Background: Proteolytic enzymes perform post-translational, processing and digestion of proteins and peptides. Six catalytic types have already been recognized, all of them peptidases cleaving substrates by hydrolysis.
Results: A seventh catalytic type has now been identified and ten families have been assembled.
Conclusion: The newly identified enzymes are not hydrolases but lyases utilizing asparagine as a nucleophile.
Significance: Not all proteolytic enzymes are peptidases.
Keywords: Peptidases, Protease, Protein Processing, Proteolytic Enzymes, Viral Protease, Asparagine, Autotransporter, Catalytic Type, Intein, Lyase
Abstract
The terms “proteolytic enzyme” and “peptidase” have been treated as synonymous, and all proteolytic enzymes have been considered to be hydrolases (EC 3.4). However, the recent discovery of proteins that cleave themselves at asparagine residues indicates that not all peptide bond cleavage occurs by hydrolysis. These self-cleaving proteins include the Tsh protein precursor of Escherichia coli, in which the large C-terminal propeptide acts as an autotransporter; certain viral coat proteins; and proteins containing inteins. Proteolysis is the action of an amidine lyase (EC 4.3.2). These proteolytic enzymes are also the first in which the nucleophile is an asparagine, defining the seventh proteolytic catalytic type and the first to be discovered since 2004. We have assembled ten families based on sequence similarity in which cleavage is thought to be catalyzed by an asparagine.
Introduction
Proteolytic enzymes release amino acids, peptides, and proteins from larger peptides and proteins. The term has often been assumed to be synonymous with “peptidase” (and to encompass the terms “proteinase” and “protease”). Peptidases have been defined as “hydrolases acting on peptide bonds” (1). However, it has recently become clear that enzymes other than hydrolases exist that also cleave peptide bonds.
Proteolytic enzymes can be broadly grouped into six catalytic types based upon the nature of the nucleophile in the reaction. These six catalytic types have been applied only to peptidases. In 1993, Rawlings & Barrett (2) recognized four catalytic types: serine, cysteine, aspartic, and metallo, a concept that could be traced back to the ideas of Hartley (3). In serine and cysteine peptidases, the nucleophile is the hydroxyl on the side chain of the active site serine or the thiol on the side chain of the active site cysteine. In both aspartic and metallopeptidases, the nucleophile is a water molecule, which in aspartic peptidases is activated by two aspartates and in metallopeptidases by one or two metal ions (usually zinc, but also cobalt, manganese, nickel, copper, and iron). In 1995, a fifth catalytic type was discovered when the structure of the proteasome was solved, and it was found that three of the fourteen different subunits were peptidases, possessing an N-terminal threonine that acted as the nucleophile (4). The sixth catalytic type was identified in 2004 when certain fungal endopeptidases now known as eqolysins were discovered to be glutamate peptidases (5).
The usefulness of the concept of the catalytic type is that proteolytic enzymes of the same catalytic type tend to share the same general inhibitors. For example, most metallopeptidases are inhibited by chelating agents such as EDTA or 1,10-phenanthroline; most serine peptidases are inhibited by diisopropyl fluorophosphate or phenylmethane sulfonylfluoride; and most cysteine peptidases are inhibited by iodoacetate (6). However, catalytic type bears little if any relationship to the evolution of the enzymes. Enzymes of the same catalytic type can be unrelated: for example, trypsin and subtilisin are unrelated serine peptidases; papain and caspases are unrelated cysteine peptidases; and methionyl aminopeptidase and thermolysin are unrelated metallopeptidases On the other hand, peptidases of different catalytic types can be evolutionarily related. For example, the poliovirus picornain 3C is a cysteine peptidase but has a similar structure to trypsin, a serine peptidase (7). Consequently, proteolytic enzymes are also classified by structure and sequence similarity (2), where proteolytic enzymes and related proteins with homologous sequences are grouped into families, and families with related structures are grouped into clans. Three clans of mixed catalytic type exist. Clan PA includes serine and cysteine peptidases with a structure similar to trypsin. The threonine peptidases that are the catalytic components of the proteasome complex are Ntn-hydrolases (clan PB), a large grouping that includes peptidases with an N-terminal serine nucleophile, such as the penicillin G acylase precursor (8), and an N-terminal cysteine nucleophile, such as the penicillin V acylase precursor (9). Clan PC includes cysteine peptidases such as gamma-glutamyl hydrolase (10) and PfpI peptidase (11) as well as the serine peptidase dipeptidase E (12). It is clear that at the clan level, catalytic type is not conserved. However, at the family level there are no known families containing peptidases of more than one catalytic type. (Though there are families that contain non-peptidase homologues that are enzymes with a different catalytic machinery, such as family S9, which includes other alpha/beta hydrolases such as lipases (13)). The classification of peptidases and their homologues is presented in the MEROPS database (14).
The recent crystal structure of the self-cleaving precursor of the Tsh autotransporter from Escherichia coli has demonstrated that a seventh catalytic type exists in which the nucleophile is an asparagine (15). Some viral capsid proteins, previously thought to be atypical aspartic peptidases, have a similar catalytic mechanism, as do intein-containing proteins. Asparagine can form a stable, five-membered succinimide ring with its own carbonyl carbon, which when induced under the right circumstances leads to cleavage of its own peptide bond. The proteolytic role of the cyclic imine derived from an asparagine residue as an alternative to peptide degradation by hydrolysis was studied by Dehart & Anderson (16). In Enzyme Nomenclature “enzymes cleaving C-C, C-O, C-N and other bonds by elimination, leaving double bonds or rings” are termed “lyases” (1). The cleavages mentioned above are not hydrolysis but the action of amidine lyases and would belong in EC subclass 4.3. These peptide lyases are thus proteolytic enzymes but not peptidases, and belong to a different enzyme class. In this report, we report the identification of ten families of asparagine peptidase lyases.
EXPERIMENTAL PROCEDURES
Assembly of Protein Domain Families
Families were assembled as follows. The scientific literature was searched for instances where autolytic processing of asparaginyl bonds had been reported. The corresponding protein sequence in the UniProt database was identified by a text search. This sequence was used in a BlastP search (17) of the entire protein sequence library at UniProt. Homologous sequences were those hits with an E-value of less than 0.001 and which overlapped the autolytic processing site in the BlastP sequence alignment.
Comparison of Tertiary Structures
Where a tertiary structure had been solved for a member of a family, it was possible to assign the family to a clan. The DALI algorithm (18) was used to find structures related to the structure of an asparagine peptide lyase by submitting the corresponding Protein Data Bank (PDB)2 database entry coordinates to the DALI server. A match was considered significant if the z score was 6.00 or greater. Where structures from peptidases in different families were significantly similar, the families could be included in the same clan.
RESULTS
Six clans and ten families of asparagine peptide lyases have been assembled (see Table 1). Clan and family names begin with the letter N. Some of the new clans and families were included in release 9.3 (September 7, 2010) and the remainder in 9.4 (January 31, 2011) of the MEROPS database. The new families are discussed below beginning with the autotransporters, then the viral capsid proteins and finally the intein-containing proteins.
TABLE 1.
Clans and families of asparagine peptide lyases
| Clan | Family | Peptidase |
|---|---|---|
| NA | N1 | Nodavirus coat protein |
| N2 | Tetravirus coat protein | |
| N8 | Picornavirus capsid protein VP0 | |
| NB | N6 | YscU protein |
| NC | N7 | Reovirus type 1 coat protein |
| ND | N4 | Tsh-associated self-cleaving domain |
| NE | N5 | Picobirnavirus capsid protein |
| PD | N9 | Intein-containing V-type proton ATPase catalytic subunit A |
| N10 | Intein-containing DNA gyrase subunit A precursor | |
| N11 | Intein-containing chloroplast ATP-dependent endopeptidase |
Autotransporter Families
Family N4
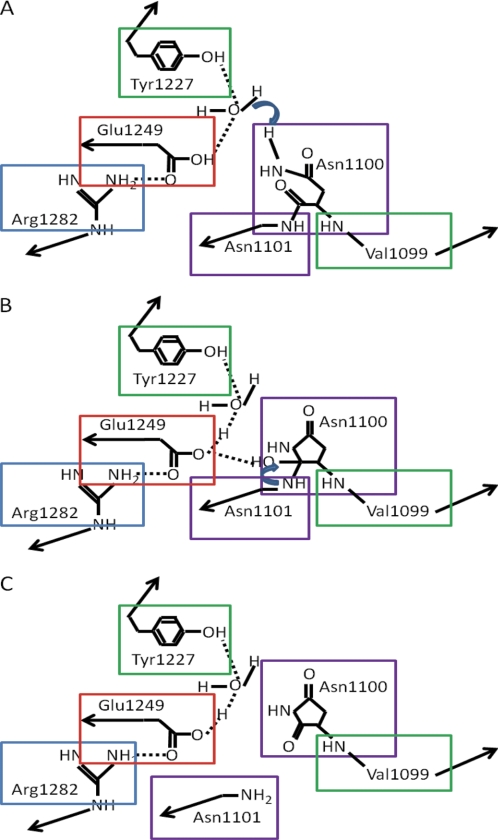
Autotransporters are virulence factors secreted by pathogenic, Gram-negative bacteria, an example being the precursor of the serine peptidase Tsh from Escherichia coli. The precursor has a signal peptide and a large C-terminal propeptide that forms a pore in the outer membrane through which the serine peptidase domain can pass. Autolytic cleavage then occurs at Asn-1100 to release the serine peptidase. The crystal structure of the autotransporter domain (15) supports the hypothesis first put forward by Dautin et al. (19) that in the E. coli EspP protein there is a catalytic dyad consisting of aspartic and asparagine residues, where the asparagine attacks and cleaves its own carbonyl carbon bond. From the crystal structure of Tsh, the residues other than Asn-1100 thought to be important are: Lys-1201 and Tyr-1227, which form hydrogen bonds with Asn-1100 holding it in place while Arg-1121 positions the side chain for nucleophilic attack. In addition, Glu-1249 assists nucleophilic attack by protonating the carbonyl and forms a salt bridge with Arg-1282, and water acts as a general base (see Fig. 1) (15).
FIGURE 1.
Mechanism for the proteolytic processing of the Tsh precursor. Residues important for the mechanism are boxed. Black arrows indicate continuing peptide chains. Hydrogen bonds are indicated by dotted lines. Blue arrows indicate nucleophilic attack. A, when the N-terminal domain passes through the outer membrane pore formed by the C-terminal autotransporter domain, the hydroxyl group of Glu-1249 in the autotransporter domain interacts with Asn-1100. B, Asn-1100 cyclizes to form a succinimide. C, cyclization leads to cleavage of the Asn-1100—Asn-1101 bond, and the N-terminal domain is released to the surrounding milieu. The image is based upon Fig. 6 of Tajima et al. (15).
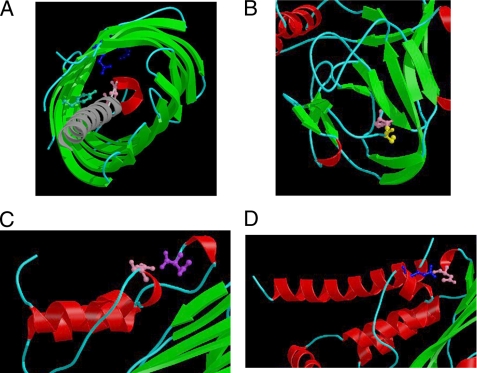
Fig. 2A shows the structure, in which the N-terminal domain was replaced by a single helix. The structure is unlike that of any other known peptidase and is therefore the type structure for clan ND.
FIGURE 2.
Richardson images (39) of selected examples from asparagines peptidase families where more than one active site residue is known are shown. A helix is shown as a coil in red, a strand is shown as an arrow in green and random coil, and loops are shown as wires in cyan. Active site residues are shown in ball-and-stick representation. Only close-ups of the regions around the active site residues are shown. A, Tsh precursor from E. coli (PDB: 3AEH) is shown (family N4). The 12-stranded beta barrel domain which forms the pore is shown in green, with the N-terminal helix (shown in gray) protruding from the center of the pore. Catalytic residues are shown in ball-and-stick representation: Asn-1100 (mutated to Asp) in pink, Tyr-1227 in green, Glu-1249 in blue, and Arg-1282 in dark blue. B, intein from the V-type proton ATPase catalytic subunit A (PDB: 1VDE) is shown (family N9). One monomer of the dimer is shown. Catalytic residues are shown: Cys-1 in yellow and Asn-454 in pink. C, coat protein from black beetle virus (PDB: 2BBV) is shown (family N1). One molecule of the trimer is shown. Catalytic residues are shown: Asp-75 in pink and Asn-363 in purple. D, coat protein from Nudaurelia capensis omega virus (PDB: 1OHF) is shown (family N2). One monomer of the tetramer is shown. Catalytic residues are shown: Glu-103 in blue and Asn-570 in pink.
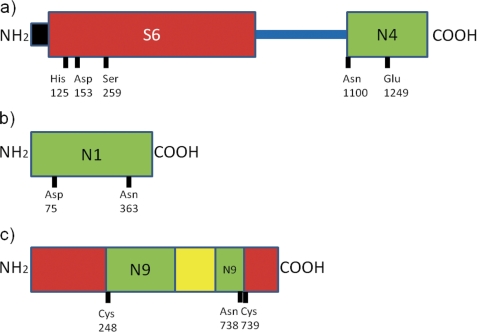
The domain organization is shown in Fig. 3a. The secreted Tsh peptidase is the N-terminal domain box, and the domain responsible for the autolytic cleavage is at the C terminus.
FIGURE 3.
The domain structures of (a) the Tsh precursor from Escherichia coli (family N4), (b) the coat protein from flock house virus (family N1), and (c) the intein-containing V-type proton ATPase catalytic subunit A from S. cerevisiae (family N9) are shown. Domains are shown as rectangles on a cyan string. The signal peptide is shown in black; the asparagine peptide lyase domain in green and other domains in red (the serine peptidase domain for the Tsh precursor and the extein domains for the RecA protein) and yellow (the homing endonuclease within the ATPase intein). Active site residues are indicated along the bottom edge of each domain.
Over thirty members of the family were found, of which 29 are predicted to be proteolytically active because all four of the essential residues mentioned above are conserved. The family is restricted to bacteria, and the majority of homologues are from the Proteobacteria. Almost all homologues are from the Gammaproteobacteria, with two from Epsilonproteobacteria; neither of which are thought to be active enzymes, and two from bacteriophages (P-EibA and P-EibC). The Epsilonproteobacteria members are Campylobacter jejuni C8J_0201 protein, where Tyr1227 is replaced with Ile and Arg-1282 with Lys; and Helicobacter mustelae major ring-forming surface antigen precursor, where Asn-1100 is replaced by Ser, Tyr-1227 by Thr, and Glu-1249 by Gln. The homologues in bacteriophages were presumably acquired by horizontal gene transfer from the host.
Family N6
Pathogenic and symbiotic Gram-negative bacteria can influence some host cells to their advantage by secreting proteins into the host cell cytoplasm. These proteins do not have signal peptides and are secreted by a special mechanism known as the type III secretion system or injectisome. The secretion is mediated by a structure similar to a flagellum, with a basal body and a multimeric filamentous protein structure which protrudes from the bacterial surface and forms a pore in the host cell membrane. The basal body sits in the bacterial cell membrane but opens to the cytoplasm, allowing unfolded proteins to pass along the hollow filament. The FlhB transmembrane protein is important when the protein being secreted changes. The FihB protein undergoes autoproteolysis at an Asn↓Pro-Thr-His motif (20), which is essential for mediating the switch in the secretion of proteins. Tertiary structures have been determined for the C-terminal domain of several homologous proteins, including the YscU precursor from Yersinia pestis which is the type structure for clan NB. Each structure shows a four-strand beta sheet surrounded by four helices, which is unique to this family. Over 180 homologues were found, all of them from bacteria. Over 120 of these are predicted to be proteolytically active. Most members are from the phyla Firmicutes and Proteobacteria, but homologues are known from almost all bacterial phyla. A widespread non-peptidase homologue is the flagellar biosynthetic protein in which the essential Asn is replaced.
Viral Coat Proteins
Family N1
In the black beetle Nodavirus, proteins beta and gamma are derived from protein alpha by cleavage of an Asn↓Ala bond. The cleavage is autolytic and occurs within the assembled virion, and both fragments remain in the virion. The active site residues are Asp-75 and Asn-363, the latter the site of cleavage (see Fig. 3b). The tertiary structure of the black beetle virus coat protein has been solved (21) and cleaved form is shown in Fig. 2C. The fold is similar to those of other viral coat proteins, including coat proteins of tetraviruses (family N2, see below) and protein VP0 of picornaviruses (family N8, see below), and all three families are included in clan NA (see supplemental Table S2). Only five members of the family were found, all of them predicted to be proteolytically active, and all of them from single-stranded RNA viruses of the family Nodaviridae. The family N1 enzymes were formerly considered to be unusual aspartic peptidases.
Family N2
A cleavage similar to that in family N1 occurs in the coat protein of Nadaurelia capensis omega virus, a Tetravirus. The residues that form the catalytic dyad in the active site are Glu-103 and Asn-570. These enzymes were also formerly considered to be unusual aspartic peptidases. The tertiary structure has been solved and shown to be similar to that of the black beetle virus coat protein (see Fig. 2D and supplemental Table S2). Consequently, family N2 is included in clan NA. Seven members of the family were found, all of them from single-stranded RNA viruses of the family Tetraviridae. Only four homologues have the catalytic dyad conserved.
Family N8
The VP0 protein from poliovirus (a Picornavirus) is cleaved into two fragments, VP2 and VP4, and its structure (22) resembles that of the Nodavirus and Tetravirus coat proteins (supplemental Table S2). Cleavage occurs at Asn-69, but the other member of the catalytic dyad has not been identified. Cleavage occurs near the N terminus, whereas in Tetravirus and Nodavirus coat proteins it is near the C terminus.
Asn-69 is replaced by glutamine in some VP0 proteins from Rhinovirus, and cleavage is known to occur in the coat protein from Rhinovirus 16 from the solved tertiary structures (23). Glutamine cyclizes less readily than asparagine, but a glutarimide intermediate can promote self-cleavage (24). It is not known if cleavage of this Gln-Ser bond is autolytic, or performed by another peptidase. This cleavage site would fit the specificity of picornain 3C (from peptidase family C3), which is present in the same polyprotein. In some enteroviruses Asn-69 is replaced by Lys; cleavage of the enterovirus VP0 protein has been assumed but not demonstrated and no cleavage occurred when enterovirus 71 polyprotein was expressed in insect cells (25). Foot-and-mouth disease virus VP0 protein is also cleaved, but at an Ala↓Asp bond (26), presumably by a host peptidase. Nearly 200 homologues were found, all from the Picornaviridae family of single-stranded RNA viruses. In 168 of these, the asparagine is conserved or replaced by glutamine.
Family N5
Picobirnaviruses are double-stranded RNA viruses. The viral particle is organized around a central icosahedral core capsid consisting of 120 identical subunits. Each subunit undergoes an autoproteolytic cleavage, releasing a peptide that remains in the capsid associated with the RNA. Cleavage occurs at Asn-45, and this is assumed to be the nucleophile. No other active site residue has been identified. The tertiary structure of the cleaved rabbit picobirnavirus coat protein has been solved (27), the structure is unrelated to that of any other peptidase (supplemental Table S2) and the rabbit picobirnavirus coat protein is the type structure for clan NE. Only one other homologue was found, from the human picobirnavirus.
Family N7
Reoviruses are double-stranded DNA viruses. The viral particle consists of two concentric, icosahedral protein capsids surrounding a ten segment, double-stranded RNA genome. One of the capsid proteins, known as mu-1, undergoes an autoproteolytic cleavage, releasing a peptide. known as mu-1N. Cleavage occurs at Asn-42, and this is assumed to be the nucleophile, but no other active site residue has been identified. Because mu-1 is myristoylated at its N terminus, mu-1N is also myristoylated and it associates with host erythrocyte membranes where it and another fragment known as phi contribute to pore formation and ultimately to membrane penetration by viral particles. The tertiary structure of the cleaved mu-1 protein has been solved (supplemental Table S2) and shows a fold unrelated to that of any other peptidase (28). The structure of the mu-1 protein is therefore the type structure for clan NC. Eleven homologues have been found, all of them from double-stranded DNA viruses of the family Reoviridae, and all predicted to be proteolytically active.
Inteins: Families N9, N10, and N11
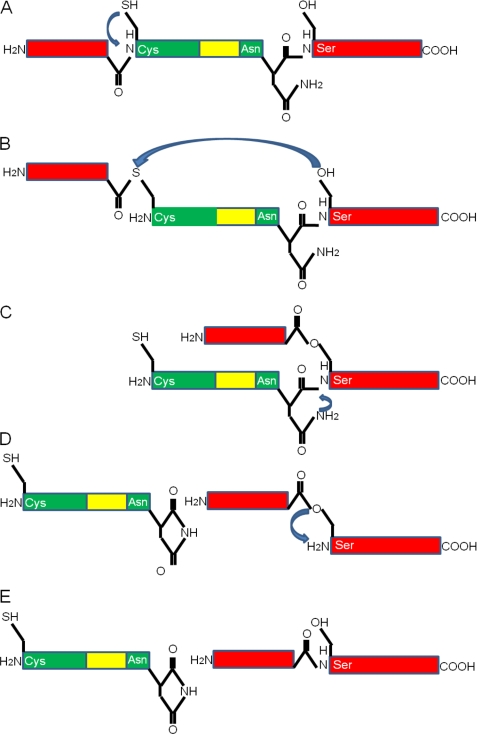
Inteins are derived from a particular form of parasitic DNA. The DNA is inserted into an existing gene. Typically, the translated protein consists of an intein domain inserted within two extein domains. The newly synthesized protein undergoes self-cleavage, releasing the intein, with simultaneous formation of a peptide bond between the remaining portions of the protein. This self-splicing reaction results in the cleavage of two peptide bonds, at either end of the intein domain. Thus, inteins are analogous to self-splicing introns (29). The first residue of the intein domain must be cysteine, serine, or threonine and the last residue asparagine. The first residue of the second extein domain must be cysteine, serine, or threonine. In the V-type proton ATPase catalytic subunit A from Saccharomyces cerevisiae, the first extein domain is residues 2–283, the intein domain is residues 284–737, and the second extein domain is residues 738–1071. The mechanism proceeds in four steps (see Fig. 4). First, a linear thioester is formed at the junction of the first extein and the intein by a rearrangement of the peptide bond involving the amino group of Cys-284. Second, a transesterification reaction occurs between the thioester and Cys-738, forming a branched thioester intermediate. During this reaction the peptide bond to Cys-284 is broken. In the third stage, this intermediate is resolved by cyclization of Asn-737 followed by peptide bond cleavage. The intein is excised and Asn-737 becomes an aminosuccinimide residue. The extein domains are linked by a thioester bond. Finally, an acyl rearrangement of the thioester bond linking the extein domains occurs (29). Neither peptide bond is broken by hydrolysis.
FIGURE 4.
The mechanism of excision of an intein is shown. The extein domains are shown in red; the intein is shown in green, and the homing endonuclease is shown in yellow. The intein-containing V-type proton ATPase catalytic subunit A from S. cerevisiae (family N9) is used as an example, and domain lengths are drawn to scale. Arrows show the direction of nucleophilic attack. A, unprocessed precursor is shown. The first residue of the intein (shown here as Cys, but it may be Ser or Thr in other inteins) attacks the carbonyl carbon atom of the preceding residue creating a linear thioester (or ester) bond. B, the thiolester intermidate is shown. The first residue of the second extein domain (shown here as Ser, but it may be Thr or Cys) attacks the thioester bond and a transesterification occurs to form a branched intermediate. C, the branched intermediate is shown. During this reaction a peptide bond is broken releasing the N terminus of the intein. The Asn at the C terminus of the intein attacks its own carbonyl-carbon bond, cyclizing to form a succinimide and breaking its own peptide bond. The intein is thus released. D, the released intein and the thiolester-linked extein domains are shown. The two portions of the extein are spliced together when the thioester bond is converted to a normal peptide bond by the action of the Ser, and the mature proteins are shown in E). The image is based upon Fig. 2 from Raghavan & Minnick (29).
The intein includes an endonuclease known as a “homing” endonuclease which propagates the intein by recognizing a specific (and rare) nucleotide sequence, cleaving it to form staggered ends. This initiates DNA repair which uses the DNA encoding the intein as a template, thus propagating the intein gene. The nucleotide sequence that the homing endonuclease recognizes has been determined for the V-type proton ATPase catalytic subunit A from S. cerevisiae. The DNA sequence occurs in the gene for the ATPase subunit, encoding the final eleven bases of the first extein domain (corresponding to residues Ile-279 to Gly-283) and the first twenty-five bases of the second extein domain (corresponding to residues Cys-738 to Met-745) (30). The self-splicing inteins in families N9, N10, and N11 are analogous to self-splicing introns (29).
Inteins have been grouped into three families. Family N9 is typified by the intein from the V-type proton ATPase catalytic subunit A of S. cerevisiae. Family N10 is built around the intein from the replicative DNA helicase of Synechocystis sp. PCC 6803. The type example for family N11 is the intein from the chloroplast ATP-dependent endopeptidase of Chlamydomonas eugametos. In both families N9 and N11, the first residue of the intein is always Cys, whereas the first residue of the second extein can by Cys, Ser, or Thr. In family N10, the first residue of the intein or the first residue of the second extein can be Cys, Ser, or Thr. There are some inteins from this family where the Asn is replaced by Gln. An example of an intein where the last residue is Gln is that from the DNA polymerase II large subunit of the archaean Haloarcula marismortui (family N10). Another unusual intein in family N10 occurs in catalytic subunit alpha of DNA polymerase III from Synechocystis sp. PCC6803. The N- and C-terminal halves of the subunit are encoded by two separate genes (dnaE-n and dnaE-c), which are far apart on the chromosome and on opposite strands. An intein is also present and split between the two genes, so that dnaE-n gene also encodes the first extein and a portion of the intein, whereas dnaE-c encodes the remainder of the intein and the second extein. Once the proteins are translated, a heterodimer forms, the intein is released and the exteins spliced together (31).
Structures have been determined for members of family N9 (see Fig. 2B) and N10. The structures of the intein self-cleaving domains are similar to each other and similar to that of the Drosophila melanogaster hedgehog protein C-terminal domain (see supplemental Table S2). A structural comparison between the intein from the V-type proton ATPase catalytic subunit A from S. cerevisiae and the D. melanogaster hedgehog protein autoprocessing domain shows conservation of all the beta-strands. The hedgehog protein consists of two domains, the N-terminal effector domain and the C-terminal autoprocessing domain. The N-terminal domain has a fold similar to that of zinc d-Ala-d-Ala carboxypeptidases in clan MD, but is not known to have any peptidase activity (32). The C-terminal domain consists of two tandem intein-like repeats derived from an ancient duplication event (33). Each intein repeat contains six beta strands arranged in an arc, and the active site sits at the junction of the two repeats. Processing occurs on the amino side of the nucleophilic Cys-208. The cysteine attacks the carbonyl carbon atom of the preceding Gly and an N-S acyl rearrangement results in the formation of a thioester bond. Then the 3-beta hydroxyl group of cholesterol reacts with the thioester, resulting in cleavage, release of the N-terminal effector domain and esterification of the new C-terminal Gly-207 (34). The similarity in structure, function, and mechanism between the intein and hedgehog self-processing domains implies common ancestry, and the four families are included in a new clan. Because this clan includes proteolytic enzymes of different catalytic types, it is named PD.
Over thirty members of family N9 have been found, with most occurring in fungi from the family Saccharomycetaceae. There are two bacterial homologues (inteins from the DnaB proteins of Salinibacter ruber and Sulfurovum sp. NBC37–1). Family N10 is the most widespread of the intein families, with over 450 homologues in archaea, bacteria, double-stranded DNA viruses, and eukaryotes (Rhodomonas salina, Ectocarpus siliculosus, Fucus vesiculosus, Porphyra purpurea, Porphyra yezoensis, and Tetrahymena thermophila). Over thirty members of family N11 have been found, with a wide distribution in double-stranded DNA viruses, archaea, bacteria, fungi (Coelomomyces stegomyiae, Phaeosphaeria nodorum, and Spiromyces aspiralis) and plants (Floydiella terrestris, Stigeoclonium helveticum, and Chlamydomonas species).
DISCUSSION
The inclusion of proteins that process themselves at asparagine residues among proteolytic enzymes might be seen as controversial. It is debatable whether proteins that cleave themselves can be considered as enzymes because the enzyme is destroyed by the enzymatic activity. Among the 256 families of proteolytic enzymes, over thirty include proteins where the only known catalytic activity is self-cleavage. This total includes all ten families of asparagine peptide lyases. The asparagine peptide lyases are not the only proteins where cleavage occurs in cis (i.e. where the enzyme and substrate are the same molecule), other examples being the hedgehog proteins (family C46), some viral polyprotein processing peptidases such as the pestivirus NS2 protein (family C74) (35), and the precursor proteins of N-terminal nucleophile (Ntn) hydrolases such as the glutamine PRPP amidotransferase (family C44), penicillin G acylase (family S45), glycosylasparaginase (family T2), gamma-glutamyl transferase (family T3), and polycystin-1 (family T6) (36). The catalytic proteasome components also process their own precursors in cis (37) but unlike most of the other precursors mentioned above continue to possess proteolytic activity. Togavirin (family S3) releases itself from the togavirus non-structural polyprotein by cleavage in cis but then has no further catalytic activity. The tertiary structure has been determined and shown to be similar to that of trypsin (38). It is clear from all these examples that self-processing, even in cis, is the action of a proteolytic enzyme, even though the enzyme is not recoverable from the reaction.
Peptidases hydrolyze peptide bonds, but the self cleavages performed by asparagine peptide lyases do not involve hydrolysis. The release of the hedgehog effector domain from its precursor also does not involve hydrolysis, instead cholesterol reacts with the newly formed thioester, resulting in cleavage and esterification of the new C-terminal Gly (34). A number of other peptidases also act as transferases, adding moieties to the newly exposed N and C termini. Peptidases from families M48 and M79 cleave proteins where a cysteine residue near the C terminus has been prenylated and methyl esterification of the new carboxyl also occurs (40). The prepilin peptidase (family A24) processes bacterial type 4 pilin precursor proteins to their mature forms by removal of a leader peptide and methylation of the new N terminus, which is generally Phe (41). However, in none of these examples is it clear that the same catalytic site performs both the cleavage and the transferase reactions. Water is not necessarily a requirement for catalytic activity of peptidases; for example the serine peptidase chymotrypsin (family S1) is able to perform transesterification of N-acetyl-l-phenylalanine ethyl ester in butanol (42).
There are very few enzymes in which asparagine is a nucleophile. Examples include a methyltransferase and purine nucleoside phosphorylase, both of which require a catalytic dyad consisting of an acidic residue (aspartate or glutamate) and asparagine, and with water acting as the general base (43). This catalytic mechanism is very similar to that of the asparagine peptide lyases. Asparagine peptide lyases are not the only lyases capable of breaking peptide bonds. Another example is peptidyl-glycine alpha-amidating monooxygenase, which is a bifunctional enzyme with two catalytic activities, which together remove the C-terminal glycine from neuropeptides such as vasopressin. First the glycine is converted to a hydroxyglycine intermediate. This unstable intermediate is then converted to a peptidyl amide and glyoxylate. The second reaction is that of a peptidylamidoglycolate lyase, and has been assigned the EC number 4.3.2.5 (44). The lyase activity is metal-dependent, but the catalytic mechanism is unknown.
Besides the autotransporters in families N4 and N6, there are other families containing self-cleaving autotransporters. The catalytic mechanism is unknown for the AIDA-I self-cleaving autotransporter protein from E. coli (the type peptidase for family U69), but self-processing occurs at a Ser-846–Ala bond (45), which would be inconsistent with this protein being an asparagine peptide lyase. Not all autotransporters process themselves. The SphB1 peptidase is a subtilisin homologue (and a member of family S8) which is known to process the autotransporter precursors of adhesin in Bordetella pertussis (46) and the IgA protease and App in Neisseria meningitides (47). The SphB1 peptidase is itself an autotransporter, processing its own precursor, for which the active site Ser in the subtilisin domain is essential (48).
In conclusion, a new type of proteolytic enzyme has been recognized, that of a peptide lyase. These enzymes are not peptidases because breaking the peptide bonds does not involve hydrolysis. Thus the terms “proteolytic enzyme” and “peptidase” are not synonymous. The nucleophile in the reaction is an asparagine, which means that this also represents a new proteolytic catalytic type bringing the total number of catalytic types to seven. Ten families of asparagine peptide lyases have been identified. There are two families of autotransporter proteins, five families of viral coat proteins, and three families of intein-containing proteins. From structural comparisons these ten families can be grouped into six clans, which implies that this activity has arisen independently on several occasions. One of these clans (PD) in addition to the intein-containing proteins, also includes the structurally-related hedgehog protein processing domain, which is a cysteine peptidase. All of the asparagine peptide lyases perform only self-cleavages, with cleavage occurring immediately C-terminal to the active site asparagine. Cleavage occurs when this asparagine forms a succinimide ring, which happens when a second active site residue, an aspartate or glutamate, is brought into close proximity. Cleavage is intramolecular in cis. No examples of asparagine peptide lyases have been identified in plants and animals, and homologues are only known from viruses, archaea, bacteria, single-celled eukaryotes and some algae. The ten families of peptide lyases have been included in the MEROPS database (14). The identification of a seventh catalytic type of proteolytic enzyme, and a new mechanism of cleavage, leads us to wonder if they are any other catalytic types left to discover.
Supplementary Material
Acknowledgment
We thank Dr. Gemma Holliday for helpful discussions.
This work was supported by the Wellcome Trust Grant Number WT077044/Z/05/Z.
This article was chosen was a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S2.
- PDB
- Protein Data Bank.
REFERENCES
- 1. NC-IUBMB (Nomenclature Committee of the International Union of Biochemistry and Molecular Biology) (1992) Enzyme Nomenclature 1992. Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes, Academic Press, Orlando [Google Scholar]
- 2. Rawlings N. D., Barrett A. J. (1993) Biochem. J. 290, 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hartley B. S. (1960) Annu. Rev. Biochem. 29, 45–72 [DOI] [PubMed] [Google Scholar]
- 4. Seemüller E., Lupas A., Stock D., Löwe J., Huber R., Baumeister W. (1995) Science 268, 579–582 [DOI] [PubMed] [Google Scholar]
- 5. Fujinaga M., Cherney M. M., Oyama H., Oda K., James M. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3364–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barrett A. J. (1994) Methods Enzymol. 244, 1–15 [DOI] [PubMed] [Google Scholar]
- 7. Allaire M., Chernaia M. M., Malcolm B. A., James M. N. (1994) Nature 369, 72–76 [DOI] [PubMed] [Google Scholar]
- 8. McDonough M. A., Klei H. E., Kelly J. A. (1999) Protein Sci. 8, 1971–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suresh C. G., Pundle A. V., SivaRaman H., Rao K. N., Brannigan J. A., McVey C. E., Verma C. S., Dauter Z., Dodson E. J., Dodson G. G. (1999) Nat. Struct. Biol. 6, 414–416 [DOI] [PubMed] [Google Scholar]
- 10. Li H., Ryan T. J., Chave K. J., Van Roey P. (2002) J. Biol. Chem. 277, 24522–24529 [DOI] [PubMed] [Google Scholar]
- 11. Du X., Choi I. G., Kim R., Wang W., Jancarik J., Yokota H., Kim S. H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14079–14084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Håkansson K., Wang A. H., Miller C. G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14097–14102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Holmquist M. (2000) Curr. Protein Pept. Sci. 1, 209–235 [DOI] [PubMed] [Google Scholar]
- 14. Rawlings N. D., Barrett A. J., Bateman A. (2010) Nucleic Acids Res. 38, D227-D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tajima N., Kawai F., Park S. Y., Tame J. R. (2010) J. Mol. Biol. 402, 645–656 [DOI] [PubMed] [Google Scholar]
- 16. Dehart M. P., Anderson B. D. (2007) J. Pharm. Sci. 96, 2667–2685 [DOI] [PubMed] [Google Scholar]
- 17. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 18. Holm L., Sander C. (1997) Nucleic Acids Res. 25, 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dautin N., Barnard T. J., Anderson D. E., Bernstein H. D. (2007) EMBO J. 26, 1942–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Björnfot A. C., Lavander M., Forsberg A., Wolf-Watz H. (2009) J. Bacteriol. 191, 4259–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wery J. P., Reddy V. S., Hosur M. V., Johnson J. E. (1994) J. Mol. Biol. 235, 565–586 [DOI] [PubMed] [Google Scholar]
- 22. Zhang Y., Simpson A. A., Ledford R. M., Bator C. M., Chakravarty S., Skochko G. A., Demenczuk T. M., Watanyar A., Pevear D. C., Rossmann M. G. (2004) J. Virol. 78, 11061–11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hadfield A. T., Lee W., Zhao R., Oliveira M. A., Minor I., Rueckert R. R., Rossmann M. G. (1997) Structure 5, 427–441 [DOI] [PubMed] [Google Scholar]
- 24. Amitai G., Dassa B., Pietrokovski S. (2004) J. Biol. Chem. 279, 3121–3131 [DOI] [PubMed] [Google Scholar]
- 25. Chung Y. C., Huang J. H., Lai C. W., Sheng H. C., Shih S. R., Ho M. S., Hu Y. C. (2006) World J. Gastroenterol. 12, 921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Curry S., Fry E., Blakemore W., Abu-Ghazaleh R., Jackson T., King A., Lea S., Newman J., Stuart D. (1997) J. Virol. 71, 9743–9752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duquerroy S., Da Costa B., Henry C., Vigouroux A., Libersou S., Lepault J., Navaza J., Delmas B., Rey F. A. (2009) EMBO J. 28, 1655–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang X., Ji Y., Zhang L., Harrison S. C., Marinescu D. C., Nibert M. L., Baker T. S. (2005) Structure 13, 1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raghavan R., Minnick M. F. (2009) J. Bacteriol. 191, 6193–6202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moure C. M., Gimble F. S., Quiocho F. A. (2002) Nat. Struct. Biol. 9, 764–770 [DOI] [PubMed] [Google Scholar]
- 31. Wu H., Hu Z., Liu X. Q. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9226–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hall T. M., Porter J. A., Beachy P. A., Leahy D. J. (1995) Nature 378, 212–216 [DOI] [PubMed] [Google Scholar]
- 33. Hall T. M., Porter J. A., Young K. E., Koonin E. V., Beachy P. A., Leahy D. J. (1997) Cell 91, 85–97 [DOI] [PubMed] [Google Scholar]
- 34. Mann R. K., Beachy P. A. (2000) Biochim. Biophys. Acta 1529, 188–202 [DOI] [PubMed] [Google Scholar]
- 35. Lackner T., Thiel H. J., Tautz N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1510–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei W., Hackmann K., Xu H., Germino G., Qian F. (2007) J. Biol. Chem. 282, 21729–21737 [DOI] [PubMed] [Google Scholar]
- 37. Jäger S., Groll M., Huber R., Wolf D. H., Heinemeyer W. (1999) J. Mol. Biol. 291, 997–1013 [DOI] [PubMed] [Google Scholar]
- 38. Choi H. K., Tong L., Minor W., Dumas P., Boege U., Rossmann M. G., Wengler G. (1991) Nature 354, 37–43 [DOI] [PubMed] [Google Scholar]
- 39. Richardson J. S. (1985) Methods Enzymol. 115, 359–380 [DOI] [PubMed] [Google Scholar]
- 40. Romano J. D., Michaelis S. (2001) Mol. Biol. Cell 12, 1957–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Strom M. S., Nunn D. N., Lory S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 2404–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eckstein M., Sesing M., Kragl U., Adlercreutz P. (2002) Biotechnol. Lett. 24, 867–872 [Google Scholar]
- 43. Doukov T. I., Hemmi H., Drennan C. L., Ragsdale S. W. (2007) J. Biol. Chem. 282, 6609–6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Satani M., Takahashi K., Sakamoto H., Harada S., Kaida Y., Noguchi M. (2003) Protein Expr. Purif. 28, 293–302 [DOI] [PubMed] [Google Scholar]
- 45. Charbonneau M. E., Janvore J., Mourez M. (2009) J. Biol. Chem. 284, 17340–17351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dé E., Saint N., Glinel K., Meli A. C., Lévy D., Jacob-Dubuisson F. (2008) Mol. Membr. Biol. 25, 192–202 [DOI] [PubMed] [Google Scholar]
- 47. van Ulsen P., van Alphen L., ten Hove J., Fransen F., van der Ley P., Tommassen J. (2003) Mol. Microbiol. 50, 1017–1030 [DOI] [PubMed] [Google Scholar]
- 48. Turner D. P., Wooldridge K. G., Ala'Aldeen D. A. (2002) Infect. Immun. 70, 4447–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.