Background: Snake venom of Notechis scutatus contains notecarin D, a coagulation factor Xa homolog.
Results: Notecarin D binds with extraordinarily high affinity to factor V/Va in the absence of membranes.
Conclusion: Notecarin D lacks the restricted membrane dependence of factor Xa for binding factor V/Va.
Significance: Notecarin D likely bypasses the membrane requirement for high affinity factor Va binding, gaining control of blood coagulation after envenomation of mammalian prey.
Keywords: Blood Coagulation Factors, Enzyme Kinetics, Fluorescence, Protein-Protein Interactions, Prothrombin, Serine Protease, Snake Venom, Factor V, Factor Xa
Abstract
Notecarin D (NotD) is a prothrombin (ProT) activator in the venom of the tiger snake, Notechis scutatus, and a factor Xa (FXa) homolog. NotD binds specifically to the FXa binding site expressed on factor V (FV) upon activation to factor Va (FVa) by thrombin. NotD active site-labeled with 5-fluorescein ([5F]FFR-NotD) binds FV and FVa with remarkably high affinity in the absence of phospholipids (KD 12 and ≤ 0.01 nm, respectively). In the presence of membranes, the affinity of [5F]FFR-NotD for FVa is similar, but increased ∼55-fold for FV. Binding of FXa active site-labeled with Oregon Green to FV and FVa in the presence of phospholipids is ∼5,000- and ∼80-fold weaker than [5F]FFR-NotD, respectively. NotD reports FVa and not FV binding by a 3-fold increase in tripeptide substrate hydrolysis, demonstrating allosteric regulation by FVa. The NotD·FVa·membrane complex activates ProT with Km(app) similar to prothrombinase, and ∼85-fold weaker without membranes. Active site-blocked NotD exhibits potent anticoagulant activity in plasma thrombin generation assays, representing inhibition of productive prothrombinase assembly and possible disruption of FXa inhibition by the tissue factor pathway inhibitor. The results show that high affinity binding of NotD to FVa is membrane-independent, unlike the strict membrane dependence of FXa for high affinity FVa binding.
Introduction
Activation of prothrombin (ProT)4 by prothrombinase to generate the clot-forming proteinase α-thrombin is a critical reaction in blood coagulation. Prothrombinase is comprised of the cofactor factor Va (FVa) bound to factor Xa (FXa) on phospholipid membranes containing phosphatidylserine in the presence of Ca2+. Assembly of the complex on membranes increases the rate of thrombin formation by six orders of magnitude (1–3). ProT cleavage by prothrombinase is sequential, with Arg320 first to generate the active intermediate meizothrombin, followed by cleavage at Arg271 to generate thrombin and fragment 1.2 (4, 5). A 100-fold decrease in the apparent Km for ProT is attributed to membrane binding and reduction in dimensionality, whereas a 3,000-fold increase in apparent kcat is assigned to FVa (6–8). Prothrombinase function is critically dependent on membrane binding, although the precise mechanisms involved are not completely understood.
Factor V (FV) circulates as a single chain 330,000-Da molecule with the domain structure of A1-A2-B-A3-C1-C2 (9). Thrombin activation of FV liberates the B domain and generates a Ca2+-stabilized heterodimer with a heavy chain (A1-A2) and light chain (A3-C1-C2). Thrombin-catalyzed FV activation is facilitated by exosite I and II interactions, resulting in a kinetically preferred order of cleavage where exosite I-mediated Arg709 is first, followed closely by Arg1018, and exosite I and II-mediated cleavage at Arg1545 is last (10–13). Cleavage at Arg1545 is required for full FVa cofactor activity (14). The single cleavage at Arg1545 by the Russell viper venom FV-activating proteinase (RVV-V) is sufficient for generation of FVa cofactor activity (15, 16).
The most toxic snake venom in the world belongs to Australian snakes from the Elapidae family (17). A member of this family is Notechis scutatus, which has a group D ProT activator in its venom called notecarin D (NotD) that is homologous to FXa (18–20). NotD is a ∼47-kDa protein, and like FXa, contains a heavy chain serine proteinase domain, a light chain consisting of two epidermal growth factor-like domains and a Gla domain with 11 γ-carboxyglutamic acid residues. NotD cleaves bovine ProT at Arg274 and Arg323 through a meizothrombin intermediate in the presence of FVa and phospholipid vesicles (18, 19, 21). Like FXa, NotD is capable of cleaving ProT in the absence of FVa, which proceeds through a predominantly prethrombin-2·fragment 1.2 pathway (18).
In the present studies, venom-derived NotD was examined to determine the functional properties that underlie its potent procoagulant, FXa-like activity. The results demonstrate for the first time that NotD binds with very high affinity to FV and FVa in the absence of phospholipid vesicles. NotD binds to the FXa site expressed on activation of FV to FVa with a ∼80-fold higher affinity than FXa. NotD binds FV in the absence and presence of membranes with lower affinity than FVa, but still binds significantly tighter than FXa. The NotD·FVa complex activates ProT in the absence and presence of phospholipid vesicles with reduced catalytic efficiency compared with prothrombinase. NotD lacks the membrane dependence of FXa for binding to FV and FVa and, presumably without this constraint, gains unregulated control of blood coagulation during N. scutatus envenomation.
EXPERIMENTAL PROCEDURES
Phospholipid Vesicles
Large and small unilamellar phospholipid vesicles (LUVs and SUVs) were prepared with minor modifications of the published method (22). Vesicles were prepared with either synthetic 60% (w/w) 1,2-dioleoyl-sn-glycero-3-phosphocholine, 20% 1,2-dioleoyl-sn-glycero-3-phospho-l-serine, and 20% 1,2-dioleoyl-sn-glycero-3-phospho-l-ethanolamine (PC/PS/PE containing synthetic dioleoyl lipids) or 75% (w/w) hen egg l-α-phosphatidylcholine and 25% porcine brain l-α-phosphatidylserine (PC/PS vesicles containing natural lipids) (Avanti Polar Lipids) and were mixed and dried under a stream of N2 and lyophilized for ∼15 h. To prepare LUVs, the lipids were dispersed in 50 mm HEPES, 125 mm NaCl, pH 7.4, and extruded through two back-to-back 0.4-μm filters. The vesicles were centrifuged (Beckman TL-100 or Optima MAX-XP) at 93,000 × g for 45 min and the top 70% of the mixture was used as LUVs. SUVs were prepared similarly, except the reconstituted lipid mixture was extruded through two back-to-back 0.4-μm filters followed by two back-to-back 0.1-μm filters. The lipids were sonicated (Branson Sonifier 250) continuously in an ice bath for 1 h at 30% duty cycle and 3-output control under a stream of N2. The vesicles were centrifuged initially at ∼130,000 × g for 30 min, followed by ∼206,000 × g for 4 h, and the top 20% of the mixture was used as SUVs. LUVs and SUVs were stored at 4 and 22 °C, respectively. The concentration of vesicles was determined by Stewart assay (23). Dynamic light scattering measurements on representative lipid preparations gave diameters of 500 ± 300 Å for SUVs and 2000 ± 580 Å for LUVs, similar to previously published values (24, 25).
Protein Purification and Characterization
Human ProT, α-thrombin, and FXa were purified as described earlier (26, 27). NotD was purified from lyophilized venom of N. scutatus (Latoxan). All chromatography matrices were regenerated in 0.1 m NaOH, 2.0 m NaCl, followed by buffer, and treated with 1 μm d-Phe-Phe-Arg-CH2Cl (FFR-CH2Cl), 1 μm d-Phe-Pro-Arg-CH2Cl (FPR-CH2Cl), and 100 μm phenylmethylsulfonyl fluoride in their respective buffers prior to purification to prevent NotD degradation. Venom (250 mg) was reconstituted in 50 mm MES, 150 mm NaCl, 10 mm benzamidine, 0.02% (w/v) NaN3, pH 6.0, dialyzed against the same buffer for ∼15 h to remove excess salts, and chromatographed on a 5-ml HiTrap heparin-Sepharose column (GE Healthcare) in the same buffer. Bound protein was eluted with a 50-ml gradient from 0.15 to 0.8 m NaCl. The activity of the fractions was determined by measuring the initial rate of 200 μm CH3SO2-d-Leu-Gly-Arg-p-nitroanilide (CH3SO2-LGR-pNA; Diagnostica Stago) hydrolysis at 405 nm and 25 °C using a microtiter plate reader. Fractions were pooled based on activity and NaCl was added to a final concentration of 1.5 m. The pooled fractions were concentrated by ultrafiltration to <2.5 ml with a YM10 membrane and loaded onto a Sephacryl-200 (Amersham Biosciences) gel filtration column (2.5 cm x 39 cm) equilibrated with 50 mm HEPES, 1.5 m NaCl, 10 mm benzamidine, 0.02% NaN3, pH 7.4. Fractions were pooled and 10% (v/v) 1 m MES, pH 6.0, was added before concentrating to <2.5 ml by ultrafiltration using a YM10 membrane. The protein was desalted using a Superdex 200 (10/300; GE Healthcare) FPLC column equilibrated with 5 mm MES, 150 mm NaCl, pH 6.0, snap-frozen in liquid N2 in PEG 20,000-coated tubes, and stored at −80 °C. The purity of the protein was evaluated by SDS-PAGE. The concentration of NotD was determined from the absorbance at 280 nm, using a calculated and validated (28) absorption coefficient of 1.10 mg·ml−1 cm−1 and molecular mass of 46,805 Da calculated from the amino acid sequence. A ratio of active sites to NotD protein concentration of 0.97 was determined by active site titration with p-nitrophenyl-p′-guanidinobenzoate (29).
Expression and Purification of ProTR271Q
HEK 293 cells expressing ProTR271Q were a gift from Dr. Sriram Krishnaswamy (University of Pennsylvania). ProTR271Q was purified by a modified method based on the published procedure (24). Cells were grown to confluence in a 10-story cell factory (Nunc) pretreated with poly-d-lysine. Two liters of expression medium (Dulbecco's modified Eagle's medium/F12 without phenol red containing 2 mm l-glutamine, 100 units/ml of penicillin, 100 μg/ml of streptomycin, 10 μg/ml of vitamin K, and 5 μg/ml of insulin/transferrin/sodium selenite supplement) were collected daily and frozen at −80 °C until 20 liters was obtained. The thawed media was centrifuged at 4,500 × g for 30 min at 4 °C, applied to a Q-Sepharose (GE Healthcare) column (5 cm × 13 cm) equilibrated with 20 mm HEPES, pH 7.4, and the ProTR271Q was eluted with a 3-liter gradient from 0 to 1 m NaCl. Na3 citrate was added at 0.425 g/100 ml of pooled protein, stirred on ice for 30 min at 4 °C, and precipitated with 1 m BaCl2 (80 ml/liter of pooled protein) over 20 min. The precipitate was centrifuged at 12,800 × g for 30 min at 4 °C, dissolved in 20 ml of 0.5 m EDTA, 5 mm benzamidine, pH 8.0, and dialyzed ∼15 h at 4 °C against 20 mm HEPES, 1 mm benzamidine, 1 mm EDTA, pH 7.4. Recovered ProTR271Q was chromatographed on a 6-ml Resource Q column (GE Healthcare) equilibrated with 20 mm HEPES, pH 7.4, and eluted with a 120-ml gradient from 0 to 1 m NaCl in the same buffer. Pooled protein was dialyzed ∼15 h at 4 °C against 1 mm NaPi, pH 6.8. ProTR271Q was loaded onto a 5-ml hydroxyapatite column (Bio-Rad) equilibrated with dialysis buffer and eluted with a 50-ml gradient from 1 to 500 mm NaPi. The pooled protein was concentrated and further purified by gel filtration on a Superdex 200 column (10 × 300 mm; Amersham Biosciences) equilibrated with 5 mm MES, 150 mm NaCl, pH 6.0. The final protein was concentrated and stored at −80 °C.
Purification of Factor V
FV was purified from human plasma (30) and activated to FVa by addition of 100 nm thrombin to 12 μm FV and incubation at 37 °C for 40 min. Thrombin was inactivated with 10 μm FPR-CH2Cl and excess inhibitor was removed by dialysis. The concentration of FV and FVa was determined from the 280-nm absorbance using the 330,000 Da molecular mass and the published absorption coefficient of 0.89 mg ml−1 cm−1 (15) for FV. An absorption coefficient for the mixture of thrombin-generated activation products (FVa) was determined. The 280-nm absorbance of 0.7 μm FV in 50 mm HEPES, 110 mm NaCl, 5 mm CaCl2, 1 mg/ml of PEG 8000, pH 7.4 (Buffer A), was measured before and after full activation by 100 nm thrombin at 37 °C for 1 h, and the ratio (0.88) was applied to the absorption coefficient of FV to obtain 0.78 mg ml−1 cm−1 for FVa (the mixture of the two-subunit form of FVa and other activation products). Terminal activation products of FV, the two-subunit form of FVa (FVa-h·FVa-l), FVa heavy chain (FVa-h), FVa light chain (FVa-l), B710–1018, and B1019–1545 were isolated as described (31).
Preparation of Active Site-labeled NotD and FXa Analogs
Nα-[(Acetylthio)acetyl]-FFR-CH2Cl (ATA-FFR-CH2Cl) and Nα-[(acetylthio)acetyl]-Glu-Gly-Arg-CH2Cl (ATA-EGR-CH2Cl) were prepared as described (26, 32). NotD active site-labeled with 5-(iodoacetamido)fluorescein ([5F]FFR-NotD) was prepared by inactivation of 18 μm NotD with 360 μm ATA-FFR-CH2Cl in 0.1 m 2,2′-(1,3-diyldiimino)bis[2-(hydroxymethyl)propane-1,3-diol]propane, 0.3 m NaCl, 1.6 mm CaCl2, 0.7 mm MgCl2, 1 mg/ml of PEG 8000, pH 7.0, at 25 °C for 3 h, until the initial rate of CH3SO2-LGR-pNA hydrolysis was <0.01% of the initial activity. The free thiol group on the incorporated inhibitor was generated by addition of 1 m NH2OH in 0.1 m 2,2′-(1,3-diyldiimino)bis[2-(hydroxymethyl)propane-1,3-diol]propane, 0.1 m NaCl, 5 mm CaCl2, pH 7.0, to a 0.1 m final concentration and labeled with 600 μm 5-(iodoacetamido)fluorescein in the same buffer. Excess inhibitor and probe were removed by Sephadex G-25 chromatography and final dialysis against 50 mm HEPES, 110 mm NaCl, 4.3 mm CaCl2, 0.7 mm MgCl2, 1 mg/ml of PEG 8000, pH 7.4. Preparations of [5F]FFR-NotD contained 0.8 mol of 5F/mol of NotD, measured as previously described (26, 33). Labeling of FXa ([OG]EGR-FXa) was achieved by inactivation with ATA-EGR-CH2Cl and labeling of the NH2OH generated a free thiol group with Oregon Green 488-maleimide (OG) following procedures as described earlier (34). Probe incorporation was 0.92 mol of OG/mol of FXa. Active site-blocked NotD (FFR-NotD) was prepared by incubating 18 μm NotD with 360 μm FFR-CH2Cl for ∼15 h at 25 °C in Buffer A. Inactivation (<0.1% active) was confirmed from the initial rate of CH3SO2-LGR-pNA hydrolysis, and excess inhibitor was removed by extensive dialysis against Buffer A.
Effects of FV, FVa, and FVa Terminal Activation Products on Chromogenic Substrate Hydrolysis by NotD
Initial rates of CH3SO2-LGR-pNA hydrolysis by NotD at 405 nm were obtained by incubating 5 nm NotD in Buffer A with various concentrations of FV/FVa terminal activation products for 1–2 min and initiating the assays with 200 μm CH3SO2-LGR-pNA. Assays were performed at 25 °C in polystyrene cuvettes coated with PEG 20,000. The fractional increase in the initial rates of substrate hydrolysis by NotD, (vobs − vo)/vo, as a function of the total FVa or FVa-h·FVa-l concentration was analyzed by nonlinear least squares fitting of the quadratic binding equation to obtain the initial velocity (vo), maximum velocity (vmax), stoichiometric factor (n), and apparent dissociation constant (KD(app)). Effects of FV and the other FVa terminal activation products were fit by linear least squares.
FV Activation by Thrombin in the Presence of Thrombin Exosite I and II Inhibitors Using the NotD Activity Assay
FV (500 nm) was activated with 0.2 nm thrombin in Buffer A in the absence and presence of 10 μm thrombin exosite I DNA aptamer (GGTTGGTGTGGTTGG; Integrated DNA Technologies), 100 nm thrombin exosite II DNA aptamer (AGTCCGTGGTAGGGCAGGTTGGGGTGACT, or 10 μm Hir54–65(SO3−) at 37 °C. Aliquots from the FV/FVa reaction mixtures were removed at various time points and the initial rates of 200 μm CH3SO2-LGR-pNA hydrolysis by 6 nm NotD were measured in Buffer A containing 1 mg/ml of BSA at 25 °C. The fractional increase in initial rates, (vobs − vo)/vo, was converted to the FVa concentration using the linear portion of a reference titration obtained by titrating 5 nm NotD with FVa in Buffer A plus BSA. FVa concentrations were obtained by interpolation of the data fit by the quadratic binding equation. FVa used in the reference titration was generated by activating the same preparation of FV (500 nm) with thrombin (1 nm) for 45 min at 37 °C and quenching with 25 units/ml of hirudin (Sigma). FV activation by thrombin in the absence of inhibitors was analyzed by nonlinear least squares fitting of a single exponential and results in the presence of inhibitors were fit by linear least squares.
Fluorescence Equilibrium Binding
Fluorescence anisotropy measurements were performed with a QuantaMaster 30 spectrofluorometer (Photon Technology International) at 25 °C using acrylic cuvettes coated with PEG 20,000. Titrations were performed in Buffer A with 10 μm FPR-CH2Cl added to prevent proteolysis. Anisotropy measurements were corrected for associated total intensity changes using a rearranged form of the equation described by Lakowicz (35) (Equation 1) to give the final anisotropy measurements (robs).
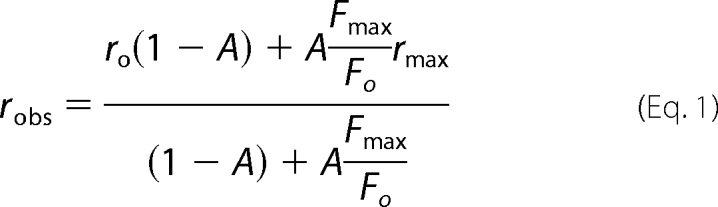 |
In Equation 1, robs is the intensity corrected anisotropy, A is fractional saturation ([PL]/n[P]o) of n equivalent and independent sites on the probe-labeled protein (P) by an unlabeled ligand (L) obtained from fitting the quadratic binding equation to the data, Fmax/Fo is the ratio of the maximum fluorescence intensity at saturation to the initial intensity (Fo) of the probe protein alone. Before fitting Equation 1, the quadratic binding equation was fit to the intensity titration expressed as (Fobs − Fo)/Fo = ΔFobs/Fo, where ΔFmax/Fo obtained from the fit is (Fmax/Fo) − 1.
[5F]FFR-NotD was titrated with FV or FVa in the presence or absence of 50 μm PC/PS SUVs with excitation at 500 nm and emission at 521 nm and band passes ranging from 4 to 10 nm depending on [5F]FFR-NotD concentration. Measurements of robs and total intensity as a function of total FV or FVa concentrations were analyzed by the quadratic binding equation (36) to obtain the dissociation constant (KD ligand), maximum fluorescence intensity (Fmax/Fo), stoichiometric factor (n), initial (ro), and maximal (rmax) anisotropies, and the change in anisotropy (Δr = rmax − ro). [OG]EGR-FXa (10 nm) was titrated with FV in the presence of 350 μm PC/PS/PE SUVs. Changes in anisotropy were measured with 488 nm excitation and 520 nm emission with 4- and 8-nm band passes, respectively. Data were analyzed as described above.
Competitive binding of [5F]FFR-NotD and active NotD to FVa was performed by titrating [5F]FFR-NotD (9 nm) with FVa in the absence and presence of two fixed concentrations of native NotD (9 and 102 nm). The three curves were analyzed simultaneously by nonlinear least squares fitting of the cubic equation (36) for tight competitive binding to obtain the dissociation constant for FVa binding to [5F]FFR-NotD (KD ligand) and for FVa binding to native NotD (KD competitor), maximum fluorescence intensity (Fmax/Fo), initial (ro) and maximal (rmax) anisotropies, the change in anisotropy (Δr = rmax − ro) and stoichiometry (m) for active NotD binding to FVa. To determine the effect of NotD on binding of FXa to FVa in the presence of SUVs (50 μm PC/PS), [OG]EGR-FXa (1 nm) was titrated with FVa in the absence and presence of two different concentrations of active site-blocked FFR-NotD (1.1 and 4.4 nm). Changes in anisotropy were measured and analyzed as described above. The solution of the cubic equation for tight competitive binding also yields A = [PL]/n[P]o (36), allowing Equation 1 to be applied to correct anisotropy data for fluorescence intensity changes. Nonlinear least squares fitting was performed in SCIENTIST (MicroMath) and error estimates represent the 95% confidence interval.
Kinetics of ProT Activation by the NotD·FVa or FXa·FVa Complex
Initial rates of native ProT or ProTR271Q activation by NotD or FXa with FVa in the presence or absence of PC/PS LUVs were expressed as the concentration of thrombin or meizothrombin, respectively, formed with time, determined from the initial rates of hydrolysis of 100 or 200 μm d-Phe-l-pipecolyl-l-Arg-pNA (FPR-pNA) measured at 405 nm. All rates of substrate hydrolysis experiments were normalized to a 1-cm path length by a correction factor of 1.46 for reaction volumes of 300 μl. Initial reference rates were measured for hydrolysis of 100 μm FPR-pNA by 1 nm thrombin or 200 μm FPR-pNA by 1 nm ecarin-generated meizothrombin from ProTR271Q. Reaction mixtures containing FVa (20 nm), ProT (0–1 μm), and 30 μm PC/PS LUVs in Buffer A were equilibrated at 25 °C for 10 min before initiation by addition of FXa (1 pm) or NotD (8.5 pm). Reactions with FXa contained 2 mg/ml of BSA, and reactions with NotD did not. The reactions were quenched by the addition of 10:1 (v/v) Buffer A and 50 mm HEPES, 125 mm NaCl, 1 mg/ml of PEG 8000, 125 mm EDTA, 2 mg/ml of soybean trypsin inhibitor (type IIS; Sigma), pH 8.2 (Buffer B), and the assays were initiated by addition of substrate. Rates of substrate hydrolysis were measured in triplicate in PEG 20,000-coated microtiter plates in a microtiter plate reader and converted to thrombin or meizothrombin formed with time using the reference rates described above. Reactions in the absence of LUVs were performed similarly with 50 pm NotD and quenched with Buffer B containing 500 mm EDTA.
To determine KD(app) for FVa binding in either the prothrombinase or NotD·FVa complexes, conditions were as described, except that reaction mixtures containing FVa (0–2 nm), PC/PS LUVs (30 μm), and ProTR271Q (1 μm) in Buffer A were equilibrated at 25 °C for 10 min before initiation by addition of 10 pm FXa or NotD. Quenching was done with Buffer B containing 300 mm EDTA and 2.5 mg/ml of soybean trypsin inhibitor. Reactions in the absence of LUVs were performed as described above using FVa (0–20 nm) and ProTR271Q (1 μm).
Inhibition of Thrombin Generation in Plasma by FFR-NotD
The effect of FFR-NotD on thrombin generation in normal human plasma was measured by calibrated automated thrombography at 37 °C (37). Assays were performed in 96-well plates coated with PEG 20,000. Corn trypsin inhibitor (50 μg/ml; Sigma) and 415 μm fluorogenic substrate, Cbz-Gly-Gly-Arg-7-amido-4-methylcoumarin (Diagnostica Stago), were added to human plasma (0.5% of total volume) (38). For each experiment two sets of readings were collected, one set for the thrombin generation reactions and the second set for the calibration reactions. During thrombin generation, the fluorescence change accompanying substrate hydrolysis was calibrated to correct for inner filter effects and substrate depletion. The plasma mixture (80 μl) was added to each well. For thrombin generation reactions, 20 μl of Tyrode buffer, pH 7.4, containing various final concentrations of FFR-NotD, 25 μm PC/PS SUVs, and either 1.6 pm tissue factor (TF) or 2 nm FXa (20 μl) were added to the wells. For calibration reactions, 20 μl of calibrator (thrombin-α2-macroglubulin; Diagnostica Stago) was added to the wells. The thrombin generation and calibration reactions were initiated by automatic dispensing of 20 mm HEPES, 100 mm CaCl2, and 60 mg/ml of BSA, pH 7.4 (20 μl), to a final reaction volume of 120 μl (1.5-fold plasma dilution). Fluorescence measurements (excitation at 390 nm and emission at 460 nm) were collected for 50 min and all measurements were done in triplicate. Data were analyzed with ThrombinoscopeTM software to obtain the endogenous thrombin potential (ETP; the integrated area under the thrombogram), peak height (maximum concentration of thrombin generated), and the lag time (time to the beginning of thrombin generation). To confirm the role of FV/FVa in binding to FFR-NotD, additional FV (25 nm) was added to normal plasma.
RESULTS
NotD Homology to FXa
Sequence alignment of NotD and α-FXa showed 55% identity and 71% conservation in the heavy chain and 53% identity and 76% conservation in the light chain (supplemental Fig. S1) (39). The COOH terminus of the NotD heavy chain ends at Lys245, similar to an autocatalytic α-FXa cleavage site (40). Secondary structure prediction showed a high degree of structural similarity between NotD and FXa (not shown). NotD has moderate conservation with FXa in exosite II (61%), involved in FVa binding (41–45), and exosite I (64%) (46). The 27-residue activation peptide of NotD is only 27% conserved with the 52-residue activation peptide of FXa. NotD contains a 9-residue insertion, FVPPNYYYV, after Lys90 in the Gly82–Arg116 loop of FXa. These comparisons support the classification of NotD as a structural homolog of FXa.
NotD Activity in the Presence of FV, FVa, and FVa Activation Fragments
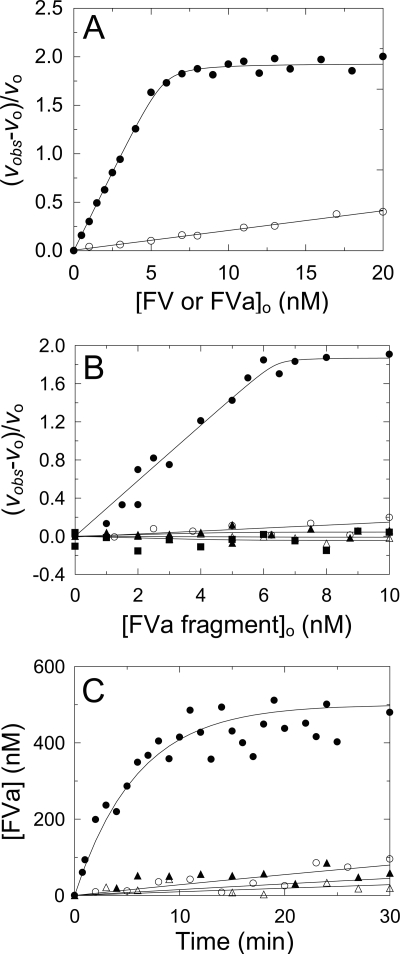
Previous studies showed that NotD exhibits an increase in the rate of CH3SO2-LGR-pNA hydrolysis by bovine FVa (18). On this basis, kinetic studies using the same substrate were performed to determine the effect of human FV and FVa on the activity of NotD. FVa titrations of NotD activity showed a saturating dependence with a stoichiometry of 1.17 ± 0.07 mol of FVa/mol of NotD, a KD ∼0.08 nm, and a 2.9 ± 0.1-fold maximum increase in the rate of CH3SO2-LGR-pNA hydrolysis (Fig. 1A). By contrast, titrations of NotD activity with FV were linear and much lower. As will be shown, the low activity of NotD was not due to low affinity of FV for NotD, as the KD for FV binding to active site-labeled NotD was 12 nm. The same tripeptide-pNA substrate was chosen for these experiments and further studies because it has a >5 mm Km, which means that the assays done at 200 μm (<0.04 × Km) reflect the specificity constant (kcat/Km), and substrate concentration did not affect the activity titrations (not shown). Comparison of the initial slopes of the FVa and FV titrations showed that ∼7% contamination of FV with FVa would be required to account for the difference, which was not detected by SDS-PAGE. It was also observed that in NotD activity titrations with FV, the rates curved upward slightly, suggesting slow activation of FV by NotD. This was taken into account in experiments with FV by fitting a parabola to the initial part of the progress curves and taking the first-order term as the initial velocity. An array of tripeptide-pNA substrates was screened with NotD in the absence and presence of a small excess of FVa with the following fold-increases in the initial rates of hydrolysis: MeO-d-cyclohexylglycyl-Gly-Arg-pNA, 4.0; d-(γ-carbobenzoxyl)-Lys-Pro-Arg-pNA, 4.3; benzyloxycarbonyl-d-Arg-Gly-Arg-pNA, 4.1; l-pyro-Glu-Pro-Arg-pNA, 1.7; and FPR-pNA, 2.7.
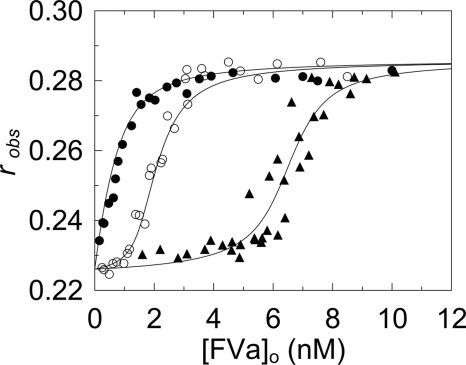
FIGURE 1.
Effect of FV, FVa, and FV activation products, and exosite I and II ligands on FV activation measured with NotD. A, fractional change in initial velocity, (vobs − vo)/vo, of 200 μm CH3SO2-LGR-pNA hydrolysis by 4.8 nm NotD as a function of total concentration of FVa (●) or FV (○) ([FVa or FV]o). The solid line in A represents the least squares fit by the quadratic equation with the parameters given under “Results.” B, initial rates (vobs) of 200 μm CH3SO2-LGR-pNA hydrolysis by 5 nm NotD as a function of the total concentration of FV activation products ([FVa fragment]o), Va-h·Va-l (●), Va-l (○), Va-h (▴), B1019–1545 (△), and B710–1018 (■). C, progress curves of 500 nm FV activation by 0.2 nm thrombin ([FVa]) determined from a stoichiometric titration of the fractional change in rate as in A, in the absence (●) or presence (○) of 100 nm exosite II aptamer, 10 μm Hir54–65(SO3−) (▴), or 1 μm exosite I aptamer (△). The solid line for the reaction in the absence of inhibitors represents the nonlinear least squares fit to the data by a single exponential and the solid lines for the inhibitors represent the linear least squares fits to the data. Activity measurements were performed and analyzed as described under “Experimental Procedures.”
To determine which of the thrombin-catalyzed FV activation terminal products was responsible for the rate increase, activity titrations with the isolated products were performed. Titrations of NotD with the purified two-subunit form of FVa, FVa-h, FVa-l, and fragments B710–1018 and B1019–1545 showed that the minimum FVa terminal activation product that enhanced NotD activity was the two-subunit form of FVa. Neither the isolated subunits, nor the other activation products had significant effects on NotD activity (Fig. 1B). FXa requires both subunits of FVa for binding (47), which was also the case for NotD.
The roles of thrombin exosites I and II in FV activation by thrombin, measured by the increase in the initial rate of NotD substrate hydrolysis in the linear portion of the stoichiometric titration (Fig. 1A), were assessed from the effects of previously characterized exosite-specific ligands (Fig. 1C). The exosite I DNA aptamer, Hir54–65(SO3−), and exosite II DNA aptamer inhibited the rate of FV activation by ≥92%. These results were compatible with previous findings that exosite I is involved in the initial cleavage of FV at Arg709, that both exosites are involved in cleavage at Arg1545, and the specificity of NotD for the two-subunit form of FVa, which requires the cleavage at Arg1545 to generate FVa-l (47).
Progress curves for FV activation by thrombin were compared with FV activation by RVV-V, which cleaves selectively only at Arg1545, yielding FVa-l with the remainder of the B domain connected to FVa-h (13–16) (supplemental Fig. S2). Analysis of these progress curves as single exponentials showed slower activation of FV by RVV-V. Interestingly, using thrombin-activated FV for the reference titration, RVV-V had a maximum amplitude ∼19% lower than that obtained with thrombin (supplemental Fig. S2C).
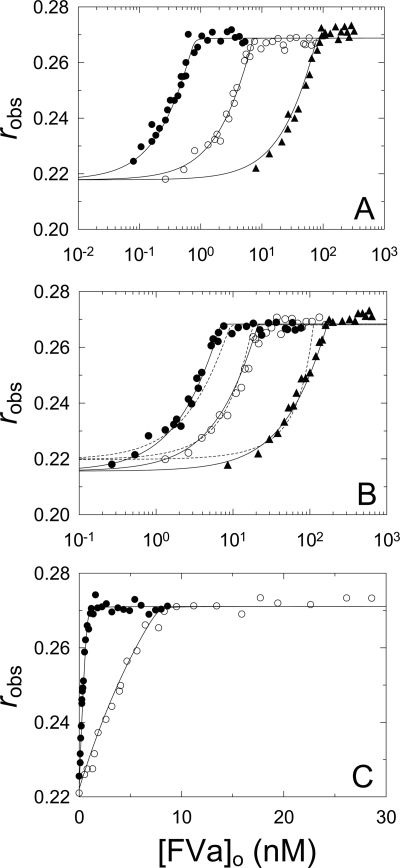
NotD Binding to FVa
Equilibrium binding studies were performed to quantitate NotD binding to FVa. Fluorescence anisotropy titrations with FVa were performed at three [5F]FFR-NotD concentrations, and simultaneous analysis of the data revealed that FVa bound tightly to NotD with KD of ≤ 10 pm, stoichiometry of 0.79 mol of FVa/mol of [5F]FFR-NotD, and a change in anisotropy (Δr) of 0.05 (Fig. 2A, Table 1). To quantitate binding of native NotD to FVa, [5F]FFR-NotD was used as a probe in competitive binding titrations with FVa and native NotD. Titrations of [5F]FFR-NotD with FVa were done in the absence and presence of two fixed concentrations of native NotD (Fig. 2B). Addition of native NotD did not affect the anisotropy of [5F]FFR-NotD. Fitting the titrations simultaneously by the competitive cubic binding equation gave a dissociation constant of <40 pm and a stoichiometric factor of 1.6 mol of native NotD/mol of FVa, with other parameters similar to those for [5F]FFR-NotD binding to FVa (Table 1). These results indicated indistinguishable affinities of native and [5F]FFR-NotD for FVa.
FIGURE 2.
[5F]FFR-NotD binding to FVa in the absence and presence of phospholipid vesicles. A, measured anisotropy (robs) as a function of total FVa concentration ([FVa]o) at 1 (●), 9 (○), and 100 nm (▴) [5F]FFR-NotD. Solid lines represent the simultaneous fit by the quadratic equation with the parameters given in Table 1. B, [5F]FFR-NotD (9 nm) in the absence (●) or presence of 9 (○) or 102 nm (▴) native NotD as a function of total FVa concentration ([FVa]o). The stoichiometric factors (n and m) were fit (solid lines) or fixed at 1 (dotted lines). Solid lines represent the simultaneous fit by the cubic binding equation with the parameters given in Table 1. C, titrations as in A in the presence of 50 μm PC/PS SUVs at 1 (●) and 9 nm (○) [5F]FFR-NotD. Solid lines represent the simultaneous fit by the quadratic binding equation with the parameters given in Table 1. Anisotropy titrations were performed and analyzed as described under “Experimental Procedures.”
TABLE 1.
Parameters for binding of FV and FVa to [5F]FFR-NotD and [OG]EGR-FXa
Dissociation constants (KD ligand), stoichiometric factors (n), initial anisotropy (ro), maximum anisotropy (rmax), and maximum fluorescence intensity (Fmax/Fo) are listed from direct titrations of [5F]FFR-NotD or [OG]EGR-FXa with FV or FVa. Experiments were performed in either the absence or presence of 50 μm PC/PS, 350 μm PC/PS/PE, or 50 μm PC/PS/PE SUVs. Results of the competitive binding studies for the indicated ligand and competitor were analyzed by simultaneous fitting of the titration in the absence and presence of two fixed concentrations of competitor to obtain the dissociation constant for the ligand (KD ligand) and competitor (KD competitor), with the stoichiometric factor for the ligand interaction (n) and competitor interaction (m) experimentally determined. For analysis of [OG]EGR-FXa binding to FV and [OG]EGR-FXa competition with FFR-NotD for FVa, n was fixed at 1. Experimental error in the parameters represents ± 2 S.D. Binding studies were performed and the data were analyzed as described under “Experimental Procedures.”
| Probe | Ligand | Competitor | SUVs | KD ligand | n | KD competitor | m | ro | rmax | Fmax/Fo |
|---|---|---|---|---|---|---|---|---|---|---|
| nm | mol/mol | nm | mol/mol | |||||||
| [5F]FFR-NotD | FVa | 0.002 ± 0.01 | 0.79 ± 0.02 | 0.22 ± 0.002 | 0.27 ± 0.008 | 1.38 | ||||
| [5F]FFR-NotD | FVa | Native NotD | 0.002 ± 0.04 | 0.76 ± 0.06 | 0.002 ± 0.04 | 1.6 ± 0.01 | 0.22 ± 0.002 | 0.27 ± 0.0008 | 1.34 | |
| [5F]FFR-NotD | FVa | +a | 0.01 ± 0.02 | 0.85 ± 0.06 | 0.22 ± 0.002 | 0.27 ± 0.001 | 1.43 | |||
| [5F]FFR-NotD | FV | 12 ± 1 | 0.79 ± 0.08 | 0.21 ± 0.002 | 0.27 ± 0.001 | 1.24 | ||||
| [5F]FFR-NotD | FV | +a | 0.22 ± 0.06 | 0.69 ± 0.06 | 0.22 ± 0.002 | 0.28 ± 0.001 | 1.31 | |||
| [OG]EGR-FXa | FV | +b | 1110 ± 250 | 1 | 0.23 ± 0.005 | 0.38 ± 0.001 | 1.06 | |||
| [OG]EGR-FXa | FVa | FFR-NotD | +a | 0.3 ± 0.2 | 1 | 0.011 ± 0.008 | 1.4 ± 0.08 | 0.23 ± 0.004 | 0.29 ± 0.004 | 1.50 |
| [OG]EGR-FXa | FVa | FFR-NotD | +c | 0.4 ± 0.3 | 1 | 0.007 ± 0.007 | 1.37 ± 0.07 | 0.23 ± 0.004 | 0.29 ± 0.004 | 1.54 |
| [OG]EGR-FXa | FVa | +c | 0.8 ± 0.5 | 1.26 ± 0.07 | 0.22 ± 0.004 | 0.28 ± 0.002 | 1.44 |
a 50 μm PC/PS.
b 350 μm PC/PS/PE.
c 50 μm PC/PS/PE.
To assess the effect of phospholipid vesicles on NotD binding to FVa, two concentrations of [5F]FFR-NotD were titrated with FVa in the presence of PC/PS SUVs (Fig. 2C). The dissociation constant was ∼11 pm, with a stoichiometry of 0.85 mol/mol, and a Δr of 0.05 (Table 1). The combined data showed an exceptionally high affinity of [5F]FFR-NotD for FVa that was not significantly affected by phospholipid vesicles (Table 1). Previous studies reported a KD of 0.7 nm for human FVa binding to [OG]EGR-FXa (48), indistinguishable from the KD of 0.8 ± 0.5 nm and the same Δr of 0.06 obtained in titrations of [OG]EGR-FXa in the presence of PC/PS/PE vesicles (supplemental Fig. S3, Table 1), and similar values have been determined (49–51). KD values of 0.8 and 2.7 μm for active site-labeled [rhodamine-X]EGR-FXa and [dansyl]EGR-FXa binding to FVa in the absence of phospholipid vesicles was determined for the bovine proteins by analytical ultracentrifugation (52) and fluorescence titrations (53), respectively. Compared with labeled human and bovine FXa, NotD binding to human FVa was enhanced ∼70-fold in the presence of phospholipid vesicles and 80,000–270,000-fold in their absence.
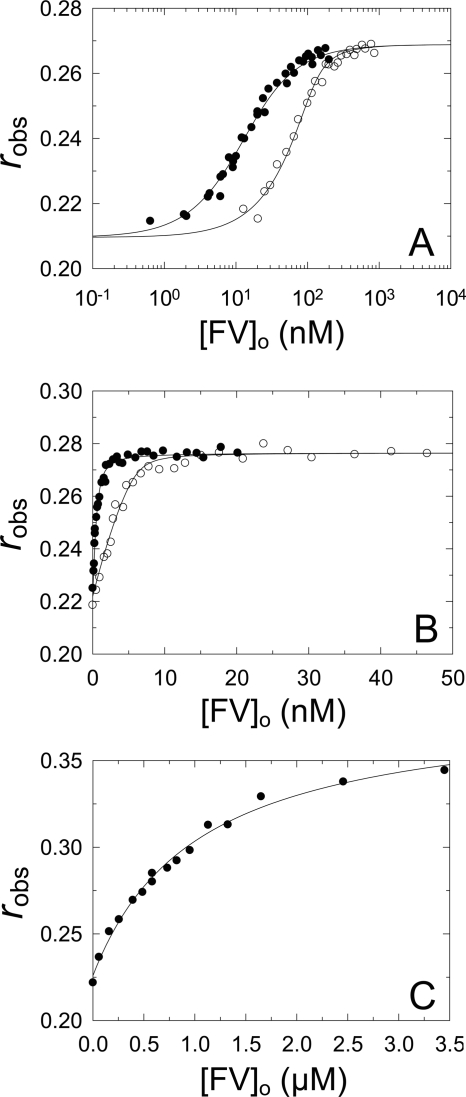
FV Binding to [5F]FFR-NotD and [OG]EGR-FXa
Similar anisotropy titrations were performed to quantitate binding of FV to [5F]FFR-NotD (Fig. 3A). Simultaneous analysis of the data collected at two fixed concentrations of [5F]FFR-NotD gave a KD of 12 nm and Δr of 0.06 (Table 1). Similar titrations were performed in the presence of PC/PS SUVs (Fig. 3B). The affinity of FV for [5F]FFR-NotD in the presence of SUVs was enhanced ∼55-fold, with a KD of 0.22 nm and Δr of 0.06 (Table 1). For comparison, [OG]EGR-FXa binding to FV in the presence of PC/PS/PE SUVs (Fig. 3C) gave a KD of 1.1 μm for FV binding with a Δr of 0.15 (Table 1). The results showed that in the presence of phospholipid vesicles there was ∼5,000-fold weaker affinity for FV binding to [OG]EGR-FXa compared with [5F]FFR-NotD.
FIGURE 3.
Binding of FV to [5F]FFR-NotD and [OG]EGR-FXa. A, observed anisotropy (robs) of 9 (●) or 150 nm (○) [5F]FFR-NotD as a function of total concentration of FV ([FV]o). B, observed anisotropy (robs) as in A in the presence of 50 μm PC/PS SUVs and 1 (●) or 9 nm (○) [5F]FFR-NotD. C, observed anisotropy (robs) of 10 nm [OG]EGR-FXa in the presence of 350 μm PC/PS/PE SUVs as in A. The solid line represents the fit by the quadratic binding equation with the parameters given in Table 1. Anisotropy titrations were performed and analyzed as described under “Experimental Procedures.”
Kinetics of ProT Activation by the NotD·FVa and Prothrombinase Complexes
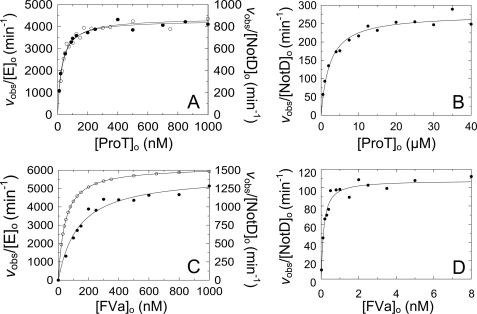
NotD activates bovine ProT by cleavages at Arg274 and Arg323 (18). Human ProT activation was measured from the rate of thrombin formation by the NotD·FVa complex or prothrombinase in the absence and presence of PC/PS LUVs and saturating FVa concentration. ProT activation by prothrombinase as a function of ProT concentration in the presence of PC/PS had a Km(app) of 31 ± 6 nm and kcat(app) of 73 ± 3 s−1 (Fig. 4A). Reactions under the same conditions with NotD-FVa showed an indistinguishable Km(app) of 27 ± 4 nm, but a 5-fold lower kcat(app) of 14.3 ± 0.4 s−1 (Fig. 4A). ProT activation by NotD-FVa in the absence of phospholipids had a Km(app) of 2.3 ± 0.6 μm and kcat(app) of 4.6 ± 0.2 s−1 (Fig. 4B). Similarly to FXa, NotD also activated ProT in the absence of FVa (not shown). The results showed that the NotD·FVa complex activated ProT in solution, but in the presence of phospholipid vesicles was more efficient with a ∼85-fold lower Km(app) and ∼3-fold higher kcat(app).
FIGURE 4.
Kinetics of ProT activation. A, initial rates of thrombin generation divided by the total enzyme concentration (vobs/[E]o; nm thrombin per min per nm Eo) for 8.5 pm NotD (●) or 1 pm FXa (○), 20 nm FVa, and 30 μm PC/PS LUVs as a function of total ProT concentration ([ProT]o). B, initial rates of thrombin generation as in A for 50 pm NotD and 20 nm FVa as a function of total ProT concentration ([ProT]o). C, initial rates as in A of 1 μm ProTR271Q activation by 10 pm NotD (●) or 10 pm FXa (○) in the presence of 30 μm PC/PS LUVs as a function of total FVa concentration ([FVa]o). NotD analysis includes FVa concentrations from 0 to 2 nm. D, initial rates as in A of 1 μm native ProT activation by 50 pm NotD as a function of total FVa concentration ([FVa]o). NotD analysis includes FVa concentrations from 0 to 20 nm. Solid lines represent the nonlinear least squares fits of the quadratic binding equation. Reactions were performed and analyzed as described under “Experimental Procedures.”
ProT activation by NotD and FXa was studied further as a function of FVa concentration in the presence or absence of PC/PS LUVs. To simplify this comparison, the single cleavage site mutant ProTR271Q was used, which can only be cleaved at Arg320 to yield meizothrombin. ProTR271Q activation in the presence of membranes by a limiting concentration of FXa and increasing FVa concentrations gave an KD(app) of 130 ± 32 pm and kcat(app) of 96 ± 7 s−1 (Fig. 4C). Similar reactions with NotD gave a ∼4-fold lower kcat(app) of 26 ± 2 s−1, but yielded a ∼3-fold tighter KD(app) of 44 ± 14 pm. The kinetic parameters for NotD-catalyzed native ProT activation in the absence of phospholipid vesicles were an KD(app) of 120 ± 30 pm for FVa and kcat(app) of 1.8 ± 0.5 s−1 (Fig. 4D). The results agreed with those of the equilibrium binding experiments, showing that FVa bound tightly to NotD even in the absence of membranes.
Competition of NotD with FXa for FVa Binding
Competitive binding experiments were performed to determine whether NotD binds to the FXa binding site on FVa. [OG]EGR-FXa was titrated with FVa in the presence of PC/PS SUVs at two fixed concentrations of FFR-NotD (Fig. 5). Simultaneous analysis of the anisotropy titrations gave KD for [OG]EGR-FXa and FVa of 0.3 nm and KD of 11 pm for FVa binding to FFR-NotD (Table 1). The KD of 11 pm was indistinguishable from that obtained for direct binding of FVa to [5F]FFR-NotD (Table 1). FVa binding to [OG]EGR-FXa had a Δr of 0.06 and Fmax/Fo of 1.5, which differed significantly from FV binding (Δr of 0.15 and Fmax/Fo of 1.06; Table 1), suggesting that the FV used in these studies did not contain traces of FVa. The results showed that NotD binds competitively with FXa for binding to FVa, providing evidence that the venom proteinase binds to the FXa binding site on FVa.
FIGURE 5.
Competitive binding of [OG]EGR-FXa and FFR-NotD to FVa. Observed anisotropy (robs) of 1 nm [OG]EGR-FXa in the absence (●) or presence of 1 (○) or 4 nm (▴) FFR-NotD and 50 μm PC/PS SUVs as a function of total FVa concentration ([FVa]o). Solid lines represent the simultaneous fit by the cubic binding equation with the parameters given in Table 1. Anisotropy titrations were performed and analyzed as described under “Experimental Procedures.”
Effect of FFR-NotD on Thrombin Generation in Human Plasma
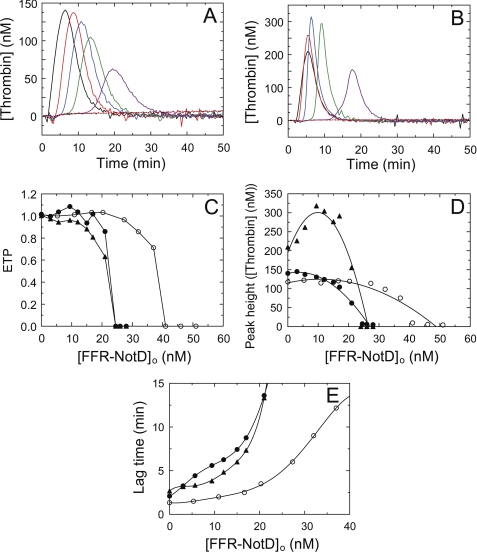
Calibrated automated thrombography was used to examine the effect of active site-blocked FFR-NotD on thrombin generation in human plasma. An abrupt decline in the ETP was observed when FFR-NotD concentrations were above 21 nm with either FXa or TF used as the trigger (Fig. 6C). The approximate concentration of FV in human plasma is 21–25 nm (54), which after correcting for dilution resulted in final FV concentrations of 14–17 nm. Thus, no detectable thrombin was generated when FFR-NotD concentrations exceeded 1.2–1.5-fold the concentration of FV in normal plasma. When 25 nm FV was added to normal plasma, the sharp decline in the ETP occurred at a higher concentration of FFR-NotD of 41 nm, 0.95–1.02-fold of the total FV concentration, which indicated that FFR-NotD affected thrombin generation by binding to FV or FVa (Fig. 6C). Increasing concentrations of FFR-NotD delayed thrombin generation as shown by the increase in lag time with both FXa and TF as triggers, which was shifted at high concentrations of FFR-NotD in the presence of 25 nm additional FV (Fig. 6E). Peak height decreased upon addition of FFR-NotD when FXa was used as a trigger (Fig. 6D). By contrast, when TF was used as the trigger, the lower concentrations of FFR-NotD (<20 nm) increased peak heights (Fig. 6D), yielded sharper peaks (Fig. 6B), and decreased lag times (Fig. 6E) without inhibiting the ETP.
FIGURE 6.
Effect of FFR-NotD on thrombin generation in normal plasma and plasma containing added FV. A, thrombin generation curves triggered by 2 nm FXa in the absence and presence of total concentrations of FFR-NotD ([FFR-NotD]o) at 0 (black), 5.7 (red), 12 (blue), 17 (green), 21 (purple), and 24.5 nm (dotted black), and 26 nm (dotted red). B, thrombin generation curves triggered by 1.6 pm TF as in A except 24 (dotted black) and 28 nm (dotted red) FFR-NotD concentrations were used. C, ETP as a function of total FFR-NotD concentration ([FFR-NotD]o). D, maximum thrombin concentration generated ([Peak height]) as a function of total FFR-NotD concentration ([FFR-NotD]o). E, lag time as a function of total FFR-NotD concentration ([FFR-NotD]o). Thrombin generation initiated with 2 nm FXa (●), 2 nm FXa with 25 nm FV added (○), or 1.6 pm TF (▴). Calibrated automated thrombography was performed as described under “Experimental Procedures.”
Inhibition of Prothrombinase by FFR-NotD
To confirm that FFR-NotD was a potent prothrombinase inhibitor, a kinetic titration of ProT activation at a low concentration (100 nm), accelerated by 5 nm FVa, was performed (supplemental Fig. S4). The results expressed as fractional inhibition were analyzed by fitting of the cubic equation for tight competitive binding of FXa and FFR-NotD to FVa. The results showed the anticipated initial plateau at concentrations of FFR-NotD <5 nm, followed by a near vertical drop to zero, corresponding to a stoichiometric factor of 1.07 ± 0.05 mol of FFR-NotD/mol of FVa and an indeterminately low dissociation constant.
DISCUSSION
The most significant conclusion of the present work is that NotD binds with extraordinarily high affinity to both human FV and FVa in the absence of membranes, with KD values of 12 nm and ≤ 10 pm, respectively. NotD, nonetheless, shows a ≥1,200-fold higher specificity for binding to FVa over FV. By contrast, active site-labeled bovine FXa, the only species for which the KD for FVa has been determined in the absence of membranes, binds with 0.8 or 2.7 μm dissociation constants, 80,000–270,000 lower affinity than NotD (52, 53). As shown here, human FXa achieves a ∼1,400-fold preference for binding to FVa compared with FV only in the presence of phospholipid membranes (48–50). [5F]FFR-NotD, FFR-NotD, and native NotD in the presence of membranes bind FVa with KD values of ≤10 pm, indistinguishable within the significant experimental error of such high affinity binding. Labeled NotD binding to FV is enhanced ∼55-fold (KD 0.22 nm) compared with the absence of membranes, reducing the difference in affinity of labeled NotD for FVa compared with FV to ∼22-fold. Collectively, the results suggest that NotD has evolved a largely membrane-independent specificity for FVa and high absolute affinity for free FV, in contrast to the stringent membrane dependence of the affinity of FXa for FV and FVa.
The results also support the conclusions that NotD binds to the FXa binding site expressed upon activation of FV, and that FVa allosterically affects the NotD active site. Initial circumstantial evidence shows that FXa has electropositive exosites in topologically similar locations as thrombin exosites I and II, of which FXa exosite II binds FVa (41). The sequence comparison with FXa indicates that NotD shares 61% sequence conservation with FXa exosite II, suggesting that this site may mediate NotD binding to FVa. Next, the specificity constant (kcat/Km) for NotD hydrolysis of CH3SO2-LGR-pNA is enhanced 3-fold and is correlated with NotD binding to FVa with high affinity in the absence of phospholipids (KD ≤ 10 pm). Six additional tripeptide substrates also reported FVa-enhanced NotD activity in the absence of membranes, indicating that unlike the active site of FXa, which is minimally affected (≤2-fold) by membrane-bound FVa in reactivity toward a tripeptide chromogenic substrate (55), the active site of NotD is allosterically regulated by FVa.
The observed increase in NotD activity requires the presence of both associated subunits of FVa and is consistent with binding to the FXa site on FVa as FXa has also been shown to require both FVa subunits for binding (47). Comparatively, NotD binding to FV is not associated with a significant change in chromogenic substrate activity. This implies that the NotD active site is affected by structural changes accompanying conversion of FV to FVa. Further evidence in support of allosteric regulation of NotD by FVa comes from RVV-V-activated FV, produced by a single cleavage at Arg1545 to generate the FVa-l subunit and FVa-h attached to the B domain (15, 16). Comparison of this form of FVa with thrombin-activated FVa showed a 19% lower stimulating effect on NotD activity, suggesting that NotD activity reports a structural difference between these FVa forms. Finally, competitive binding of FFR-NotD with [OG]EGR-FXa to FVa provides direct support for the conclusion that NotD binds to the FXa site on FVa, and thus is a probe of the FV/FVa structural and functional mechanisms modulating this essential binding site.
The NotD·FVa complex in the presence of phospholipids functions similarly to the prothrombinase complex, with an equivalent Km(app) of ∼30 nm and a 5-fold lower kcat(app) of 14.3 s−1. In the absence of membranes, the Km(app) of the NotD·FVa complex for ProT is increased ∼74-fold and has a 16-fold lower kcat(app) compared with prothrombinase. Initially, the kinetically determined KD(app) for assembly of NotD-FVa-membranes based on FVa titration of the rate increase of native ProT activation by NotD was 20 ± 10 pm, indistinguishable from the value of 30 ± 10 pm obtained for FVa and 1 pm FXa (not shown). Subsequently, the KD(app) was determined using the ProTR271Q mutant to simplify the activation mechanism to a single cleavage at Arg320 to form meizothrombin. Comparison of the FXa- and NotD-catalyzed reactions gave KD(app) values of 130 and 44 pm, respectively, showing a ∼3-fold tighter affinity for NotD compared with FXa. It should be recognized that dissociation constants obtained from kinetic experiments involving multiple interactions often cannot be unequivocally interpreted in terms of the binding equilibria of the different components involved. Bovine, and likely human, prothrombinase assembly is thermodynamically linked through membrane, FXa, and FVa interactions (2, 56). Whether this is the case for NotD is unknown, but such linkage may explain the high affinities observed with both native ProT and the mutant. Moreover, published KD values obtained from kinetic studies of ProT activation employing SUVs range from 0.083 to 3 nm whereas a value as low as 0.025 nm was reported for LUVs (43, 50, 56–58). LUVs were used here because SUVs contain few binding sites per vesicle for assembly of prothrombinase at low FXa concentrations and for ProT substrate delivery, resulting in slow assembly and rate-limiting ProT binding to prothrombinase (24, 56, 59, 60). In addition, kinetic titration of FXa with FVa measured by thrombin formation involves transition of the pathway of ProT activation from prethrombin 2-fragment 1.2 as the intermediate, to meizothrombin, which has activity measured with FPR-pNA similar to that of thrombin. On this basis, the kinetic titration using native ProT may or may not reflect the true KD. The change in pathway was not possible with ProTR271Q, but may have affected the values obtained with native ProT. Compared with the divergent estimates of FXa-FVa binding affinity inferred from kinetics, equilibrium binding studies with human [OG]EGR-FXa and FVa in the presence of membranes have reported more consistent KD values ranging from 0.7 to 4.3 nm with a mean of 1.6 nm (24, 48–50, 55). These considerations illustrate the difficulty of interpreting complex constants inferred from kinetic studies in terms of discrete molecular events, compared with equilibrium binding studies, which are less problematic. Further kinetic studies of ProT activation by NotD-FVa will be required to resolve some of these questions.
The potent anticoagulant effects of FFR-NotD in plasma involve at least two distinct modes of inhibition, distinguished in thrombin generation initiated by FXa compared with TF. Inhibition of FXa-initiated thrombin generation as a function of FFR-NotD concentration was fairly uniform in showing a progressive increase in lag time. This was followed by a precipitous decrease to zero in the maximum concentration of thrombin generated (peak height) and the peak area (ETP) at approximately equivalent concentrations of FFR-NotD to the concentration of FV in human plasma. The most likely mechanism for the total decrease in thrombin generation is FFR-NotD binding to thrombin-generated FVa in competition with FXa. This would result in the formation of a nonproductive FFR-NotD·FVa·membrane complex, facilitated by the high affinity of NotD for FVa compared with FXa. When the FV concentration in plasma was increased by addition of 25 nm purified FV to FXa-triggered thrombin generation, the lag time, peak height, and ETP were shifted to proportionally higher concentrations of FFR-NotD compared with untreated plasma. This is consistent with the competitive formation of inhibited prothrombinase as the underlying mechanism. The mechanism of inhibition was confirmed in a titration with FFR-NotD of the FVa-accelerated rate of ProT activation in the presence of membranes. The results paralleled the drop to zero in the ETP seen in the FXa-initiated plasma thrombin generation experiments as a function of FFR-NotD concentration, validating the mechanism of inhibition.
A target upstream of prothrombinase is presumably responsible for the increase in peak height at FFR-NotD concentrations below the abolishment of the ETP during TF-initiated thrombin generation. The FVIIa·TF complex activates FX and FIX and is inhibited by the tissue factor pathway inhibitor (TFPI) (61), which may be the target of FFR-NotD. The FVIIa·TF complex is inhibited by active site-dependent binding to the Kunitz-1 domain of TFPI (62, 63). TFPI acts as a slow, tight-binding inhibitor of FXa by the Kunitz-2 domain of TFPI, through a mechanism that enhances overall affinity from the nanomolar to picomolar range (62, 64). Rigorous kinetic studies of the TFPI mechanism of inhibition of FXa and the FVIIa·TF complex support the conclusion that the transiently formed product complex, FVIIa·TF·FXa is the primary target of TFPI inhibition, yielding a quaternary TFPI·FVIIa·TF·FXa complex (65, 66). This is based on the findings that FVIIa·TF binding to the TFPI Kunitz-1 domain occurs at a very rapid, near diffusion controlled rate, and that free FXa is rapidly inactivated by the high physiologic concentration of antithrombin (65, 66). Based on these studies, FXa in the product complex may be rapidly displaced by FFR-NotD as an alternate product, freeing FXa and inhibiting TFPI. Although this scenario may account for the observed effects of FFR-NotD on thrombin generation, it is not the only possibility, and will require further investigation to elucidate.
Fully active NotD in the venom of the mainland tiger snake is presumably necessary for defensive and/or predatory purposes. Envenomation of humans and other mammals by N. scutatus causes consumptive coagulopathy, hypotension, cerebral hemorrhage, severe fibrinogen depletion, and thrombocytopenia (67–70). This work demonstrates how the tiger snake has adapted NotD for extremely high affinity recognition of FV and FVa, and compared with prothrombinase, somewhat poor activity as a ProT activator in the absence and presence of membranes. The 5- and 16-fold lower catalytic rate constants for ProT activation by NotD compared with FXa in the presence and absence of membranes, respectively, is paradoxical. On the one hand, formation of the NotD·FVa complex can compete with prothrombinase and thereby, inhibit thrombin generation, but on the other hand, the unique features of NotD also allow the snake to bypass the requirement of membranes for high affinity FVa binding and ProT activation, usurping the blood coagulation system after envenomation.
Supplementary Material
Acknowledgments
We thank Dr. Heather K. Kroh for excellent technical assistance with the figures, Miranda Nolan for improving the purification procedure for NotD, and Christian Disney and Dr. Samrat Saha for performing some of the chromogenic substrate experiments. We thank Dr. Sriram Krishnaswamy for the dynamic light scattering measurements and for supplying the mammalian cells expressing ProTR271Q.
This work was supported, in whole or in part, by National Institutes of Health Grant HL038779 (to P. E. B.) from the NHLBI.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- ProT
- prothrombin
- NotD
- notecarin D
- pNA
- para-nitroanilide
- ProTR271Q
- recombinant ProT with Arg271 substituted with Gln
- Hir54–65(SO3−)
- Tyr63-sulfated hirudin 54–65
- FPR
- d-Phe-Pro-Arg
- FFR
- d-Phe-Phe-Arg
- EGR
- Glu-Gly-Arg
- ATA-FFR
- Nα-[(acetylthio)acetyl]-FFR
- ATA-EGR
- Nα-[(acetylthio)acetyl]-EGR
- RVV-V
- Russell viper venom factor V-activating proteinase
- FXa
- factor Xa
- TFPI
- tissue factor pathway inhibitor
- TF
- tissue factor
- FV
- factor V
- FVa
- factor Va
- ETP
- endogenous thrombin potential
- SUV
- small unilamellar vesicle
- LUV
- large unilamellar vesicle
- PS
- phosphatidylserine
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- FPR-pNA
- d-Phe-l-pipecolyl-l-Arg-pNA
- CH3SO2-LGR-pNA
- CH3SO2-d-Leu-Gly-Arg-pNA
- [5F]FFR-NotD
- NotD inactivated with Nα-[(acetylthio)acetyl]-d-Phe-Phe-Arg-CH2Cl and labeled with 5-(iodoacetamido)fluorescein
- [OG]EGR-FXa
- FXa inactivated with Nα-[(acetylthio)acetyl]-Glu-Gly-Arg-CH2Cl and labeled with Oregon Green 488-maleimide
- dansyl
- 5-dimethylaminonaphthalene-1-sulfonyl.
REFERENCES
- 1. Mann K. G., Jenny R. J., Krishnaswamy S. (1988) Annu. Rev. Biochem. 57, 915–956 [DOI] [PubMed] [Google Scholar]
- 2. Krishnaswamy S. (1990) J. Biol. Chem. 265, 3708–3718 [PubMed] [Google Scholar]
- 3. Krishnaswamy S., Jones K. C., Mann K. G. (1988) J. Biol. Chem. 263, 3823–3834 [PubMed] [Google Scholar]
- 4. Rosing J., Zwaal R. F., Tans G. (1986) J. Biol. Chem. 261, 4224–4228 [PubMed] [Google Scholar]
- 5. Krishnaswamy S., Mann K. G., Nesheim M. E. (1986) J. Biol. Chem. 261, 8977–8984 [PubMed] [Google Scholar]
- 6. Rosing J., Tans G., Govers-Riemslag J. W., Zwaal R. F., Hemker H. C. (1980) J. Biol. Chem. 255, 274–283 [PubMed] [Google Scholar]
- 7. Nesheim M. E., Taswell J. B., Mann K. G. (1979) J. Biol. Chem. 254, 10952–10962 [PubMed] [Google Scholar]
- 8. Krishnaswamy S., Church W. R., Nesheim M. E., Mann K. G. (1987) J. Biol. Chem. 262, 3291–3299 [PubMed] [Google Scholar]
- 9. Jenny R. J., Pittman D. D., Toole J. J., Kriz R. W., Aldape R. A., Hewick R. M., Kaufman R. J., Mann K. G. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 4846–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mann K. G., Kalafatis M. (2003) Blood 101, 20–30 [DOI] [PubMed] [Google Scholar]
- 11. Kane W. H. (2001) in Hemostasis and Thrombosis: Basic Principles and Clinical Practice (Colman R. W., Hirsh J., Marder V. J., Clowes A. W., Geroge J. N. eds) pp. 157–169, Lippincott, Williams and Wilkins, Philadelphia [Google Scholar]
- 12. Esmon C. T., Lollar P. (1996) J. Biol. Chem. 271, 13882–13887 [DOI] [PubMed] [Google Scholar]
- 13. Segers K., Dahlbäck B., Bock P. E., Tans G., Rosing J., Nicolaes G. A. (2007) J. Biol. Chem. 282, 33915–33924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thorelli E., Kaufman R. J., Dahlbäck B. (1998) Thromb. Haemost. 80, 92–98 [PubMed] [Google Scholar]
- 15. Kane W. H., Majerus P. W. (1981) J. Biol. Chem. 256, 1002–1007 [PubMed] [Google Scholar]
- 16. Kalafatis M., Beck D. O., Mann K. G. (2003) J. Biol. Chem. 278, 33550–33561 [DOI] [PubMed] [Google Scholar]
- 17. Broad A. J., Sutherland S. K., Coulter A. R. (1979) Toxicon 17, 661–664 [DOI] [PubMed] [Google Scholar]
- 18. Tans G., Govers-Riemslag J. W., van Rijn J. L., Rosing J. (1985) J. Biol. Chem. 260, 9366–9372 [PubMed] [Google Scholar]
- 19. Rao V. S., Joseph J. S., Kini R. M. (2003) Biochem. J. 369, 635–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jobin F., Esnouf M. P. (1966) Nature 211, 873–875 [DOI] [PubMed] [Google Scholar]
- 21. St Pierre L., Masci P. P., Filippovich I., Sorokina N., Marsh N., Miller D. J., Lavin M. F. (2005) Mol. Biol. Evol. 22, 1853–1864 [DOI] [PubMed] [Google Scholar]
- 22. Higgins D. L., Mann K. G. (1983) J. Biol. Chem. 258, 6503–6508 [PubMed] [Google Scholar]
- 23. Stewart J. C. (1980) Anal. Biochem. 104, 10–14 [DOI] [PubMed] [Google Scholar]
- 24. Orcutt S. J., Krishnaswamy S. (2004) J. Biol. Chem. 279, 54927–54936 [DOI] [PubMed] [Google Scholar]
- 25. Anderson P. J., Nesset A., Dharmawardana K. R., Bock P. E. (2000) J. Biol. Chem. 275, 16435–16442 [DOI] [PubMed] [Google Scholar]
- 26. Bock P. E. (1992) J. Biol. Chem. 267, 14974–14981 [PubMed] [Google Scholar]
- 27. Bock P. E., Craig P. A., Olson S. T., Singh P. (1989) Arch. Biochem. Biophys. 273, 375–388 [DOI] [PubMed] [Google Scholar]
- 28. Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chase T., Jr., Shaw E. (1969) Biochemistry 8, 2212–2224 [DOI] [PubMed] [Google Scholar]
- 30. Dharmawardana K. R., Bock P. E. (1998) Biochemistry 37, 13143–13152 [DOI] [PubMed] [Google Scholar]
- 31. Dharmawardana K. R., Olson S. T., Bock P. E. (1999) J. Biol. Chem. 274, 18635–18643 [DOI] [PubMed] [Google Scholar]
- 32. Bock P. E. (1993) Methods Enzymol. 222, 478–503 [DOI] [PubMed] [Google Scholar]
- 33. Panizzi P., Friedrich R., Fuentes-Prior P., Kroh H. K., Briggs J., Tans G., Bode W., Bock P. E. (2006) J. Biol. Chem. 281, 1169–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nicolaes G. A., Bock P. E., Segers K., Wildhagen K. C., Dahlbäck B., Rosing J. (2010) J. Biol. Chem. 285, 22890–22900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lakowicz J. R. (2006) Principles of Fluorescence Spectroscopy, 3rd Ed., Springer, New York [Google Scholar]
- 36. Bock P. E., Olson S. T., Björk I. (1997) J. Biol. Chem. 272, 19837–19845 [DOI] [PubMed] [Google Scholar]
- 37. Hemker H. C., Giesen P., Al Dieri R., Regnault V., de Smedt E., Wagenvoord R., Lecompte T., Béguin S. (2003) Pathophysiol. Haemost. Thromb. 33, 4–15 [DOI] [PubMed] [Google Scholar]
- 38. Kravtsov D. V., Matafonov A., Tucker E. I., Sun M. F., Walsh P. N., Gruber A., Gailani D. (2009) Blood 114, 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 40. Talbot K., Meixner S. C., Pryzdial E. L. (2010) Biochim. Biophys. Acta 1804, 723–730 [DOI] [PubMed] [Google Scholar]
- 41. Bock P. E., Panizzi P., Verhamme I. M. (2007) J. Thromb. Haemost. 5, Suppl. 1, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yegneswaran S., Mesters R. M., Griffin J. H. (2003) J. Biol. Chem. 278, 33312–33318 [DOI] [PubMed] [Google Scholar]
- 43. Rudolph A. E., Porche-Sorbet R., Miletich J. P. (2001) J. Biol. Chem. 276, 5123–5128 [DOI] [PubMed] [Google Scholar]
- 44. Rezaie A. R. (2000) J. Biol. Chem. 275, 3320–3327 [DOI] [PubMed] [Google Scholar]
- 45. Rezaie A. R., Kittur F. S. (2004) J. Biol. Chem. 279, 48262–48269 [DOI] [PubMed] [Google Scholar]
- 46. Page M. J., Macgillivray R. T., Di Cera E. (2005) J. Thromb. Haemost. 3, 2401–2408 [DOI] [PubMed] [Google Scholar]
- 47. Guinto E. R., Esmon C. T. (1984) J. Biol. Chem. 259, 13986–13992 [PubMed] [Google Scholar]
- 48. Buddai S. K., Layzer J. M., Lu G., Rusconi C. P., Sullenger B. A., Monroe D. M., Krishnaswamy S. (2010) J. Biol. Chem. 285, 5212–5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Betz A., Krishnaswamy S. (1998) J. Biol. Chem. 273, 10709–10718 [DOI] [PubMed] [Google Scholar]
- 50. Toso R., Camire R. M. (2004) J. Biol. Chem. 279, 21643–21650 [DOI] [PubMed] [Google Scholar]
- 51. Toso R., Camire R. M. (2006) J. Biol. Chem. 281, 8773–8779 [DOI] [PubMed] [Google Scholar]
- 52. Pryzdial E. L., Mann K. G. (1991) J. Biol. Chem. 266, 8969–8977 [PubMed] [Google Scholar]
- 53. Boskovic D. S., Giles A. R., Nesheim M. E. (1990) J. Biol. Chem. 265, 10497–10505 [PubMed] [Google Scholar]
- 54. Duckers C., Simioni P., Spiezia L., Radu C., Gavasso S., Rosing J., Castoldi E. (2008) Blood 112, 3615–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Buddai S. K., Toulokhonova L., Bergum P. W., Vlasuk G. P., Krishnaswamy S. (2002) J. Biol. Chem. 277, 26689–26698 [DOI] [PubMed] [Google Scholar]
- 56. Ye J., Esmon C. T. (1995) Biochemistry 34, 6448–6453 [DOI] [PubMed] [Google Scholar]
- 57. Nicolaes G. A., Tans G., Thomassen M. C., Hemker H. C., Pabinger I., Varadi K., Schwarz H. P., Rosing J. (1995) J. Biol. Chem. 270, 21158–21166 [DOI] [PubMed] [Google Scholar]
- 58. Bukys M. A., Blum M. A., Kim P. Y., Brufatto N., Nesheim M. E., Kalafatis M. (2005) J. Biol. Chem. 280, 27393–27401 [DOI] [PubMed] [Google Scholar]
- 59. Billy D., Willems G. M., Hemker H. C., Lindhout T. (1995) J. Biol. Chem. 270, 26883–26889 [DOI] [PubMed] [Google Scholar]
- 60. Lu Y., Nelsestuen G. L. (1996) Biochemistry 35, 8201–8209 [DOI] [PubMed] [Google Scholar]
- 61. Broze G. J., Jr. (1995) Annu. Rev. Med. 46, 103–112 [DOI] [PubMed] [Google Scholar]
- 62. Petersen L. C., Bjørn S. E., Olsen O. H., Nordfang O., Norris F., Norris K. (1996) Eur. J. Biochem. 235, 310–316 [DOI] [PubMed] [Google Scholar]
- 63. Broze G. J., Jr., Warren L. A., Novotny W. F., Higuchi D. A., Girard J. J., Miletich J. P. (1988) Blood 71, 335–343 [PubMed] [Google Scholar]
- 64. Huang Z. F., Wun T. C., Broze G. J., Jr. (1993) J. Biol. Chem. 268, 26950–26955 [PubMed] [Google Scholar]
- 65. Baugh R. J., Broze G. J., Jr., Krishnaswamy S. (1998) J. Biol. Chem. 273, 4378–4386 [DOI] [PubMed] [Google Scholar]
- 66. Lu G., Broze G. J., Jr., Krishnaswamy S. (2004) J. Biol. Chem. 279, 17241–17249 [DOI] [PubMed] [Google Scholar]
- 67. Tibballs J. (1998) Anaesth. Intensive Care 26, 536–547 [PubMed] [Google Scholar]
- 68. Tibballs J. (1998) Anaesth. Intensive Care 26, 529–535 [DOI] [PubMed] [Google Scholar]
- 69. Francis B., Williams E. S., Seebart C., Kaiser I. I. (1993) Toxicon 31, 447–458 [DOI] [PubMed] [Google Scholar]
- 70. Tibballs J., Henning R. D., Sutherland S. K., Kerr A. R. (1991) Med. J. Aust. 154, 275–276 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.