Background: Dipeptidyl peptidases (DPPs) are required for protein metabolism in Porphyromonas gingivalis.
Results: Asp/Glu-specific novel DPP (DPP11) was discovered and characterized.
Conclusion: DPP11 ensures efficient degradation of oligopeptide substrates in Gram-negative anaerobic rods.
Significance: This observation suggests further variation of substrate specificity in the DPP members.
Keywords: Aspartate, Glutamate, Metabolism, Peptidases, Periodontal Disease, Serine Protease, Porphyromonas endodontalis, Porphyromonas gingivalis, Butylate, Dipeptidyl Peptidase
Abstract
Porphyromonas gingivalis and Porphyromonas endodontalis, asaccharolytic black-pigmented anaerobes, are predominant pathogens of human chronic and periapical periodontitis, respectively. They incorporate di- and tripeptides from the environment as carbon and energy sources. In the present study we cloned a novel dipeptidyl peptidase (DPP) gene of P. endodontalis ATCC 35406, designated as DPP11. The DPP11 gene encoded 717 amino acids with a molecular mass of 81,090 Da and was present as a 75-kDa form with an N terminus of Asp22. A homology search revealed the presence of a P. gingivalis orthologue, PGN0607, that has been categorized as an isoform of authentic DPP7. P. gingivalis DPP11 was exclusively cell-associated as a truncated 60-kDa form, and the gene ablation retarded cell growth. DPP11 specifically removed dipeptides from oligopeptides with the penultimate N-terminal Asp and Glu and has a P2-position preference to hydrophobic residues. Optimum pH was 7.0, and the kcat/Km value was higher for Asp than Glu. Those activities were lost by substitution of Ser652 in P. endodontalis and Ser655 in P. gingivalis DPP11 to Ala, and they were consistently decreased with increasing NaCl concentration. Arg670 is a unique amino acid completely conserved in all DPP11 members distributed in the genera Porphyromonas, Bacteroides, and Parabacteroides, whereas this residue is converted to Gly in all authentic DPP7 members. Substitution analysis suggested that Arg670 interacts with an acidic residue of the substrate. Considered to preferentially utilize acidic amino acids, DPP11 ensures efficient degradation of oligopeptide substrates in these Gram-negative anaerobic rods.
Introduction
Porphyromonas gingivalis, a Gram-negative black-pigmented anaerobe, is a major causative organism of aggressive forms of chronic periodontitis (1, 2) which leads to loss of permanent teeth (3–5). Recently, much attention has been paid to this bacterium because of its close relationship with systemic diseases, such as cardiovascular diseases (6), decreased kidney function (7), and rheumatoid arthritis (8). An important relative of the bacterium is Porphyromonas endodontalis, which is predominantly isolated from periapical periodontitis sites, i.e. infected root canals with acute symptoms such as pain, swelling, and drainage (9–11).
Both Porphyromonas species are asaccharolytic in principal and do not ferment glucose, cellobiose, lactose, or sucrose (12) and require proteinaceous substrates as carbon and energy sources. P. gingivalis possesses extracellular arginine aminopeptidase activity, which is mediated by Arg-gingipains (Rgps)3 isoforms A and B of Arg-X-specific cysteine proteinase (X is any amino acid), whereas other aminopeptidase activities are not present (13, 14). Lys-specific gingipain (Kgp), another potent cysteine proteinase, does not exhibit aminopeptidase activity (15). In accord with the lack of predominant aminopeptidase activities, it has been demonstrated that P. gingivalis mainly incorporates nutritional amino acids as forms of di- and tripeptides, not as single amino acids, and produces metabolic end products such as ammonia, acetate, propionate, and butyrate (16, 17), which are considered to be virulence factors of this bacterium, causing host tissue damage (18, 19). Accordingly, the cell-surface and extracellular peptidases of P. gingivalis that produce di- and tri-peptides are considered to play critical roles in cell growth as well as its pathogenicity.
Although entire genome sequencing has annotated 72 peptidase genes in P. gingivalis W83 (20) as well as ATCC 33277 (21), at present only the several peptidases that have been well characterized allow us to explain their proteinaceous substrate utilization. First, the predominant proteolytic activities of Kgp and Rgps (22–25) are believed to digest nutritional proteins into oligopeptides. Subsequently, oligopeptides are processed by dipeptidyl peptidase IV (EC 3.4.14.5, DPPIV) (26), DPP7 (27) and prolyl tripeptidyl peptidase-A (PTP-A) (28, 29). P. gingivalis DPPIV preferentially cleaves NH2-XP↓Xn and less potently NH2-XA↓Xn (26, 30). DPP7 (PgDPP7) cleaves NH2-XY↓Xn, where Y is an aliphatic or aromatic amino acid (27). When Pro is located at the third position from the N terminus, neither DPPIV nor DPP7 cleaves the peptides, and instead, PTP-A cleaves an NH2-XXP↓Xn bond (29, 31). Therefore, the integrated actions of DPPIV, DPP7, and PTP-A may be responsible for utilization of peptides from scarce resources in the oral cavity. On the other hand, although the growth of a DPPIV-, DPP7-, and PTP-A-triple knock-out P. gingivalis strain was demonstrated to be retarded, the mutant showed growth (32), which may indicate the presence of a complementary mechanism for supplying substances for the metabolic pathway. Moreover, Asp/Asn and Glu/Gln are the most intensively consumed nutritional amino acids in Tryptone-based medium (16). Therefore, the existence of P. gingivalis DPP that hydrolyzes Asp and Glu at the P1 position is reasonably surmised. In the case of P. endodontalis, which does not have the proteolytic activity equivalent to gingipains (10, 33), the mechanism on oligopeptide metabolism is further obscure.
In the present study, we started from the analysis of a DPP activity of P. endodontalis, an important pathogenic organism in periapical lesions, because substantial DPP activities were observed in the extracellular fraction obtained from a dialysis membrane culture on an agar plate, whereas DPPs seemed completely cell-associated in P. gingivalis. In addition, because P. endodontalis does not have marked gingipain-like activities, this microorganism has a great advantage over P. gingivalis in handling of the activities without care about degradation or modification by gingipain-like proteinases (33).
We isolated a gene of P. endodontalis using a PCR method with degenerated primers designed based on the sequence homology of DPP7-family genes and subsequently found the existence of its P. gingivalis orthologue, PGN0607, in a homology search that had already been proposed as an isoform of DPP7 (27). Recombinant proteins of the cloned gene and PGN0607 expressed in Escherichia coli did not show DPP7-like activity, indicating their identity distinct from DPP7. Because I-VI and 6–10 have been already allocated to DPPs with various specificities and their subtypes, we designated this novel DPP as DPP11. The enzymatic and biochemical analyses on of DPP11 from P. endodontalis (PeDPP11) and P. gingivalis (PgDPP11) revealed that they possessed the activity to hydrolyze NH2-X(D/E)↓Xn. We also found Arg670 that is critical for the substrate specificity of DPP11. Furthermore, a MEROPS data base (34) search indicated that DPP11 orthologues are widely distributed in anaerobic Gram-negative species in the genera Porphyromonas, Bacteroides, and Parabacteroides.
EXPERIMENTAL PROCEDURES
Materials
The expression and cloning vectors used were pTrcHisTOPO from Invitrogen, pQE60 from Qiagen, and pGEM-Teasy from Promega. DEAE-Sephacel, Sephacryl S200HR, CNBr-Sepharose, low molecular weight molecular markers, and rainbow markers were obtained from GE Healthcare. Restriction enzymes and DNA-modifying enzymes came from TAKARA BIO and New England Biolabs, respectively, whereas KOD Plus DNA polymerase came from Toyobo (Tokyo, Japan). MCA peptides and neuromedin B were obtained from the Peptide Institute Inc. (Osaka, Japan). Variants of neuromedin B with an amino acid substitution to Asp at the first, second, third, or fifth position were synthesized by BEX (Tokyo, Japan), and LD-, LE-, acetyl (ac)-LD-, and benzyloxycarbonyl (Z)-LLQ-MCA were synthesized by Thermo Fisher Scientific (Ulm, Germany) and TORAY (Tokyo, Japan). A Genome Walker Universal kit and Talon metal affinity resin came from Clontech Laboratories. Thermolysin from Bacillus thermoproteolyticus rokko and trifluoroacetic acid were obtained from Sigma, and polyvinylidene difluoride membranes and ZipTipμ-C18 came from Millipore. α-Cyano-4-hydroxycinnamic acid was purchased from Applied Biosystems, oligonucleotide primers came from FASMAC (Atsugi, Japan), and alkaline phosphatase-conjugated rabbit anti-mouse Ig(G+A+M) was from Zymed Laboratories Inc.. Recombinant Glu-specific endopeptidase from Staphylococcus aureus V8 strain (GluV8) was expressed using a method reported previously (35).
Bacterial Strains and Growth Conditions
P. endodontalis ATCC 35406 and P. gingivalis ATCC 33277 were obtained from American Type Culture Collection. They were cultured in anaerobic bacteria culture media (ABCM) (Eiken Chemical, Tokyo, Japan) in the presence of 1 μg/ml menadione (Sigma) at 35 °C under anaerobic conditions (90% N2, 5% H2, 5% CO2). To separate the cellular and soluble extracellular fractions, bacterial cells grown to the early stationary phase were inoculated onto ABCM agar plates covered with a sterilized dialysis membrane (cutoff, 6 kDa) and further cultured for 2 days. The cells were then suspended with 10 ml of phosphate-buffered saline (PBS) (pH 7.4) and centrifuged at 10,000 × g for 20 min at 4 °C. The resultant supernatant was filtered with a 0.45-μm membrane and used as the extracellular fraction. The cell pellet was washed once with PBS, and the resultant pellet was resuspended in 10 ml of PBS and used as the cellular fraction. CDC anaerobe blood agar plates (Nippon Becton Dickinson, Tokyo, Japan) were used to examine black pigmentation. E. coli XL-1 Blue was grown at 37 °C in Luria-Bertani broth and on agar plates supplemented with 75 μg/ml ampicillin.
PCR Cloning of the Gene Encoding a DPP-family Member from P. endodontalis
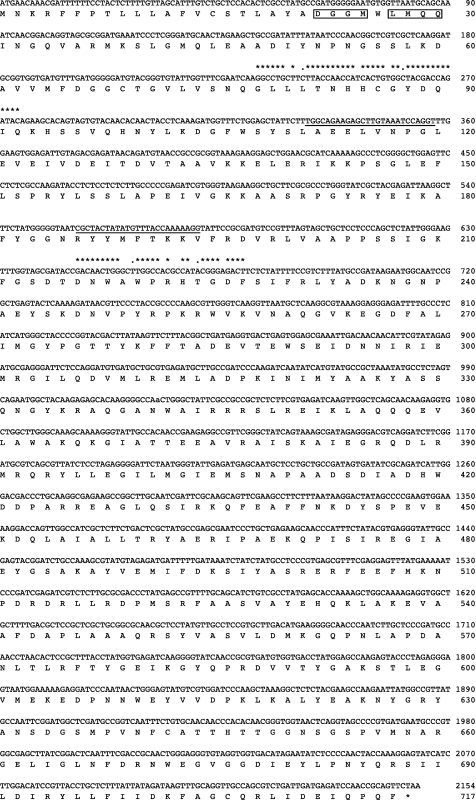
Genomic DNA from P. endodontalis and P. gingivalis (0.1 μg) was prepared as described previously (36) and used as templates. Degenerate 5′- and 3′-oligonucleotide primers (DPP7deg244–276 and DPP7deg676–643) (supplemental Table S1) were synthesized in consideration of the homology of DPP7-related peptidases from P. gingivalis (PG0491/Q7MWU6 and PG1283/BAG33126), Xyella fastidiosa (ZP00652070), and Shewanella putrefaciens (YP001182286 and YP001182445) (27). Degenerate PCR was performed with 7 cycles at 94 °C for 3 s and 72 °C for 3 min then 25 cycles at 94 °C for 3 s and 60 °C for 3 min using a Genome Walker Universal kit. The resultant 448-bp DNA fragment was amplified, inserted into a pGEM-T Easy vector, and sequenced. Genomic DNA of P. endodontalis (2 μg) was digested with DraI, EcoRV, PvuII, or StuI, then ligated with an adaptor nucleotide, and the obtained DNA fragments were used as a template for genome walking. PCR was performed using one of the gene specific primers (8Fcomo127–102 and 8Fcomp326–350, Fig. 1 and supplemental Table S1) in combination with the adaptor primer (5′-GTAATACGACTCACTATAGGGC-3′), resulting in amplification of 1- and 2-kbp DNA fragments from DraI- and EcoRV-digested genome DNA, respectively. Consequently, a 2303-bp nucleotide containing an open reading frame composed of 2154 bp was identified. The gene was designated as P. endodontalis DPP11 (PeDPP11, registered as AB610284 in DNA Data Bank of Japan).
FIGURE 1.
Nucleotide and deduced amino acid sequences of PeDPP11. The DNA sequence of DPP11 from P. endodontalis and its deduced amino acid sequence are shown. Matching the nucleotide sequence with those of the degenerate primers DPP7deg244–276 and DPP7deg676–643 (supplemental Table S1) is indicated by asterisks (completely matched) or dots (matched with one of the degenerate bases). The nucleotide sequences (8Fcomp127–102 and 8Fcomp326–350) used for genome walking are underlined. The N-terminal amino acid sequence determined with an immuno-purified 75-kDa protein is boxed (see Fig. 3C).
Construction of Expression Plasmids and in Vitro Mutagenesis
Because the N terminus of the soluble form of PeDPP11 was Asp22 (see Fig. 3), a DNA fragment encoding Asp22-Phe717 was amplified by PCR with genomic DNA and a set of primers (5PeDPP11D22Bgl, 3PeDPP11F717Bgl) containing BglII sites (supplemental Table S1), then the BglII-digested 2.2-kDa PCR fragment was cloned into the BamHI site of pTrcHisTOPO (designated pTrcHis-PeDPP11). A DNA fragment encoding from Asp22 to the C-terminal Pro720 of PGN0607, the P. gingivalis gene (PgDPP11) most homologous to PeDPP11, was amplified by PCR with a set of primers (5PgDPP11D22Bam/3PgDPP11P720Bam, supplemental Table S1) and genomic DNA, then cloned into a BamHI site of pQE60 to generate pQE-PgDPP11. In vitro mutagenesis was performed using a PCR technique with primers to introduce an amino acid substitution (supplemental Table S1). The substitutions of S652A and S655A were introduced into plasmids encoding pTrcHis-PeDPP11 and pQE-PgDPP11, respectively, whereas substitution of Arg670 to Gly, Lys, His, Gln, or Asp was introduced to pTrcHis-PeDPP11. All mutations were confirmed by nucleotide sequencing.
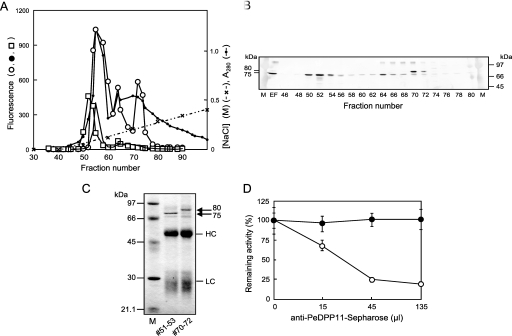
FIGURE 3.
Detection and identification of native PeDPP11 separated by DEAE-Sephacel anion-exchange chromatography. A, a soluble extracellular fraction from P. endodontalis was separated using DEAE-Sephacel chromatography with a 0–0.6 m linear gradient of NaCl in buffer A, as described under “Experimental Procedures.” Hydrolysis of KA (open circle)-, suc-AAA (closed circle)-, and ac-DNLD-MCA (square) of even fractions (10 μl) was measured. B, proteins at fractions 46–80 (even numbers) were separated on SDS-PAGE, then subjected to immunoblotting with anti-PeDPP11 serum (105-diluted). EF, extracellular fraction (10 μl). M, rainbow marker. C, fractions 51–53 and 70–72 (3 ml each) were immunoadsorbed to the PeDPP11 antibody resin, as described under “Experimental Procedures.” Proteins bound to the resin were extracted with SDS-sample buffer, separated on SDS-PAGE, and subjected to N-terminal sequencing. LC and HC, light and heavy chains, respectively, of immunoglobulins. D, fraction 52 or 56 (100 μl) of DEAE-chromatography (panel A) was incubated with 15, 45, or 135 μl of a 50% suspension of a PeDPP11-antibody resin. After rotation at 0 °C for 8 h, samples were centrifuged, and the remaining activity in the supernatant was determined with ac-DNLD-MCA for the sample from fraction 52 (open circle) and suc-AAA-MCA from fraction 56 (closed circle). Values are expressed as percent (mean ± S.D., n = 3) of the control incubated with Sepharose 4B. M, molecular mass markers.
Expression of DPP in E. coli and Production of Polyclonal Antibodies
Recombinant PeDPP11 and PgDPP11 were expressed in E. coli by induction with 0.2 mm isopropyl-β-thiogalactopyranoside at 30 °C for 4 h. Recombinant proteins were purified by Talon affinity chromatography as reported previously (35). For usage as antigens for developing antibodies, Talon affinity column-purified PeDPP11 and PgDPP11 were further separated by size-exclusion gel chromatography with a Sephacryl S200HR column (1 × 86 cm) equilibrated with 20 mm ammonium bicarbonate (pH 8.0). The peak fraction was lyophilized and used for immunization of rabbits.
Construction of P. gingivalis Strain with Disrupted PGN0607
A DNA fragment encoding Asp22–Pro720 of PGN0607 was amplified with a primer set (5PgDPP11D22Bam and 3PgDPP11-P720Bam) and cloned into a pGEM-T Easy vector (pGEM-T Easy-PGN0607). An erythromycin-resistant gene fragment from Bacteroides fragilis was amplified by PCR from pUC19-Em with 5ErmF-AM-Cla and 3ErmF-AM-Cla primers (37, 38), digested with ClaI, and inserted into a ClaI site of pGEM-T Easy-PGN0607 (designated pGEM-T Easy-PGN0607-Em). Electro-transformation of P. gingivalis ATCC 33277 was carried out with SalI-linearlized pGEM-T Easy-PGN0607-Em, and PGN0607-disrupted strains were selected on ABCM agar supplemented with 1 μg/ml menadione and 10 μm erythromycin, according to a previously reported method (39).
Hydrolyzing Activity toward MCA Peptides
Generally, various fractions of native DPP11 at 10 μl or 2 ng of recombinant DPP11 were used for measurement of proteolytic activity in 200 μl of reaction solution composed of 50 mm sodium phosphate (pH 7.0) and containing 5 mm EDTA. The reaction was started with an addition of 20 μm MCA-peptides and continued at 37 °C for 1 h. Fluorescence intensity was measured with excitation at 380 nm and emission at 460 nm with a Fluorescence Photometer F-4000 (Hitachi). In some experiments, appropriate buffers (50 mm) from pH 4 to 10.5 or NaCl concentrations from 0 to 1.6 m were used. Dipeptidyl-MCA substrates not commercially available were prepared from tri- or tetrapeptidyl-MCA (0.4 mm) through digestion with 0.3 μg of GluV8 in 50 mm Tris-HCl (pH 8.0) containing 5 mm EDTA (100 μl) or 0.3 μg of thermolysin in 10 mm sodium borate (pH 8.0) and 2 mm CaSO4 containing 0.005% (v/v) Triton-X100 (100 μl) at 37 °C for 4 h (35). The resultant protease-pretreated MCA peptides were used as substrates for DPP11 at 20 μm as described above.
Ion-exchange Chromatography of P. endodontalis Extracellular Fraction
Chromatographic procedures were performed at 4 °C. Soluble extracellular fraction (100 ml) obtained from dialysis membrane cultures of P. endodontalis was dialyzed against 20 mm Tris-HCl (pH 8.0) containing 1 mm EDTA (Buffer A) and subjected to anion-exchange chromatography on a DEAE-Sephacel column (1.5 × 19 cm) at a flow rate of 2 ml/min. After washing with Buffer A, bound proteins were eluted with a linear gradient of 0–0.6 m NaCl in Buffer A (300 ml). Eighty-drop fractions (∼4 ml) were collected, then the proteolytic activities toward ac-DNLD-, KA-, and succinyl (suc)-AAA-MCA were determined as described above.
Mass Spectrometry
Neuromedin B and its derivatives (1 nmol) were separately incubated with recombinant PeDPP11 (50 nm) or ion-exchange-fractionated fractions (10 μl) in 50 μl of 50 mm Tris-HCl (pH 8.0) at 30 °C for 1 h. The reaction was stopped by the addition of trifluoroacetic acid (final 0.1%), then hydrolysates were adsorbed to ZipTipμ-C18, washed with 0.1% trifluoroacetic acid, and eluted with 50% of acetonitrile containing 5 mg/ml α-cyano-4-hydroxycinnamic under an isocratic condition. The molecular masses of the products were determined by mass spectrometry using a Voyager DE-PRO (Applied Biosystems).
SDS-PAGE, Immunoblotting, and N-terminal Sequencing
Proteins were separated by PAGE in the presence of 0.1% (w/v) of SDS with a polyacrylamide concentration of 11% (w/v), then stained with Coomassie Brilliant Blue or transferred to a polyvinylidene difluoride membrane. Immunoblotting was performed using anti-PeDPP11 or anti-PgDPP11 serum (104–106-fold dilutions), and the blots were visualized with alkaline phosphatase-conjugated anti-rabbit Ig(G+A+M) using 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium (Promega). For N-terminal sequencing, separated proteins were transferred to a Sequi-Blot membrane (Bio-Rad) and stained with Coomassie Brilliant Blue. The N-terminal sequences of stained bands were determined with a model Procise 49XcLC protein sequencer (Applied Biosystems) as previously described (40).
Immunoadsorption of PeDPP11
Recombinant PeDPP11-coupled Sepharose was prepared using a CNBr-Sepharose 4B (GE Healthcare) according to the manufacturer's protocol. A PeDPP11-specific antibody was purified from rabbit antiserum using PeDPP11-Sepharose affinity chromatography, then PeDPP11 antibody-conjugated Sepharose was prepared as above. For immunoadsorption assays, aliquots (100 μl) of DEAE-Sephacel-separated fractions were incubated with 15, 45, or 135 μl of PeDPP11-antibody resin (50% suspension). After rotation at 0 °C for 8 h, samples were centrifuged, and the activity remaining in the supernatant was determined. To determine the N-terminal sequence, fractions (3 ml) obtained by DEAE-Sephacel chromatography were immunoadsorbed to anti-PeDPP11 antibody resin (0.4 ml as 50% suspension), then the resin was washed with 1 ml of PBS 4 times. After centrifugation, materials bound to the resin were extracted with SDS-sample buffer (100 μl) at 94 °C for 10 min, and aliquots were separated on SDS-PAGE.
RESULTS
Degenerate PCR Cloning of Potential DPP
We first attempted to purify DPP7-like activities from extracellular fraction of P. endodontalis, which exhibited higher activity than those of P. gingivalis (33); however, they were not purified to homogeneity, and thus their N-terminal sequences were not determined. Then gene cloning by degenerate PCR was undertaken with a set of primers (DPP7deg244–276 and DPP7deg676–643, supplemental Table S1) designed in consideration of the amino acid homology of DPP7s among P. gingivalis, X. fastidiosa, and S. putrefaciens. Using a combination of degenerate PCR and subsequent genome walking, we cloned a 2303-bp nucleotide sequence carrying a gene of 2154-bp ORF, which encoded a 717-amino acid protein with a deduced molecular mass of 81,090 Da (Fig. 1). This gene was designated as DPP11. The amino acid sequence identity of PeDPP11 to DPP7 from P. gingivalis ATCC 33277 (PGN1479) (21) was 38.3% (Table 1). The arrangement of potential catalytic triad for the serine proteases His84, Asp197, and Ser652 in PeDPP11 was identical to that in PgDPP7 (Fig. 2), and the amino acid sequence at around Ser652 shared significant homology with the Glu-specific serine protease GluV8 (Fig. 2). However, the arrangement of the catalytic triad was distinct from that of P. gingivalis DPPIV (Ser593, Asp668, and His700). Interestingly, a BLAST search identified PGN0607 as having the highest homology (57.9%) to the PeDPP11 gene (Table 1) and the corresponding gene of P. gingivalis W83 (PG1283), which has only four amino acid substitutions with PGN0607 and has been annotated to be an isoform of authentic DPP7 in P. gingivalis, whereas its enzymatic property was not directly examined (27).
TABLE 1.
Amino acid sequence identity among DPPIV, DPP7, and DPP11 of P. gingivalis and P. endodontalis
| DPPs | PeDPP11 | PgDPP11 | PgDPP7 | PgDPPIV | PeDPPIV |
|---|---|---|---|---|---|
| % | % | % | % | % | |
| P. endodontalis DPP11 (AB610284a) | 100 | 57.9 | 38.3 | 13.8 | 12.4 |
| P. gingivalis DPP11 (PGN0607b/MER034628c) | 100 | 38.7 | 11.1 | 11.4 | |
| P. gingivalis DPP7 (PGN1479b/MER014366c) | 100 | 11.6 | 12.6 | ||
| P. gingivalis DPPIV (PGN1469b) | 100 | 55.1 | |||
| P. endodontalis DPPIV (MER192286c/ZP04390817d) | 100 |
FIGURE 2.
Alignment of amino acid sequences of PeDPP11, PgDPP11, and PgDPP7. The amino acid sequence of PeDPP11 was compared with those of PGN0607/PgDPP11 and PGN1479/PgDPP7 from P. gingivalis and residues 232–250 of GluV8 carrying an essential Ser237 (60). Hyphens represent gaps introduced for maximal matching. Common amino acid residues are marked with asterisks, whereas those matched in two DPPs are indicated by dots. Amino acids corresponding to the regions used for amplification of PeDPP11 with degenerate primers are underlined. Amino acid residues to form the active triad of PeDPP11 predicted by comparison with those of PGN0607 and PgDPP7 are indicated by arrows.
When the PeDPP11 gene and PGN0607 were expressed in E. coli, 85-kDa proteins were purified in accordance with their estimated molecular masses (supplemental Fig. S1). However, neither of those proteins showed hydrolyzing activity toward KA-MCA, a typical substrate for DPP7. Furthermore, none of 76 commercially available MCA substrates was cleaved by the recombinant proteins. These findings could be interpreted in the following three ways; they are pseudogenes, they are recombinant proteins that require maturation processing to achieve the activity, or they have novel substrate specificity that cannot be detected with commercial MCA substrates. Although the possibility of pseudogenes cannot be completely excluded, the maintenance of the ORF of 717 amino acids comparable to the 712 amino acid ORF of PgDPP7 and the similarity to PG0607 suggest its integrity. To achieve in vitro maturation, recombinant proteins were incubated with trypsin, chymotrypsin, thermolysin, papain, or GluV8, as reported on recombinant GluV8 (35). However, they noted no activity for KA-MCA (data not shown). Hence, based on the third assumption, we attempted to reveal the peptidase activity of DPP11. To identify and purify native DPP11, antibodies against recombinant PeDPP11 and PgDPP11 were developed. Immunoblotting (supplemental Fig. S1B) and enzyme-linked immunosorbent assay (data not shown) demonstrated that these antibodies reacted with 7–8-fold higher affinities for the original antigens than the counterparts.
Peptidase Activities in Extracellular Fraction of P. endodontalis
Initially, we arranged 76 commercially available mono- to octapeptidyl MCA substrates with or without N-terminal modification in which the P1 position was 14 amino acids other than Cys, His, Ile, Gln, Ser, and Thr, then we determined the peptidase activities of extracellular and cell-associated fractions from P. endodontalis as well as P. gingivalis. As a result, KA-MCA was predominantly hydrolyzed with the P. endodontalis extracellular fraction, whereas the Rgp and Kgp activities were negligible (supplemental Fig. S2, Ref. 33). In addition, we detected activities hydrolyzing suc-AAA- and ac-DNLD-MCA in P. endodontalis, and the hydrolysis was significantly increased by prolonged incubation, suggesting a multiple step reaction. These activities were also present in the P. endodontalis cellular fraction (data not shown). Noticeably, the hydrolyzing activities toward KA-MCA, suc-AAA-MCA, and ac-DNLD were very low in the extracellular fraction of P. gingivalis (supplemental Fig. S2). Based on these observations, we assumed the presence of two kinds of DPPs in P. endodontalis, one that is responsible for the cleavage of KA-MCA and another that cleaves the NH2-X(D/N)↓Xn bonds. Considering the suc group as an N-terminal amino acid, suc-AAA-MCA could be cleaved by the DPP responsible for the cleavage of KA-MCA, most likely to DPP7.
Separation of DPP11 Activity from Extracellular Fraction of P. endodontalis by Anion-exchange Gel Chromatography and Immunochemical Techniques
To determine whether the hydrolyzing activity toward the NH2-X(D/N)↓Xn bonds was related to DPP11, soluble extracellular proteins from P. endodontalis were separated by DEAE-Sephacel chromatography (Fig. 3). The DPP7-like activity hydrolyzing KA-MCA was split into 3 peaks at fractions 56, 64, and 74. Among them, the first major KA-MCA hydrolyzing activity was co-eluted with that for suc-AAA-MCA, further suggesting their identity. The activity for ac-DNLD-MCA was eluted as a major peak at around fraction 52, which was separate from the DPP7-like Ala-specific activity. In addition, the 75-kDa band immunoblotted with the PeDPP11 antibody was co-eluted with the peak of hydrolyzing activity toward ac-DNLD-MCA. Moderate staining was found at 80 kDa at around fraction 70 and weak staining at 75 kDa at fraction 64. These 75- and 80-kDa substances were purified by immunoadsorption, and their N-terminal amino acid sequences were determined. The N-terminal sequence of the 75-kDa molecule, DGGMXLMQQ, coincided with the deduced sequence of PeDPP11 (22DGGMWLMQQ30), with an estimated molecular mass of 78,706 Da (Asp22-Phe717). Although the N-terminal sequence of the 80-kDa species was not determined because of impurity, it was speculated to be a full-length form (Met1–Phe717) with a calculated molecular mass of 81,095 Da. Furthermore, the ac-DNLD-MCA-hydrolyzing activity of fraction 52 was decreased by incubation with PeDPP11-antibody-Sepharose in a dose-dependent manner (Fig. 3), whereas the DPP7-like activity of fraction 56 toward KA-MCA did not change. Thus, the ac-DNLD-MCA hydrolyzing activity was tightly associated with the 75-kDa PeDPP11.
Identification of Peptidase Activity in Recombinant DPP11
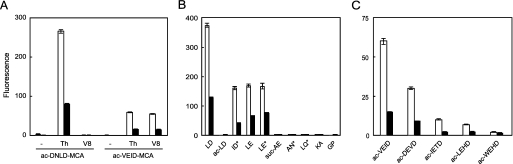
Despite the prominent activity of native PeDPP11, recombinant PeDPP11 did not exhibit activity for ac-DNLD-MCA. Apparently, because tetrapeptidyl-MCA is suboptimal to measure DPP activity, ac-DNLD-MCA was pretreated by thermolysin, which was expected to produce LD-MCA. Consequently, PeDPP11 was shown for the first time to exhibit peptidase activity, whereas hydrolysis was not detected when ac-DNLD-MCA was preincubated with GluV8, which did not produce dipeptidyl-MCA (Fig. 4A). Furthermore, by use of ac-VEID-MCA pretreated with thermolysin or GluV8, both of which could produce ID-MCA, the activity was evidently demonstrated. Similar results were obtained with PgDPP11 (Fig. 4A). The P1-position specificity of DPP11 was further examined using synthesized dipeptidyl substrates, i.e. LD-, ac-LD-, and LE-MCA, and additional dipeptidyl-MCA substrates, such as ID-, LE-, AN-, LQ-MCA, which were enzymatically prepared from ac-VEID-, Z-LLE-, Z-AAN-, Z-LLQ-MCA, respectively, and commercially available suc-AE-, KA-, and GP-MCA. We found that both PeDPP11 and PgDPP11 most efficiently hydrolyzed LD-MCA, although ID- and LE-MCA did so moderately, whereas they did not hydrolyze ac-LD-, suc-AE-, AN-, LQ-, KA-, or GP-MCA (Fig. 4B). These findings demonstrate that DPP11 cleaves peptides with penultimate N-terminal Asp and Glu but does not cleave those either with Asn, Gln, Ala, and Pro at the position or N-terminal modification.
FIGURE 4.
Determination of P1 specificity and P2 preference of DPP11. A, Ac-DNLD- and ac-VEID-MCA were preincubated without (−) or with thermolysin (Th) and GluV8 (V8), as described under “Experimental Procedures.” The hydrolyzing activities of recombinant PeDPP11 (open column) and PgDPP11 (closed column) were determined with the pretreated substrates. B, the activities of PeDPP11 (open column) and PgDPP11 (closed column) hydrolyzing peptidyl MCA substrates were determined. Asterisks indicate enzymatically prepared materials. C, the activities of PeDPP11 (open column) and PgDPP11 (closed column) were determined with acetyl tetrapeptidyl MCAs preincubated with GluV8. All values are shown as the mean ± S.D. (n = 3).
The enzymatic activity of DPP11 was also studied using a bombesin-related human neuropeptide, neuromedin B, and its derivatives, in which Asp residue was located at the first, second, third, or fifth position (Asp1-, Asp2-, Asp3- or Asp5-neuromedin B, respectively) from the N terminus. Asp2-neuromedin B was solely cleaved to the two peptides, NH2-GD-COOH and NH2-LWATGHFM-NH2 (Table 2, data not shown). Moreover, DPP11 did not cleave at NH2-GN↓L, NH2-DG↓L, or NH2-GS↓(D/L) nor at NH2-LW↓A. Therefore, our results explicitly show that DPP11 has genuine Asp- and Glu-specific DPP activity.
TABLE 2.
Peptidase activities of recombinant PeDPP11 and fraction 52 from DEAE-Sephacel chromatography
Neuromedin B and its derivatives were separately incubated with recombinant PeDPP11 or fraction 52 for 1 h at 30 °C. Amounts of the peptide products were semiquantitatively analyzed by MALDI-TOF MS. Substituted amino acid residues from neuromedin B are underlined.
| Substrate | Sequence | Specimen | Degradationa |
|---|---|---|---|
| % | |||
| Neuromedin B | GNLWATGHFM-NH2 | PeDPP11 | 0 |
| Asp1-neuromedin B | DGLWATGHFM-NH2 | PeDPP11 | 0 |
| Asp2-neuromedin B | GDLWATGHFM-NH2 | PeDPP11 | 100 |
| Asp3-neuromedin B | GSDWATGHFM-NH2 | PeDPP11 | 0 |
| Asp5-neuromedin B | GSLWDTGHFM-NH2 | PeDPP11 | 0 |
| Neuromedin B | GNLWATGHFM-NH2 | Fraction 52b | 5 |
| Asp2-neuromedin B | GDLWATGHFM-NH2 | Fraction 52b | 100 |
a Expressed as percent of fragment products losing N-terminal moiety.
b Fraction 52 was from DEAE-Sephacel chromatography (Fig. 3A).
We also tested the activity of fraction 52 from ion-exchange chromatography (see Fig. 3) and found that Asp2-neuromedin B was completely cleaved at NH2-GD↓L, and neuromedin B was partially hydrolyzed at NH2-GN↓L (Table 2). These results suggested that the hydrolysis of ac-DNLD-MCA by fraction 52 as well as the culture supernatant was probably achieved through a 2-step reaction mediated by a co-existing Asn-targeting peptidase and DPP11.
Next, we examined the P2-position preference of DPP11. The dipeptidyl MCA substrates ID-, VD-, and TD-, and HD-MCA were prepared by pretreatment with GluV8 from ac-VEID-, ac-DEVD-, ac-IETD-, and ac-LEHD-/ac-WEHD-MCA, respectively. After pretreatment, these substrates were hydrolyzed by DPP11 with the efficiency in the order of ID> VD > TD > HD-MCA (Fig. 4C). Taken the data of Fig. 4A together, the order of the substrates suitable for DPP11 was aligned to LD > ID > VD > TD > HD-MCA, suggesting a hydrophobic residue preference at the P2 position.
Biochemical Properties of DPP11
The optimum pH of both PeDPP11 and PgDPP11 was 7.0 (supplemental Fig. SA3). This pH was slightly lower than those of the other members, i.e. pH 7.5 for DPPIV, pH 6.5–9.0 for DPP7, and pH 7.0–8.0 for PTP-A from P. gingivalis (26–28). Inhibitor analysis demonstrated that the activity of PeDPP11 was completely blocked by diisopropyl fluorophosphates, moderately by phenylmethylsulfonyl fluoride (PMSF) and 4-(2-methyl)benzenesulfonyl fluoride, and slightly by pepstatin (data not shown), whereas PgDPP11 showed a similar tendency. All of the potent inhibitors were serine protease inhibitors, except for pepstatin, an aspartyl protease inhibitor. The substitution of Ser652 in PeDPP11 and Ser655 in PgDPP11 to Ala completely abolished their hydrolyzing activities toward LD- and LE-MCA (supplemental Fig. S3B). Therefore, Ser652/655 is likely the essential amino acid forming the catalytic triad of the chymotrypsin-superfamily serine proteases, as proposed in Fig. 2, and the target of diisopropyl fluorophosphate, PMSF, and 4-(2-methyl)benzenesulfonyl fluoride.
The activity was maximal without NaCl and consistently declined in a concentration-dependent manner (supplemental Fig. S3C), which was in contrast to PgDPP7, which had a maximal activity at 0.5 m NaCl in 100 mm HEPES buffer (pH 7.0) (27). To confirm the salt effect, the activities hydrolyzing GP-, KA-, and LD-MCA, specific for DPPIV, DPP7, and DPP11, respectively, were compared with the cellular fraction of P. gingivalis (supplemental Fig. S3C). Activities specific for native DPPIV and DPP11 were maximal without NaCl and then decreased in a concentration-dependent manner. In contrast, the activity of DPP7 was maximal at 0.1 m NaCl, confirming that NaCl at appropriate concentrations enhances the activity of DPP7.
Although we attempted to determine their enzymatic parameters, it was impossible to directly measure Km and Vmax for these MCA substrates, because the Km values seemed much higher than the maximal concentration (20 μm) that could be employed in the present study. Hence, kcat/Km toward LD- and LE-MCA was determined by nonlinear regression analysis for fitting the Michaelis-Menten equation using GraphPad Prism software. kcat/Km (s−1m−1) values of PeDPP11 for LD- and LE-MCA were 126,300 ± 4,600 and 55,000 ± 4,400 (mean ± S.D., n = 4), respectively, whereas those of PgDPP11 were 36,100 ± 1,100 and 33,900 ± 300, respectively. Therefore, we concluded that PeDPP11 had kcat/Km values higher than PgDPP11 for the 2 substrates and that LD-MCA was a more preferable substrate as compared with LE-MCA.
Localization of DPP11 and Effect of Gene Disruption on P. gingivalis
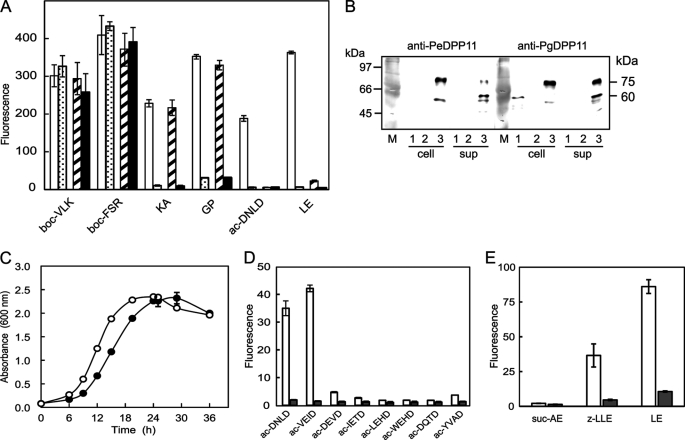
A PgDPP11 gene (PGN0607) disruptant was prepared with P. gingivalis ATCC 33277 by homologous recombination, whereas the same attempt with P. endodontalis was not successful due to feebleness of the bacterium. Potent activities corresponding to Kgp and Rgps were observed in both cell-associated and extracellular forms in the disruptant as in the wild type (Fig. 5A). The hydrolyzing activities toward KA-MCA of DPP7 and GP-MCA of DPPIV, which were exclusively cell-associated, were also demonstrated in both the wild type and disruptant strains. Similarly, the hydrolyzing activities toward ac-DNLD- and LE-MCA were exclusively cell-associated but were abolished in the disruptant, suggesting that PgDPP11 was solely responsible for the hydrolysis in P. gingivalis. Furthermore, these results demonstrated that the DPP11-gene defect did not affect other major proteolytic activities. In addition, the absence of a soluble form of PgDPP11 in P. gingivalis was distinguishable from its presence in P. endodontalis, thus the lack of hydrolyzing activity toward ac-DNLD as shown in supplemental Fig. S2 could be explained by PgDPP11 localization on the cells.
FIGURE 5.
Localization of DPP11 and effect of DPP11 gene disruption in P. gingivalis. A, the hydrolyzing activities toward MCA peptides were determined using aliquots (10 μl) of cellular (open column) and extracellular (dotted column) fractions of P. gingivalis wild type and cellular (hatched column) and extracellular (closed column) fractions of a DPP11 disrupted mutant. boc, t-butyloxycarbonyl-[(2S)-2-amino-3-(benzyloxycarbonyl)propionyl]. B, aliquots (10 μl) of the cellular factions (cell) and culture supernatants (sup) of P. gingivalis wild type (lane 1), disruptant (lane 2), and P. endodontalis (lane 3) were separated on SDS-PAGE and subjected to immunoblotting with anti-PeDPP11 (105-diluted) or anti-PgDPP11 (104-diluted) serum. C, P. gingivalis wild type (open circle) and the disruptant (closed circle) were cultured in ABCM broth supplemented with 1 μg/ml menadione under an anaerobic condition. Cell growth was monitored by measuring the absorbance at 600 nm. A representative result of three separate experiments in triplicates is presented. D and E, the hydrolyzing activities of the cellular fraction (10 μl) from P. gingivalis wild type (open column) and the disruptant (closed column) were determined with MCA peptides carrying Asp (D) or Glu (E) at the P1 position. All values are shown as the mean ± S.D. (n = 3).
Differential localization of DPP11 between P. gingivalis and P. endodontalis was reproduced by immunoblotting (Fig. 5B). A 75-kDa species was observed in the soluble and cell-associated fractions from P. endodontalis, and a truncated 55–60-kDa species was increased in the extracellular fraction. In contrast, a relatively low amount of a 60-kDa species was detected in the cell-associated fraction of the P. gingivalis wild type strain. In the DPP11 gene disruptant, this species disappeared. Exclusive cellular localization was not specific for PgDPP11, as it was also seen in DPPIV and DPP7 (Fig. 5). Hence, the lack of soluble forms of these DPPs may be caused by degradation with gingipains predominantly produced in P. gingivalis.
The growth of the DPP11 gene disruptant was retarded in ABCM medium containing 10 μg/ml erythromycin (Fig. 5C). These results suggest that, although not essential, DPP11 is involved in nutrition metabolism. Growth retardation was also reported in a DPPIV, DPP7, and PTP-A triple disruptant (29). Incomplete arrest in growth of the DPP11 disruptant may be explained by compensation with the remaining DPPs and vice versa. In addition, black pigmentation on sheep blood agar was observed with both the wild type and mutant strains (data not shown).
Complete loss of hydrolysis for ac-DNLD- and LE-MCA by the disruptant suggested an absence of Asp- and Glu-cleaving peptidases other than DPP11. In fact, none of the eight acetylated tetrapeptidyl MCAs carrying Asp at the P1 position were cleaved by the disruptant (Fig. 5D). Moreover, suc-AE-, Z-LLE-, and LE-MCA were scarcely hydrolyzed by the disruptant (Fig. 5E). Taken together, we concluded that DPP11 accounted for all Asp-specific and most Glu-specific cleavages of environmental peptide substrates in P. gingivalis.
Arg670 of DPP11 as an Essential Residue
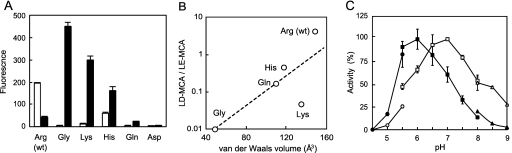
Because Arg670 (PeDPP11 numbering) is a unique amino acid residue that is conserved in all DPP11 orthologues and is thoroughly converted to Gly in all DPP7 orthologues (details described under “Discussion”), we examined whether Arg670 is indispensable for Asp- and Glu-specific DPP11 activities by its substitutions. The LD-MCA hydrolyzing activity became negligible in the mutants with the substitution from Arg670 to Gly, Lys, His, Gln, or Asp, indicating that Arg670 was truly important for DPP11 activity. Moreover, when a 250-fold excess of mutant protein was subjected to the assay, qualitative alterations in the remaining activities among the substituted amino acids emerged (Fig. 6A). Among the mutants, the hydrolysis of LD-MCA was maximal with substitution by His followed by Lys. On the other hand, that of LE-MCA was maximal with the substitution by Gly, followed by Lys, His, and Gln. Again, no hydrolysis was observed in the mutant substituted to Asp. Importantly, in contrast to the wild type of PeDPP11, the hydrolysis of LE-MCA exerted by the mutations was consistently higher than those of LD-MCA, whereas the ratio of hydrolysis of LD-MCA to that of LE-MCA was correlated to the size of the amino acid residue (Fig. 6B). It seems important to note that the van der Waals volume of Asp (91 Å3) is smaller than that of Glu (109 Å3), which reasonably explains why the size reduction from Arg670 (148 Å3) to other amino acids (48–135 Å3) more significantly affected the activity for LD-MCA than for LE-MCA.
FIGURE 6.
Essential role of Arg670 in hydrolyzing activity of DPP11. A, the hydrolyzing activities of wild type PeDPP11 (2 ng) and with substitution of Gly, Lys, His, Gln, or Asp (0.5 μg) were measured with LD-MCA (open column) or LE-MCA (closed column). B, the ratio of hydrolysis of LD-MCA to that of LE-MCA was plotted against the van der Waals volumes of the amino acids at position 670. Data for the Asp substitution were not plotted, as there were no activities for either substrate. C, the hydrolyzing activities of wild type PeDPP11 (open symbol) and R670H (closed symbol) were determined with LE-MCA in 50 mm sodium acetate (circle), sodium phosphate (square), and Tris-HCl (triangle).
Next we investigated whether the basic charge of Arg670 is indispensable for the activity. For this purpose, we determined the pH profile of the R670H mutant. If the positive charge is indispensable for the activity, the pH optimum of the mutant was expected to shift to acidic side. Truly, the R670H mutant possessed the pH optimum one unit lower than that of the wild type (Fig. 6C). Taken together, it is proposed that the guanidinium group of Arg670 directly interacts with the carboxyl group of Asp or Glu in a peptide substrate. Along this line, complete loss of the activity to LD- and LE-MCA observed in the R670D species can be explained by the repulsion between Asp670 and Asp/Glu.
Cooperative Actions of DPP11 and Other Peptidases on Protein Metabolism
The efficiency of peptide utilization of proteinaceous nutrients in the wild type and DPP11 disruptant was calculated under the following hypothetical culture conditions containing human albumin (pI 4.7) and human hemoglobin α12β2 (pI 7.2) as the most probable substrates for P. gingivalis. (i) Rgps and Kgp cleave all Arg-X and Lys-X bonds, respectively, of protein substrates; (ii) subsequently, DPPIV and DPP7 liberate dipeptides with any residues, except for Asp and Glu at the second position, and Pro at the third position; (iii) when Pro is located at the third position from the N terminus, PTP-A liberates tripeptides; (iv) DPP11 liberates dipeptides with Asp and Glu at the second position, except that Pro is located at the third position because its incapability is reasonably postulated by analogy to the properties of DPPIV and DPP7; (v) the resultant di- and tripeptides are incorporated into the bacterium, but free amino acids may not be; (vi) the influence of the disulfide bond is not taken into consideration in this calculation. Consequently, the efficiencies of incorporation of human albumin and hemoglobin in the mutant strain were calculated to be 53.0 and 82.3%, respectively, of the wild type (100%). Therefore, we concluded that DPP11 greatly enhances peptide utilization efficiency, especially for acidic proteins.
DISCUSSION
In this study, we identified a novel Asp- and Glu-specific DPP, DPP11, from P. gingivalis and P. endodontalis. DPP11 possesses essential Ser652 and Arg670 for its activity, the former of which constitutes an active triad with His84 and Asp197 conserved in the serine protease superfamily. It is curious that, although DPP11 has a similar sequence around the essential Ser of GluV8 (Fig. 2), DPP11 is more than double the size of GluV8. Currently, we suspect that DPP11 as well as DPP7 possesses a region, which is associated with other DPPs and PTP-A for co-operative degradation.
PeDPP11 was observed in both cell-associated and soluble extracellular forms, of which the 75-kDa form starting from Asp22 was increased in the soluble extracellular fraction. In contrast, PgDPP11 was solely observed as cell-associated, the same as DPPIV and DPP7 (Figs. 5 and supplemental Fig. S2). The existence of DPP11 is compatible with both experimental observation utilizing Tryptone-based medium (16) and the entire metabolic network model of P. gingivalis to achieve maximal growth (41), whereas Glu and Asp are most intensively consumed from nutritional peptides. These previous studies also indicated that production of butyrate and propionate, estimated end products of Glu and Asp metabolism (16, 20, 41), could become secured under a condition that includes consumption of Glu and Asp in P. gingivalis. Therefore, DPP11 may be one of the pathogenic factors of P. gingivalis.
To date, DPPI-III (42–44), eukaryotic (45) and bacterial (26) DPPIV, DPPV (46), DPPVI (bacterium) (47, 48), DPP6 (eukaryote) (49), DPP7 (bacterial and eukaryote types) (27, 50), DPP8–10 (51–53), fibroblast activation protein α (54), and peptidyl dipeptidase (EC 3.4.15.5) (55) have been reported to be DPP members (supplemental Table S2). Peptidyl dipeptidase, which liberates dipeptides from the C terminus of polypeptides, is localized in an intracellular compartment; thus, it seems to be driven for the breakdown of intracellular proteins (54). Among DPPs excluding peptidyl dipeptidase, eukaryotic DPP7 belongs to the DPPII family (50), and fibroblast activation protein α, DPP6 (eukaryote), and DPP8–10 belong to the DPPIV family in respect to substrate specificity and sequence homology (49, 51–53, 56). In summary, DPPI-DPPV are expressed in eukaryotic organisms, including fungi, mammals, fishes, and plants, whereas DPPIV, DPPVI, and DPP7 are known as bacterial enzymes. DPPVI is expressed in the genera Bacillus and Oceanobacillus and has specificity for d-amino acid-containing peptide moieties of some peptidoglycans (47, 48). Thus, DPPIV, DPP7, and new member DPP11 are involved with dipeptide liberation from extracellular oligopeptides consisting of l-amino acids in bacteria.
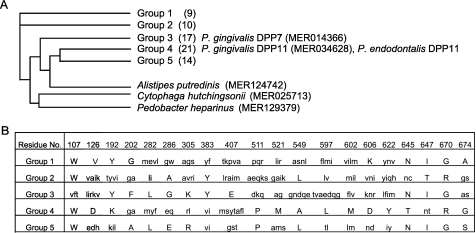
DPP11 from P. gingivalis is classified into the S46.001 subfamily of the S46/DPP7 family in the MEROPS data base (34). Based on sequence homology, S46.001 is divided into 5 major groups (71 members) and an additional 3 members (Fig. 7A and supplemental Table S3). The present findings suggest that Group 3 is genuine DPP7, and Group 4 is DPP11. In studies of a Group 4 species (MER217397) from Porphyromonas uenonis, a bacterium isolated from human urogenital or intestinal tracts (57, 58), the homology was found to be 53.5% to PgDPP11 and 39.7% to PgDPP7, indicating its attribution to DPP11. In addition to the genus Porphyromonas, Group 4/DPP11 consists of members from the genera Bacteroides and Parabacteroides (supplemental Table S3). Based on the present results, it is likely that the substrate specificities of the Group 1, 2, and 5 members are also different from that of DPP7. In particular, Group-5 members are distributed only in the genus Bacteroides, in which at least 9 species simultaneously carry Group-3 (DPP7) and Group-4 (DPP11) genes (supplemental Table S3). Hence, it is possible that Group-5 DPP possesses substrate specificity distinct from DPP7 and DPP11. To address these issues, the proteolytic activities of several members in the S46.001 subfamily are now being examined in our laboratory.
FIGURE 7.
Group-specific amino acid residues in S46.001-subfamily members. A, shown is the phylogenetic tree of the S46.001 subfamily of S46/DPP7 family. Numbers in parentheses indicate number of members (supplemental Table S3). B, 20 amino acids of PeDPP11 completely conserved in Group-3 members and simultaneously not present in any member of Group 4 and vice versa were compared among Group 1–5 members. Amino acids unique to each group are shown in capital letters, whereas others are shown together in each box with small letters. Note that the amino acid at position 670 is completely conserved among all members of Groups 1, 3, and 5 as Gly and Groups 2 and 4 as Arg.
The amino acid residues that define the substrate specificity of DPP11 were considered likely to be altered from those of DPP7 but highly conserved within every group of the S46.001 subfamily. Hence, we selected 20 amino acid residues that are perfectly conserved within all members of either DPP7/Group 3 or DPP11/Group 4 and not simultaneously present in even one member of the other group (Fig. 7B). Among them, Arg670 was the sole residue that was found to be completely conserved in all Group 4 and Group 2 members and was also shown to be completely converted to Gly670 in all Group 3 as well as Group 1 and 5 members. Indeed, Arg670 of PeDPP11 could not be substituted by other amino acids (Fig. 6). Furthermore, the present results suggest that the guanidinium side chain of Arg670 directly interacts with the carboxyl group of target Asp and Glu of a substrate. In turn, Gly670 is absolutely located in all DPP7 family members; thus, a tiny and hydrophobic side chain (hydrogen) of Gly may be indispensable for the acceptance of bulky aliphatic and aromatic residues of a substrate for DPP7. The enhancing effect of NaCl on the activity of DPP7 and its negative effect on DPP11 (supplemental Fig. S3, C and D) were consistent with their substrate specificities, because ions generally strengthen hydrophobic interactions and weaken hydrophilic interactions.
Takahashi and Sato (59) reported on the metabolic efficiency of dipeptides in P. gingivalis and other periodontopathic bacteria, i.e. Prevotella intermedia, Prevotella nigrescens, and Fusobacterium nucleatum. Ammonia production from P. gingivalis was significantly increased in the presence of aspartylaspartate and glutamylglutamate, whereas that production was limited with Asp4–100 and Glu5–100. This observation is now reasonably explained by the P2-position preference of DPP11 to hydrophobic residues (Fig. 4C), as polyglutamic and polyaspartic peptides are not suitable substrates for DPP11. Their study also demonstrated that (Glu)4 enhanced ammonia production in P. nigrescens, suggesting DPP11 expression in this bacterium, although the DPP11 gene has yet to be identified (supplemental Table S3). Therefore, we speculate that DPP11 is distributed in the genus Prevotella as well as the genera Porphyromonas, Bacteroides, and Parabacteroides.
CONCLUSION
We cloned, identified, and characterized a novel type of DPP, designated as DPP11, from P. endodontalis and P. gingivalis, that specifically cleaves at the NH2-(Y(D/E)↓Xn bond, in which hydrophobic residues are preferred at the Y position. The substrate specificity of DPP11 expands the repertoire of substrates covered by DPPIV, DPP7, and PTP-A and provides advantage to these asaccharolytic oral pathogens for survival in an oligotrophic oral environment. P. gingivalis DPP11 is likely keenly involved in the metabolism of abundant acidic residues in proteins and production of end products harmful to host tissues. Hence, DPP11 may become a therapeutic target of chronic periodontitis. Our findings also suggested the distribution of DPP11 in the genera Bacteroides, Parabacteroides, and Prevotella and a further variation of substrate specificity in the S46.001 DPP members.
Supplementary Material
Acknowledgments
We greatly appreciate T. Kobayakawa and F. Tetsuo (Nagasaki University) for technical assistance. We also gratefully acknowledge the generous gift of a plasmid carrying an erythromycin-resistant cassette and helpful advice from Drs. K. Nakayama, M. Naito, M. Shoji, and K. Sato (Nagasaki University).
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture of Japan (to Y. O.-N., T. K. N., and S. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S3.
- Rgp
- Arg-gingipains
- Rgp
- Arg-specific gingipain
- Kgp
- Lys-specific gingipain
- DPP
- dipeptidyl peptidase
- PeDPP and PgDPP
- DPP from P. endodontalis and P. gingivalis, respectively
- PTP-A
- prolyl tripeptidyl peptidase A
- GluV8
- glutamyl endopeptidase from S. aureus
- Z-
- benzyloxycarbonyl-
- MCA
- 4-methylcoumaryl-7-amide
- suc
- succinyl
- ABCM
- anaerobic bacteria culture media.
REFERENCES
- 1. Kastelein P., van Steenbergen T. J., Bras J. M., de Graaff J. (1981) Antonie Van Leeuwenhoek 47, 1–9 [DOI] [PubMed] [Google Scholar]
- 2. van Steenbergen T. J. M., van Winkelhoff A. J., Mayrand D., Grenier D., de Graaff J. (1984) Int. J. Syst. Bacteriol. 34, 118–120 [Google Scholar]
- 3. White D., Mayrand D. (1981) J. Periodontal Res. 16, 259–265 [DOI] [PubMed] [Google Scholar]
- 4. Moore W. E., Holdeman L. V., Smibert R. M., Hash D. E., Burmeister J. A., Ranney R. R. (1982) Infect. Immun. 38, 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Loesche W. J., Syed S. A., Schmidt E., Morrison E. C. (1985) J. Periodontol. 56, 447–456 [DOI] [PubMed] [Google Scholar]
- 6. Iwai T., Inoue Y., Umeda M., Huang Y., Kurihara N., Koike M., Ishikawa I. (2005) (2005) J. Vasc. Surg. 42, 107–115 [DOI] [PubMed] [Google Scholar]
- 7. Kshirsagar A. V., Offenbacher S., Moss K. L., Barros S. P., Beck J. D. (2007) Blood Purif. 25, 125–132 [DOI] [PubMed] [Google Scholar]
- 8. Detert J., Pischon N., Burmester G. R., Buttgereit F. (2010) Arthritis Res. Ther. 12, 218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Steenbergen T. J. M., de Graaff J. (1986) FEMS Microbiol. Lett. 33, 219–222 [Google Scholar]
- 10. van Winkelhoff A. J., Kippuw N., de Graaff J. (1987) J. Dent. Res. 66, 1663–1667 [DOI] [PubMed] [Google Scholar]
- 11. Sundqvist G., Johansson E., Sjögren U. (1989) J. Endod. 15, 13–19 [DOI] [PubMed] [Google Scholar]
- 12. Citron D. M., Poxton I. R., Baron E. J. (2007) Manual of Clinical Microbiology, 9th Ed., Vol. 1, pp. 911–932, American Society for Microbiology, Washington, D. C [Google Scholar]
- 13. Suido H., Nakamura M., Mashimo P. A., Zambon J. J., Genco R. J. (1986) J. Dent. Res. 65, 1335–1340 [DOI] [PubMed] [Google Scholar]
- 14. Grenier D., Gauthier P., Plamondon P., Nakayama K., Mayrand D. (2001) Oral Microbiol. Immunol. 16, 212–217 [DOI] [PubMed] [Google Scholar]
- 15. Curtis M. A., Kuramitsu H. K., Lantz M., Macrina F. L., Nakayama K., Potempa J., Reynolds E. C., Aduse-Opoku J. (1999) J. Periodontal Res. 34, 464–472 [DOI] [PubMed] [Google Scholar]
- 16. Takahashi N., Sato T., Yamada T. (2000) J. Bacteriol. 182, 4704–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takahashi N., Sato T. (2001) J. Dent. Res. 80, 1425–1429 [DOI] [PubMed] [Google Scholar]
- 18. Mayrand D., Holt S. C. (1988) Microbiol. Rev. 52, 134–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holt S. C., Kesavalu L., Walker S., Genco CA. (1999) Periodontol 2000 20, 168–238 [DOI] [PubMed] [Google Scholar]
- 20. Nelson K. E., Fleischmann R. D., DeBoy R. T., Paulsen I. T., Fouts D. E., Eisen J. A., Daugherty S. C., Dodson R. J., Durkin A. S., Gwinn M., Haft D. H., Kolonay J. F., Nelson W. C., Mason T., Tallon L., Gray J., Granger D., Tettelin H., Dong H., Galvin J. L., Duncan M. J., Dewhirst F. E., Fraser C. M. (2003) J. Bacteriol. 185, 5591–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Naito M., Hirakawa H., Yamashita A., Ohara N., Shoji M., Yukitake H., Nakayama K., Toh H., Yoshimura F., Kuhara S., Hattori M., Hayashi T., Nakayama K. (2008) DNA Res. 15, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujimura S., Shibata Y., Nakamura T. (1992) Oral Microbiol. Immunol. 7, 212–217 [DOI] [PubMed] [Google Scholar]
- 23. Chen Z., Potempa J., Polanowski A., Wikstrom M., Travis J. (1992) J. Biol. Chem. 267, 18896–18901 [PubMed] [Google Scholar]
- 24. Pike R., McGraw W., Potempa J., Travis J. (1994) J. Biol. Chem. 269, 406–411 [PubMed] [Google Scholar]
- 25. Kadowaki T., Yoneda M., Okamoto K., Maeda K., Yamamoto K. (1994) J. Biol. Chem. 269, 21371–21378 [PubMed] [Google Scholar]
- 26. Banbula A., Bugno M., Goldstein J., Yen J., Nelson D., Travis J., Potempa J. (2000) Infect. Immun. 68, 1176–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Banbula A., Yen J., Oleksy A., Mak P., Bugno M., Travis J., Potempa J. (2001) J. Biol. Chem. 276, 6299–6305 [DOI] [PubMed] [Google Scholar]
- 28. Banbula A., Mak P., Bugno M., Silberring J., Dubin A., Nelson D., Travis J., Potempa J. (1999) J. Biol. Chem. 274, 9246–9252 [DOI] [PubMed] [Google Scholar]
- 29. Ito K., Nakajima Y., Xu Y., Yamada N., Onohara Y., Ito T., Matsubara F., Kabashima T., Nakayama K., Yoshimoto T. (2006) J. Mol. Biol. 362, 228–240 [DOI] [PubMed] [Google Scholar]
- 30. Kiyama M., Hayakawa M., Shiroza T., Nakamura S., Takeuchi A., Masamoto Y., Abiko Y. (1998) Biochim. Biophys. Acta 1396, 39–46 [DOI] [PubMed] [Google Scholar]
- 31. Krieger T. J., Bartfeld D., Jenish D. L., Hadary D. (1994) FEBS Lett. 352, 385–388 [DOI] [PubMed] [Google Scholar]
- 32. Oda H., Saiki K., Tonosaki M., Yajima A., Konishi K. (2009) J. Periodontal Res. 44, 362–367 [DOI] [PubMed] [Google Scholar]
- 33. Kon A. (2002) Dent. J. Iwate Med. Univ. 27, 187–196 (in Japanese) [Google Scholar]
- 34. Rawlings N. D., Barrett A. J., Bateman A. (2010) Nucleic Acids Res. 38, D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nemoto T. K., Ohara-Nemoto Y., Ono T., Kobayakawa T., Shimoyama Y., Kimura S., Takagi T. (2008) FEBS J. 275, 573–587 [DOI] [PubMed] [Google Scholar]
- 36. Ikeda Y., Ohara-Nemoto Y., Kimura S., Ishibashi K., Kikuchi K. (2004) Can. J. Microbiol. 50, 493–498 [DOI] [PubMed] [Google Scholar]
- 37. Nakayama K., Kadowaki T., Okamoto K., Yamamoto K. (1995) J. Biol. Chem. 270, 23619–23626 [DOI] [PubMed] [Google Scholar]
- 38. Shoji M., Ratnayake D. B., Shi Y., Kadowaki T., Yamamoto K., Yoshimura F., Akamine A., Curtis M. A., Nakayama K. (2002) Microbiology 148, 1183–1191 [DOI] [PubMed] [Google Scholar]
- 39. Ueshima J., Shoji M., Ratnayake D. B., Abe K., Yoshida S., Yamamoto K., Nakayama K. (2003) Infect. Immun. 71, 1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ohara-Nemoto Y., Ono T., Shimoyama Y., Kimura S., Nemoto T. K. (2008) Biol. Chem. 389, 1209–1217 [DOI] [PubMed] [Google Scholar]
- 41. Mazumdar V., Snitkin E. S., Amar S., Segrè D. (2009) J. Bacteriol. 191, 74–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McDonald J. K., Schwabe C. (1971) Proteases in Mammalian Cells and Tissues, pp. 311–391, North Holland Publishing, Amsterdam [Google Scholar]
- 43. McDonald J. K., Reilly T. J., Zeitman B. B., Ellis S. (1968) J. Biol. Chem. 243, 2028–2037 [PubMed] [Google Scholar]
- 44. Ellis S., Nuenke J. M. (1967) J. Biol. Chem. 242, 4623–4629 [PubMed] [Google Scholar]
- 45. Oya H., Harada M., Nagatsu T. (1974) Arch. Oral Biol. 19, 489–491 [DOI] [PubMed] [Google Scholar]
- 46. Beauvais A., Monod M., Debeaupuis J. P., Diaquin M., Kobayashi H., Latgé J. P. (1997) J. Biol. Chem. 272, 6238–6244 [DOI] [PubMed] [Google Scholar]
- 47. Vacheron M. J., Guinand M., Françon A., Michel G. (1979) Eur. J. Biochem. 100, 189–196 [DOI] [PubMed] [Google Scholar]
- 48. Guinand M., Vacheron M. J., Michel G., Tipper D. J. (1979) J. Bacteriol. 138, 126–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wada K., Yokotani N., Hunter C., Doi K., Wenthold R. J., Shimasaki S. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Maes M. B., Lambeir A. M., Gilany K., Senten K., Van der Veken P., Leiting B., Augustyns K., Scharpé S., De Meester I. (2005) Biochem. J. 386, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Abbott C. A., Yu D. M., Woollatt E., Sutherland G. R., McCaughan G. W., Gorrell M. D. (2000) Eur. J. Biochem. 267, 6140–6150 [DOI] [PubMed] [Google Scholar]
- 52. Dubois V., Lambeir A. M., Vandamme S., Matheeussen V., Guisez Y., Scharpé S., De Meester I. D. (2010) Biochem. Biophys. Acta 1804, 781–788 [DOI] [PubMed] [Google Scholar]
- 53. McNicholas K., Chen T., Abbott C. A. (2009) Clin. Chem. Lab. Med. 47, 262–267 [DOI] [PubMed] [Google Scholar]
- 54. Park J. E., Lenter M. C., Zimmermann R. N., Garin-Chesa P., Old L. J., Rettig W. J. (1999) J. Biol. Chem. 274, 36505–36512 [DOI] [PubMed] [Google Scholar]
- 55. Vimr E. R., Green L., Miller C. G. (1983) J. Bacteriol. 153, 1259–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Qi S. Y., Riviere P. J., Trojnar J., Junien J. L., Akinsanya K. O. (2003) Biochem. J. 373, 179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Conrads G., Citron D. M., Tyrrell K. L., Horz H. P., Goldstein E. J. (2005) Int. J. Syst. Evol. Microbiol. 55, 607–613 [DOI] [PubMed] [Google Scholar]
- 58. Finegold S. M., Vaisanen M. L., Rautio M., Eerola E., Summanen P., Molitoris D., Song Y., Liu C., Jousimies-Somer H. (2004) J. Clin. Microbiol. 42, 5298–52301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Takahashi N., Sato T. (2002) Oral Microbiol. Immunol. 17, 50–54 [DOI] [PubMed] [Google Scholar]
- 60. Carmona C., Gray G. L. (1987) Nucleic Acids Res. 15, 6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.