Background: Role of copper as an attenuator or facilitator in prion diseases is controversial.
Results: Copper-bound PrP does not aggregate at physiological temperature and shows two novel interactions between its N- and C-terminal domains.
Conclusion: Copper may act as an attenuator in prion diseases and induces novel long range inter-domain interactions in PrP.
Significance: This study might help in understanding the role of copper in prion diseases.
Keywords: Aggregation, Copper, Metals, NMR, Prions, Small Angle X-ray Scattering, Copper Prion Protein Interaction, Copper-bound Prion Protein, Prion Diseases
Abstract
Copper is reported to promote and prevent aggregation of prion protein. Conformational and functional consequences of Cu2+-binding to prion protein (PrP) are not well understood largely because most of the Cu2+-binding studies have been performed on fragments and truncated variants of the prion protein. In this context, we set out to investigate the conformational consequences of Cu2+-binding to full-length prion protein (PrP) by isothermal calorimetry, NMR, and small angle x-ray scattering. In this study, we report altered aggregation behavior of full-length PrP upon binding to Cu2+. At physiological temperature, Cu2+ did not promote aggregation suggesting that Cu2+ may not play a role in the aggregation of PrP at physiological temperature (37 °C). However, Cu2+-bound PrP aggregated at lower temperatures. This temperature-dependent process is reversible. Our results show two novel intra-protein interactions upon Cu2+-binding. The N-terminal region (residues 90–120 that contain the site His-96/His-111) becomes proximal to helix-1 (residues 144–147) and its nearby loop region (residues 139–143), which may be important in preventing amyloid fibril formation in the presence of Cu2+. In addition, we observed another novel interaction between the N-terminal region comprising the octapeptide repeats (residues 60–91) and helix-2 (residues 174–185) of PrP. Small angle x-ray scattering studies of full-length PrP show significant compactness upon Cu2+-binding. Our results demonstrate novel long range inter-domain interactions of the N- and C-terminal regions of PrP upon Cu2+-binding, which might have physiological significance.
Introduction
Prion protein (PrP)3 is involved in transmissible spongiform encephalopathies, a group of diseases such as Creutzfeldt-Jakob disease, Kuru, Fatal Familial Insomnia, and Gerstmann-Straussler-Scheinker syndrome, characterized by neurodegeneration, spongiform cerebral tissue, and amyloid plaques. Such diseases are caused by the conformational transition of α-helix-rich cellular prion protein (PrPC) to the β-sheet-rich scrapie prion protein (PrPSC) (1). Copper ions (Cu2+) appear to have a significant role in such conformational transition (2). PrP exhibits strong binding toward Cu2+ with nano- to femtomolar affinity (3–5). Cu2+-binding sites are reported in the flexible N-terminal region (residues 23–120); four of them are present in the octapeptide repeats (residues 60–91) and one located at His-96 and His-111 (6–8) (residues are numbered according to the human PrP sequence). An additional Cu2+-binding site at His-187 has also been recognized in the C-terminal region of PrP (9–12).
Although the functional implication of Cu2+-binding to PrP is not yet clear, it is believed that Cu2+ is an important factor (as a facilitator or an attenuator) in prion diseases. Cu2+-binding induces conformational changes leading to reduction in the helical content and an increase in the β-sheet content especially in the region 90–120 (13), which partially overlaps with the well established amyloidogenic stretch 106–126 (14). Cu2+ has also been shown to induce conformational changes and increased stability in the octapeptide repeats of PrP (15, 16).
Cu2+ is shown to convert PrPC from brain homogenates (2) and aged recombinant PrP (17) to protease-resistant and detergent-insoluble aggregates. However, these aggregates are structurally distinct from the scrapie prion protein (PrPSC). Expansion of octapeptide repeats has been suggested to cause an early onset of the prion disease (18, 19). In contrast, several studies show compelling evidence on the inhibitory role of Cu2+ on the amyloid formation of PrP in vitro (20–23). In vivo studies of Cu2+-binding to PrP are also not unequivocal. One study reports the delay in the onset of prion disease upon removal of Cu2+ by chelation (24), whereas a different study shows that Cu2+-depleted diet enhances prion disease (25). Thus, the nature and consequences of interaction of Cu2+ with PrP, both in vitro and in vivo, appear to be a complex phenomenon that remains to be investigated.
Conformational studies on binding of Cu2+ to PrP in vitro have been performed using peptides (26–28) or truncated variants of PrP (29–31) and at nonphysiological temperatures (2, 10, 13–17) and in water or buffers of low ionic strength (3, 9, 13, 32–34). We have used full-length PrP(23–231) to investigate conformational changes and aggregation upon Cu2+-binding at physiological temperature (37 °C). We find that Cu2+-bound PrP undergoes unusual temperature-dependent reversible aggregation. Interestingly, it aggregates at lower temperatures and resolubilizes as the temperature is raised. Our investigations using isothermal calorimetry (ITC), NMR, and small angle x-ray scattering (SAXS) provide insight into thermodynamic and conformational aspects of this process. Based on our study, we propose the involvement of the interaction of helix-2 and the octapeptide repeats region in the aggregation process. Our results reveal two novel interactions between the flexible N- and the C-terminal regions of PrP upon Cu2+-binding as follows: (i) between the N-terminal region and helix-1 (residues 144–147) and its nearby loop region (residues 139–143) and (ii) between the N-terminal region (residues 60–91) and helix-2 (residues 174–185) of PrP. We believe these observations have physiological implications.
EXPERIMENTAL PROCEDURES
All chemicals and reagents were of analytical or ultra pure grade. Isopropyl 1-thio-β-d-galactopyranoside was obtained from Bangalore Genei (India), and urea was procured from United States Biochemical (Cleveland, OH), and ampicillin was purchased from Biochem Pharmaceutical Industries Ltd. (India). Protease inhibitor mixture was from Roche Diagnostics; NaCl and glycine were from Qualigens, Fine Chemicals (India); CuSO4 was from Sisco Research Laboratories Pvt. Ltd. (Mumbai, India). Nickel-nitrilotriacetic acid matrix was obtained from Qiagen (GmbH, Hilden, Germany). Other chemicals or reagents were from Sigma. Water used in all reactions was obtained from MilliQ, Millipore (Bedford, MA).
Cloning, Expression, and Purification of Prion Protein Variants
We have cloned, expressed, and purified mouse PrP(23–231) as described earlier (35). PrP(90–231) was cloned into pET21a vector after PCR amplification from PrP(23–231)-pET21a template using gene-specific primers (forward primer TGGCATATGGGCCAAGGAGGGGGTAC and reverse primer GCTTTCGAATCAGCTGGATCTTCTCCCG). Inserts in pMOS blue vector (Novagen) were digested with NdeI and HindIII and ligated by T4-DNA polymerase (Promega) in pET21a vector. Escherichia coli DH5α strain was transfected with the plasmid constructs. Positive colonies were screened with the help of ampicillin. The sequence of this construct was verified using 3700ABI automated DNA sequencer. MoPrP(90–231) (pET21a vector) was expressed in E. coli Rosetta DE3. After 12 h of induction, cells were harvested, lysed, and centrifuged. The insoluble pellet was washed and dissolved in 8 m urea and 10 mm reduced glutathione and bound to nickel-nitrilotriacetic acid matrix. On-column oxidative folding of PrP was done by slow removal of the denaturant and the reducing agent. Protein was eluted in 50 mm imidazole. The eluted protein was extensively dialyzed against MilliQ water and then concentrated. Aliquots of protein were stored at −70 °C. The protein concentrations were estimated using the extinction coefficient of 2.70 and 1.70 at 280 nm for 1 mg/ml solutions of PrP(23–231) and PrP(90–231), respectively. The purity of the protein was checked using SDS-PAGE and found to be free of any contaminants.
PrP Preparations for Spectroscopic Measurements
Scattering experiments were performed in 20 mm sodium phosphate buffer (pH 7.4) and 100 mm NaCl, unless otherwise specified. For ITC, SAXS, dynamic light scattering, sedimentation velocity, and NMR experiments, PrP in water with pH 7.4 maintained by dilute NaOH solution was used to avoid aggregation. At low protein (∼0.25 mg/ml) concentrations, the reversible aggregation was not noticeable. However, at higher concentrations up to 2 mg/ml in water (pH 7.4), reversible aggregation was observed below 22 °C.
Cu2+-binding Studies
For all Cu2+-binding studies, including aggregation, NMR, SAXS, sedimentation velocity, and ITC experiments, Cu2+ was complexed with glycine in a 1:4 molar ratio to form Cu-(Gly)2 complex (referred to as Cu2+ everywhere in the text) to avoid any hydroxide formation. The Cu-(Gly)2 complex was maintained at pH 6.0 to avoid drastic change in pH of PrP (water, pH 7.4) upon titration in ITC experiments. After titration, no significant change in pH was observed. Except for ITC, all the binding studies were done using 6 m eq of Cu2+.
Scattering Studies
Scattering measurements were monitored using Spectrofluorimeter Fluorolog-3 model FL-3–22 from Horiba Jobin Yvon, Inc. Rayleigh scattering of solutions was recorded at any given temperature with excitation and emission monochromators fixed at 465 nm, and bandpasses for excitation and emission were set at 0.5 and 1 nm, respectively. Datamax software provided by the manufacturer was used to monitor scattering. The instrument takes 2.5 and 3 min to increase or decrease the sample temperatures from 15 to 37 °C, respectively. To measure the extent of reversibility, scattering was monitored at various temperatures between 37 and 15 °C and brought back to 37 °C with similar intervals.
Isothermal Calorimetry
Binding of Cu-(Gly)2 complex with PrP(23–231) was performed using VP-ITC microcalorimeter from MicroCal, LLC. Data acquisition and analysis were done with software provided by the manufacturer. PrP(23–231) at a concentration of 0.25 mg/ml (12 μm) in water (pH 7.4) was used and 2 mm of Cu-(Gly)2 was titrated against it. Each solution was filtered or centrifuged and degassed before binding experiments. Initial injection of 2 μl followed by 29 injections of 4 μl was used for binding reactions. To remove the heat of dilution generated because of injection of ligands to the protein solution in the binding reactions, base-line correction for raw isotherms was performed by subtracting average saturated values (obtained from six points) from its corresponding isotherms. For analysis, a sequential mode with four binding sites was used. Binding reactions were performed at 16, 20, 25, 30, and 37 °C. Three reactions were done at each temperature, and averaged values are reported.
NMR of Prion Protein
Uniformly 15N-labeled protein samples of PrP(23–231) and PrP(90–231) were prepared by growing E. coli cells in M9 minimal medium containing 15NH4Cl as the sole source of nitrogen. Protein samples (2 mg/ml) were prepared in ultra pure water (10−18 Ω−1 cm−1 resistivity) whose pH was maintained at 7.4. Two-dimensional 15N-1H HSQC spectra were recorded for PrP(23–231) and PrP(90–231) at various temperatures ranging from 16 to 37 °C. Each 15N-1H HSQC was recorded with 1024 and 128 complex points along 1H and 15N dimensions, respectively, using watergate sequence for water saturation, on a Bruker Avance 800 MHz NMR spectrometer equipped with a 5-mm triple-resonance cryogenic probe, whose temperature was calibrated using methanol (280–300 K). All the spectra were referenced using 4,4-dimethyl-4-silapentane-1-sulfonic acid as an external reference at 298 K and then adding the correction of 0.01 ppm/K for the temperature change. The spectra were processed with 4096 and 1024 points along 1H and 15N dimensions, and a window function of 60°-shifted sine square bell was used for apodization along both the dimensions using TopSpin 2.0 software. Analysis was carried out manually using computer-aided resonance assignment (CARA).
NMR Assignments
Prior to the assignment of the peaks in the two-dimensional 15N-1H HSQC of PrP(90–231), recorded in a mixed solvent of 2H2O and H2O (pH 7.4), we recorded the same spectrum at pH 7.0 for which the chemical shift information (32) is available in the BioMagResBank (code 16071) under similar buffer conditions, 5 mm phosphate buffer (pH 7.0). All the observed peaks in the pH 7.0 spectrum, totaling 128 out of 138, could be assigned by visual inspection. The missing spectral information about the 10 remaining residues belongs to the flexible part of the protein. In such an assignment, no changes were noticed in the respective chemical shifts, except for the residue Phe-141. We then assigned the 15N-1H HSQC of PrP(90–231) recorded in water at pH 7.4 by comparing it with the spectral assignments made at pH 7.0. Once again, no spectral changes were noticed except for the absence of 17 more cross-peaks belonging to the unstructured part of the protein (residues 90–120). Thus, we could unambiguously assign the spectral signatures of 111 residues. Such assignments were further used to assign 85 of the observed peaks in the 15N-1H HSQC of wild-type prion protein, recorded under identical experimental conditions. Among the residues, whose spectral signatures were absent in the spectrum, 28 belong to the unstructured polypeptide stretch (residues 90–120), and the residues Val-166–Asn-171, Arg-230, and Ser-231, all belonging to the globular domain of the protein.
Sedimentation Velocity Measurements
Sedimentation velocity measurements were performed using Optima XL-I analytical ultracentrifuge (Beckman Coulter, Fullerton, CA). Prion protein samples (2 mg/ml (pH 7.4), conditions identical to those used in NMR experiments), with and without Cu2+, were subjected to centrifugation at 58,000 rpm (∼240,000 × g) at 37 °C, using An60Ti rotor with an aluminum center piece. The protein boundary was scanned at 1-min intervals for 8 h by interference measurements using 30-milliwatt 660 nm laser diode. The sedimentation coefficient s20,w and molecular mass of the protein were calculated using the program SEDFIT (36), which uses nonlinear regression fitting of the sedimenting boundary profile with the Lamm Equation 1,
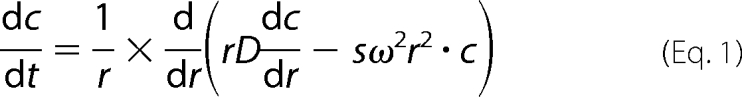 |
which describes the concentration distribution c(r,t) of a species with sedimentation coefficient s and diffusion coefficient D in a sector-shaped volume and in the centrifugal field ω2r.
SAXS Studies
SAXS measurements were performed with a HECUS S3-Micro SAXS camera attached to a Xenocs microbeam delivery system (copper target, wavelength λ = 1.54 Å) operating at a power of 50 watts. SAXS measurements of PrP samples with and without Cu2+ (2 mg/ml in water (pH 7.4)) at 37 °C for 10,800 s were recorded with a Pilatus 100k pixel detector. The two-dimensional image was reduced to a one-dimensional scattering curve using the Fit2D software and processed using PRIMUS (37). The Rg and I(0) values were obtained using the Guinier approximation (38). To obtain shape information, the scattering data were converted to P(r) function by GNOM software, and the Dmax was calculated (39). The P(r) function was taken to GASBOR, to generate ab initio models (40). This process was repeated 10 times and then averaged using DAMAVER (41). The high resolution NMR data of prion protein and the ab initio SAXS envelopes of PrP in the absence and presence of Cu2+ obtained from GASBOR program were matched using SUPCOMB (42).
RESULTS
Reversible Aggregation of PrP(23–231) upon Cu2+-binding
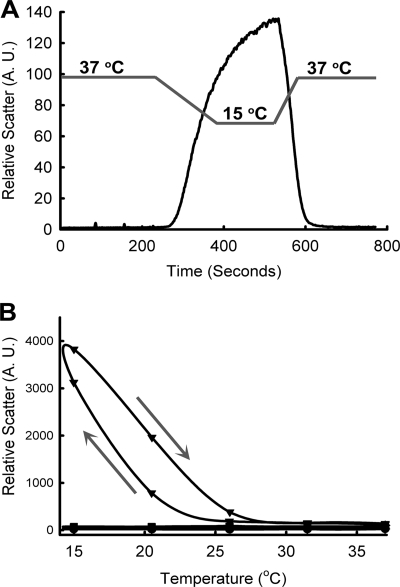
We measured aggregation of PrP upon Cu2+-binding by monitoring light scattering of the solution under physiological conditions (temperature 37 °C and pH 7.4). Six molar equivalents of Cu2+ in the form of Cu-(Gly)2 (see under “Experimental Procedures”) were added to full-length PrP (Fig. 1A). No changes in the scattering at 37 °C were observed upon Cu2+-binding to PrP(23–231), indicating the absence of aggregation at this temperature (see Fig. 1A, before 200 s). We made a serendipitous observation that as the temperature was lowered to 15 °C, there was a sudden increase in the scattering, suggesting protein aggregation (200–400 s). When the temperature was raised back to 37 °C, the observed light scattering suddenly decreased (400–600 s), indicating reversible protein aggregation. The extent of reversibility was monitored by continuous heating and cooling of the sample for 14 times (supplemental Fig. S2A). The aggregation was reversible except for a minor decrease in the observed light scattering. Temperature dependence of this process (supplemental Figs. S1, A and B, and S2A) suggests that the aggregation occurs below 20 °C. However, at higher concentrations of PrP(23–231), the protein aggregates at higher temperatures (below 25 °C). We also observed that the extent of reversible aggregation increased with increasing concentration of PrP(23–231) (supplemental Fig. S1, C and D). Although this aggregation occurs at a nonphysiological temperature, a detailed understanding of this process is likely to provide insights into the molecular basis of Cu2+/PrP interaction. We have therefore investigated the temperature-dependent, reversible aggregation of Cu2+-bound PrP.
FIGURE 1.
Temperature-dependent reversible aggregation of Cu2+-bound PrP. A, reversible aggregation of PrP(23–231) in the presence of Cu2+. Gray lines indicate temperature of the sample, and slopes of lines indicate decrease or increase in the temperature. B, reversible aggregation of similar concentrations of PrP(23–231) (circles), Cu2+-bound PrP(23–231) (triangles), PrP(90–231) (diamonds), and Cu2+-bound PrP(90–231) (squares). The circles, diamonds, and squares are overlapping. Arrows show the directions of experiments. Arrow pointing downward shows cooling, and arrow pointing upward shows heating of sample.
To examine the role of electrostatic interactions in the Cu2+-bound PrP(23–231) aggregation, we monitored the protein aggregation at varying concentrations of salt (sodium chloride, NaCl) (supplemental Fig. S1, E and F). The extent of aggregation increased with an increase in salt concentration, suggesting the involvement of nonelectrostatic interactions such as hydrogen bonds (H-bonds) and hydrophobic interactions in the PrP(23–231) aggregation. PrP(23–231) binds Cu2+ in the N-terminal region involving four octapeptide repeats (residues 60–91) and at the sites His-96 and His-111 (2–12). To probe the region(s) responsible for temperature-dependent aggregation of Cu2+-bound PrP(23–231), we have generated an N-terminal deletion mutant PrP(90–231), which lacks the four octapeptide repeats (region 23–89). Interestingly, Cu2+-bound PrP(90–231) (Fig. 1B, squares) did not show any aggregation, whereas the Cu2+-bound PrP(23–231) shows reversible aggregation as expected (Fig. 1B, triangles). This result shows that the region encompassing octapeptide repeats is responsible for the temperature-dependent reversible aggregation of Cu2+-bound PrP.
We used an amyloid-specific fluorescent probe thioflavin T (43) to further understand the nature of these aggregates (amyloid or amorphous). We did not observe any change in thioflavin T fluorescence with Cu2+-bound PrP(23–231) at various temperatures (16, 20, 25, 30, and 37 °C), indicating that the aggregates formed are amorphous in nature (supplemental Fig. S2B). This result rules out any amyloidogenic conversion of PrP(23–231) upon Cu2+-binding in the given temperature range.
Binding Affinity of Cu2+ to PrP(23–231) at Various Temperatures
It is possible that the observed temperature-dependent aggregation could be due to temperature-dependent changes in the affinity of Cu2+ for PrP. We determined the affinity of Cu2+ for PrP(23–231) at different temperatures using ITC. The apparent dissociation constants (Kd(app)) thus obtained from the curve fitting of base-line-corrected isotherms (see “Experimental Procedures”) did not show significant differences at various temperatures (statistical analysis shows p values in the range 0.11 to 0.23) (supplemental Fig. S2C). This result clearly suggests that the Cu2+-induced reversible aggregation of PrP(23–231) cannot be attributed to changes in the binding affinities.
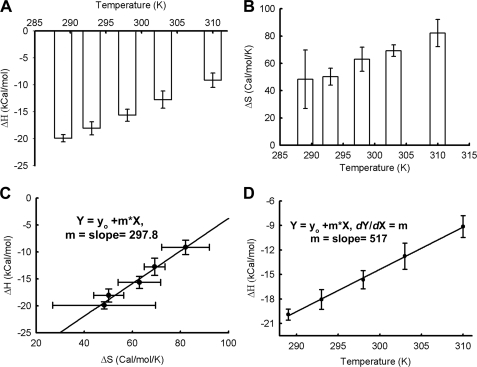
The ITC data suggest that both ΔH and ΔS show linear changes with temperature (Fig. 2, A and B); although ΔH decreased, ΔS increased with temperature, indicating conformational changes associated with Cu2+-binding to PrP(23–231). Interestingly, we observed a linear correlation between ΔH and ΔS with a slope of 297.8 K (Fig. 2C). Such a linear and positive correlation between ΔH and ΔS, with a slope of ∼300 K, has been proposed to indicate enthalpy/entropy compensation for small solutes and proteins in water solutions (44, 45). Temperature-dependent compensation requires heat capacity to remain more or less constant throughout the temperature range used (46). In the present case, the calculated molar heat capacity indeed was found to be constant (∼517 cal/mol/K, see Fig. 3D). This observation may be taken to suggest that at high temperatures the process is entropy-driven, whereas at lower temperatures it is driven by enthalpy.
FIGURE 2.
Calorimetric studies on binding of Cu2+ to PrP. A, ΔH; B, ΔS of Cu2+-binding to PrP at different temperatures. C, plot of ΔH and ΔS. Enthalpy/entropy compensation was observed in the case of Cu2+-binding to PrP at various temperatures. Data were fitted with the linear equation (y = m·x + C), yielding a slope of 297.8 K. D, plot of ΔH and T; slope yields heat capacity change (∼517 cal/mol/K). For all figures, S.E. was calculated using three sets of experiments.
FIGURE 3.
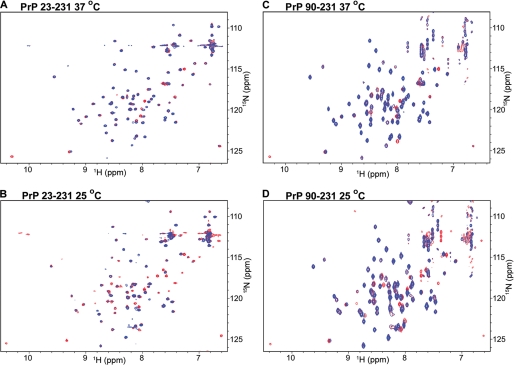
Two-dimensional 15N-1H HSQC spectra of PrP(23–231) and PrP(90–231). A and B, two-dimensional 15N-1H HSQC spectra of PrP(23–231) (red cross-peaks) were overlaid with those of Cu2+-bound PrP(23–231) (blue cross-peaks) at 37 °C (A) and 25 °C (B). C and D, two-dimensional 15N-1H HSQC spectra of PrP(90–231) (red cross-peaks) and Cu2+-bound PrP(90–231) (blue cross-peaks) were at 37 °C (C) and 25 °C (D).
Novel Interactions between the N- and C-terminal Regions upon Cu2+-binding
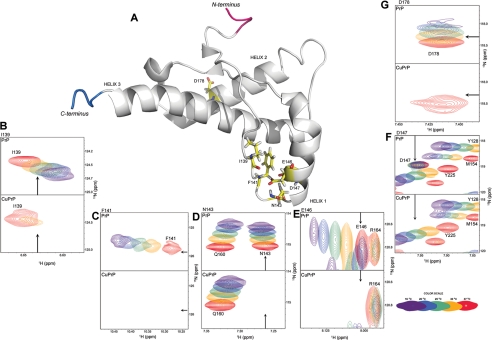
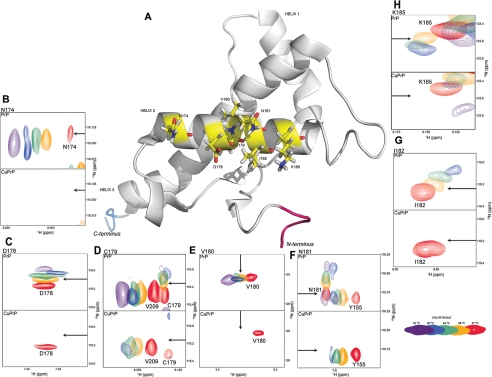
Isothermal titration calorimetry studies, inter alia, show positive and large heat capacity change upon Cu2+-binding to PrP(23–231) (∼517 cal/mol/K, see Fig. 2D). Several studies associate positive and large heat capacity change with the breaking of H-bonds or long range electrostatic interactions or loss of polar residues on the surface- or temperature-dependent conformational changes (47). In our experiments, positive and large heat capacity change suggests that PrP(23–231) undergoes conformational changes upon binding to Cu2+ at various temperatures. We have investigated the possible structural alteration using NMR and SAXS. Because PrP is a glycosylphosphatidylinositol-anchored protein, it has been suggested by Nishina et al. (48) that it is likely to exist in relatively nonpolar micro-environments with a dielectric constant similar to those of low ionic strength buffer solutions. We have therefore carried out these experiments under low ionic strength conditions (see “Experimental Procedures”). We have recorded a series of 15N-1H HSQC spectra of PrP(23–231) (full length) at different temperatures in the presence and absence of Cu2+ (Fig. 3; supplemental Fig. S3). We could monitor cross-peaks from the C-terminal region of PrP (residues 120–231). The cross-peaks from the N-terminal region (residues 23–120), which included Cu2+-binding sites such as the octapeptide repeats region (residues 60–91) and His-96/His-111, were not visible in the given spectra in the presence and absence of Cu2+ (see “Experimental Procedures”). This facilitated observation of specific conformational changes in the C-terminal region of PrP(23–231) induced by Cu2+-binding to the N-terminal region. Being paramagnetic in nature, Cu2+ is known to increase transverse (spin-spin) relaxation rates, thereby broadening the NMR spectral lines (49). Upon Cu2+ addition, we observed the line broadening of only some but not all the peaks, indicating strong binding of Cu2+ to specific sites. Several of the original peaks in the HSQC spectrum of PrP(23–231) broadened, upon addition of Cu2+, although others remained unaffected in their line widths. The intensities of these broadened peaks were below the NMR detection limit (Fig. 3). We further noted that the number of broadened cross-peaks increased with a decrease in temperature in the HSQC spectra of Cu2+-bound PrP(23–231) (compare blue cross-peaks in Fig. 3A with those in B and those in supplemental Fig. S3, A and F). However, in the absence of Cu2+, no such broadening of cross-peaks was noticed in the HSQC spectra of PrP(23–231) (compare red (without Cu2+) and blue (with Cu2+) cross-peaks in Fig. 3, A and B). At 37 °C, cross-peaks arising from Phe-141, Asn-143, and Asp-147 were found missing (Fig. 3A; Table 1; and Fig. 4, C, D, and F). As the temperature was lowered, peaks arising from Glu-146 and Ile-139 disappeared at 34 and 31 °C, respectively (Fig. 4, E and B; Table 1). Residues Ile-139 to Asn-143 correspond to the loop region preceding helix-1, whereas Glu-146 and Asp-147 are part of helix-1 (Fig. 4A). In addition to the above-mentioned residues, some of the peaks arising from the helix-2 region (residues 171–188) also showed line broadening at temperatures below 37 °C (Table 1; Fig. 5, B–H). Asn-174 and Asp-178 were the first residues whose spectral signatures disappeared at 34 °C (Fig. 5, B and C). This was followed by the disappearance of peaks of Val-180 and Asn-181 at 31 °C (Fig. 5, E and F) and also Cys-179, Ile-182, and Lys-185 at 28 °C (Fig. 5, D, G, and H; Table 1).
TABLE 1.
Lists of residues whose cross-peaks are missing upon addition of Cu2+ at various temperatures
In the two-dimensional 15N-1H HSQC spectra of Cu2+-bound PrP(23–231), cross-peaks of these residues are broadened at a particular temperature compared with that of PrP(23–231). Cross-peaks of the residues (boldface) are also found to be missing in the HSQC spectra of Cu2+-bound PrP(90–231). In the case of Cu2+-bound PrP(90–231), cross-peak arising from Ile-139 disappeared at 25 °C in contrast to that of PrP(23–231), where it disappeared at 31 °C. Below 24 °C, cross-peaks are broadening due to the aggregation process, which is prevalent at or below 22 °C (shown in supplemental Table S1). Except Asp-178, residues in boldface belong to the helix-1 region and others correspond to the helix-2 region.
| 37 °C | 34 °C | 31 °C | 28 °C |
|---|---|---|---|
| Ala-120a | Ala-120 | Ala-120 | Ala-120 |
| Phe-141 | Phe-141 | Phe-141 | Phe-141 |
| Asn-143 | Asn-143 | Asn-143 | Asn-143 |
| Asp-147 | Asp-147 | Asp-147 | Asp-147 |
| Glu-146 | Glu-146 | Glu-146 | |
| Asn-174 | Asn-174 | Asn-174 | |
| Asp-178 | Asp-178 | Asp-178 | |
| Ile-139b | Ile-139b | ||
| Cys-180 | Cys-180 | ||
| Asn-181 | Asn-181 | ||
| Cys-170 | |||
| Ile-182 | |||
| Lys-185 | |||
| Arg-208a |
a Ala-120 is present in a flexible N-terminal region, and Arg-208 is present in helix-3.
b In the case of PrP(90–231), the cross-peak arising from Ile-139 was found to disappear at 25 °C.
FIGURE 4.
Representation of the residues corresponding to missing cross-peaks in helix-1 of PrP(23–231) and PrP(90–231). A, NMR structure of PrP(121–231) (BioMagResBank code 16071 and Protein Data Bank code 1XYX) displaying residues (in sticks with yellow backbone) from helix-1 and its nearby loop region, whose cross-peaks were found missing upon addition of Cu2+ to PrP(23–231) and PrP(90–231) in the temperature range of 37–24 °C. The cross-peaks of helix-1 residues Ile-139 (B), Phe-141 (C), Asn-143 (D), Glu-146 (E), Asp-147 (F), and helix-2 residue Asp-178 (G) in the absence of Cu2+ (upper panel) are compared with the cross-peaks obtained in the presence of Cu2+ (lower panel). Each panel has five different color cross-peaks that correspond to the same residues obtained at different temperatures from 16 °C (dark purple) to 37 °C (red) as shown in the color cross-peaks bar (bottom right corner). In each panel an arrow is used to indicate the approximate position of cross-peaks in the absence and presence of Cu2+. Asp-178 is the only residue in helix-2 whose cross-peaks were found to be missing at and above 30 °C in the two-dimensional 15N-1H HSQC of Cu2+-bound PrP(90–231). Residues are numbered according to the human PrP sequence. Val-121 and Ser- 231 are the N-terminal (red) and C-terminal (blue) residues, respectively.
FIGURE 5.
Representation of the residues corresponding to missing cross-peaks in the helix-2 of PrP(23–231). A, NMR structure of PrP(121–231) (BioMagResBank code 16071 and Protein Data Bank code 1XYX) displaying residues from helix-2 whose cross-peaks were found missing upon addition of Cu2+ to PrP(23–231) in the temperature range of 37–24 °C. The cross-peaks of these residues, Asn-174 (B), Asp-178 (C), Cys-179 (D), Val-180 (E), Asn-181 (F), Ile-182 (G), and Lys-185 (H), in the absence of Cu2+ (upper panel) are compared with those obtained in the presence of Cu2+ (lower panel). Each panel has five different color cross-peaks that correspond to the same residues obtained at different temperatures from 16 °C (dark purple) to 37 °C (red) as shown in the color cross-peaks bar (bottom right corner). In each panel an arrow is used to indicate approximate position of cross-peaks in the absence and presence of Cu2+. Residues are numbered according to the human PrP sequence. Val-121 and Ser-231 are the N-terminal (red) and C-terminal (blue) residues, respectively.
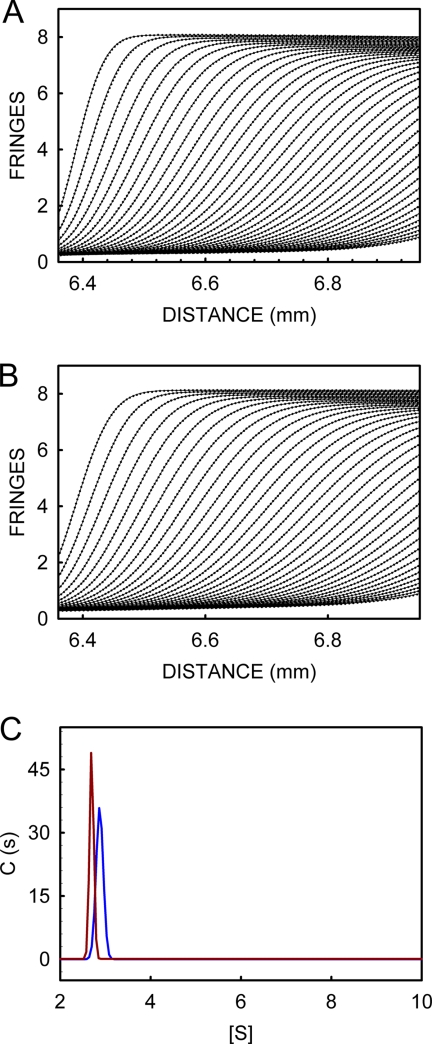
Proximity of paramagnetic Cu2+ could lead to the line broadening (disappearance of peaks) in the HSQC spectra listed in Table 1. Formation of higher order assemblies, however, can also lead to line broadening. The results from the scattering experiments (supplemental Fig. S1, A and B) show that PrP(23–231) does not undergo aggregation in the presence of Cu2+ at these temperatures. We carried out dynamic light scattering measurements (under conditions identical to NMR experiments) and found no change in the hydrodynamic radius of PrP and Cu2+-bound PrP (supplemental Fig. S2D). We also carried out sedimentation velocity measurements with PrP and Cu2+-bound PrP at 37 °C. Fig. 6, A and B, shows the movement of sedimenting boundary, represented as fringe displacement with time, of the samples of PrP and Cu2+-bound PrP, respectively. Fig. 6C shows the sedimentation coefficient profiles of PrP (s20,w, 2.694) and Cu2+-bound PrP (s20,w, 2.862) derived from curve-fitting analyses of the sedimentation profiles. Sedimentation profiles indicate that ∼98.5% of the population in the samples containing PrP and Cu2+-bound PrP are in monomeric form with a molecular mass of ∼23.4 kDa. The remaining negligible fraction (∼1.5%), which corresponds to higher oligomers (including dimers, trimers, or tetramers), is observed both in PrP and Cu2+-bound PrP, clearly showing that addition of Cu2+ to PrP samples at 37 °C does not lead to the formation of higher order assemblies. It is important to note that the sedimentation velocity and dynamic light scattering measurements were carried out under the same conditions of concentration of PrP, pH, and temperature as used in the NMR experiments. All these observations taken together clearly show that proximity of paramagnetic Cu2+, but not the formation of higher order assemblies, is responsible for the observed line broadening in HSQC spectra.
FIGURE 6.
Sedimentation velocity profiles of PrP and Cu2+-bound PrP. Movement of sedimentary boundary as monitored using interference pattern of PrP (A) and Cu2+-bound PrP (B). C, sedimentation coefficient distribution of PrP (red) and Cu2+-bound PrP (blue).
Interestingly, none of the residues in helix-1 and its nearby loop region (Ile-139–Asp-147) are implicated in Cu2+-binding to PrP. It is worth mentioning here that in the polypeptide stretch Ile-139 to Asp-147, His-140, not detected in the absence or presence of Cu2+ in the given HSQC spectrum, is the lone residue that could bind Cu2+. However, studies by Van Doorslaer and co-workers (33) have clearly ruled out the possibility of Cu2+-binding at this residue. Therefore, we believe that the reason for the disappearance of peaks from these regions can only be explained if Cu2+ bound at other sites such as the octapeptide repeats region and/or His-96/His-111 of the N-terminal region might become proximal to these residues and thus cause the paramagnetic line broadening leading to the disappearance of their spectral signatures.
To probe residues from the N-terminal regions that might interact with helix-1 and helix-2 of the C-terminal region, we have recorded 15N-1H HSQC spectra of the N-terminal deletion mutant PrP(90–231), which lacks the four octapeptide repeats (region 23–89) leaving His-96/His-111, as the only Cu2+-binding site. We noticed that the cross-peaks arising from Phe-141, Asn-143, Glu-146, Asp-147, and Asp-178 completely disappeared upon addition of Cu2+ at 37 °C (Figs. 3C and 4, C–G). Upon lowering the temperature to 25 °C, the cross-peak arising from Ile-139 also disappeared (Figs. 3D and 4B; supplemental Fig. S4B; in Table 1 see the boldface residues). Interestingly, the cross-peaks of PrP(90–231) from helix-1 and its nearby loop region (Ile-139–Asp-147), which broadened in the presence of Cu2+ at 37 °C, were similar to those of PrP(23–231), as mentioned above. PrP(90–231) did not show protein aggregation in the presence of Cu2+ at low temperatures (Fig. 1B). We therefore propose here that the C-terminal region (Ile-139 to Asp-147) of PrP is involved in a novel interaction with the Cu2+-bound N-terminal region (residues 90–120 that contains the site His-96/His-111). The role of the octapeptide repeats in this interaction is less pronounced as Cu2+-bound PrP(90–231) does not have residues from 23 to 89 that harbor octapeptide repeats. Thus, on the basis of our observations in proteins PrP(90–231) and PrP(23–231), we suggest that the region 90–120 (which contains the site His-96/His-111) becomes proximal to the region Ile-139–Asp-147 for interaction, upon binding to Cu2+.
PrP(23–231) also shows broadened cross-peaks from the helix-2 region, which is not observed in the case of PrP(90–231). A few reports suggest that Cu2+ binds to His-187 (9–12). However, we found that the cross-peak arising from His-187 broadened only below 22 °C in the HSQC spectra of Cu2+-bound PrP(23–231) but not in that of Cu2+-bound PrP(90–231) (Table S1). Because both the variants of PrP (PrP(23–231) and PrP(90–231)) have His-187, they should have shown a similar broadening phenomenon if Cu2+ binds to this site. Moreover, the broadened cross-peaks arising from the helix-2 region (residues 174–182 until 28 °C) are distal to His-187. Thus, the NMR data clearly shows that His-187 is not involved in Cu2+-binding under the chosen experimental conditions. This observation indicates the presence of Cu2+ in close proximity of the helix-2 region, which might be responsible for the observed line broadening. We believe that the reason for the disappearance of peaks from this region can only be explained by a second novel interaction of the N-terminal domain with the helix-2 region. In the case of PrP(90–231) (which lacks octapeptide repeats), we did not observe broadening of peaks in the helix-2 region suggesting the role of Cu2+-bound octapeptide repeats in the disappearance of cross-peaks from the helix-2 region. Therefore, we further propose that Cu2+-bound octapeptide repeats region (residues 60–91) interacts with the helix-2 region (residues 174–185) and is responsible for the broadening of cross-peaks arising from this region. Viles et al. (28) investigated a small region of PrP covering 51–91 residues that harbor the Cu2+-binding octapeptide repeats region and observed line broadening of some lines from this region. Because we have used full-length PrP(23–231), we were able to observe long range, intra-protein interactions upon Cu2+-binding.
Our NMR results also show that residues present in hydrophobic core were less affected until 24 °C. Upon further lowering the temperature to 22 °C and below, Cu2+-bound PrP(23–231) exhibited protein aggregation. HSQC spectra recorded at these temperatures showed general broadening of cross-peaks originating from residues of helices 1 and 2 (supplemental Table S1) and a few of the residues belonging to helix-3, which are in contact with these helices. As described earlier, the octapeptide repeats are responsible for the temperature-dependent reversible aggregation of Cu2+-bound PrP(23–231). On the basis of NMR results, we suggest the role of interaction of helix-2 and octapeptide repeats region in the temperature-dependent reversible aggregation of Cu2+-bound PrP(23–231).
Compaction of the Extended Conformation of PrP upon Cu2+-binding
Our results indicate that the line broadening of the peaks from helix-1 and helix-2 regions above 24 °C is not due to the formation of higher order assemblies but a result of the interaction of Cu2+-bound N-terminal region with the C-terminal region of PrP(23–231).
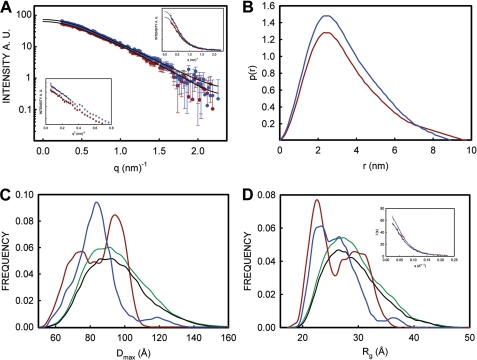
To get an insight into these intra-molecular interactions of the N-terminal region with the C-terminal counterpart, we estimated Rg and Dmax of PrP(23–231) and Cu2+-bound PrP(23–231) using SAXS. The Rg of PrP(23–231) (2.55 nm) and Cu2+-bound PrP(23–231) (2.46 nm) determined by Guinier analysis show small differences (left inset, Fig. 7A). Distance distribution function, P(r) analysis, reveals reduction in the Dmax value from 10 nm for PrP(23–231) to 9 nm for Cu2+-bound PrP(23–231) (Fig. 7B), which suggests that Cu2+-bound PrP(23–231) might have become more compact compared with PrP. The N-terminal region of PrP is unstructured and flexible in nature. Hence, we analyzed the data using ensemble optimization method (EOM), which gives useful information such as Rg and Dmax distributions (50, 51) in the case of proteins with flexible domains. Fig. 7, C and D, compare the Dmax and Rg distributions, respectively, of the ensembles of the selected conformations with the Dmax and Rg distributions of the initial pool (randomly generated) of 10,000 structures. The Dmax and Rg distributions of PrP(23–231) and Cu2+-bound PrP(23–231) showed multimodal distributions. To discern these distribution curves, we have deconvoluted the Dmax and the Rg distribution plots (supplemental Fig. S5, A–D). In the Dmax distribution of PrP(23–231), the two conformer populations (mean around 7.31 and 9.52 nm with fractional contributions of 48.7 and 49.9%, respectively) (Fig. 7C, red trace; supplemental Fig. S5B) converged into a major population with Dmax 8.39 nm (72.7%) upon Cu2+-binding (Fig. 7C, blue trace; supplemental Fig. S5D). This suggests that a significant amount of compaction of the extended conformation of PrP(23–231) occurs upon Cu2+-binding. We also found similar features in the Rg distributions of PrP(23–231) and Cu2+-bound PrP(23–231). Rg distributions of PrP(23–231) corresponding to 2.25 nm (46.4%) and 3.05 nm (34.5%) (Fig. 7D, red trace; supplemental Fig. S5A) are changed to 2.24 nm (22.8%) and 2.65 nm (73.4%) (Fig. 7D, blue trace; supplemental Fig. S5C), respectively, upon Cu2+-binding, which supports the Dmax distribution data obtained above. The results from the EOM analysis of the size distribution are in agreement with the values obtained from P(r) distribution function, where reduction of 1 nm is seen for Dmax of Cu2+-bound PrP(23–231) compared with that of PrP(23–231). The reduction in the Dmax leading to the global compactness of Cu2+-bound PrP(23–231) might be the result of a decrease in the flexibility of the N-terminal region, which exhibits interaction with the C-terminal region of PrP(23–231) upon Cu2+-binding. Study from Viles et al. (28) suggests that upon Cu2+-binding the octapeptide repeats region achieves compact structure due to four coordinating histidine residues. Studies from Takeuchi et al. (52) have reported that Cu2+ induces helix formation in the octapeptide repeat region (53). However, in both these studies short peptides comprising octapeptide or the repeat region (residues 51–91) have been used. We report here a global compaction of full-length PrP(23–231) due to inter-domain interactions upon Cu2+-binding. Thus, our SAXS results lend credence to the existence of long range interactions between the N- and C-terminal regions in the Cu2+-bound PrP(23–231). The observed marginal increase (Fig. 6C) in the sedimentation coefficient of Cu2+-bound PrP (2.86 from 2.69 for PrP) also indicates global compaction.
FIGURE 7.
SAXS measurements of PrP and Cu2+-bound PrP. A, scattering profiles of PrP (red circles) and Cu2+-bound PrP (blue circles) plotted against q (q = 4πsinθ/λ). Black traces are fitted lines obtained from P(r) distribution function. Differences in the scatter profiles of PrP and Cu2+-bound PrP are better seen in the linear plot (top right inset). Left bottom panel shows Guinier plots (ln(I(q)) versus q2) of PrP (red circle) and Cu2+-bound PrP (blue triangle). Straight lines show regions of the curve from where Rg values were calculated using Guinier plot. B, distance distribution function (P(r) function) of PrP (red trace) and Cu2+-bound PrP (blue trace). C, Dmax; D, Rg distribution function calculated from the scattering profiles of PrP(23–231) (red trace in A) and Cu2+-bound PrP (blue trace in a) using the EOM method. In each (C and D), black traces show Dmax (C) and Rg (D) distributions calculated from the pool of 10,000 conformations from PrP scattering profiles and green traces are from those of Cu2+-bound profiles. Red trace in each panel shows distribution from the selected 20 structures from PrP scattering profiles, and blue trace shows that of Cu2+-bound PrP. D, inset, EOM fitting of the PrP (black over blue trace) and Cu2+-bound PrP (black over red trace) with χ values of 0.778 and 0.775, respectively.
DISCUSSION
We have observed temperature-dependent reversible aggregation of Cu2+-bound PrP(23–231); it aggregates at lower temperatures and resolubilizes at physiological temperature. We believe that a novel interaction between helix-2 and the octapeptide repeats region of PrP(23–231) is involved in this reversible aggregation. Our study further suggests that the aggregation might proceed through the formation of hydrogen bonds (H-bonds), which might dissociate rendering the PrP(23–231) soluble at physiological temperature. We have investigated the conformational consequences of Cu2+-binding to PrP(23–231) under physiological conditions. On the basis of our results, we propose novel interactions between the N- and C-terminal regions involving the flexible region and the helices-1 and -2.
Aggregation of Cu2+-bound PrP
Cu2+ is known to play a role in prion diseases. It is shown to promote and prevent the process. We noticed that most of the Cu2+-binding experiments are either performed at nonphysiological temperatures (2, 10, 13–17) or at very high concentrations of Cu2+ (54, 55). Under physiological conditions, we find that Cu2+-bound PrP(23–231) does not aggregate. Interestingly, it starts to aggregate when the temperature is lowered. Aggregation at lower temperature has been reported earlier (2, 10, 13–17). Our results suggest that Cu2+ may not play a role in the aggregation of PrP at physiological temperature (37 °C). Our experiments with mutant protein (PrP(90–231)) show that the region encompassing octapeptide repeats (residues 60–91) is responsible for the temperature-dependent reversible aggregation of Cu2+-bound PrP(23–231) (Fig. 1). Our ITC experiments show that the observed temperature-dependent aggregation might not be due to changes in the affinity of Cu2+-binding. Binding affinity remains invariant; however, enthalpy and entropy changes show linear and reciprocal relation indicating an enthalpy-entropy-compensation phenomenon (44, 45). The observed enthalpy/entropy compensation phenomenon might be useful in understanding the temperature-dependent aggregation; due to higher entropy at 37 °C, the Cu2+-binding sites will be flexible and solvated and might disrupt any cohesive inter-molecular interactions. This facilitates Cu2+-bound PrP(23–231) to remain in solution at 37 °C. However, at lower temperatures, these sites might come together initiating cohesive interactions via H-bonds or hydrophobic interactions. Hydrophobic interactions are known to increase in strength with increasing temperatures (56). As mentioned earlier, we observed that Cu2+-bound PrP(23–231) aggregates at low temperature, but not at higher temperature (supplemental Figs. S1, A and B, and S2A ), indicating a rather lesser role for hydrophobic interactions in the process. Thus, we believe that H-bonds might play a prominent role in the temperature-dependent aggregation of Cu2+-bound PrP(23–231).
Interaction between the N- and C-terminal Regions
The possibility of interaction between the N- and C-terminal regions of PrP(23–231) has been indicated in earlier studies. Binding of antibodies specific to the N- and C-terminal regions (57), high pressure (58), and NMR studies indicates the presence of a transient interaction of the C-terminal region of helix-2 (residues 187–193) and helix-3 (residues 217–226) with the unfolded N-terminal domain (34). Our Cu2+-binding studies using ITC revealed changes in enthalpy with temperatures that resulted in large positive heat capacity change. Large heat capacity change reflects structural rearrangements upon Cu2+-binding to PrP(23–231). Our NMR studies indicate previously unknown interactions of the N-terminal region with the C-terminal region in the Cu2+-bound PrP(23–231). The NMR of full-length PrP has been reported earlier; there are also reports of NMR studies on Cu2+-binding to fragments of PrP (28, 32, 59). To the best of our knowledge, this is the first investigation of NMR of Cu2+-bound full-length PrP, which shows novel long range inter-domain interactions.
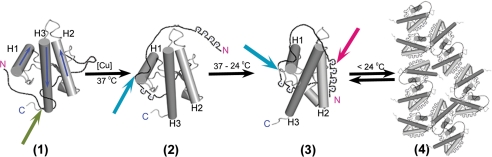
Fig. 8 schematically describes temperature-dependent reversible aggregation and long range inter-domain interactions observed with Cu2+-bound PrP(23–231). PrP (Fig. 8, panel 1) shows interaction of the N-terminal region with the C-terminal region at helix-2 and -3 (residues 187–193 and 217–226, respectively) (green arrow) as reported earlier (55–57). At 37 °C, upon Cu2+ addition, our studies suggest interaction of the N-terminal flexible domain with helix-1 and its nearby loop region (Fig. 8, panel 2, blue arrow). To the best of our knowledge, this is the first report of the interaction of helix-1 and its nearby region (residues 139–147) with the flexible N-terminal region (residues 90–120 that contains the site His-96/His-111). Cu2+-bound PrP(23–231), in the temperature range 37–24 °C, exhibits another novel interaction between the N-terminal (residues 60–91) and the C-terminal (residues 174–185) regions in the presence of Cu2+ (Fig. 8, panel 3, red arrow). Aggregation‘ at lower temperatures might be initiated via interaction of helix-2 and the Cu2+-bound octapeptide repeats region. This interaction will further result in aggregation of Cu2+-bound PrP (Fig. 8, panel 4).
FIGURE 8.
Schematic model for novel interactions and Cu2+-induced temperature-dependent aggregation of prion protein. Panel 1, helices 1–3 are represented as H1, H2, and H3, respectively. Blue arrow in each helix shows the direction of helix. N (red) and C (blue) represents the N and C terminus of PrP. Green arrow in panel 1 describes transient interaction of the N with C terminus reported earlier in the absence of Cu2+ (see “Discussion” for details). Blue arrow in panels 2 and 3 shows interaction of the N-terminal region (90–120) with helix-1 and its nearby loop region (139–147) in the presence of Cu2+ obtained from our results. Red arrow in panel 3 shows interaction of octapeptide repeat region (60–91) with helix-2(174–185) upon Cu2+ addition. Below 24 °C, Cu2+-bound PrP undergoes reversible aggregation (panel 4) via inter-molecular interaction initiated by interaction between octapeptide repeats region and helix-2. Structures are not to scale. Structural features are adopted from the NMR structure (Protein Data Bank code 1XYX) using PyMol software.
Our SAXS results lend credence to the scheme described in Fig. 8. We have generated low resolution ab initio SAXS models for PrP(23–231) and Cu2+-bound PrP(23–231) (supplemental Fig. S6). Cu2+-bound PrP(23–231) (orange envelope) shows more curved and smaller envelope compared with that of PrP(23–231) (blue envelope) (supplemental Fig. S6, A and B). It is possible that the interaction of the flexible N-terminal region with the C-terminal region, upon Cu2+-binding, leads to the observed changes in the shape of envelope of Cu2+-bound PrP(23–231) compared with that of PrP(23–231).
Functional Consequence of Interaction of Helix-1 and N-terminal Region (Residues 90–120)
We believe that the interaction of the N-terminal region (residues 90–120 that contain the site His-96/His-111) with the C-terminal region (residues 139–147) is important because of the role played by helix-1 and its nearby loop in the conversion of PrPC to PrPSC. Involvement of helix-1 in the conversion of PrPC to PrPSC was first predicted by Morrissey and Shakhnovich(60) using CHARMM energy calculation. The β-nucleation model proposed by them states that PrPSC is an aggregate with a hydrophilic core, consisting of a β-sheet-like arrangement of constituent helix-1 components (60). Helix-1 has been shown to promote aggregation (61, 62). It was also found in the dimer interface of the crystal structure of human PrP (63). Site 139–141 has been shown to be the most pressure-sensitive region (58) and found to have a much higher aggregation tendency compared with helix-1 (64). The helix-1 region Asn-143–Glu-146 also plays a prominent role in the prion conversion in vivo (65). Thus, it is evident that helix-1 and the nearby loop region play a major role in the aggregation and conversion of PrP to PrPSC. Our study shows that helix-1 and the nearby loop region interact with the N-terminal region (residues 90–120 that contain the site His-96/His-111) upon Cu2+-binding. As mentioned earlier, Cu2+ inhibits amyloid formation of PrP in vitro (22). Baskakov and co-workers (22) attributed this inhibition to binding of Cu2+ in the C-terminal region of PrP. However, even in the absence of the C-terminal region, PrP(82–146), in the presence of Cu2+, did not show amyloid fibril formation (23). It is possible that the interaction of the N-terminal region (residues 90–120) with the loop (Ile-139–Phe-141) and helix-1 (Asn-143–Arg-148) region of PrP(23–231) upon Cu2+-binding is responsible for the observed inhibition of the amyloid fibril formation in all the three cases, namely PrP(82–146) (23), PrP(89–231) (22), and PrP(23–231) (22). This interaction is not present in PrP(23–231) in the absence of Cu2+. We therefore propose an inhibitory role of Cu2+ on the conversion of PrPC to PrPSC in vitro and in vivo via the interaction between the N-terminal region and helix-1 and its nearby loop region.
Functional Consequence of Interaction of Helix-2 and Octapeptide Repeats
PrP(23–231), in contrast to PrP(90–231), undergoes temperature-dependent reversible aggregation upon Cu2+-binding possibly because of the interaction between the octapeptide repeats region (residues 60–91) and helix-2. Reversible aggregation of Cu2+-bound PrP(23–231) is only observed at lower temperature but not at 37 °C. Therefore, we believe that interaction between the octapeptide repeats region and the helix-2 region primes Cu2+-bound PrP(23–231) for inter-protein interactions at 37 °C. Such interactions might have physiological significance.
The globular C-terminal domain of PrP includes two subdomains, H1-B1 and B2-H2-H3. NMR relaxation dynamics show that H1-B1 and the connecting loop are more flexible compared with the B2-H2-H3 subdomain (66). Rare, large scale motions of the subdomains have been proposed to initiate prion protein aggregation (67). Recent studies have shown that separation of these subdomains and formation of domain-swapped dimers are a prerequisite for the aggregation of prion protein (68, 69). This implies that increased proximity between these subdomains might have an opposite effect and might delay or completely abrogate the aggregation process. In this study, we show that N-terminal region wraps around C-terminal region by interacting with helix-1 and helix-2 upon copper binding. This might lead to increased proximity of the subdomains, thus inhibiting the aggregation process. It appears that copper inhibits amyloid formation either by interfering with the region comprising H1 and the nearby loop (see above) or by decreasing separation between the two subdomains of the C-terminal region. These implications are important in the context of amyloid formation and role of copper in prion disease.
Harris and Pauly (70) proposed that conformational changes in PrPC upon Cu2+-binding might increase the affinity of the protein for a putative endocytic receptor that localizes PrPC in clathrin-coated pits. Based on our observations, we propose that the interaction between the octapeptide repeats region and the helix-2 region might be responsible for the conformational changes required for increased endocytosis.
The role of Cu2+ as an attenuator or facilitator in prion diseases is highly controversial. Our observation that PrP(23–231) in the presence of Cu2+ does not aggregate at 37 °C supports the suggestion that Cu2+ attenuates prion disease. However, we have also observed an unusual temperature-dependent aggregation of Cu2+-bound PrP(23–231). It aggregated at lower temperature and resolubilized at physiological temperature. We have investigated this phenomenon using various biophysical techniques such as ITC, NMR, and SAXS thus providing detailed thermodynamic and conformational aspects of Cu2+-binding to PrP(23–231). We observed two novel long range inter-domain interactions between the N- and C-terminal regions. Our study shows that Cu2+-bound His-96/His-111 becomes proximal to helix-1 and its nearby loop region (residues 139–147). This region is shown to have a prominent role in amyloidogenesis in vitro (22) and prion conversion in vivo (23). Our observation suggests that upon Cu2+-binding, the proximity of His-96/His-111 to Ile-139–Asp-147 and proximity between the two subdomains of the C-terminal region are involved in the inhibition of amyloid formation. In addition, we also observed interaction of the octapeptide repeats region (residues 60–91) with helix-2 (residues 174–185) upon Cu2+-binding. We speculate the involvement of this interaction in the aggregation process at lower temperatures. We believe that these interactions should prove useful in understanding the biological functions of prion protein upon Cu2+-binding in vivo such as conformational changes (71), endocytosis (70), lipid rafts localization, and Cu2+-induced signal transduction (72, 73) and in developing therapeutic strategies against prion diseases.
Supplementary Material
Acknowledgments
We thank T. Ramakrishna, T. K. Chowdary, Z. Kamal, and B. Raman for insightful discussion and critical reading of the manuscript. A. K. S. and K. V. R. C. acknowledge the facilities provided by the National Facility for High Field NMR, supported by Department of Science and Technology, Department of Biotechnology, Council of Scientific and Industrial Research, New Delhi, India, and Tata Institute of Fundamental Research, Mumbai, India.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Table S1.
- PrP
- prion protein
- SAXS
- small angle x-ray scattering
- ITC
- isothermal calorimetry
- EOM
- ensemble optimization method.
REFERENCES
- 1. Baldwin M. A., Pan K. M., Nguyen J., Huang Z., Groth D., Serban A., Gasset M., Mehlhorn I., Fletterick R. J., Cohen F. E. (1994) Philos. Trans. R. Soc. Lond. B Biol. Sci. 343, 435–441 [DOI] [PubMed] [Google Scholar]
- 2. Quaglio E., Chiesa R., Harris D. A. (2001) J. Biol. Chem. 276, 11432–11438 [DOI] [PubMed] [Google Scholar]
- 3. Thompsett A. R., Abdelraheim S. R., Daniels M., Brown D. R. (2005) J. Biol. Chem. 280, 42750–42758 [DOI] [PubMed] [Google Scholar]
- 4. Badrick A. C., Jones C. E. (2009) J. Inorg. Biochem. 103, 1169–1175 [DOI] [PubMed] [Google Scholar]
- 5. Nadal R. C., Davies P., Brown D. R., Viles J. H. (2009) Biochemistry 48, 8929–8931 [DOI] [PubMed] [Google Scholar]
- 6. Brown D. R., Qin K., Herms J. W., Madlung A., Manson J., Strome R., Fraser P. E., Kruck T., von Bohlen A., Schulz-Schaeffer W., Giese A., Westaway D., Kretzschmar H. (1997) Nature 390, 684–687 [DOI] [PubMed] [Google Scholar]
- 7. Jackson G. S., Murray I., Hosszu L. L., Gibbs N., Waltho J. P., Clarke A. R., Collinge J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8531–8535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hasnain S. S., Murphy L. M., Strange R. W., Grossmann J. G., Clarke A. R., Jackson G. S., Collinge J. (2001) J. Mol. Biol. 311, 467–473 [DOI] [PubMed] [Google Scholar]
- 9. Cereghetti G. M., Schweiger A., Glockshuber R., Van Doorslaer S. (2001) Biophys. J. 81, 516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown D. R., Guantieri V., Grasso G., Impellizzeri G., Pappalardo G., Rizzarelli E. (2004) J. Inorg. Biochem. 98, 133–143 [DOI] [PubMed] [Google Scholar]
- 11. Colombo M. C., Vandevondele J., Van Doorslaer S., Laio A., Guidoni L., Rothlisberger U. (2008) Proteins 70, 1084–1098 [DOI] [PubMed] [Google Scholar]
- 12. Watanabe Y., Hiraoka W., Igarashi M., Ito K., Shimoyama Y., Horiuchi M., Yamamori T., Yasui H., Kuwabara M., Inagaki F., Inanami O. (2010) Biochem. Biophys. Res. Commun. 394, 522–528 [DOI] [PubMed] [Google Scholar]
- 13. Jones C. E., Abdelraheim S. R., Brown D. R., Viles J. H. (2004) J. Biol. Chem. 279, 32018–32027 [DOI] [PubMed] [Google Scholar]
- 14. Tagliavini F., Prelli F., Verga L., Giaccone G., Sarma R., Gorevic P., Ghetti B., Passerini F., Ghibaudi E., Forloni G., et al. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9678–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zahn R. (2003) J. Mol. Biol. 334, 477–488 [DOI] [PubMed] [Google Scholar]
- 16. Hodak M., Chisnell R., Lu W., Bernholc J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11576–11581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qin K., Yang D. S., Yang Y., Chishti M. A., Meng L. J., Kretzschmar H. A., Yip C. M., Fraser P. E., Westaway D. (2000) J. Biol. Chem. 275, 19121–19131 [DOI] [PubMed] [Google Scholar]
- 18. Stevens D. J., Walter E. D., Rodríguez A., Draper D., Davies P., Brown D. R., Millhauser G. L. (2009) PLoS Pathog. 5, e1000390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu S., Yin S., Pham N., Wong P., Kang S. C., Petersen R. B., Li C., Sy M. S. (2008) FEBS J. 275, 5564–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giese A., Levin J., Bertsch U., Kretzschmar H. (2004) Biochem. Biophys. Res. Commun. 320, 1240–1246 [DOI] [PubMed] [Google Scholar]
- 21. Orem N. R., Geoghegan J. C., Deleault N. R., Kascsak R., Supattapone S. (2006) J. Neurochem. 96, 1409–1415 [DOI] [PubMed] [Google Scholar]
- 22. Bocharova O. V., Breydo L., Salnikov V. V., Baskakov I. V. (2005) Biochemistry 44, 6776–6787 [DOI] [PubMed] [Google Scholar]
- 23. Ricchelli F., Buggio R., Drago D., Salmona M., Forloni G., Negro A., Tognon G., Zatta P. (2006) Biochemistry 45, 6724–6732 [DOI] [PubMed] [Google Scholar]
- 24. Sigurdsson E. M., Brown D. R., Alim M. A., Scholtzova H., Carp R., Meeker H. C., Prelli F., Frangione B., Wisniewski T. (2003) J. Biol. Chem. 278, 46199–46202 [DOI] [PubMed] [Google Scholar]
- 25. Hortells P., Monleón E., Acín C., Vargas A., Vasseur V., Salomon A., Ryffel B., Cesbron J. Y., Badiola J. J., Monzón M. (2010) Zoonoses. Public Health 57, 358–366 [DOI] [PubMed] [Google Scholar]
- 26. Chattopadhyay M., Walter E. D., Newell D. J., Jackson P. J., Aronoff-Spencer E., Peisach J., Gerfen G. J., Bennett B., Antholine W. E., Millhauser G. L. (2005) J. Am. Chem. Soc. 127, 12647–12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aronoff-Spencer E., Burns C. S., Avdievich N. I., Gerfen G. J., Peisach J., Antholine W. E., Ball H. L., Cohen F. E., Prusiner S. B., Millhauser G. L. (2000) Biochemistry 39, 13760–13771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Viles J. H., Cohen F. E., Prusiner S. B., Goodin D. B., Wright P. E., Dyson H. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2042–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Di Natale G., Osz K., Nagy Z., Sanna D., Micera G., Pappalardo G., Sóvágó I., Rizzarell E. (2009) Inorg. Chem. 48, 4239–4250 [DOI] [PubMed] [Google Scholar]
- 30. Osz K., Nagy Z., Pappalardo G., Di Natale G., Sanna D., Micera G., Rizzarelli E., Sóvágó I. (2007) Chemistry 13, 7129–7143 [DOI] [PubMed] [Google Scholar]
- 31. Di Natale G., Grasso G., Impellizzeri G., La Mendola D., Micera G., Mihala N., Nagy Z., Osz K., Pappalardo G., Rigó V., Rizzarelli E., Sanna D., Sóvágó I. (2005) Inorg. Chem. 44, 7214–7225 [DOI] [PubMed] [Google Scholar]
- 32. Hornemann S., von Schroetter C., Damberger F. F., Wüthrich K. (2009) J. Biol. Chem. 284, 22713–22721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cereghetti G. M., Schweiger A., Glockshuber R., Van Doorslaer S. (2003) Biophys. J. 84, 1985–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zahn R., Liu A., Lührs T., Riek R., von Schroetter C., López García F., Billeter M., Calzolai L., Wider G., Wüthrich K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thakur A. K., Rao ChM. (2008) PLoS One 3, e2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 38. Guinier A., Fournet G. (1955) Small Angle Scattering of X-rays, pp. 5–82, John Wiley & Sons, Inc., New York [Google Scholar]
- 39. Svergun D. I. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 40. Svergun D. I., Petoukhov M. V., Koch M. H. (2001) Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Volkov V. V., Svergun D. I. (2001) J. Appl. Crystallogr. 36, 860–864 [Google Scholar]
- 42. Kozin M., Svergun D. I. (2000) J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 43. LeVine H., 3rd (1999) Methods Enzymol. 309, 274–284 [DOI] [PubMed] [Google Scholar]
- 44. Lumry R., Rajender S. (1970) Biopolymers 9, 1125–1227 [DOI] [PubMed] [Google Scholar]
- 45. Dunitz J. D. (1995) Chem. Biol. 2, 709–712 [DOI] [PubMed] [Google Scholar]
- 46. Cooper A., Johnson C. M., Lakey J. H., Nöllmann M. (2001) Biophys. Chem. 93, 215–230 [DOI] [PubMed] [Google Scholar]
- 47. Sturtevant J. M. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 2236–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nishina K., Jenks S., Supattapone S. (2004) J. Biol. Chem. 279, 40788–40794 [DOI] [PubMed] [Google Scholar]
- 49. Banci L., Pierattelli R., Vila A. J. (2002) Adv. Protein Chem. 60, 397–449 [DOI] [PubMed] [Google Scholar]
- 50. Bernadó P. (2010) Eur. Biophys. J. 39, 769–780 [DOI] [PubMed] [Google Scholar]
- 51. Bernadó P., Mylonas E., Petoukhov M. V., Blackledge M., Svergun D. I. (2007) J. Am. Chem. Soc. 129, 5656–5664 [DOI] [PubMed] [Google Scholar]
- 52. Miura T., Hori-i A., Takeuchi H. (1996) FEBS Lett. 396, 248–252 [DOI] [PubMed] [Google Scholar]
- 53. Brown D. R., Sassoon J. (2002) Mol. Biotechnol. 22, 165–178 [DOI] [PubMed] [Google Scholar]
- 54. Tsiroulnikov K., Rezaei H., Dalgalarrondo M., Chobert J. M., Grosclaude J., Haertlé T. (2006) Biochim. Biophys. Acta 1764, 1218–1226 [DOI] [PubMed] [Google Scholar]
- 55. Redecke L., von Bergen M., Clos J., Konarev P. V., Svergun D. I., Fittschen U. E., Broekaert J. A., Bruns O., Georgieva D., Mandelkow E., Genov N., Betzel C. (2007) J. Struct. Biol. 157, 308–320 [DOI] [PubMed] [Google Scholar]
- 56. Chandler D. (2005) Nature 437, 640–647 [DOI] [PubMed] [Google Scholar]
- 57. Li R., Liu T., Wong B. S., Pan T., Morillas M., Swietnicki W., O'Rourke K., Gambetti P., Surewicz W. K., Sy M. S. (2000) J. Mol. Biol. 301, 567–573 [DOI] [PubMed] [Google Scholar]
- 58. Kachel N., Kremer W., Zahn R., Kalbitzer H. R. (2006) BMC Struct. Biol. 6, 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pérez D. R., Damberger F. F., Wüthrich K. (2010) J. Mol. Biol. 400, 121–128 [DOI] [PubMed] [Google Scholar]
- 60. Morrissey M. P., Shakhnovich E. I. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11293–11298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Watzlawik J., Skora L., Frense D., Griesinger C., Zweckstetter M., Schulz-Schaeffer W. J., Kramer M. L. (2006) J. Biol. Chem. 281, 30242–30250 [DOI] [PubMed] [Google Scholar]
- 62. Solforosi L., Bellon A., Schaller M., Cruite J. T., Abalos G. C., Williamson R. A. (2007) J. Biol. Chem. 282, 7465–7471 [DOI] [PubMed] [Google Scholar]
- 63. Knaus K. J., Morillas M., Swietnicki W., Malone M., Surewicz W. K., Yee V. C. (2001) Nat. Struct. Biol. 8, 770–774 [DOI] [PubMed] [Google Scholar]
- 64. Ziegler J., Viehrig C., Geimer S., Rösch P., Schwarzinger S. (2006) FEBS Lett. 580, 2033–2040 [DOI] [PubMed] [Google Scholar]
- 65. Norstrom E. M., Mastrianni J. A. (2006) J. Virol. 80, 8521–8529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Viles J. H., Donne D., Kroon G., Prusiner S. B., Cohen F. E., Dyson H. J., Wright P. E. (2001) Biochemistry 40, 2743–2753 [DOI] [PubMed] [Google Scholar]
- 67. Schwarzinger S., Horn A. H., Ziegler J., Sticht H. (2006) J. Biomol. Struct. Dyn. 23, 581–590 [DOI] [PubMed] [Google Scholar]
- 68. Eghiaian F., Daubenfeld T., Quenet Y., van Audenhaege M., Bouin A. P., van der Rest G., Grosclaude J., Rezaei H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7414–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hafner-Bratkovic I., Bester R., Pristovsek P., Gaedtke L., Veranic P., Gaspersic J., Mancek-Keber M., Avbelj M., Polymenidou M., Julius C., Aguzzi A., Vorberg I., Jerala R. (2011) J. Biol. Chem. 286, 12149–12156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pauly P. C., Harris D. A. (1998) J. Biol. Chem. 273, 33107–33110 [DOI] [PubMed] [Google Scholar]
- 71. Leclerc E., Serban H., Prusiner S. B., Burton D. R., Williamson R. A. (2006) Arch. Virol. 151, 2103–2109 [DOI] [PubMed] [Google Scholar]
- 72. Stuermer C. A., Langhorst M. F., Wiechers M. F., Legler D. F., Von Hanwehr S. H., Guse A. H., Plattner H. (2004) FASEB J. 18, 1731–1733 [DOI] [PubMed] [Google Scholar]
- 73. Monnet C., Gavard J., Mège R. M., Sobel A. (2004) FEBS Lett. 576, 114–118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.