Background: Protein glutaminase (PG) catalyzes deamination of Gln residues in proteins.
Results: The structures of mature and pro forms and a pro form mutant reveal that the side chain of Gln-47 of mutant A47Q mimics the protein substrate of PG.
Conclusion: Gln-47 of A47Q forms an S-acyl covalent intermediate with the catalytic Cys.
Significance: PG shares a common catalytic mechanism with transglutaminase and cysteine protease.
Keywords: Enzyme Catalysis, Enzyme Mechanisms, Enzyme Mutation, Enzyme Structure, X-ray Crystallography, Covalent Intermediate, Cysteine Protease, Proenzyme, Protein Glutaminase, Transglutaminase
Abstract
Protein glutaminase, which converts a protein glutamine residue to a glutamate residue, is expected to be useful as a new food-processing enzyme. The crystal structures of the mature and pro forms of the enzyme were refined at 1.15 and 1.73 Å resolution, respectively. The overall structure of the mature enzyme has a weak homology to the core domain of human transglutaminase-2. The catalytic triad (Cys-His-Asp) common to transglutaminases and cysteine proteases is located in the bottom of the active site pocket. The structure of the recombinant pro form shows that a short loop between S2 and S3 in the proregion covers and interacts with the active site of the mature region, mimicking the protein substrate of the enzyme. Ala-47 is located just above the pocket of the active site. Two mutant structures (A47Q-1 and A47Q-2) refined at 1.5 Å resolution were found to correspond to the enzyme-substrate complex and an S-acyl intermediate. Based on these structures, the catalytic mechanism of protein glutaminase is proposed.
Introduction
Protein glutaminase (PG2; EC 3.5.1.44) can deamidate glutamine residues in proteins to glutamate residues, but not asparagine residues or free glutamines. The enzyme was discovered in culture supernatant from Chryseobacterium proteolyticum strain 9670 and purified by Yamaguchi and Yokoe (1). It was cloned and expressed in Escherichia coli (2) and investigated for its reactivity with substrate proteins such as α- and β-caseins and gluten (3), zein (4), and gliadin and α- and β-lactalbumins (5), including proteins exhibiting low solubility in aqueous solutions (6), to improve the quality of these food proteins. One of the potential applications of PG is to reduce the allergenicity of food proteins. The sequence Gln-Gln-Gln-Pro-Pro is considered the primary candidate for the IgE-binding epitope in wheat gluten (7). By reacting with PG, wheat glutens lose their allergenicity toward patients' sera in proportion to the degree of deamidation (4). PG was also demonstrated to be non-pathogenic and non-mutagenic (8).
Deamidation of proteins can improve their solubility, emulsifying activity, foaming activity, and other functional properties by increasing the number of negative charges and changing the overall structure (9). The chemical methods for protein deamidation, which include acid treatment, anion-catalyzed deamidation, dry heating under alkaline conditions, and thermal treatment, all cause collateral peptide bond cleavages (10, 11). Because of its selectivity and mildness, enzymatic protein deamidation is regarded in the food protein industry as a promising method to improve protein functionality (12). Although peptide glutaminase has been identified as an enzyme that deamidates short peptides, it had little activity against proteins (13, 14). PG can deamidate a wide range of substrates from peptides to proteins with low solubility.
Transglutaminase (TG), which is an important food-modifying enzyme, can also deamidate proteins in addition to catalyzing protein cross-linking with amine incorporation (15). TG catalyzes acyl transfer reactions in which the γ-carboxamide group of peptide-bound glutamine residues acts as the acyl donor and the ϵ-amino group of lysine residues acts as the acyl accepter. It deamidates proteins only in the absence of the acyl accepter.
Mature PG is a monomer enzyme consisting of 185 amino acid residues with a calculated molecular weight of 19,861 and is expressed as a non-active proprotein with a putative signal peptide of 21 amino acids and a prosequence of 114 amino acids. The amino acid sequence of the mature form has no obvious homology to any published sequence. No amino acid sequence motifs or domains defined in PROSITE were found (2). The solution structure of PG determined by NMR has reported recently (16).
Here, the high resolution structures of mature PG and pro-PG were determined by x-ray crystallography. The enzyme reaction intermediates were trapped by mutation of pro-PG Ala-47, which is located just above the catalytic site. We discuss the catalytic mechanism and substrate specificity of this enzyme based on these structures of mutant pro-PG.
EXPERIMENTAL PROCEDURES
Preparation of Mature PG
The expression and purification of mature PG from C. proteolyticum were performed as described previously (2).
Construction of the Pro-PG Expression System
An expression system for pro-PG was constructed in E. coli cells as follows. To introduce the insert C. proteolyticum prgA gene into the pET20b vector (Novagen), PCR was performed using KOD -Plus- polymerase (Toyobo) with previously manufactured (1) pBluescript vector (Stratagene) including the prgA gene as the template and two synthetic oligonucleotides as primers: cgtgcatATGGATTCCAACCGGGAATCAGG and ctcgctcgagAAATCCACAGCTGGATACAT (with the NdeI and XhoI sites underlined, respectively). The amplified pro-PG gene fragment was digested with NdeI and XhoI and then ligated with NdeI- and XhoI-digested pET20b. The accuracy of the gene amplification and cloning was confirmed by DNA sequencing. The resultant plasmid containing the gene for pro-PG and a C-terminal His6 tag was designated pET20b-prgA. pET20b-prgA was transformed into E. coli BL21(DE3) (Novagen) as a host cell for overexpression.
Site-directed Mutagenesis
Ala-47 was replaced with glutamine using a QuikChange site-directed mutagenesis kit (Stratagene) with pET20b-prgA as the template. The primers used were 5′-ATGTTAACTCAGCAATTCTAT-3′ and 5′-ATAGAATTGCTGAGTTAACAT-3′. E. coli BL21(DE3) cells were transformed with the mutant plasmid; the mutation was confirmed by DNA sequencing.
Expression, Processing, and Purification of Pro-PG
Wild-type pro-PG and the A47Q mutant were overexpressed in the cytoplasm of E. coli BL21(DE3) cells with a C-terminal His6 tag. For expression in E. coli, cells were aerobically cultured at 37 °C in LB medium supplemented with ampicillin (0.1 mg/ml). When the turbidity reached ∼0.5 at 600 nm, isopropyl β-d-thiogalactopyranoside (1 mm) was added to the culture, and the cells were further cultured at 20 °C for 18 h. The cultured cells were collected by centrifugation at 6000 × g for 15 min at 4 °C and washed with buffer containing 20 mm sodium phosphate, 10 mm imidazole, 0.5 m sodium chloride (pH 6.1). Cells were then resuspended in the same buffer and ultrasonically disrupted at 0 °C for 20 min at 9 kHz. The clear solution obtained after centrifugation at 15,000 × g for 20 min at 4 °C was used as the cell extract. The cell extracts were purified by nickel-immobilized metal affinity chromatography (Invitrogen) and eluted with 20 mm sodium phosphate and 0.3 m imidazole (pH 7.3).
Crystallization
Crystals of mature PG were grown by the hanging drop vapor diffusion method by mixing 5 μl of 34.0 mg/ml mature PG dissolved in 20 mm NaHEPES (pH 7.0) with 5 μl of reservoir solution (1 ml) composed of 1.2 m ammonium dihydrogen phosphate and 0.1 m sodium citrate (pH 5.6). Crystallization of pro-PG was performed by the sitting drop vapor diffusion method by mixing 2 μl of 20 mg/ml pro-PG dissolved in 20 mm NaHEPES (pH 6.5) with 2 μl of reservoir solution (0.7 ml) composed of 0.2 m ammonium citrate (pH 5.1) and 20% (w/v) PEG-3350. Crystallization of the A47Q mutant was carried out by the hanging drop vapor diffusion method in two ways. A47Q-1 was crystallized by mixing 5 μl of 20 mg/ml A47Q dissolved in 20 mm ammonium tartrate (pH 6.7) with 5 μl of reservoir solution (0.7 ml) composed of 0.2 m ammonium tartrate (pH 6.7) and 20% PEG-3350. A47Q-2 was crystallized by mixing 5 μl of 20 mg/ml A47Q dissolved in 20 mm sodium phosphate (pH 6.0) with 5 μl of reservoir solution (0.7 ml) composed of 0.2 m sodium tartrate (pH 8.6) and 20% (w/v) PEG-3350. All crystals were grown at 20 °C.
For phasing of mature PG, the crystal of mature PG was soaked in 2 mm Na[AuCl4] dissolved in reservoir solution for 10 min at 20 °C. Flash cooling was carried out in a nitrogen gas stream at 100 K after brief soaking of the crystals in 30% glycerol (mature PG and gold derivative) or 20% 2-methyl-2,4-pentanediol (pro-PG and A47Q). These crystals were stored in liquid nitrogen after a diffraction check with an in-house Bruker HI-STAR detector system.
Data Collection
All x-ray diffraction data were collected at 100 K under a nitrogen gas stream at beamline BL38B1 of the synchrotron facility SPring-8 (Hyogo, Japan) with approval from the Japan Synchrotron Radiation Research Institute (JASRI). All data were processed with the HKL2000 software package (17).
Structure Determination and Refinement
The gold sites were located with SHELXD (18), and single-wavelength anomalous diffraction phases to 3.0 Å resolution were calculated with SHELXE (19) using the data collected at the x-ray wavelength (1.0 Å) corresponding to the peak of the x-ray fluorescence spectrum. Two gold atom sites in the asymmetric unit were found. The improvement of the initial phases and peptide fragment modeling were performed using RESOLVE (20). The initial model was built manually with WinCoot (21) and refined with REFMAC5 from the CCP4 package (22). After repeated model rebuilding and refinement, the final model was refined using SHELXL (23) at 1.15 Å resolution. The B-anisotropy and hydrogen atoms except for the hydroxyl hydrogens of Tyr, Ser, and Thr were incorporated into the model. Finally, Rwork dropped to 0.0967 for all 72,390 data, and Rfree dropped to 0.1367 for all 3817 data. The estimated standard deviation plot was calculated using SHELXPRO (24).
The structure of pro-PG was determined by molecular replacement with PHASER (25) using the refined mature PG structure as the search model. The structures of pro-PG and its mutant were built with WinCoot and further refined with REFMAC5 and PHENIX (26). The structures of the two pro-PG mutants (A47Q-1 and A47Q-2) were refined with B-anisotropy. The statistics of the data collection and refinement are shown in Table 1. The secondary structure was assigned by DSSP (27). Superposition of structures was carried out by SSM Superpose implemented in WinCoot (28).
TABLE 1.
Statistics of data collection and refinement
The values of the highest resolution shells are in parentheses.
| Crystal |
|||||
|---|---|---|---|---|---|
| Mature/Gold | Mature | Pro-PG | A47Q-1 | A47Q-2 | |
| Data collection | |||||
| Device | SPring-8 BL38B1 | SPring-8 BL38B1 | SPring-8 BL38B1 | SPring-8 BL38B1 | SPring-8 BL38B1 |
| X-ray λ (Å) | 1.0 | 0.9 | 1.0 | 0.8 | 0.8 |
| Detector | Jupiter 210 CCD | R-AXIS V IP | Jupiter 210 CCD | Jupiter 210 CCD | Jupiter 210 CCD |
| Crystal system | Hexagonal | Hexagonal | Orthorhombic | Orthorhombic | Orthorhombic |
| Space group | P6522 | P6522 | P212121 | P212121 | P212121 |
| Cell length (Å) | a, b = 62.45, c = 185.84 | a, b = 62.31, c = 185.53 | a = 56.64, b = 103.29, c = 132.51 | a = 57.02, b = 104.10, c = 134.10 | a = 56.75, b = 103.74, c = 133.85 |
| Resolution (Å) | 50–2.32 (2.36–2.32) | 50–1.15 (1.19–1.15) | 50–1.73 (1.79–1.73) | 50–1.50 (1.55–1.50) | 50–1.50 (1.53–1.50) |
| Total reflections | 130,380 (1161) | 1,008,233 (41,726) | 533,046 (34,587) | 967,700 (43,806) | 944,033 (21,657) |
| Unique reflections | 17,536 (1691) | 76,489 (7451) | 80,701 (7359) | 125,864 (11,528) | 119,408 (6016) |
| Rmerge | 0.043 (0.058) | 0.058 (0.385) | 0.056 (0.314) | 0.037 (0.402) | 0.041 (0.332) |
| Completeness (%) | 99.5 (95.6) | 99.8 (99.8) | 98.2 (90.9) | 98.6 (91.5) | 93.5 (96.1) |
| Redundancy | 7.4 (6.6) | 13.2 (5.6) | 6.6 (4.7) | 4.3 (3.8) | 4.1 (3.6) |
| 〈I/σ(I)〉 | 54 (58.4) | 20.1 (5.9) | 19.6 (3.16) | 17.5 (2.7) | 21.4 (2.8) |
| Phasing | SADa | MR | |||
| Number of gold site | 2 | ||||
| Figure of merit | 0.78 | ||||
| Refinement | |||||
| Resolution (Å) | 50–1.15 (1.20–1.15) | 30.6–1.73 (1.749–1.730) | 19.9–1.502 (1.519–1.502) | 46.9–1.501 (1.520–1.501) | |
| Reflections | 76,207 (8499) | 76,594 (2302) | 125,753 (3431) | 113,344 (8232) | |
| Residues | |||||
| Amino acids | 185 | 563 | 560 | 560 | |
| Water | 448 | 722 | 907 | 838 | |
| Ligand | One Na+, one glycerol | Two citric acids | |||
| Rcryst | 0.097 (0.178) | 0.175 (0.214) | 0.140 (0.177) | 0.157 (0.180) | |
| Rfree | 0.137 | 0.206 (0.242) | 0.167 (0.208) | 0.181 (0.220) | |
| Average B-factor (Å2) | 19.0 | 29.6 | 28.5 | 26.6 | |
| r.m.s.d. bond (Å) | 0.030 | 0.007 | 0.005 | 0.008 | |
| r.m.s.d. angle | 0.032° | 1.067° | 1.020° | 1.091° | |
a SAD, single-wavelength anomalous diffraction; MR, molecular replacement; r.m.s.d., root mean square deviation.
Amino Acid Sequencing
The N-terminal protein sequence of mature PG was analyzed by Edman degradation in a pulsed liquid phase protein sequencer (PROCISE 492, Applied Biosystems) after blotting 5.1 nmol of PG onto a polyvinylidene difluoride membrane (ProSorb, Applied Biosystems). The amino acid sequences determined by the protein sequencer were compared with the protein sequences of PG in the NCBI Protein Database (BAB21508).
RESULTS
Structures Determined
We obtained four crystal structures of mature PG, pro-PG, and pro-PG mutants A47Q-1 and A47Q-2. The crystal structure of mature PG was determined by single-wavelength anomalous diffraction using a gold derivative and refined at 1.15 Å resolution. The crystal structures of pro-PG and its two mutants (A47Q-1 at pH 6.7 and A47Q-2 at pH 8.7) were refined at 1.73, 1.50, and 1.50 Å resolution, respectively. The data collection and refinement statistics are shown in Table 1.
The mature PG model is composed of 185 amino acid residues, 448 water molecules, one glycerol molecule, and one sodium ion. A Ramachandran plot analysis by PROCHECK (29) revealed that 91.4% of the residues are in the most favored regions and 8.6% in additionally allowed regions; there are no non-glycine residues in the disallowed regions. The estimated standard deviation plot of the final model shows that the absolute positional error is <0.04 Å in the most ordered region (B < 10 Å2) and <0.16 Å in the rest of the molecule (B < 60 Å2). Thus, we can discuss the precise features of the hydrogen bonds in the active site of mature PG usually within 0.1 Å deviation.
The asymmetric unit of crystals of pro-PG, A47Q-1, and A47Q-2 contains two monomers of 282, 280, and 280 amino acid residues with 722, 907, and 838 water molecules, respectively. Two citric acids are found in the pro-PG crystal. N-terminal residues 1–17 in pro-PG and 1–19 in the mutants are invisible because of disorder. Ramachandran plot analyses show that 92.7–92.8% of the residues are in the most favored regions, 6.7–6.8% in additionally allowed regions, and 0.4% (one residue/chain) in disallowed regions. This sole exception at Leu-45 is described below.
Overall Structure of Mature PG
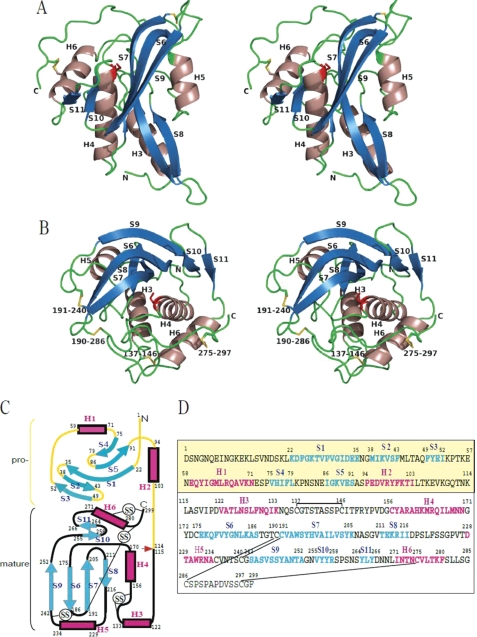
The structure of mature PG (residues 115–299) is composed of a central four-stranded antiparallel β-sheet (S6–S9) with four α-helices (H3–H6) and a two-stranded antiparallel β-sheet (S10-S11) that surrounds the central sheet (Fig. 1). The amino acid residue positions and the secondary structure elements are designated in common with pro-PG so that the N terminus of mature PG starts at residue 115 and the element from H3 (residues 122–133) (Fig. 1, B and D). The structure of mature PG is stabilized by four disulfide bonds of 11 cysteine residues (positions 137–146, 190–286, 191–240, and 275–297). The active site cleft is formed on one side of the central β-sheet together with H4, H6, and the other β-sheet. One glycerol molecule is found on this cleft.
FIGURE 1.
Overall structures of the mature PG molecule. A and B, ribbon diagrams drawn in left and right stereo from different directions. S–S bonds are drawn as yellow sticks with labeled residue numbers. The putative catalytic residue, Cys-156, is drawn as a red stick. These images were drawn using PyMOL Version 0.99. C, topology diagram of the secondary structures of mature PG and pro-PG. Boxes and arrows represent α-helices and β-strands, respectively. D, amino acid sequences of mature PG and pro-PG with assigned secondary structure elements. The first 114 residues in yellow belong to the proregion. α-Helices and β-strands are colored in red and blue, respectively.
A structural similarity search using the VAST server (30) suggested that PG does not belong to any reported superfamilies. The highest VAST structure similarity score is 8.4 with TG2 from Homo sapiens (Protein Data Bank code 2Q3Z) (31). This can be inferred from the similarity of their catalytic reactions, and the similarity is restricted only around the active center. The root mean square deviation is 1.21 Å for 62 Cα atoms, including only H4 and S7 of PG.
Active Cleft of PG
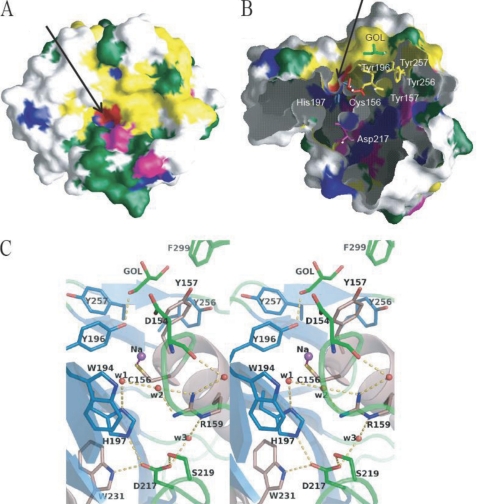
One shallow cleft (25 × 5 Å) is located on the protein surface surrounded by S6, S7, S10, S11, H4, H6, and loop 290–299 as shown in Fig. 1 (A and B). PG has several aromatic or hydrophobic amino acid residues (yellow and green in Fig. 2, A and B) on this cleft. In particular, five aromatic residues, Tyr-157, Tyr-196, Tyr-256, Tyr-257, and Phe-299, cover half of the cleft (Fig. 2B). In the other half of this cleft, there is one narrow pocket (4 × 4 × 6 Å) surrounded by S7, H4, and a loop before H4, which is connected to Cys-156 and His-197 in the bottom of the pocket indicated by the arrow in Fig. 2A. Thus, the cleft is divided into two parts: half of the aromatic region and the rest of the half of the catalytic pocket.
FIGURE 2.
Active cleft and pocket of mature PG. A and B, molecular surface models of PG drawn from different directions. The molecular surface is colored according to the amino acid residues: aromatic in yellow, hydrophobic in green, basic in blue, acidic in magenta, and Cys-156 in red. A is viewed from the same orientation as in Fig. 1B. The bound glycerol is colored green in B. The active pocket is indicated by arrows. These images were drawn using GRASP (40). C, stereoview of the active site of mature PG. The Four water molecules are indicated by red balls. Hydrogen bonds are designated as dashed yellow lines. This image was drawn using PyMOL.
One glycerol molecule, which had been added as cryoprotectant, was found to bind to the aromatic surface of the cleft of PG (Fig. 2, B and C). This glycerol forms C–C contacts with Asn-272, Leu-265, Phe-299, Tyr-157, Tyr-256, and Tyr-257, making a hydrogen bond with Tyr-196 (O1 … Oη of Tyr-196; 2.66 Å with an estimated deviation of 0.12 Å). The binding of glycerol suggests that this aromatic region is important for interaction with the protein substrate. The ability of PG to catalyze protein substrates with little solubility may be attributed to this aromatic region.
Catalytic Pocket
The putative catalytic residues of PG are Cys-156, His-197, and Asp-217 (Fig. 2C). This triad (Cys-His-Asp) is conserved in transglutaminases such as TG factor XIII (32, 33), human TG2 (31), and fish transglutaminase (34). A similar triad (Cys-His-Asn) is also shared by some cysteine proteases such as papain (35) and actinidin (36).
The present high resolution structure of mature PG reveals the precise conformation of the active site, which is shown in Fig. 2C. The side chain of Asp-217 is situated at a position where it can form hydrogen bonds with a water molecule (W3), His-197 Nϵ, Trp-231 Nϵ, Ser-219 Oγ, and Ser-219 nitrogen (supplemental Table S1). This indicates that the position of Asp-217 is strictly fixed in the active site. On the other hand, His-197 and Arg-159 form hydrogen bonds with W1 and W2, respectively. W1 and W2 form hydrogen bonds with each other (supplemental Table S1). These water molecules are situated 3.06 and 4.08 Å from Cys-156 Sγ, respectively, and are suggested to be involved in the enzyme reaction as described below.
There was a small spherical electron density in the vicinity of Cys-156 Sγ (2.17 Å apart), which is tentatively assigned to be a sodium ion (Fig. 2C). It is also within the ligand distance from W1 (supplemental Table S1). Although there has been no report on the metal-bound form of the active cysteine residue in transglutaminases, the presence of this metal ion represents the deprotonated thiol ion of Cys-156 in PG. The distances of Cys-156 Sγ from His-197 Nδ1 and Arg-159 Nη2 are 3.82 and 5.48 Å, respectively. This suggests that the positive charges of His-197 and Arg-159 decrease the pKa of Cys-156 to 3.9 as calculated using PROPKA (37) and increase its nucleophilic reactivity.
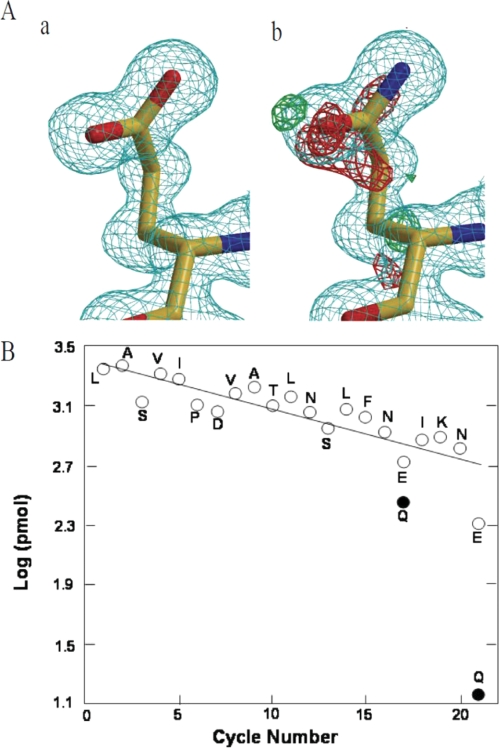
Self-deamidation
A previous study of PG suggested that some Gln residues were self-deamidated to Glu residues (2). The present 1.15 Å resolution model demonstrates that Gln-135 is converted to Glu based on the electron density maps (Fig. 3A). The direct amino acid sequencing also demonstrated that part of Gln-131 (cycle 17) and most of Gln-135 (cycle 21) have been converted to Glu (Fig. 3B). In the crystal structure of PG, both residues are located on the surface of the enzyme and thus may be easily self-deamidated.
FIGURE 3.
Self-deamidation of mature PG. A, comparison of the electron density maps at 1.15 Å resolution modeled as Glu (panel a) and Gln (panel b) for residue 135. The σ levels of the |Fo| − |Fc| maps are 3.0 (green) and −3.0 (red), respectively, and that of the 2|Fo| − |Fc| map is 1.2 (cyan). These images were drawn using MOLSCRIPT (41), BOBSCRIPT (42), and Raster3D (43). B, graph of the results of N-terminal sequencing of mature PG. The straight line shows the theoretical yield calculated with a repetitive yield of 0.978. The detected Glu and Gln residues are plotted with open and closed circles for Gln-131 (cycle 17) and Gln-135 (cycle 21), respectively.
Structure of Pro-PG
The overall structure of pro-PG shows that there is no noteworthy difference between the mature region of pro-PG and the mature PG structure (Fig. 4). The proregion is folded to form two helices (H1 and H2) and two three-stranded antiparallel β-sheets (S1/S5/S4 and S1/S2/S3) connected by long S1 and H1 located between S3 and S4. After S4, H2 and a long C-terminal loop continue to the N terminus of mature region at residue 115 (Fig. 1, B and C). The proregion belongs to the superfamily of bacterial enterotoxins based on the results of the VAST search. Heat-labile enterotoxin type IIb (Protein Data Bank code 1QB5) has the highest homology with a root mean square deviation of 1.77 Å for 76 residues from positions 19 to 94 of pro-PG.
FIGURE 4.
Structure of recombinant pro-PG. A, overall structure of pro-PG. The mature region is shown in gray, and the proregion is shown in rainbow colors. Cys-156 and Ala-47 are drawn as sticks. B, superposition of pro-PG (shown in purple) and mature PG (shown in green) drawn by Cα lines. C, interaction between the active site (gray) and a short loop between S2 and S3 in the proregion (cyan) drawn by stereo view. The hydrogen bonds are designated by dashed yellow lines. These images were drawn using PyMOL.
The proregion interacts with the mature region through several hydrogen bonds and many van der Waals contacts (supplemental Table S2). The solvent-accessible surface area of 1216 Å2 is buried by the interface between the proregion and mature region, which reaches 14.1% of the accessible surface of mature PG. The major contacts occur in the loop between S2 and S3 (residues 45–48) and the C-terminal loop (residues 105–111) of the proregion. It is especially remarkable that residues 45–49 cover and interact with the active site of mature PG. As shown in Fig. 4C and supplemental Table S2, Leu-45 oxygen, Ala-47 nitrogen, and Ala-47 oxygen form hydrogen bonds with Tyr-196 Oη, Asp-154 Oϵ2, and Ser-195 nitrogen, respectively. The side chain atoms of Thr-46 Oγ and Gln-48 Oϵ also form hydrogen bonds with Asp-154 Oδ1 and Ala-291 nitrogen, respectively. Additionally, Phe-49 nitrogen makes a water-mediated hydrogen bond with Ala-193 oxygen. A Ramachandran plot analysis shows that Leu-45 in the loop is in disallowed regions (φ and ψ = 72.8° and −59.7°). This distortion may be attributed to formation of a hydrogen bond between Leu-45 oxygen and Tyr-196 Oη. The position of glycerol found in mature PG is occupied by Leu-45. These results indicate that at least three or four residues with a near-extended form of substrate protein can be recognized by PG.
The side chain of Ala-47 is located in the entrance of the catalytic pocket, indicating that this proregion loop mimics a substrate protein bound to PG. Therefore, we substituted Ala-47 with a true substrate, glutamine, by site-directed mutagenesis.
Structures of A47Q-1 and A47Q-2
As a result of the crystallization screening of A47Q, two different structures of Gln-47 were obtained in different pH solutions and named A47Q-1 (pH 6.7) and A47Q-2 (pH 8.6).
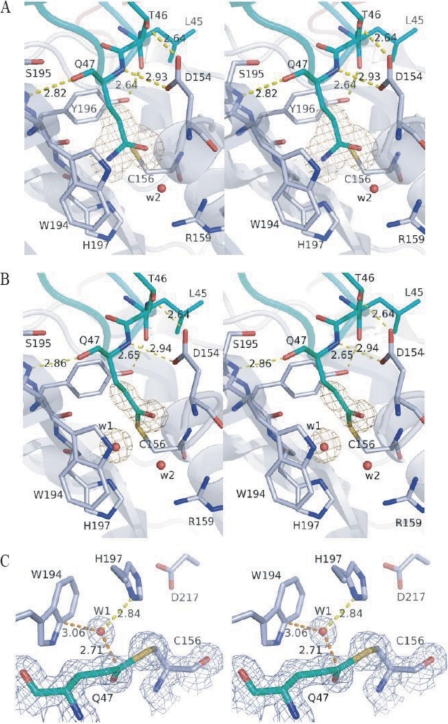
The structure model of A47Q-1 shows that the side chain of Gln-47 is inserted in the catalytic pocket just like a substrate (Fig. 5A). The position of Gln-47 is fixed by the hydrogen bonds formed between Gln-47 nitrogen and Asp-154 Oδ2 and between Gln-47 oxygen and Ser-195 nitrogen, as is the case for pro-PG. The side chain of Gln-47 is surrounded by the side chains of Cys-156, Trp-194, Tyr-196, His-197, Gly-155, and Asp-154. The distance between Gln-47 Cδ and Cys-156 Sγ is within the sum of van der Waals radii (3.0 Å). The distances from Gln-47 Nϵ to Trp-194 Cδ, Tyr-196 oxygen, and His-197 Nδ1 are 3.3, 2.9, and 3.2 Å, respectively, and those from Gln-47 Oϵ to Cys-156 nitrogen and Gly-155 nitrogen are 3.1 and 3.4 Å, respectively. The last two main chain nitrogen atoms form a oxyanion hole that stabilizes the tetrahedral intermediate, as found in serine and cysteine proteases. Thus, the A47Q-1 structure shows a strict fitness of the glutamine residue to the PG catalytic pocket just like the peptide main chain binds to cysteine proteases. The possibility that residue 47 of A47Q-1 is not Gln but Glu was examined by refinement after conversion, which showed that Gln is more suitable as judged by the |Fo| − |Fc| maps.
FIGURE 5.
Structures of A47Q mutants of pro-PG. The active sites of A47Q-1 (A) and A47Q-2 (B) are drawn in stereo view. The |Fo| − |Fc| omit maps calculated after changing their Gln-47 to Ala are shown in yellow mesh at 3.5σ. C, stereoview of the density of A47Q-2 with the 2|Fo| − |Fc| map of Gln-47 and Cys-156 shown in gray mesh at 1.2σ. These images were drawn using PyMOL.
The structure model of A47Q-2 is the same as that of A47Q-1 except for the deamidated Gln-47 residue (Fig. 5, B and C). The distance between Cys-156 Sγ and Gln-47 Cδ is 1.89 Å, so close that they nearly form a covalent bond (Fig. 5C). This A47Q-2 structure corresponds to the S-acyl intermediate formed after ammonia leaves the side chain of Gln-47. The superposition of A47Q-2 with A47Q-1 (Fig. 6A) shows that two water molecules (W1 and W2) found in mature PG exist also in the structure of A47Q-2. W1 is close to Gln-47 Cδ (2.62 Å) and makes a hydrogen bond with His-197 Nδ1 (2.81 Å) and Cys-156 Sγ (3.29 Å). In the structure of A47Q-1, W1 is replaced with Gln-47 Nϵ. W1 and W2 make hydrogen bonds with each other with distance of 3.0 Å. The side chain of Gln-47 in A47Q-2 rotates around χ2 (−20°) and χ3 (50.5°) from A47Q-1 to form the covalent intermediate. It is remarkable that the position of Gln-47 Oϵ1 is close to that of the presumed sodium ion found in mature PG (0.5 Å).
FIGURE 6.
A, stereoview of the superposition of A47Q-1 and A47Q-2 colored in pink and green, respectively. Water molecules are represented as pink balls (A47Q-1) and red balls (A47Q-2). B, stereoview of the superposition of the active site of A47Q-2 (colored in green) with that of TG2 (Protein Data Bank code 2Q3Z; colored in gray). Cys-156–His-197–Asp-217 of PG (green) corresponds to Cys-277–His-335–Asp-358 of TG2 (blue). These images were drawn using PyMOL.
DISCUSSION
Catalytic Mechanism of PG
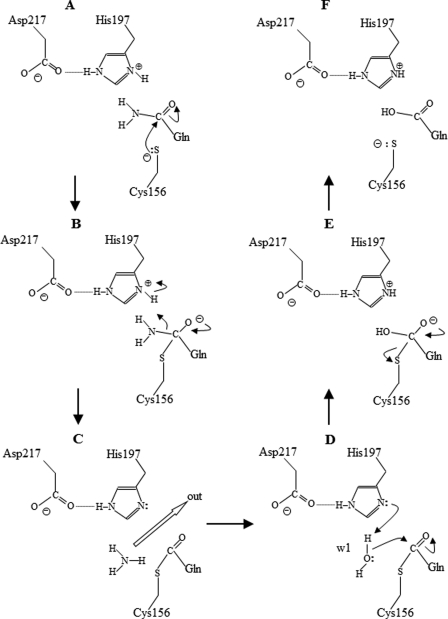
On the basis of the non-bonded distances of atoms in the active site pocket of mature PG (supplemental Table S1) and the structures of A47Q-1 and A47Q-2, we can postulate the catalytic mechanism of PG. As shown in Fig. 7A, nucleophilic Sγ of Cys-156 approaches Cδ of the amide group of the substrate glutamine within 3 Å distance from above the plane of the carboxamide group as found in the structure of A47Q-1 (Fig. 5A and supplemental Table S2). Nϵ2 of Gln-47 occupies the position of W1 (Fig. 6A). Cys-156 Sγ attacks Cδ of the substrate, and they form a tetrahedral intermediate as shown in Fig. 7B. The distances of Cδ-Oδ1 and Cδ-Oδ2 of Asp-217 in the mature PG structure are 1.28 and 1.24 Å, respectively, indicating a deprotonated (negatively charged) state of the side chain. The primary role of Asp-217 seems to be to activate the side chain of His-197 to a protonated (positively charged) state. The proton at His-197 Nδ1 can attack the tetrahedral intermediate to release an ammonium molecule and to form the S-acyl intermediate observed in A47Q-2 (Fig. 7C). Following the release of the ammonium, W1 attacks the S-acyl intermediate (Fig. 7D) to form a glutamate side chain, which is coupled by the regeneration of the positive charge of His-197. The conformational change in the side chain of A47Q, which flips to face its plane of the S-acyl group toward W1 in A47Q-2 from that of the carboxamide group toward Cys-156 Sγ in A47Q-1, is important for this step (Fig. 6A) because the reactivities of the carboxamide and S-acyl groups against the nucleophile are controlled by their lowest unoccupied molecular orbitals that are located on the carbon atom perpendicular to the O=C–N and O=C–S planes, respectively. The hydrolytic water found in the acyl-enzyme complex of porcine pancreatic elastase is located almost in the same arrangement of W1 in A47Q-2 (38). The final step is the cleavage of the S-acyl bond to form a free glutamate side chain (Fig. 7, E and F). The regained positive charge of His-197 accelerates the cleavage by stabilizing the carboxyl anion intermediate. The formation of the negatively charged glutamate side chain facilitates the release of the product from the catalytic pocket by an unfavorable interaction with the thiol anion of Cys-156 in the bottom of the pocket and with the carboxyl anion of Asp-154 at the entrance of the pocket.
FIGURE 7.
Postulated catalytic mechanism of PG. Gln refers to the residue of a substrate protein. The details in each step are elaborated under “Discussion.”
As the Gln-47 side chain of A47Q mutants of pro-PG is fixed and cannot be released from the catalytic pocket, the enzyme reaction ceased on the way of the whole reaction. The lower pH (pH 6.7) and the presence of ammonium ion in the crystallization medium of A47Q-1 prevent the release of ammonium from Gln-47, keeping the enzyme-substrate complex prior to nucleophilic attack (Fig. 7A). The lower pH decreases the nucleophilicity of Cys-156 Sγ. The side chain is fixed by three hydrogen bonds (Nϵ2–Tyr-196 oxygen, 2.9 Å; Oϵ1–Cys-156 nitrogen, 3.1 Å; and Oϵ1–Gly-155 nitrogen, 3.3 Å) except for the interaction with Cys-156 Sγ.
The S-acyl covalent intermediate of Gln-47 was attained using the crystallization medium of A47Q-2 in the absence of ammonium ion at higher pH (pH 8.6). This S-acyl intermediate seems relatively stable. A similar but artificial thioether adduct is also found in the complex of human TG2 with its peptide inhibitor (31). The higher pH may also suppress the nucleophilicity of His-197 so that the activation of W1 (which attacks the S-acyl intermediate) is lowered. It is interesting that the Bürgi-Dunitz angles found in A47Q-1 (Cys-156 Sγ–C=O, 92.9°) and in A47Q-2 (W1–C=O, 90.1°) are slightly different from that predicted to be optimal for nucleophilic attack of the carbonyl compounds (105 ± 5°) (39), suggesting that the trapped PG intermediates have insufficient orbital overlap to form tetrahedral intermediates.
Comparison with TG
The partial similarity between mature PG and human TG2 is restricted only around the active center. TG has longer intervening loops or domains among the secondary structure components compared with PG. However, the superposition of the catalytic Cys-His-Asp triads showed a good agreement (Fig. 6B). In PG and factor XIII-like TG, it is common that the catalytic Cys residue is located at the N terminus of an α-helix, His on a β-strand, and Asp on a loop. This commonality is also found in papain-like cysteine proteases without the folding analogy found in PG and TGs. Thus, PG is evolutionally related to TG, probably as an ancestor of TG, suggesting that the structure comparison of PG and TG provides information about their catalysis. As the structure of TG complexed with an acyl acceptor such as lysine has not yet been reported, it is worth mentioning that the position of W1 in the A47Q-2 structure (Fig. 6B) should be the same as that of Nζ of the lysine residue occupied in the cross-link reaction catalyzed by TG2.
Beside the triad, PG has Arg-159 as the holder of a water molecule (W2). The primary role of Arg-159 seems to be to decrease the pKa of Cys-156 Sγ. In contrast to PG, factor XIII-like TG does not have water molecules (W1 and W2) in the vicinity of catalytic Cys Sγ as judged from published Protein Data Bank data. In factor XIII-like TG, phenylalanine is in the position corresponding to Arg-197 in PG (Fig. 6B) with a wider cleft around this residue. This difference may account for the variation in enzyme reactions and implies that factor XIII-like TG activity is inclined to cross-link rather than deamidation.
Conclusion
In this study, we have reported, for the first time, the crystal structures of mature PG, pro-PG, and pro-PG mutants with two different reaction intermediates (A47Q-1 and A47Q-2). A47Q-1 and A47Q-2 provide the first examples of reaction intermediates (the enzyme-substrate complex and S-acyl covalent intermediate, respectively) with a natural substrate in PG and TG. On the basis of these structures, we could postulate the catalytic mechanism of this enzyme.
The overall folding of mature PG has a partial homology to factor XIII-like TG, especially its Cys-His-Asp catalytic triad. Comparison of the active site structures of PG and TG demonstrates their similarities and differences in enzyme reactions. Increased thermal or oxidative stabilities of this enzyme based on the present structural information are needed for industrial use of PG.
Supplementary Material
Acknowledgments
We thank Drs. K. Hasegawa, H. Sakai, and S. Baba (Japan Synchrotron Radiation Research Institute) for valuable help with data collection.
This work was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
The atomic coordinates and structure factors (codes 2ZK9, 3A54, 3A55, and 3A56) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PG
- protein glutaminase
- TG
- transglutaminase
- W
- water.
REFERENCES
- 1. Yamaguchi S., Yokoe M. (2000) Appl. Environ. Microbiol. 66, 3337–3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yamaguchi S., Jeenes D. J., Archer D. B. (2001) Eur. J. Biochem. 268, 1410–1421 [DOI] [PubMed] [Google Scholar]
- 3. Yong Y. H., Yamaguchi S., Matsumura Y. (2006) J. Agric. Food Chem. 54, 6034–6040 [DOI] [PubMed] [Google Scholar]
- 4. Yong Y. H., Yamaguchi S., Gu Y. S., Mori T., Matsumura Y. (2004) J. Agric. Food Chem. 52, 7094–7100 [DOI] [PubMed] [Google Scholar]
- 5. Gu Y. S., Matsumura Y., Yamaguchi S., Mori T. (2001) J. Agric. Food Chem. 49, 5999–6005 [DOI] [PubMed] [Google Scholar]
- 6. Zayas J. F. (1997) Functionality of Proteins in Food, pp. 6–75, Springer-Verlag, Berlin [Google Scholar]
- 7. Tanabe S., Arai S., Yanagihara Y., Mita H., Takahashi K., Watanabe M. (1996) Biochem. Biophys. Res. Commun. 219, 290–293 [DOI] [PubMed] [Google Scholar]
- 8. Scheuplein R. J., Mizutani A., Yamaguchi S. (2007) Regul. Toxicol. Pharmacol. 49, 79–89 [DOI] [PubMed] [Google Scholar]
- 9. Hamada J. S. (1992) J. Agric. Food Chem. 40, 719–723 [Google Scholar]
- 10. Riha W. E., 3rd, Izzo H. V., Zhang J., Ho C. T. (1996) Crit. Rev. Food Sci. Nutr. 36, 225–255 [DOI] [PubMed] [Google Scholar]
- 11. Schwenke K. D. (1997) in Food Proteins and Their Applications (Damodaran S., Paraf A. eds) pp. 393–424, Marcel Dekker, New York [Google Scholar]
- 12. Hamada J. S. (1994) Crit. Rev. Food Sci. Nutr. 34, 283–292 [DOI] [PubMed] [Google Scholar]
- 13. Hamada J. S., Shih F. F., Frank A. W., Marshall W. E. (1988) J. Food Sci. 53, 671–672 [Google Scholar]
- 14. Gill B. P., O'Shaughnessey A. J., Henderson P., Headon D. R. (1985) J. Food Sci. Technol. 9, 33–41 [Google Scholar]
- 15. Larre C., Chiarello M., Blanloeli Y., Chenu M., Gueguen J. (1993) J. Food Biochem. 17, 267–282 [Google Scholar]
- 16. Kumeta H., Miwa N., Ogura K., Kai Y., Mizukoshi T., Shimba N., Suzuki E., Inagaki F. (2010) J. Biomol. NMR 46, 251–255 [DOI] [PubMed] [Google Scholar]
- 17. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 18. Usón I., Sheldrick G. M. (1999) Curr. Opin. Struct. Biol. 9, 643–648 [DOI] [PubMed] [Google Scholar]
- 19. Sheldrick G. M. (2002) Z. Kristallogr. 217, 644–650 [Google Scholar]
- 20. Terwilliger T. C. (2002) Acta Crystallogr. D 59, 34–44 [Google Scholar]
- 21. Emsley P., Cowtan K. (2004) Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22. Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D 50, 760–763 15299374 [Google Scholar]
- 23. Sheldrick G. M., Schneider T. R. (1997) Methods Enzymol. 277, 319–343 [PubMed] [Google Scholar]
- 24. Sheldrick G. (1984) Acta Crystallogr. A 40, C440 [Google Scholar]
- 25. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 27. Kabsch W., Sander C. (1983) Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 28. Krissinel E., Henrick K. (2004) Acta Crystallogr. D 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 29. Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 30. Madej T., Gibrat J. F., Bryant S. H. (1995) Proteins 23, 356–369 [DOI] [PubMed] [Google Scholar]
- 31. Pinkas D. M., Strop P., Brunger A. T., Khosla C. (2007) PLoS Biol. 5, e327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yee V. C., Pedersen L. C., Le Trong I., Bishop P. D., Stenkamp R. E., Teller D. C. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7296–7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pedersen L. C., Yee V. C., Bishop P. D., Le Trong I., Teller D. C., Stenkamp R. E. (1994) Protein Sci. 3, 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Noguchi K., Ishikawa K., Yokoyama K., Ohtsuka T., Nio N., Suzuki E. (2001) J. Biol. Chem. 276, 12055–12059 [DOI] [PubMed] [Google Scholar]
- 35. Kamphuis I. G., Kalk K. H., Swarte M. B., Drenth J. (1984) J. Mol. Biol. 179, 233–256 [DOI] [PubMed] [Google Scholar]
- 36. Kamphuis I. G., Drenth J., Baker E. N. (1985) J. Mol. Biol. 82, 317–329 [DOI] [PubMed] [Google Scholar]
- 37. Bas D. C., Rogers D. M., Jensen J. H. (2008) Proteins 73, 765–783 [DOI] [PubMed] [Google Scholar]
- 38. Katona G., Wilmouth R. C., Wright P. A., Berglund G. I., Hajdu J., Neutze R., Schofield C. J. (2002) J. Biol. Chem. 277, 21962–21970 [DOI] [PubMed] [Google Scholar]
- 39. Bürgi H. B., Dunitz J. D., Shefter E. (1973) J. Am. Chem. Soc. 95, 5065–5067 [Google Scholar]
- 40. Nicholls A., Sharp K. A., Honig B. (1991) Proteins Struct. Funct. Genet. 11, 281–296 [DOI] [PubMed] [Google Scholar]
- 41. Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 946–950 [Google Scholar]
- 42. Esnouf R. M. (1999) Acta Crystallogr. D 55, 938–940 [DOI] [PubMed] [Google Scholar]
- 43. Merritt E. A., Murphy M. E. (1994) Acta Crystallogr. D 50, 869–873 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.