three families of carotenoid ketolases, CrtW, CrtO, and CrtA, had been identified in chlorophototrophic bacteria prior to this study, but no close homologs of these ketolases were encoded in the genomes of the three okenone-producing PSB strains examined in this study. CrtW and CrtO introduce keto groups on β-rings at the 4- and/or 4′-positions in cyanobacteria, marine bacteria, and some other organisms (29–32). The first cDNAs or genes encoding β-carotene ketolases were isolated from the green alga Haematococcus pluvialis (56, 57) and from bacteria Agrobacterium aurantiacum and Alcaligenes sp. strain PC-1 (58). These CrtW-type ketolases, which have been suggested to be strictly oxygen-dependent (30), belong to the membrane fatty acid desaturase superfamily of di-iron-containing dioxygenases. However, Mukoyama et al. (35) reported that CrtW from Paracoccus sp. MBIC1143 was active when expressed in the marine purple bacterium Rhodovulum sulfidophilum grown under anoxic, phototrophic conditions. This observation suggests that at least some CrtW enzymes may not be oxygen-dependent.
CrtO is structurally unrelated to CrtW and is a member of the pyridine nucleotide-disulfide oxidoreductase superfamily, which includes flavin-containing oxidoreductases such as phytoene desaturase (COG1233). CrtO from the cyanobacterium Synechocystis sp. PCC 6803 is dioxygen-dependent and requires NADPH, which suggests that the introduction of the keto group may occur by a monooxygenation/dehydrogenation mechanism (29). CrtA is found in purple bacteria and a few other non-phototrophic bacteria, and under oxic conditions, it ketolates the C-2 position of the ψ-end group (23, 33). The oxygen of the keto group at C-2 is derived from O2 under these conditions in R. sphaeroides (23, 59). However, about 10 and 40% of the total carotenoids in R. capsulatus and Rhodobaca bogoriensis, respectively, were ketocarotenoids under anoxic, phototrophic growth conditions (60); ketocarotenoids were also produced in R. sphaeroides during phototrophic growth (61). For R. sphaeroides grown phototrophically, water was the source of oxygen for the methoxy group of spheroidenone, but neither water nor CO2 appeared to be the source of the oxygen for the keto group under anoxic condition (61). The authors speculated that unknown oxygen-containing compounds derived from cellular metabolites provided the oxygen for the keto group under anoxic conditions. However, the ketolation reaction was reportedly dependent upon CrtA because no ketocarotenoids were observed under oxic or anoxic conditions in a crtA mutant (61). These results suggest that CrtA may catalyze the ketolation reaction by different mechanisms in the presence and absence of oxygen. A recent study reported that CrtA is a non-P450 heme protein and that the enzyme in E. coli can introduce a hydroxyl group at C-1 and C-1′ in addition to a keto group at the C-2 and C-2′ position in various substrates (34).
In Thiodictyon sp. CAD16 and T. marina, the genomic region encoding genes for carotenoid biosynthesis included two homologs of CrtD desaturases (see Fig. 1). In contrast to the reactivities of known CrtD enzymes, which introduce a double bond at the 3,4-position and thereby extend the conjugated double bond system of the substrates like lycopene and neurosporene, the CrtD1 homolog, redesignated here as CruO, was shown to be a 4/4′-ketolase. CruO is also a member of the pyridine nucleotide-disulfide oxidoreductase superfamily of flavin-containing oxidoreductases (COG1233), but unlike CrtO to which it is only very distantly related, CruO was able to introduce a keto group into its substrates under anoxic conditions. This demonstrated that CruO does not require dioxygen as a substrate and showed that the introduced oxygen atom is probably derived from water. The absorption spectra of the ketocarotenoid and its borohydride reduction product in combination with results from mass spectroscopy confirmed that the keto group is formed at the C-4′ position. CruO can also modify the C-4 position of some substrates, such as mono- or dihydroxy derivatives of lycopene and neurosporene, leading to compounds containing two keto groups. Therefore, the CruO ketolase has several unique characteristics: it can ketolate the ψ-end of carotenoids at C-4′ under anoxic conditions, a combination of properties that have not previously been described for enzymes of carotenogenesis. Accordingly, the gene encoding this enzyme was thus given a unique locus tag, cruO.
Because the cruS gene is not present in the genome of M. purpuratum, an okenone-producing PSB, it was clear that CruS does not play a role in okenone biosynthesis. The crtD2 gene encoded a bifunctional enzyme that was only active in the presence of oxygen, and because this enzyme also performs a unique reaction, it has similarly been given a unique gene locus designation, cruS. One of the functions of this probable flavoprotein was consistent with the desaturation reactions catalyzed by other, previously characterized CrtD enzymes. CrtD from R. gelatinosus performs a 3,4-desaturation reaction with 1-hydroxy carotenes or with 1,1′-dihydroxy derivatives with a double bond at the C-3′,4′ position (48). However, 1′-hydroxyneurosporene and 1,1′-dihydroxyneurosporene were the only substrates for CruS identified in this study. When the carotenoids produced by CruS were reduced with sodium borohydride, the resulting products had chromophores with 10 conjugated double bounds (absorption maxima, 430, 456, and 487 nm (38)). To extend the chromophore of 1′-hydroxyneurosporene and 1,1′-dihydroxyneurosporene to 10 conjugated double bonds, the C-3,4 bond must be desaturated. Reduction of the carotenoid product also revealed the presence of a keto group. The mass of the carotenoids and the similarity of the absorption spectra of the products to spheroidenone (62) were all consistent with the presence of a keto group at the C-2 position. This second activity corresponds to that of CrtA of purple non-sulfur bacteria, and like the reaction catalyzed by CrtA, this activity of CruS only occurred under oxic conditions. Thus, CruS alone performs the same reactions as those catalyzed by CrtD and CrtA in the biosynthesis of spheroidenone in purple non-sulfur bacteria (13, 23, 33). Based on our results from heterologous expression in E. coli, Thiodictyon sp. CAD16, T. marina, and other PSB harboring a cruS gene should in principle be able to synthesize spheroidenone in the presence of oxygen provided that the cruS and crtC genes can be transcribed and translated to produce functional proteins under these conditions and that neurosporene is available as a substrate. Because spheroidenone has not yet been detected in PSB even when cells were grown under microoxic conditions and because of the extensive overlap of the cruS gene with the crtC gene in the same operon, it is possible that cruS may no longer be active in PSB, and it may represent a pseudogene or relic of some past capability that was present in the genome even though the recombinant product is still active in E. coli. These issues will be addressed in future studies.
An important question concerning okenone is whether this compound is uniquely produced by PSB (15–17). Among microorganisms whose genomes have been sequenced thus far, the CruO and CruS genes appear to be uniquely associated with PSB. Only one additional CruO and one additional CruS sequence could be identified in GenBank, and both were encoded by the same contig assembled as part of a marine, metagenome sequencing project. The source of these sequences appears to be an organism distantly related to T. marina (Fig. 8). Therefore, CruO catalyzes a reaction that is uniquely required to produce okenone, and CruO orthologs have only been found in PSB that synthesize okenone (assuming that this is true of the marine organism(s) present in the metagenomic study).
Geochemists have proposed that okenane is a biomarker indicative of sediments deposited from aquatic environments that were anoxic and that harbored PSB (15–17). However, as we have shown here, okenone biosynthesis is not dependent upon oxygen, nor is it inhibited by oxygen. Although some PSB have the genetic potential to synthesize methoxyspheroidenone, conditions for the production of this carotenoid have not yet been identified, and those PSB grown under dark, microoxic conditions still produced only okenone. Taken together with recent findings on χ-ring formation in PSB (21), one must be very cautious in inferring the physicochemical properties of ancient environments as well as the biochemical and physiological properties of ancient microorganisms on the basis of the properties of extant microorganisms.
Supplementary Material
Acknowledgments
We thank James R. Miller of the Proteomics and Mass Spectrometry Core Facility, The Huck Institutes of the Life Sciences, The Pennsylvania State University, for technical expertise for the mass spectrometry analyses and Dr. C. Neil Hunter and Dr. Daniel Canniffe of the University of Sheffield for providing aerobically grown R. sphaeroides cells for the isolation of spheroidenone.
This work was supported by United States Department of Energy Grant DE-FG02-94ER20137 (to D. A. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S8.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) JN206680 and JN206681.
Z. Liu, K. Vogl, N.-U. Frigaard, L. P. Tomsho, S. C. Schuster, and D. A. Bryant, unpublished results.
N. Storelli, M. Tonollo, and N.-U. Frigaard, personal communication.
- PSB
- purple sulfur bacteria.
REFERENCES
- 1. Imhoff J. (2006) in The Prokaryotes (Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. eds) pp. 874–886, Springer, New York [Google Scholar]
- 2. Imhoff J. (2006) in The Prokaryotes (Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. eds) pp. 41–64, Springer, New York [Google Scholar]
- 3. Imhoff J. (2006) in The Prokaryotes (Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. eds) pp. 593–601, Springer, New York [Google Scholar]
- 4. Imhoff J. F. (2006) in The Prokaryotes (Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. eds) pp. 846–873, Springer, New York [Google Scholar]
- 5. Madigan M. T., Jung D. O. (2009) in The Purple Phototrophic Bacteria (Hunter C. N., Daldal F., Thurnauer M. C., Beatty J. T. eds) pp. 1–15, Springer, Dordrecht, The Netherlands [Google Scholar]
- 6. Yurkov V. V., Beatty J. T. (1998) Microbiol. Mol. Biol. Rev. 62, 695–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Imhoff J. (2004) in Anoxygenic Photosynthetic Bacteria (Blankenship R., Madigan M., Bauer C. eds) pp. 1–15, Springer, Dordrecht, The Netherlands [Google Scholar]
- 8. Stackebrandt E., Murray R. G., Truper H. G. (1988) Int. J. Syst. Bacteriol. 38, 321–325 [Google Scholar]
- 9. McEwan A. G. (1994) Antonie Van Leeuwenhoek 66, 151–164 [DOI] [PubMed] [Google Scholar]
- 10. Hoff A. J., Deisenhofer J. (1997) Phys. Rep. 287, 2–247 [Google Scholar]
- 11. Blankenship R. E. (1994) Antonie Van Leeuwenhoek 65, 311–329 [DOI] [PubMed] [Google Scholar]
- 12. Willows R. D., Kriegel A. M. (2009) in The Purple Phototrophic Bacteria (Hunter C. N., Daldal F., Thurnauer M. C., Beatty J. T. eds) pp. 57–79, Springer, Dordrecht, The Netherlands [Google Scholar]
- 13. Takaichi S. (2009) in The Purple Phototrophic Bacteria (Hunter C. N., Daldal F., Thurnauer M. C., Beatty J. T. eds) pp. 97–117, Springer, Dordrecht, The Netherlands [Google Scholar]
- 14. Polli D., Cerullo G., Lanzani G., De Silvestri S., Hashimoto H., Cogdell R. J. (2006) Biophys. J. 90, 2486–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brocks J. J., Schaeffer P. (2008) Geochim. Cosmochim. Acta 72, 1396–1414 [Google Scholar]
- 16. Brocks J. J., Banfield J. (2009) Nat. Rev. Microbiol. 7, 601–609 [DOI] [PubMed] [Google Scholar]
- 17. Brocks J. J., Love G. D., Summons R. E., Knoll A. H., Logan G. A., Bowden S. A. (2005) Nature 437, 866–870 [DOI] [PubMed] [Google Scholar]
- 18. Schmidt K., LiaaenJensen S., Schlegel H. G. (1963) Arch. Mikrobiol. 46, 117–126 [PubMed] [Google Scholar]
- 19. Aasen A. J., Jensen S. L. (1967) Acta Chem. Scand. 21, 970–982 [Google Scholar]
- 20. Peduzzi S., Tonolla M., Hahn D. (2003) FEMS Microbiol. Ecol. 45, 29–37 [DOI] [PubMed] [Google Scholar]
- 21. Vogl K., Bryant D. A. (2011) Geobiology, in press [DOI] [PubMed] [Google Scholar]
- 22. Maresca J. A., Graham J. E., Bryant D. A. (2008) Photosynth. Res. 97, 121–140 [DOI] [PubMed] [Google Scholar]
- 23. Scolnik P. A., Walker M. A., Marrs B. L. (1980) J. Biol. Chem. 255, 2427–2432 [PubMed] [Google Scholar]
- 24. Steiger S., Mazet A., Sandmann G. (2003) Arch. Biochem. Biophys. 414, 51–58 [DOI] [PubMed] [Google Scholar]
- 25. Steiger S., Takaichi S., Sandmann G. (2002) J. Biotechnol. 97, 51–58 [DOI] [PubMed] [Google Scholar]
- 26. Badenhop F., Steiger S., Sandmann M., Sandmann G. (2003) FEMS Microbiol. Lett. 222, 237–242 [DOI] [PubMed] [Google Scholar]
- 27. Pinta V., Ouchane S., Picaud M., Takaichi S., Astier C., Reiss-Husson F. (2003) Arch. Microbiol. 179, 354–362 [DOI] [PubMed] [Google Scholar]
- 28. Graham J. E., Bryant D. A. (2009) J. Bacteriol. 191, 3292–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fernández-González B., Sandmann G., Vioque A. (1997) J. Biol. Chem. 272, 9728–9733 [DOI] [PubMed] [Google Scholar]
- 30. Fraser P. D., Miura Y., Misawa N. (1997) J. Biol. Chem. 272, 6128–6135 [DOI] [PubMed] [Google Scholar]
- 31. Maresca J. A., Braff J. C., Delong E. F. (2009) Environ. Microbiol. Rep. 1, 524–534 [DOI] [PubMed] [Google Scholar]
- 32. Zhu Y., Graham J. E., Ludwig M., Xiong W., Alvey R. M., Shen G., Bryant D. A. (2010) Arch. Biochem. Biophys. 504, 86–99 [DOI] [PubMed] [Google Scholar]
- 33. Gerjets T., Steiger S., Sandmann G. (2009) Biochim. Biophys. Acta 1791, 125–131 [DOI] [PubMed] [Google Scholar]
- 34. Lee P. C., Holtzapple E., Schmidt-Dannert C. (2010) Appl. Environ. Microbiol. 76, 7328–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mukoyama D., Takeyama H., Kondo Y., Matsunaga T. (2006) FEMS Microbiol. Lett. 265, 69–75 [DOI] [PubMed] [Google Scholar]
- 36. Cunningham F. X., Jr., Sun Z., Chamovitz D., Hirschberg J., Gantt E. (1994) Plant Cell 6, 1107–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Britton G. (1985) Methods Enzymol. 111, 113–149 [DOI] [PubMed] [Google Scholar]
- 38. Britton G. (1995) in Carotenoids, Volume 1B: Spectroscopy (Britton G., Liaaen-Jensen S., Pfander H. eds) pp. 13–62, Birkhäuser, Basel, Switzerland [Google Scholar]
- 39. Frigaard N. U., Sakuragi Y., Bryant D. A. (2004) Methods Mol. Biol. 274, 325–340 [DOI] [PubMed] [Google Scholar]
- 40. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 41. Felsenstein J. (1989) Cladistics 5, 164–166 [Google Scholar]
- 42. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 43. Caumette P., Guyoneaud R., Imhoff J. F., Süling J., Gorlenko V. (2004) Int. J. Syst. Evol. Microbiol. 54, 1031–1036 [DOI] [PubMed] [Google Scholar]
- 44. Imhoff J. F., Trüper H. G. (1980) Zentralbl. Bakteriol. Mikrobiol. Hyg. C 1, 61–69 [Google Scholar]
- 45. Frigaard N. U., Maresca J. A., Yunker C. E., Jones A. D., Bryant D. A. (2004) J. Bacteriol. 186, 5210–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Graham J. E., Bryant D. A. (2008) J. Bacteriol. 190, 7966–7974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maresca J. A., Romberger S. P., Bryant D. A. (2008) J. Bacteriol. 190, 6384–6391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Steiger S., Astier C., Sandmann G. (2000) Biochem. J. 349, 635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Britton G., Liaaen-Jensen S., Pfander H. (2004) in Carotenoids Handbook (Britton G., Liaaen-Jensen S., Pfander H. eds) Birkhäuser, Basel, Switzerland [Google Scholar]
- 50. Maresca J. A., Graham J. E., Wu M., Eisen J. A., Bryant D. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11784–11789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yu Q., Schaub P., Ghisla S., Al-Babili S., Krieger-Liszkay A., Beyer P. (2010) J. Biol. Chem. 285, 12109–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Andrewes A. G., Liaaen-Jensen S. (1972) Acta Chem. Scand. 26, 2194–2204 [DOI] [PubMed] [Google Scholar]
- 53. Bryant D. A., Klatt C. G., Frigaard N.-U., Liu Z., Li T., Zhao F., Garcia Costas A. M., Overmann J., Ward D. M. (2011) in Advances in Photosynthesis and Respiration, Vol. 33, Functional Genomics and Evolution of Photosynthetic Systems (Burnap R. L., Vermaas W. eds) pp. 47–102, Springer, Dordrecht, The Netherlands [Google Scholar]
- 54. Garcia Costas A. M., Liu Z., Tomsho L., Schuster S. C., Ward D. M., Bryant D. A. (2011) Environ. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- 55. Sun Z., Shen S., Wang C., Wang H., Hu Y., Jiao J., Ma T., Tian B., Hua Y. (2009) Microbiology 155, 2775–2783 [DOI] [PubMed] [Google Scholar]
- 56. Kajiwara S., Kakizono T., Saito T., Kondo K., Ohtani T., Nishio N., Nagai S., Misawa N. (1995) Plant. Mol. Biol. 29, 343–352 [DOI] [PubMed] [Google Scholar]
- 57. Lotan T., Hirschberg J. (1995) FEBS Lett. 364, 125–128 [DOI] [PubMed] [Google Scholar]
- 58. Misawa N., Satomi Y., Kondo K., Yokoyama A., Kajiwara S., Saito T., Ohtani T., Miki W. (1995) J. Bacteriol. 177, 6575–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shneour E. A. (1962) Biochim. Biophys. Acta 65, 510–511 [DOI] [PubMed] [Google Scholar]
- 60. Takaichi S., Jung D. O., Madigan M. T. (2001) Photosynth. Res. 67, 207–214 [DOI] [PubMed] [Google Scholar]
- 61. Yeliseev A. A., Kaplan S. (1997) FEBS Lett. 403, 10–14 [DOI] [PubMed] [Google Scholar]
- 62. Liaaen-Jensen S. (1963) Acta Chem. Scand. 17, 303–312 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.