Abstract
Subunit “a” is associated with the membrane-bound (VO) complex of eukaryotic vacuolar H+-ATPase (V-ATPase) acidification machinery. It has also been shown recently to be involved in diverse membrane fusion/secretory functions independent of acidification. Here, we report the crystal structure of the N-terminal cytosolic domain from the Meiothermus ruber subunit “I” homolog of subunit a. The structure is composed of a curved long central α-helix bundle capped on both ends by two lobes with similar α/β architecture. Based on the structure, a reasonable model of its eukaryotic subunit a counterpart was obtained. The crystal structure and model fit well into reconstruction densities from electron microscopy of prokaryotic and eukaryotic V-ATPases, respectively, clarifying their orientations and interactions and revealing features that could enable subunit a to play a role in membrane fusion/secretion.
Keywords: A/V-type ATPase, EM fit, membrane fusion/secretion, protein structure, proton pumping
V-ATPase is a ubiquitous multi-subunit proton pump that acidifies a wide variety of intracellular compartments, which in turn affects many biological processes, including membrane trafficking, protein degradation and coupled transport of small molecules and pH homeostasis.1, 2 The subunits are organized into the peripheral, soluble V1 and membrane-bound VO sectors. The V1 sector hydrolyzes ATP, providing the energy to pump protons via VO. V-ATPases are found in archaea and eubacteria (also identified as A-ATPases) and in eukaryotic cells, where they reside in both intracellular organelles (e.g., golgi, endosomes, lysosomes and secretory vesicles) and plasma membranes of certain cells (e.g. renal intercalated cells, osteoclasts and macrophages).3 Reversible dissociation of the V1 and VO sectors has been observed in yeast and other mammalian cells.4, 5 The composition, mass, stoichiometry in either V1 or VO sector and proposed organizations of the subunits of the eukaryotic and prokaryotic rotatory V-ATPases are shown in Supplementary Table S1 and Fig. S1.
Subunit “a” of the VO sector is highly conserved across eukaryotic species and exists in multiple isoforms with unique intracellular localizations.6 It is the largest subunit of V-ATPases and partitioned almost equally into an N-terminal cytosolic (hereafter named acyt) domain and a C-terminal integral membrane (amem) domain with six to nine transmembrane α-helices. The same is true of its bacterial homolog subunit “I”. The cytosolic domain of subunit a or I is believed to participate in linking VO with components of the V1 complex, particularly the peripheral stalks,1, 2 and the membrane-bound domain is implicated to play an important role in the mechanism of proton transfer.7 Recently, subunit a homologs have been demonstrated to be involved in membrane fusion or secretion, independent of the V-ATPase proton pumping activity in numerous systems. This includes vacuolar fusion in yeast,8 synaptic vesicle exocytosis in Drosophila,9 secretion of hedgehog related proteins in worm,10 secretion of insulin from pancreatic islets in mice11 and phagosomal-endosomal fusion in microglial-mediated neuronal degradation in zebrafish.12 The role and regulation of subunit a in these multiple functions are largely unknown. As a first step in understanding the structure-function relationship of subunit a in the two basic independent cellular functions of acidification and membrane fusion, we here report the crystal structure of the homologous subunit I cytosolic N-terminal domain (Icyt) from the thermophilic bacterium Meiothermus ruber and its high-fidelity fit, as well as that of an acyt model, to EM reconstructions of intact prokaryotic and eukaryotic V-ATPases.
Icyt crystal structure
The recombinant M. ruber Icyt (residues 1-344) was obtained and its crystal structure determined by single wavelength anomalous dispersion technique and refined to 2.6 Å resolution (see Supplementary Methods and Table 1). A portion of the electron density is shown in Supplementary Fig. S2. The C-terminal 43 residues show no electron density and likely indicate a flexible segment (Supplementary Methods).
With a long central α-helix bundle capped on both ends by two similar lobes, the Icyt structure has the shape of a barbell, with the bar/linker bent (Figs. 1a-b). The central linker is formed by a bundle of two long tandems of α-helices; the first and longest consists of αIII-αIV-αV, and the second, which is anti-parallel to the first, consists of αVIII-IX-αX. This topological arrangement creates three approximate pairs of α-helices III-X, IV-IX, and V-VIII. The curvature of the bar is caused by four kinks between α-helices, two in each of the tandem of helices (Supplementary Table S2). Another α-helix (II), which is close to the proximal lobe, is associated with the linker by interacting with segments of α-helices III, IV and X.
Fig. 1.
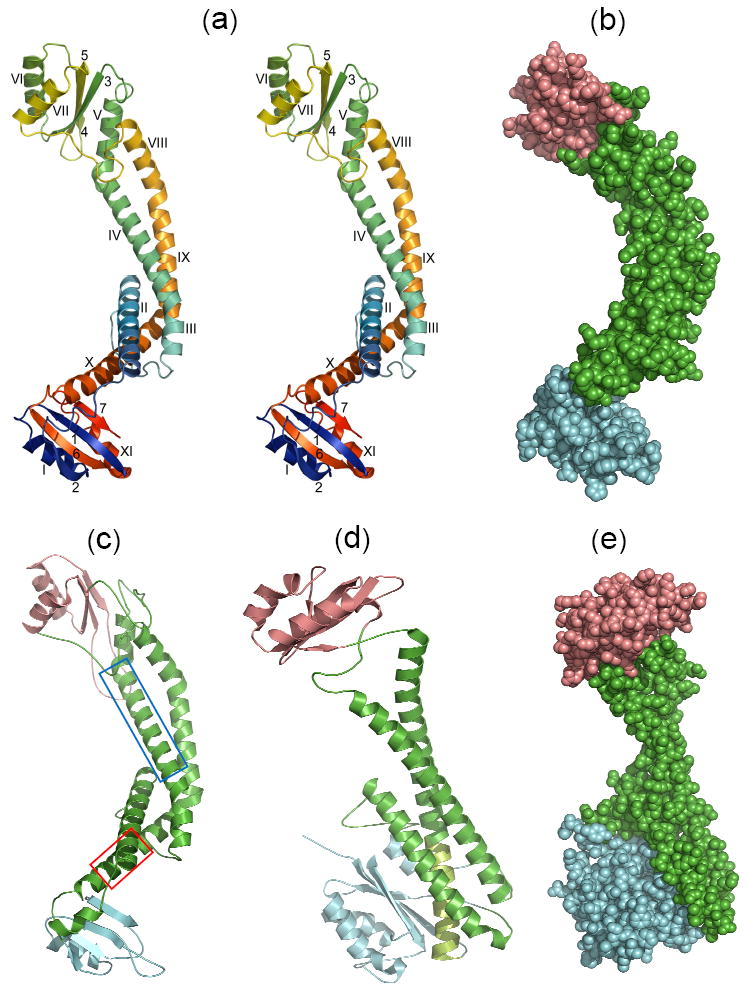
X-ray and modeled structures in similar orientations. (a) Stereoview of the ribbon backbone trace of the Icyt crystal structure color ramped from the N- to the C-terminal ends, and α-helices and β-sheet strands identified with Roman and Arabic numbers, respectively. Superposition of the distal (top) and proximal (bottom) lobes showed an rmsd of 5.3 Å over 23 α-carbons. (b) Space-filling representation of Icyt structure. The long linker region of anti-parallel pairs of α-helices is colored green, and the distal and proximal lobes are distinguished by salmon and cyan colors, respectively. These distinctions are adopted in c to e. (c) Model of acyt of the neuronal zebrafish subunit a1 obtained as described in the Supplementary Methods and Table S3. The α-helices boxed in blue (αV) and red (αX) in the sequence alignment which are equivalent to Icyt’s αIV and αX, respectively, have been implicated as sites for binding of t-SNARE protein syntaxin or SNAP259 and Ca2+-calmodulin, correspondingly30 (described further in Supplementary Fig. S3). (d) Ribbon trace and (e) space-filling representation, of the yeast subunit C (Vma5p) crystal structure (PDB 1U7L).14 The match from the DALI database13 between Icyt and subunit C structure gave Z score of 6.8 and rmsd of 10.1 Å over 216 aligned residues and sequence identity of 7%. A large substructure is formed by extensive hydrophobic interactions of the C-terminal segment of the longest α helix (light green) of the linker with the inner face of β-sheet of the foot domain (blue).
The two lobes, similar in size, adopt comparable α/β architecture (Fig. 1a). The lobe at one (distal) end is made from one continuous segment near midpoint of the polypeptide chain. The β-sheet is composed of three anti-parallel β-strands (3, 4 and 5) and flanked on one side by α-helices VI and VII and on the opposite side by the αV helix of the central helix bundle. In contrast, the second lobe at the opposite (proximal) end is folded from the N- and C-termini segments, each contributing equal numbers of β-sheet strands (1 and 2 and 6 and 7, respectively) and α-helices (I and XI, respectively) which stack against one face of the β-sheet. The opposite side of the β-sheet is devoid of a helix.
Structural comparison of Icyt with the yeast V-ATPase subunit C
The search for structural similarity of Icyt structure against the DALI database13 revealed a fairly good match with the structure of the yeast V-ATPase V1 subunit C (Vma5p),14 despite no apparent sequence similarity. Subunit C also has a also a central bundle of anti-parallel pair of α-helices capped on both ends by domains (identified as “head” and “foot”), although one is much larger than the other (Fig. 1d). There are several additional major differences. First, the linker of α-helices is curved, mediated by kinks in Icyt, whereas that in subunit C is continuous and displays smooth bends (Figs. 1a and d). Second, the entire length of the linker in Icyt is fairly uniform in size, whereas that in subunit C is described as a “neck” coupled to a large substructure “below” it (Figs. 1b, d-e). Third, the proximal lobe in Icyt is loosely connected at a right angle (like in a foot) to the end of the linker, suggesting possible rigid-body rotation or articulation between them (Fig. 1a). In contrast, the foot domain in subunit C, the counterpart of the proximal lobe, is integrated into a larger substructure through extensive non-polar interactions of its large β-sheet in parallel with the last third of the longer of the two α-helices and neighboring residues below the neck (Figs. 1d-e). Similar but substantially fewer interactions occur in the Icyt distal lobe, between the smaller β-sheet and αV helix of the linker (Figs. 1a-b). Fourth, although more similar in sizes, the packing of the distal lobe of Icyt to the linker tip is completely different from that of the head domain of subunit C (Figs. 1a-b and d-e). It has been suggested that eukaryotic subunit C and prokaryotic Icyt are functionally identical due to their ability to cross-link two peripheral stalks, which is composed of heterodimer of subunits E and G (EG for short) in V1 (briefly summarized in reference 15), but given the considerable differences described above, the geometries and molecular nature of the cross-links via the lobes in Icyt and domains in subunit C would be very different (discussed further below).
Fits of the N-terminal domains of subunits I and a into EMs of intact V-ATPases
The fits that are described as follows allow the Icyt or acyt domain to be visualized for the first time in both intact prokaryotic or eukaryotic V-ATPases, respectively, providing insights into its interactions and major role. With the exception of subunit I, the crystal structures of all of the subunits of prokaryotic V-ATPases have been determined (Supplementary Table S1) and used to fit into mass density of reconstructions of cryo- and negative stain-EMs of intact prokaryotic and eukaryotic V-ATPases, thereby generating overall architectures of V-ATPases in both taxonomic systems (e.g., see Supplementary Fig. S1; further reviewed in ref. 2). The associations of Icyt are believed to differ from those of its eukaryotic counterpart acyt in their respective V-ATPase due entirely to the presence of V1 subunits H and C that are unique in eukaryotes. The rod-like Icyt density in the reconstructions of prokaryotic V-ATPase (e.g., T. thermophilus15 or Pyrococcus furiosus16) is predicted to lie parallel to the cytoplasmic surface of the membrane. The elongated Icyt crystal structure fits very well into the 16 Å cryo-EM density of the T. thermophilus V-ATPase reconstruction17 (Supplementary Methods) in this parallel orientation with its two flanking lobes in close proximity to the N-termini of the equally well-fitted EG heterodimer structure15 of the peripheral stalks (Fig. 2a). Furthermore, the proximal lobe, at right angle to the linker, is near the density connecting Icyt to Imem. The inner curvature of the long central helical bundle (Figs. 1a and b) interfaces with the funnel-shaped, all helix-containing subunit C, the prokaryotic homolog of eukaryotic subunit d that serves as a socket to attach the V1 central stalk to the L-ring. Although the two lobes of Icyt exhibit a similar motif, there are enough differences between them, especially in the relative orientations with respect to the linker (Figs. 1a-b and 2a) and the somewhat low sequence conservation (23% identity), to indicate that the molecular nature of the coupling with the identical stalks will be largely dissimilar. Notwithstanding the differences, the picture that has emerged supports the notion that Icyt substitutes the exclusively eukaryotic subunits C and H (discussed below), limiting to only two EG peripheral stalks in prokaryotic V-ATPases.
Fig. 2.
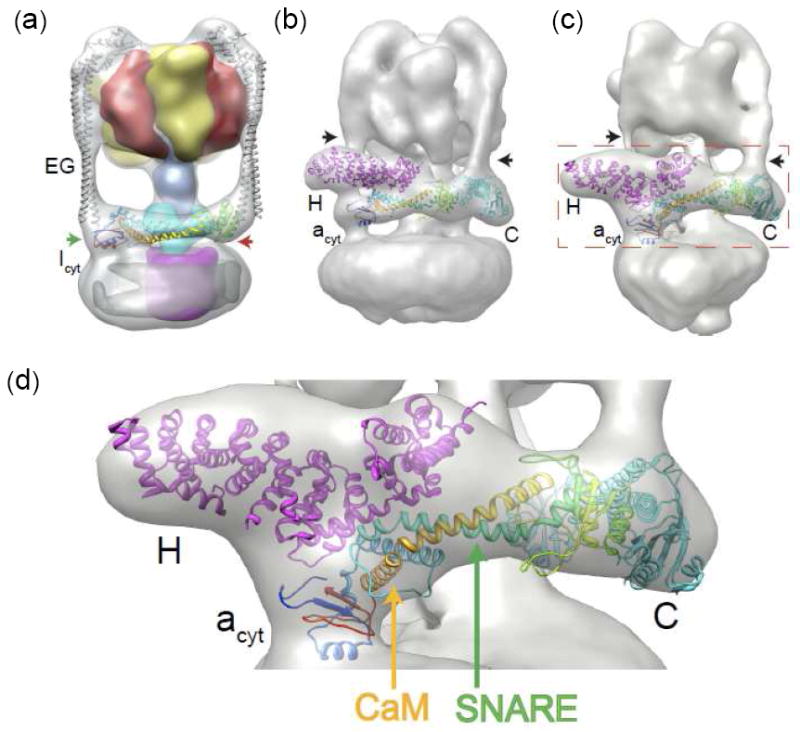
Fitting of the Icyt crystal structure and acyt model to respective EM densities. The methods are described in the Supplementary Information. All the fits are unique as assessed by e3fhstat.py in EMAN231 which indicated 99th percentile for volume inclusion and real-space correlation (for additional statistics see Supplementary Methods). (a) Fit of the crystal structures of Icyt (color ramped ribbon trace) and the T. thermophilus heterodimer of subunits EG15 (gray trace) into the 16 Å cryo-EM density of the T. thermophilus V-ATPase.17 The surface density (available from ref. 15) of subunits A is colored yellow; B, red; D, blue; C, cyan; and c, magenta. The green and red arrows indicate the proximal and distal lobes of Icyt, respectively. (b) Fit of the zebrafish acyt model (Fig. 1c) color ramped trace and crystal structures of yeast subunits H20 and C14 into the 25 Å negative-stain EM reconstruction density of yeast V-ATPase.18 The black arrows in this figure and that in panel c indicate the vertical column of density of two of the three peripheral stalks. (c) Fit of the identical components identified in b into the 17 Å cryo-EM reconstruction density of M. sexta V-ATPase.19 Red dashed rectangle delineates an area of the V-ATPase fit that has been magnified in (d). In both Figs. b and c, the third peripheral stalk is in the “rear” and obstructed from view. Compared to yeast V-ATPase, in M. sexta, acyt and subunit H are rotated clockwise by ~25° and ~30°, respectively, about a horizontal axis, whereas subunit C is rotated clockwise by ~ 10° about an axis normal to the plane of the figure. This level of difference may be attributed to the differences in the imaging methods and the resulting resolutions. (d) A magnified region of the zebrafish acyt fit delineated in panel c. Potential binding sites for SNARE and Ca2+-calmodulin (CaM) are indicated by green and orange arrows, respectively.
Unlike for Icyt, there is at present no consensus for the cytoplasmic side membrane orientation of the acyt density. We therefore sought to assess the fit of acyt to two EM reconstructions of intact eukaryotic V-ATPases, one 25 Å negative stain EM from yeast18 and the other to a 17 Å cryo-EM of Manduca sexta19 which portrayed two significantly different orientations. The assumed acyt density in the yeast reconstruction is portrayed in a parallel orientation and in a position to bridge the fitted crystal structures of subunits H 20 and C14. In sharp contrast, in M. sexta it is assigned to a large protrusion or a “nose” which projects above the membrane surface (or vertical orientation) and is in contact with only subunit H which in turn is in close proximity to subunit C. The nose (or Arm 2) is also present in the yeast reconstruction, but is assigned to subunit H. The two different orientations lead to two significantly different arrangements of subunits H and C which, together with the acyt domain, form the “collar” around the mid-section of the V-ATPase.18, 19 They also have ramifications in the role of acyt in membrane fusion (discussed below). Yet a third geometry has been proposed in a very recent review article by fitting subunit H to the nose density in the M. sexta cryo-EM, which in turn contacts the acyt density at an assumed alternate location and also subunit C.2 To undertake the fitting, we obtained fairly reasonable and very similar acyt models of subunit a from fly, zebrafish and yeast by threading each of their sequences into the Icyt crystal structure (Supplementary Methods and Table S3). The zebrafish acyt model (Fig. 1c), which gave somewhat better modeling statistics (Supplementary Table S3), together with the crystal structures of yeast subunits H and C, was fitted first into the reconstruction of the yeast V-ATPase and then M. sexta (Supplementary Methods). In similar fashion, all three components fit well into respective densities in both reconstructions (Figs. 2b, c; see also Supplementary Methods). Both fits form a consensus which shows that the acyt model lies parallel to the plane of the membrane and acts as a bridge between H and C subunits, all three components forming the expansive collar. This also indicates that subunits H and C are far apart and devoid of intermolecular contacts, contrary to the two previous fits into the M. sexta reconstruction which place them in close proximity.2, 19 It further resolves other conflicting and inconsistent portrayals of acyt, particularly its association within the collar and with other components of V1 and VO. Obviously, the fits of subunit H and C and the acyt density deduced in the yeast V-ATPase reconstruction18 (also portrayed in Supplemental Fig. S1b) more closely matches the consensus fit.
The following are other new finer features of the involvements of acyt revealed by the consensus fit as typified in Figs. 2c and d. First, the proximal lobe, which is roughly at right angle to the linker (Fig. 1a and b) and just below the segment connecting the N- and C-terminal domains of subunit H, is directed towards the density linking the acyt and amem domains. Second, the interfacing of acyt with subunit d resembles those observed in Icyt with the equivalent prokaryotic subunit C (Fig. 2a and Supplementary Table S1). Third, parts of the linker, mostly the α-helices at the proximal half, and the proximal lobe make contacts with α-helices in both the N- and C-terminal domains of subunit H. The larger N-terminal domain of subunit H, playing a greater role, together with the proximal lobe of acyt, anchors one EG peripheral stalk (or Stator 1). Fourth, the exposed α-helices in the distal lobe interface with those flanking the β-sheet of the broad foot of subunit C (Fig. 1d), and together serve to dock another peripheral stalk (Stator 2). The smaller head domain of subunit C contacts the third stalk or Stator 3 (not visible in Figs. 2b and c). Fifth, based on the features described above, the strengths of the interactions of acyt with subunits H and C are likely to vary, stronger with H since they cover more area involving the proximal lobe and the linker helices and somewhat weaker with subunit C since they entail mainly the distal lobe. Briefly, the molecular picture emerging depicts acyt playing important roles by tethering subunits H and C and, in collaboration with the two subunits, by docking two peripheral stalks.
A variety of experimental techniques (e.g., cross-linking, yeast two hybrid, pull down, etc.) provide evidence in support of the interactions described above, including those of subunit a with subunits H,21, 22, C23, E and G,24, 25, as well as subunit H with E and G26 and subunit C with E and G.23, 27, 28 The interactions, particularly the likelihood of a stronger affinity of acyt with subunit H than with C, are consistent with the regulation of the V-ATPases by reversible disassembly and reassembly of the two sectors which is initiated by undocking of subunit C.4 Moreover, negative stain EM reconstruction of VO in isolation indicated that acyt remains in the parallel orientation.29
The fit of Icyt shows a simpler and shorter collar confined solely to Icyt with its two lobes interacting with the N-terminal ends of the peripheral stalks. In sharp contrast, the consensus fit of acyt demonstrates a more expansive, intricate and nonuniform collar as a result of acyt coupling subunit H to C. It is also noteworthy that the two or three docking sites on the collar of the prokaryotic or eukaryotic V-ATPase, respectively, for equivalent peripheral stalks in each kingdom are dissimilar. These observations elicit three key questions that remain to be elucidated. How do the different docking sites recognize and bind presumably with different affinities the stalk, how are the dramatic differences between the eukaryotic and propkaryotic geometries of the collars and consequently their interactions with the V1 and VO components reflected in the detailed molecular mechanism of proton pumping and how did the collar in the eukaryotic V-ATPase evolve?
SNARE and calmodulin binding domains of subunit a
Presumably V0 plays a role in membrane fusion and secretion in the absence of the V1 sector. In this case, the increase in accessibility and conformational flexibility, the potential rigid body rotations between the linker and proximal or distal lobe or both, the presumed disordered segment in the C-terminal end of the domain and the potential pliability of the linker are features that would enable acyt to play a role in membrane fusion and secretion through its interactions with partner proteins. We envision the acyt in the VO state sampling between the horizontal and semi-vertical orientations. The conserved segments of acyts implicated to interact with target-SNARE proteins in synaptic vesicle exocytosis9 and Ca2+- calmodulin,30 whose signal transduction function remains to be elucidated, map to α-helices V and X, respectively (Figs. 1c and 2d and Supplementary Fig. S3). However, since both α-helices are a part of the linker of a long anti-parallel pair of helices, acyt would have to undergo a partner protein-induced conformational change to fully accommodate either SNAREs or Ca2+- calmodulin. Target-SNARE proteins and Ca2+-calmodulin have been demonstrated to bind to their respective isolated acyt binding segments.9, 30 The sequences of these segments are highly conserved in homologs of eukaryotic subunit a (e.g., see Supplementary Fig. S3, Table S3 and ref. 30). Lack of sequence conservation between these specific binding segments and Icyt (Supplementary Fig. S3) suggests that the SNARE and calmodulin binding properties of acyts for membrane fusion/secretion have emerged later in evolution.
Our work presented here indicates that the cytosolic acyt domain is a hub for functions important for the two basic cellular functions of acidification and membrane fusion. It advances molecular understanding of the roles of the cytoplasmic N-terminal domain of subunit a or I in V-ATPases, as well as provides a framework for further functional and structural dissections of the function and biology of subunit a proteins in eukaryotic membrane fusion and secretion.
Protein Data Bank accession number
The coordinates and structure factors of Icyt have been deposited with accession code 3rrk.
Supplementary Material
Acknowledgments
We are grateful to Jing He and Jennifer Fallon for technical assistance. We also thank the staff members for assistance in data collection on the APS beam line 19-ID. We thank Katherine Sippel for critical reading of the manuscript. M.L.B. is supported by a grant from NIH National Center for Research Resources (P41RR002250). The work was supported by grants from NIH (5R01-GM088803) and The Welch Foundation (Q-581) to F.A.Q.
Abbreviations used
- EM
electron microscopy
- SNARE
soluble N-ethyl maleimide sensitive fusion protein receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 2.Muench SP, Trinick J, Harrison MA. Structural divergence of the rotary ATPases. Q Rev Biophys. 2011:1–46. doi: 10.1017/S0033583510000338. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida M, Muneyuki E, Hisabori T. ATP synthase-a marvellous rotary engine of the cell. Nat Rev Mol Cell Biol. 2001;2:669–77. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 4.Kane PM. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J Biol Chem. 1995;270:17025–32. [PubMed] [Google Scholar]
- 5.Sumner JP, Dow JA, Earley FG, Klein U, Jager D, Wieczorek H. Regulation of plasma membrane V-ATPase activity by dissociation of peripheral subunits. J Biol Chem. 1995;270:5649–53. doi: 10.1074/jbc.270.10.5649. [DOI] [PubMed] [Google Scholar]
- 6.Toei M, Saum R, Forgac M. Regulation and isoform function of the V-ATPases. Biochemistry. 2010;49:4715–23. doi: 10.1021/bi100397s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawasaki-Nishi S, Nishi T, Forgac M. Arg-735 of the 100-kDa subunit a of the yeast V-ATPase is essential for proton translocation. Proc Natl Acad Sci U S A. 2001;98:12397–402. doi: 10.1073/pnas.221291798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters C, Bayer MJ, Buhler S, Andersen JS, Mann M, Mayer A. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature. 2001;409:581–8. doi: 10.1038/35054500. [DOI] [PubMed] [Google Scholar]
- 9.Hiesinger PR, Fayyazuddin A, Mehta SQ, Rosenmund T, Schulze KL, Zhai RG, Verstreken P, Cao Y, Zhou Y, Kunz J, Bellen HJ. The v-ATPase V0 subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila. Cell. 2005;121:607–20. doi: 10.1016/j.cell.2005.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liegeois S, Benedetto A, Garnier JM, Schwab Y, Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J Cell Biol. 2006;173:949–61. doi: 10.1083/jcb.200511072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun-Wada GH, Toyomura T, Murata Y, Yamamoto A, Futai M, Wada Y. The a3 isoform of V-ATPase regulates insulin secretion from pancreatic beta-cells. J Cell Sci. 2006;119:4531–40. doi: 10.1242/jcs.03234. [DOI] [PubMed] [Google Scholar]
- 12.Peri F, Nusslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133:916–27. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- 13.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drory O, Frolow F, Nelson N. Crystal structure of yeast V-ATPase subunit C reveals its stator function. EMBO Rep. 2004;5:1148–52. doi: 10.1038/sj.embor.7400294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee LK, Stewart AG, Donohoe M, Bernal RA, Stock D. The structure of the peripheral stalk of Thermus thermophilus H+-ATPase/synthase. Nat Struct Mol Biol. 2010;17:373–8. doi: 10.1038/nsmb.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vonck J, Pisa KY, Morgner N, Brutschy B, Muller V. Three-dimensional structure of A1A0 ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus by electron microscopy. J Biol Chem. 2009;284:10110–9. doi: 10.1074/jbc.M808498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau WC, Rubinstein JL. Structure of intact Thermus thermophilus V-ATPase by cryo-EM reveals organization of the membrane-bound V(O) motor. Proc Natl Acad Sci U S A. 2010;107:1367–72. doi: 10.1073/pnas.0911085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diepholz M, Venzke D, Prinz S, Batisse C, Florchinger B, Rossle M, Svergun DI, Bottcher B, Fethiere J. A different conformation for EGC stator subcomplex in solution and in the assembled yeast V-ATPase: possible implications for regulatory disassembly. Structure. 2008;16:1789–98. doi: 10.1016/j.str.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Muench SP, Huss M, Song CF, Phillips C, Wieczorek H, Trinick J, Harrison MA. Cryo-electron microscopy of the vacuolar ATPase motor reveals its mechanical and regulatory complexity. J Mol Biol. 2009;386:989–99. doi: 10.1016/j.jmb.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 20.Sagermann M, Stevens TH, Matthews BW. Crystal structure of the regulatory subunit H of the V-type ATPase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2001;98:7134–7139. doi: 10.1073/pnas.131192798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landolt-Marticorena C, Williams KM, Correa J, Chen W, Manolson MF. Evidence that the NH2 terminus of vph1p, an integral subunit of the V0 sector of the yeast V-ATPase, interacts directly with the Vma1p and Vma13p subunits of the V1 sector. J Biol Chem. 2000;275:15449–57. doi: 10.1074/jbc.M000207200. [DOI] [PubMed] [Google Scholar]
- 22.Diab H, Ohira M, Liu M, Cobb E, Kane PM. Subunit interactions and requirements for inhibition of the yeast V1-ATPase. J Biol Chem. 2009;284:13316–25. doi: 10.1074/jbc.M900475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue T, Forgac M. Cysteine-mediated cross-linking indicates that subunit C of the V-ATPase is in close proximity to subunits E and G of the V1 domain and subunit a of the V0 domain. J Biol Chem. 2005;280:27896–903. doi: 10.1074/jbc.M504890200. [DOI] [PubMed] [Google Scholar]
- 24.Xu T, Vasilyeva E, Forgac M. Subunit interactions in the clathrin-coated vesicle vacuolar (H(+))-ATPase complex. J Biol Chem. 1999;274:28909–15. doi: 10.1074/jbc.274.41.28909. [DOI] [PubMed] [Google Scholar]
- 25.Norgett EE, Borthwick KJ, Al-Lamki RS, Su Y, Smith AN, Karet FE. V1 and V0 domains of the human H+-ATPase are linked by an interaction between the G and a subunits. J Biol Chem. 2007;282:14421–7. doi: 10.1074/jbc.M701226200. [DOI] [PubMed] [Google Scholar]
- 26.Lu M, Vergara S, Zhang L, Holliday LS, Aris J, Gluck SL. The amino-terminal domain of the E subunit of vacuolar H(+)-ATPase (V-ATPase) interacts with the H subunit and is required for V-ATPase function. J Biol Chem. 2002;277:38409–15. doi: 10.1074/jbc.M203521200. [DOI] [PubMed] [Google Scholar]
- 27.Jones RP, Durose LJ, Findlay JB, Harrison MA. Defined sites of interaction between subunits E (Vma4p), C (Vma5p), and G (Vma10p) within the stator structure of the vacuolar H+-ATPase. Biochemistry. 2005;44:3933–41. doi: 10.1021/bi048402x. [DOI] [PubMed] [Google Scholar]
- 28.Oot RA, Wilkens S. Domain characterization and interaction of the yeast vacuolar ATPase subunit C with the peripheral stator stalk subunits E and G. J Biol Chem. 2010;285:24654–64. doi: 10.1074/jbc.M110.136960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkens S, Forgac M. Three-dimensional structure of the vacuolar ATPase proton channel by electron microscopy. J Biol Chem. 2001;276:44064–8. doi: 10.1074/jbc.M106579200. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W, Wang D, Volk E, Bellen HJ, Hiesinger PR, Quiocho FA. V-ATPase V0 sector subunit a1 in neurons is a target of calmodulin. J Biol Chem. 2008;283:294–300. doi: 10.1074/jbc.M708058200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


