Abstract
Time of flight secondary ion mass spectrometry 2D images and molecular depth profiles of human HeLa cells treated with bromodeoxyuridine (BrdU) were acquired in the dual beam mode (Bi3+ analysis beam, C60+ etching beam). Several preparation protocols were investigated and were compared to a simple wash-and-dry method. The feasibility of using C60 to clean the samples prior to imaging with Bi was also investigated quantitatively by calibrating full depth profiles of the cells using atomic force microscopy. BrdU was used as a marker for the cell nucleus, facilitating identification and localization of sub-cellular features during depth profiling. Results show that C60 can be used to remove the surface contamination and to access different layers within the cells for 2D imaging. For a 1 nA, 10 keV C60+ beam incident at 45° and rastered over a 500 × 500 μm2 area, ~1 nm of biological material was sputtered every second. Our results show that HeLa cells were completely removed after etching with 1.3×1015 C60+ ions per cm2, giving an average etching rate of 3.9 nm for every 1013 C60 per cm2 at 10 keV and 45° incidence.
Keywords: HeLa cells, imaging, ToF-SIMS, C60, depth profiling, dual beam, bromodeoxyuridine, BrdU
1. Introduction
Cluster ion beams offer new opportunities for 2D and 3D time-of-flight secondary ion mass spectrometry (ToF-SIMS) analysis of biological samples, providing enhanced secondary ion yields with minimal sample damage1-2. In particular, C60 ions have been shown to allow molecular depth profiling, thus providing 3D images of single cells with subcellular resolutions3-6. The clusters can also be used to “clean” the sample surface7, which subsequently allows high resolution imaging with LMIG sources that can be focused down to 100 nm (e.g., Bi3+, Au+). When analyzing biological samples, however, two concerns need to be addressed. First, the heterogeneity of biological samples, especially cells, may lead to variable etching rates during depth profiling or cleaning. For example, most published etching rate measurements for biological samples assume a constant etching rate throughout the entire depth of the cell or tissue, but this may not always be true3-5. Thus, a better understanding of the etching rates for the different components of biological samples is necessary to establish optimal depth profiling parameters. Second, sample preparation is a critical aspect to acquiring relevant images since the chemical and spatial integrity of biological samples must be preserved when analyzed under a vacuum environment. This area has been investigated extensively and numerous protocols have been reported in the literature, ranging from an easy wash-anddry method to a more tedious freeze-fracture method after a plunge-freeze in a cryogen7-14. The latter is considered to be the gold standard for obtaining relevant images of highly diffusible ions and molecules, since the fast freezing of the sample instantly arrests all biological activity. However, the emergence of cluster ion sources has fundamentally changed the way biological samples can be prepared, since the removal of surface contamination with minimal damage can be done for samples with less than pristine surfaces7. Also, this allows sample preparation methods that were not adequate when using monoatomic species to be revisited.
In this work, results for 2D imaging and depth profiling of human HeLa cells are reported. Different sample preparations were investigated and were compared to the results obtained using a simple wash-and-dry method with ammonium acetate. The use of C60 to clean the sample prior to imaging with a Bi3+ beam was also investigated. Finally, results of ToF-SIMS depth profiles of HeLa cells treated with bromodeoxyuridine (BrdU) were combined with atomic force microscopy (AFM) measurements to determine the etching rate of various cellular components using C60+ at 10 keV and to investigate the sensitivity of the technique for 3D imaging of biomolecules. For this purpose, the cell thickness was measured by AFM before and after etching with various ion fluences, and the erosion rate was calculated by dividing these thicknesses by the corresponding C60+ fluence needed to reach the Si substrate. BrdU, a well known nucleus marker, was also used to help identify and delimit the different sub-cellular regions in the ToF-SIMS depth profiles (i.e., nucleus, top and bottom membranes, and cytoplasm).
2. Experimental
HeLa cells, human cervical carcinoma cells (ATCC CCL-2), were maintained in minimum essential media (MEM) containing L-glutamine (Gibco), 1% penicillin-streptomycin (Gibco), and 10% fetal bovine serum (FBS, Invitrogen) at 37 °C and 5% CO2. After trypsinizing, the HeLa cells (12,000 cells/cm2) were seeded onto Si surfaces that had been cleaned by sequential sonications in DI water, dichloromethylene, acetone, and methanol. Then the cells were allowed to adhere overnight to the Si surface. The cells were then prepared for ToF-SIMS analysis in the following ways (as shown in the figures):
Figure 1. Wash-and-dry with ammonium acetate8: The cells were washed for 30 seconds in ammonium acetate and dried in air before introduction into the instrument.
Figure 2a. Cells were washed in ammonium acetate, fixed in 4% phosphate-buffered paraformaldehyde, rinsed with double distilled H2O (ddH2O), and dried with N2.
Figure 2b. Cells were rinsed with ammonium acetate, frozen in liquid nitrogen, and freeze-dried for 24 hours.
Figure 2c. Trehalose was added to the culture media13. After several hours of incubation, cells were rinsed first with PBS buffer containing 100 mM trehalose then with a 100 mM trehalose, 0.3% glycerol water solution. The samples were freeze-dried.
Figure 2d. Cells were washed in ammonium acetate and introduced hydrated into the ToF-SIMS loadlock, then the sample was cooled to -150°C. Pumping in the chamber was started when the sample's temperature was -90°C. Analysis was performed in the main chamber at -90°C to minimize ice deposition.
Figure 2e. Cells were washed in ammonium acetate for 60 seconds, then snap-frozen in liquid ethane and freeze-dried for 24 hours.
Figure 3. Cells were incubated in the presence of 20 μM bromodeoxyuridine (BrdU) for 48 hours prior to seeding. BrdU, which is incorporated into DNA during cell division, is a good label for cell depth profiling. BrdU can be detected using the 81Br- signal (79Br- overlaps with PO3-), as well as C4H2N2O2Br- at m/z 189 and 191 (79Br- and 81Br- isotopes). After seeding and incubation overnight, the cells were washed in PBS, fixed in 4% phosphate-buffered paraformaldehyde, and rinsed with ammonium acetate.
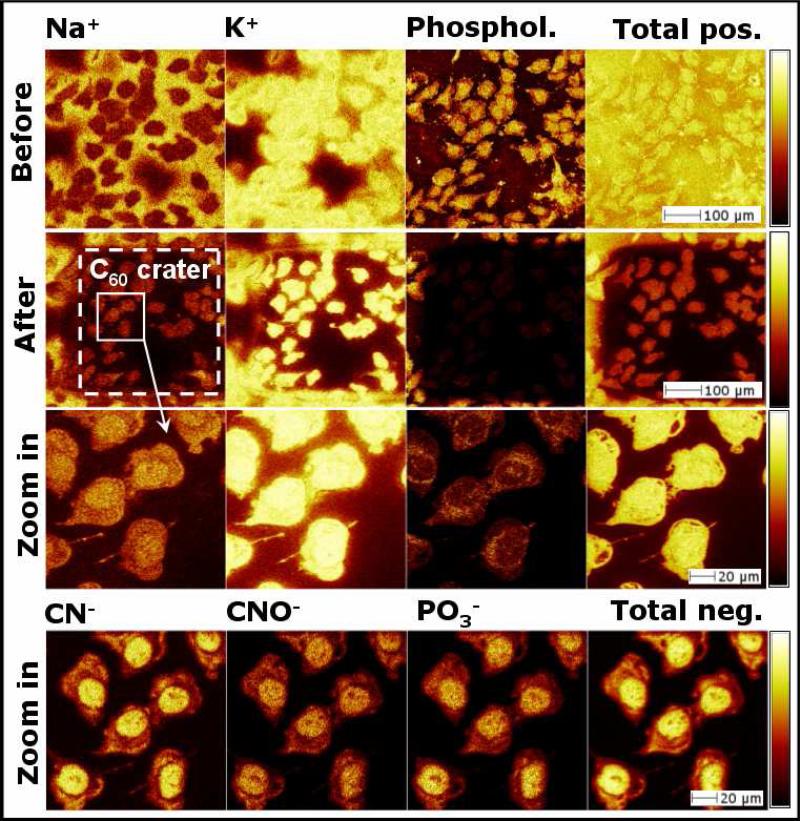
Figure 1.
Positive and negative ion images of HeLa cells prepared by a wash-anddry method. The images in the first row were acquired before C60++ etching, the second, the third, and the fourth rows were acquired after etching with 1.03×1014 C60++ per cm2.
Figure 2.
Positive ion images of phospholipids signals (sum of m/z = 58, 86 and 184) before (first row) and after etching (second row). The methods used to prepare the cells are used to label the images. K+ to Na+ ratios shown in the bottom row were measured inside the cells after etching.
Figure 3.
Depth profiles of HeLa cells acquired in the dual beam mode with 25 keV Bi3+ for analysis and 10 keV C60+ for etching. These depth profiles were reconstructed from two regions of interest (ROI). The first ROI was chosen inside a single cell nucleus region (a) and the second one was chosen inside the cytoplasm region (b). The depth scales were calibrated using AFM. The overlaid positive and negative ion signals were obtained from different cells on the same sample.
ToF-SIMS experiments were performed using an ION-TOF TOF.SIMS 5-100 (ION-TOF GmbH, Münster, Germany) equipped with a LMIG analysis gun and an electron impact gun. The analysis beam used for this study was a pulsed 25 keV Bi3+ whereas the etching beam was 10 keV C60q+ (q = 1, 2). Both beams strike the target at an angle of 45°. Images were collected in the non-interlaced mode (i.e., analysis and sputtering are active in different TOF-cycles) while depth profiles were obtained in the interlaced mode (i.e., analysis and sputtering are done within one TOF-cycle). Bi was set in the burst alignment mode for imaging (long pulses, nominal mass) and in the bunched high current mode for depth profiling (short pulses, high mass resolution). Bi3+ was typically rastered over a 200 × 200 μm2 area, centered inside a 500 × 500 μm2 C60q+ crater. Target currents were measured separately before each measurement, with Bi3+ ranging between 0.03 and 0.3 pA and C60q+ between 0.15 nA and 1 nA, depending on the species used and on the beam parameters.
AFM (Dimension 3100, Veeco Metrology Inc., Santa Barbara, CA) was used to measure the height of the cells and their morphology before and after sputtering with differing C60 fluences. The AFM was equipped with a 315 kHz, 42 N/m PointProbe Plus silicon tip (Nanosensors, Neuchâtel, Switzerland), and operated in the intermittent contact mode in air. At least four locations of each cell at a particular area were scanned to determine the average height. These measurements were used to calibrate the depth scale of molecular depth profiles. The cell nucleus and cytoplasm regions were each considered, as a first approximation, to be uniform, flat regions.
3. Results and discussion
Since the qualitative evaluation of a cell is complex, some criteria for cell samples have been reported in the literature9, 15. Due to the high concentration of sodium in the nutrient media and the presence of sodium pumps in the cell's plasma membrane, the Na+ signal is typically 10 times higher than K+ extracellularly, and vice versa intracellularly. Phospholipids and other characteristic signals such as cholesterol and carbohydrates should be detected only around the cell surface and not from the surrounding areas. Detection of these species outside the cell surface indicates leakage and damage to the cell's membrane.
Figures 1 and 2 show ToF-SIMS images of HeLa cells prepared according to the protocols described earlier. The images were acquired before and after C60 etching. The wash-and-dry method shown in Figure 1 was used as a control to compare the quality of other protocols. Interestingly, ammonium acetate proved to be the most efficient solution for salt removal when compared to ddH2O, PBS and ammonium formate (unpublished studies): the first row of Figure 1 shows that Na+ is detected outside the cells, K+ is primarily detected inside the cells, and the phospholipid signals (sum of m/z = 58, 86 and 184) are detected on the cells surface. The second row shows the same cells after etching with 1.03×1014 C60++ per cm2. Na+ and K+ signals detected around the cells have now been removed, and the phospholipids and total positive ion signals are seen to decrease significantly inside the crater. Also, the K/Na ratio increases from 4.8 to 5.5 inside the cells after etching, which supports the claim that the K/Na ratio is higher inside the cells. At higher magnification, both the positive (third row) and negative (fourth row) ion images show that subcellular features can be imaged more clearly after C60 etching. The positive images display phospholipid signals around the cells’ nuclei originating from the endoplasmic reticulum and/or the nuclear envelope, and the negative images show strong CN-, CNO- and PO3- signals indicative of nucleic acids contained within the cell.
Figure 2 shows the ToF-SIMS phospholipid images before and after C60 etching of HeLa cells prepared using the different protocols described in the experimental section. Compared to the wash-and-dry method, the morphology of the cells in Figure 2 was seen to be better preserved. However, morphology alone is not a good indicator of a well-preserved cell. For example, paraformaldehyde fixation (a) and trehalose vitrification (c) damaged the cells membrane, as can be seen from the discharge of phospholipids signals from the cells. The best result was obtained when the short ammonium rinse was followed by plunge-freezing in liquid ethane and freeze-drying (e). This method resulted in an optimal K/Na ratio inside the cells, as well as the preservation of morphology with minimal membrane damage. More importantly, it should be noted that C60 “cleaning” always resulted in the removal of surface contamination, which made imaging of subcellular features possible.
The capability of C60 ions as a tool to both “clean” and etch through biological samples requires an understanding of its rate of erosion through extracellular and intracellular components. To address this issue, depth profiles of HeLa cells treated with BrdU were acquired in the dual beam mode with 25 keV Bi3+ (analysis) and 10 keV C60+ (etching). Figure 3 shows the depth profiles reconstructed from a single HeLa cell nucleus (a) and cytoplasm (b), fixed in paraformaldehyde on a Si substrate.
The depth profile of the cell's nucleus region in Figure 3a shows that the phospholipid signals (sum of the peaks at m/z 58, 86, 125, 166, 184 and 224) first decrease by one order of magnitude at the beginning of the profile, likely due to chemical damage and surface contamination removal. The phospholipid signals exhibit two local maxima near 45 nm and 450 nm, marking the top and bottom cell membrane positions, as well as delimiting the cell nucleus location. The sum of most intense amino acid peaks observed by Apte16 (i.e., Σ m/z 43, 44, 60, 61, 70, 72, 110, 120 and 130) and the sum of the most intense DNA peaks observed by May17 (i.e., Σ m/z 112, 127, 136 and 152) were seen to decrease with increasing C60 fluence, probably due to material removal and/or to chemical damage. Figure 3a also shows that the BrdU signals (Br- and C4H2N2O2Br-) first increase as the contamination and the top membrane are removed, then reach steady state inside the cell nucleus between 90 nm and 390 nm. A thickness of 300 nm for the cell nucleus without the membranes is in good agreement with the value reported by Fletcher et al. for HeLa cells fixed in ethanol/acetic acid18. Figure 3b also shows that BrdU is not detected in the cytoplasm region, as expected.
It can be seen in the profiles that the cell's nucleus region (~508 nm by AFM, including the membranes and the surface contamination) is completely removed after etching with 1.3 × 1015 C60 per cm2, which indicate an etching rate of 3.9 nm for every 1013 C60 per cm2. The cell's cytoplasm region (~85 nm by AFM) is removed after 0.27×1015 C60 per cm2, indicating an etching rate of 3.2 nm for every 1013 C60 per cm2. The same experiments performed with a higher energy C60 beam at 20 keV resulted in an average etching rate of 8.7 nm for every 1013 C60 per cm2, showing a linear increase of etching rate with C60 beam energy.
Conclusions
Results for 2D imaging and depth profiling of human HeLa cells were reported and discussed. Several sample preparation protocols were investigated and were compared to a reference wash-and-dry method using ammonium acetate. The results show that increasing the complexity of the sample preparation does not always improve the quality of ToF-SIMS images. Moreover, it was shown that etching with C60 allows removal of the surface contamination, providing subsequent high resolution imaging of subcellular features. To investigate the rate of etching through different cellular components, full depth profiles of HeLa cells treated with BrdU were acquired. For a 1 nA, 10 keV C60+ beam incident at 45° and rastered over 500 × 500 μm2, 20 seconds of etching was necessary to remove the surface contamination (~20 nm). After approximately 75 seconds of etching, the top membrane was completely removed (~54 nm), with intracellular components visible. After 100 seconds of etching, the cytoplasm of the cells was completely sputtered away (~85 nm), and only the nucleus remained. Finally, the bottom membrane was reached after 403 seconds of etching, and the cell was completely sputtered after 515 seconds (~508 nm). Both the atomic and molecular species from BrdU were detected inside the cell's nucleus after the surface contamination and the top membrane were removed by C60 etching.
Acknowledgment
This research was supported by NIH grants EB-002027 and EB-006163. Danielle S.W. Benoit is a Merck Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1948-07). The AFM experiments were conducted at the University of Washington NanoTech User Facility, a member of the NSF National Nanotechnology Infrastructure Network (NNIN).
References
- 1.Gillen G, Roberson S. Rapid Commun. Mass Spectrom. 1998;12:1303–1312. doi: 10.1002/(SICI)1097-0231(19981015)12:19<1303::AID-RCM330>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 2.Kotter F, Benninghoven A. Appl. Surf. Sci. 1998;133:47–57. [Google Scholar]
- 3.Breitenstein D, Rommel CE, Möllers R, Wegener J, Hagenhoff B. Angew. Chem. Int. Ed. 2007;46:5332–5335. doi: 10.1002/anie.200604468. [DOI] [PubMed] [Google Scholar]
- 4.Fletcher JS, Lockyer NP, Vaidyanathan S, Vickerman JC. Anal. Chem. 2007;79:2199–2206. doi: 10.1021/ac061370u. [DOI] [PubMed] [Google Scholar]
- 5.Fletcher JS, Vickerman JC. Anal. Bioanal. Chem. 2010;396:85–104. doi: 10.1007/s00216-009-2986-3. [DOI] [PubMed] [Google Scholar]
- 6.Nygren H, Hagenhoff B, Malmberg P, Nilsson M, Richter K. Micros. Res. Tech. 2007;70:969–974. doi: 10.1002/jemt.20502. [DOI] [PubMed] [Google Scholar]
- 7.Kurczy ME, Piehowski PD, Parry SA, Jiang M, Chen G, Ewing AG, Winograd N. Appl. Surf. Sci. 2008;255:1298–1304. doi: 10.1016/j.apsusc.2008.05.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berman ESF, Fortson SL, Checchi KD, Wu L, Felton JS, Wu KJJ, Kulp KS. J. Am. Soc. Mass Spectrom. 2008;19:1230–1236. doi: 10.1016/j.jasms.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Chandra S. Appl. Surf. Sci. 2008;255:1273–1284. [Google Scholar]
- 10.Chandra S, Morrison GH. Biol. Cell. 1992;74:31–42. doi: 10.1016/0248-4900(92)90006-m. [DOI] [PubMed] [Google Scholar]
- 11.Malm J, Giannaras D, Riehle MO, Gadegaard N, Sjövall P. Anal. Chem. 2009;81:7197–7205. doi: 10.1021/ac900636v. [DOI] [PubMed] [Google Scholar]
- 12.Nygren H, Eriksson C, Malmberg P, Sahlin H, Carlsson L, Lausmaa J, Sjövall P. Colloid Surf. B-Biointerfaces. 2003;30:87–92. [Google Scholar]
- 13.Parry SA, Kurczy ME, Fan X, Halleck MS, Schlegel RA, Winograd N. Appl. Surf. Sci. 2008;255:929–933. doi: 10.1016/j.apsusc.2008.05.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sjovall P, Lausmaa J, Nygren H, Carlsson L, Malmberg P. Anal. Chem. 2003;75:3429–3434. doi: 10.1021/ac0207675. [DOI] [PubMed] [Google Scholar]
- 15.Arlinghaus HF, Kriegeskotte C, Fartmann M, Wittig A, Sauerwein W, Lipinsky D. Appl. Surf. Sci. 2006;252:6941–6948. [Google Scholar]
- 16.Apte J, Cheng F, Michel R, Castner DG. in preparation.
- 17.May CJ, Canavan HE, Castner DG. Anal. Chem. 2004;76:1114–1122. doi: 10.1021/ac034874q. [DOI] [PubMed] [Google Scholar]
- 18.Fletcher JS, Henderson A, Biddulph GX, Vaidyanathan S, Lockyer NP, Vickerman JC. Appl. Surf. Sci. 2008;255:1264–1270. [Google Scholar]