Abstract
The Hsp90 cycle depends on the coordinated activity of a range of cochaperones, including Hop, Hsp70 and peptidyl-prolyl isomerases such as FKBP52. Using mass spectrometry, we investigate the order of addition of these cochaperones and their effects on the stoichiometry and composition of the resulting Hsp90-containing complexes. Our results show that monomeric Hop binds specifically to the Hsp90 dimer whereas FKBP52 binds to both monomeric and dimeric forms of Hsp90. By preforming Hsp90 complexes with either Hop, followed by addition of FKBP52, or with FKBP52 and subsequent addition of Hop, we monitor the formation of a predominant asymmetric ternary complex containing both cochaperones. This asymmetric complex is subsequently able to interact with the chaperone Hsp70 to form quaternary complexes containing all four proteins. Monitoring the population of these complexes during their formation and at equilibrium allows us to model the complex formation and to extract 14 different KD values. This simultaneous calculation of the KDs from a complex system with the same method, from eight deferent datasets under the same buffer conditions delivers a self-consistent set of values. In this case, the KD values afford insights into the assembly of ten Hsp90-containing complexes and provide a rationale for the cellular heterogeneity and prevalence of intermediates in the Hsp90 chaperone cycle.
Keywords: dissociation constants, interaction network
Hsp90 selectively interacts with and regulates the activation of many client proteins involved in cellular signalling pathways such as kinases and steroid hormone receptors (1–3). The activation of Hsp90 clients relies on interaction with its cochaperones which facilitate different stages of the cycle (4–6). In human cells, Hsp90 cochaperones include Hsp70, Hop, Aha1, Cdc37, p23, and peptidyl-propyl isomerases (PPIases) such as FKBP51, FKBP52, and CyP40. The exact complexes that are formed between Hsp90 and cochaperones depend on the client protein. Cdc37 appears to be specific towards kinase clients whilst p23 is associated with the binding and activation of steroid hormone receptors. Despite a wealth of research, the exact complexes formed during the cycle are not well established and their stoichiometry remains controversial (6, 7).
Hsp90 is a homodimer containing three structural domains. The N-terminal domain contains the ATP-binding site and binds cochaperones including p23. The middle domain is suggested as the client protein binding region. The C-terminal domain encompasses the dimerization interface and also contains a C-terminal MEEVD motif which binds tetratricopeptide repeat (TPR)-containing cochaperones (8). The ATPase activity of Hsp90 is essential for its function and ATP hydrolysis is linked with a conformational rearrangement in which the homodimer changes from an open conformation to a closed state in which the two N-terminal domains interact (5, 6, 9–12). A general model was proposed for the activation of steroid hormone receptors (SHR) by the Hsp90 chaperone machinery (13, 14) in which the SHR binds to the Hsp40/Hsp70 chaperones and is then transferred to Hsp90 through the action of Hop which binds simultaneously to Hsp70 and Hsp90 through its different TPR domains (15). Subsequent binding of PPIases such as FKBP52 and cochaperone p23 leads to a mature complex in which the client protein is capable of being activated by binding of ligands and cofactors (13).
The Hsp90 dimer has two C-terminal MEEVD motifs which are the primary binding sites for TPR-containing cochaperones such as Hop and FKBP52 (16, 17). Hop has been established as a crucial component of the Hsp90 cycle forming a link that facilitates client protein transfer from Hsp70 to Hsp90 (15, 18). Hop binding is also known to inhibit the ATPase activity of Hsp90 by stabilizing the open conformation (6, 19). When bound to the Hsp90 dimer however there is conflicting evidence showing Hop binds as a dimer (6, 7, 20, 21) or a monomer (22). Some molecular details of how the TPR domains in FKBP52 and Hop bind to the C-terminal MEEVD motifs of Hsp90 are known from crystal structures of isolated TPR domains in complex with short MEEVD-containing peptides (17). However, many details of their binding in the context of the full-length Hsp90 and cochaperones remain poorly understood. In particular, as Hsp90 is a homodimer containing two MEEVD motifs, little is known about the ternary complexes that might form with the different TPR cochaperones. There is a general view that Hop/FKBP52 may compete for available binding sites in the Hsp90 dimer (13, 16, 23, 24). The lack of reliable dissociation constants means that it is often difficult to rationalize competitive binding and to predict the complexes that form in the cellular environment.
Mass spectrometry (MS) is making significant inroads as an adjunct to established structural biology tools by defining the stoichiometry and interactions of protein complexes (25). Particularly significant have been recent applications of nanoflow electrospray to polydisperse molecular chaperone-substrate complexes wherein a snapshot of all complexes present at a given time can be obtained within a single mass spectrum (26). This MS approach should therefore enable investigation of the dynamics of a protein system complicated by the formation of multiple species in solution. By correlating the intensity of the MS peaks, assigned to proteins and their complexes, with their concentration in solution we show here that it is possible to calculate KD values for coexisting species. In this study, we investigate the heterogeneity and dynamics of the Hsp90 complexes formed in the presence of Hop, FKBP52, and Hsp70. Although exemplified with part of the Hsp90 reaction cycle, the approach detailed here is applicable to other protein systems and is particularly suited to assemblies that are complicated by heterogeneity and polydispersity.
By manipulating the ratio of Hsp90 and Hop, we find that the predominant complex is the (Hsp90)2(Hop)1 although up to two molecules of Hop can bind in excess of Hop. By preforming complexes of the type (Hsp90)2(Hop)n=1 or 2 and adding FKBP52, we observe how FKBP52 competes with Hop for available Hsp90 binding sites to form the asymmetric ternary complex (Hsp90)2(Hop)1(FKBP52)1. Similarly, this complex can be assembled by addition of Hop to a preformed (Hsp90)2(FKBP52)n complex. By monitoring the formation of these complexes in real time, we calculate KD values for all ten complexes using eight different datasets. Additionally, in the presence of Hsp70 we identify complexes that are competent to bind Hsp70. Overall by monitoring these heterogeneous chaperone assemblies we reveal a self-consistent set of KD values which together with their established cellular concentrations, allows us to predict the multiple Hsp90 complexes existing in a cellular environment.
Results
Hsp90 Binds Hop with a 2∶1 or 2∶2 Stoichiometry.
We first recorded mass spectra of all four proteins individually (Hsp90, Hsp70, Hop, and FKBP52) under nondenaturing MS conditions (27). Hsp90 revealed well resolved charge state series corresponding to monomeric and predominantly dimeric Hsp90, in line with previous MS studies (28, 29) (SI Appendix, Fig. S1A). The MS of Hop revealed a mass corresponding to a monomer and no higher oligomers (SI Appendix, Fig. S1B). For Hop, FKBP52, and Hsp70 measured masses corresponded in each case to a monomer and no higher oligomers were detected (SI Appendix, Fig. S1 B–D). All calculated and experimental masses, with the exception of Hsp90, confirmed these proteins are monomeric under the conditions used here (SI Appendix, Table S1).
To determine the binding stoichiometry of Hop to Hsp90, we varied the concentration of both components. First the Hop concentration was kept at 1 μM and the concentration of the Hsp90 dimer (Hsp90)2 varied from 1 to 5 μM. At equimolar concentrations of Hop and (Hsp90)2, the dominant complex was (Hsp90)2(Hop)1 as well as Hsp90 monomer (Hsp90)1 and unbound Hsp90 dimer (Hsp90)2 (SI Appendix, Fig. S1E). No free Hop was seen, implying that all Hop molecules were bound in the complex. Higher Hsp90 concentrations triggered no changes in stoichiometry, the (Hsp90)2(Hop)1 remained the prevalent and only complex formed. The reverse experiment was also performed using a constant (Hsp90)2 concentration of 1 μM and increasing the Hop concentration from 1 to 5 μM. We found that with excess Hop (2 μM) an additional complex of (Hsp90)2(Hop)2 formed; concomitantly the concentration of free Hsp90 was depleted (SI Appendix, Fig. S1F). A further increase in Hop concentration resulted in the (Hsp90)2(Hop)2 complex becoming more dominant than the (Hsp90)2(Hop)1 complex. No other higher-order complexes were observed.
Because the Hsp90 dimer adopts an open conformation in the presence of ADP and converts to a closed state when ATP is bound (5), we examined how nucleotides affected these complexes. Addition of 100 μM ADP to the Hop and Hsp90 complexes showed no changes in the stoichiometry or ratios of the complexes. Given that the KDs for ATP and AMPpNp are reported to be higher than for ADP (30, 31), we also examined binding at higher nucleotide concentration (300 μM). At this nucleotide concentration and using a KD value of 148 μM (30) and an Hsp90 monomer concentration of 2 μM we calculate that > 65% of the Hsp90 binding sites will be occupied. While the peaks are significantly broader, due to multiple nonspecific attachments of nucleotide, there is no change in the ratio of the complexes formed (SI Appendix, Fig. S2 A–F). We conclude that while both Hsp90 subunits within the dimer can bind a Hop monomer, the stoichiometry and ratios of the complexes formed is nucleotide independent.
Hsp90 Can Bind Simultaneously to FKBP52 and Hop.
We established the interactions of (Hsp90)2 with FKBP52, by incubating equimolar concentrations of (Hsp90)2 and FKBP52. The spectra recorded revealed peaks that corresponded to the (Hsp90)2(FKBP52)1 and (Hsp90)2(FKBP52)2 complexes (SI Appendix, Fig. S3A). We also observed minor peaks for both FKBP52 and Hsp90 monomers and an unexpected complex of (Hsp90)1(FKBP52)1. It is noteworthy that the analogous complex (Hsp90)1(Hop)1 was not observed for Hop binding to Hsp90.
We then examined competitive binding of Hop and FKBP52 to the Hsp90 dimer by forming either the (Hsp90)2(FKBP52)n or (Hsp90)2(Hop)n complexes respectively at different molar concentrations as follows: In the first set of experiments, we preformed complexes of (Hsp90)2(FKBP52)n at equimolar concentrations of FKBP52 and Hsp90 dimer (both 1 μM) and subsequently added an equimolar amount of Hop (SI Appendix, Fig. S3A). In the second experiment, a binary complex of (Hsp90)2(Hop)1, was preformed at equimolar concentrations and challenged with an equimolar amount of FKBP52 (SI Appendix, Fig. S3B). In the third set of experiments, we formed complexes of (Hsp90)2(Hop)1 and (Hsp90)2(Hop)2 with 1 μM Hsp90 dimer and a twofold excess of Hop (2 μM) and then challenged these with 1 μM FKBP52 (Fig. 1). In all three experiments, spectra were recorded at regular time intervals up to 1 h after addition of the competing protein.
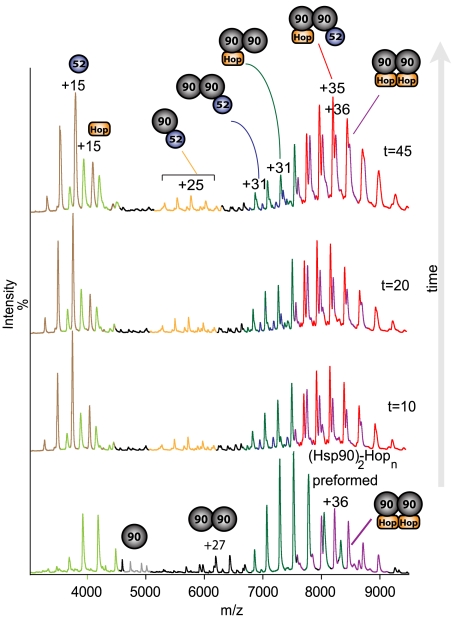
Fig. 1.
Hsp90, Hop, and FKBP52 interact to form binary and ternary complexes. MS recorded after t = 0 to t = 45 min showing the formation of the asymmetric complex containing both Hop and FKBP52. (Hsp90)2(Hop)1 and (Hsp90)2(Hop)2 were formed with twofold excess of Hop prior to the addition of FKBP52. Charge states colored as follows: Hop (light green), Hsp90 monomer and dimer (gray and black respectively), (Hsp90)2(Hop)1 dark green, (Hsp90)2(Hop)2 purple, (Hsp90)1(FKBP52)1 yellow, (Hsp90)2(FKBP2)1 blue, and (Hsp90)2(Hop)1(FKBP52)1 red. Same colors used throughout.
In the first experiment, the asymmetric ternary complex (Hsp90)2(FKBP52)1(Hop)1 was clearly visible after 5 min (SI Appendix, Fig. S3A). Concomitantly with the formation of this complex, the relative abundance of the (Hsp90)2(Hop)1 complex was also dominant compared to the (Hsp90)2(FKBP52)1. In the second experiment, addition of equimolar FKBP52 to (Hsp90)2(Hop)n complexes preformed at equimolar concentrations revealed peaks corresponding (Hsp90)2(FKBP52)n and the asymmetric ternary complex (SI Appendix, Fig. S3B). Again we observed the (Hsp90)1(FKBP52)1 complex. If we compare the abundance of the Hsp90 dimer with only one binding partner, we find the (Hsp90)2(Hop)1 is more prevalent than (Hsp90)2(FKBP52)1 complex throughout the time course of both experiments. In the third experiment, addition of FKBP52 to complexes containing (Hsp90)2(Hop)1 and (Hsp90)2(Hop)2 similarly leads to formation of the asymmetric ternary complex, as well as the same complexes observed in the first two experiments. In this case, the (Hsp90)2(Hop)2 complex persists throughout the time course showing Hop was not displaced, but rather FKBP52 binds on to the already formed (Hsp90)2(Hop)1 complex (Fig. 1, SI Appendix, Fig. S4A). To test stability of these complexes formed in the third experiment, we also recorded spectra after 3 h (SI Appendix, Fig. S4B). The same complexes prevailed allowing us to conclude that the asymmetric complex forms readily from a variety of solution conditions and is stable for at least 3 h. As a control, we also tested for an interaction between FKBP52 and Hop alone. No binding was observed validating this control experiment and our observation of specific complexes for Hsp90 with Hop or Hsp90 with FKBP52.
To compare the proportion of complexes formed as a function of the total protein components including individual proteins and complexes, we employed a deconvolution algorithm (Massign) (32, 33). An example is shown for a spectrum recorded 15 min after (Hsp90)2(Hop)n complexes were challenged with an equimolar ratio of FKBP52 (Fig. 2A). After 15 min the asymmetric ternary (Hsp90)2(FKBP52)1(Hop)1 and (Hsp90)2(FKBP52)1 complexes represent 19% and 16% of the total population, respectively. Similarly, we calculated the relative abundance of all other components and plotted their populations as a function of time (Fig. 2B). We find the asymmetric ternary complex forms rapidly after addition of FKBP52. We also find that the homodimeric (Hsp90)2 becomes depleted over time implying that more of the Hsp90 dimer is forming complexes with the TPR-containing cochaperones. The concentration of the (Hsp90)2(Hop)1 complex is also depleted over time while the (Hsp90)2(Hop)2 remains constant throughout implying that Hop is not displaced. The heterodimer complex (Hsp90)1(FKBP52)1 forms while Hop is unable to form an (Hsp90)1(Hop)1 complex in the same fashion. Our data shows both TPR-containing proteins readily bind to an available site on the Hsp90 dimer, despite prior binding of another, to form the asymmetric ternary complex.
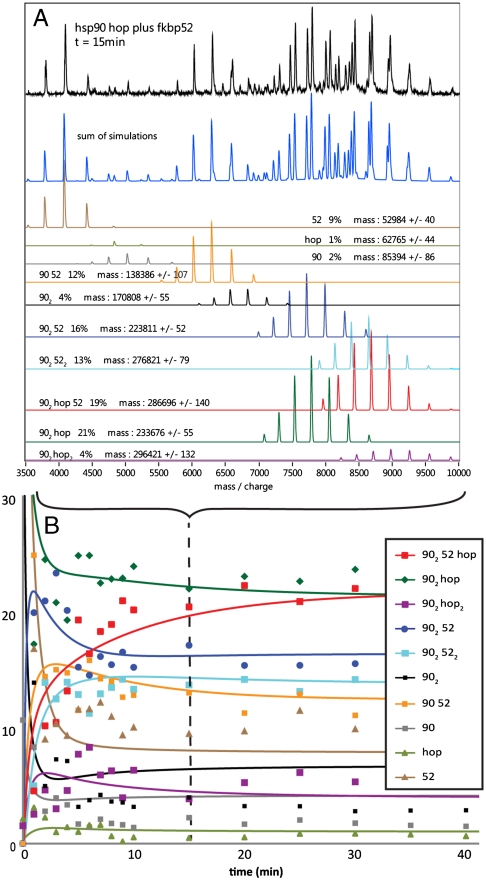
Fig. 2.
Monitoring the formation of Hsp90 complexes in real-time (A) MS recorded 15 min after addition of an equimolar ratio of FKBP52 to (Hsp90)2(Hop)n (black) is compared with a simulated spectrum (blue) formed by summing the deconvoluted spectra of all components (SI Appendix, Fig. S3). The composition, relative intensity (%), and mass of each component are given. (B) Kinetic model established from the relative intensity of all components plotted against time (SI Appendix). The simulated kinetics (lines) are shown together with the experimental data (symbols) from 0 to 40 min. The data with dashed lines is t = 15 min.
Hsp90-Hop Complexes Promote Subsequent Binding of Hsp70.
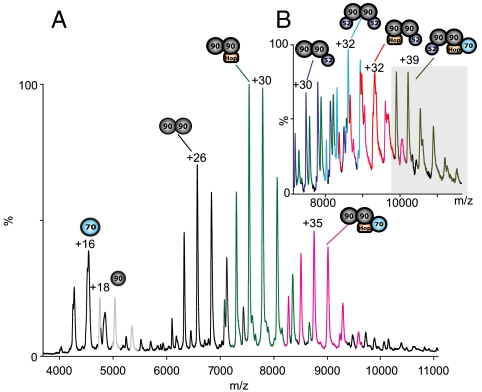
The presence of multiple coexisting complexes prompts investigation of complexes that are competent to bind Hsp70. Initially, we investigated binary interactions between Hsp70 and Hop or Hsp70 and Hsp90. At equimolar or higher concentrations of Hop, no interaction was seen between Hsp70 and Hop in the absence of nucleotides. The presence of 100 μM ADP however induced formation of a weak 1∶1 complex of the Hsp70∶Hop heterodimer (SI Appendix, Fig. S5A). Subsequently, we also examined whether there was any direct interaction between Hsp90 and Hsp70 but no binding was observed (SI Appendix, Fig. S5B). In contrast, on addition of Hsp90 to Hsp70 in the presence of equimolar amount of Hop, we observed a species with a measured mass corresponding to an intermediate complex containing (Hsp90)2(Hop)1(Hsp70)1 (Fig. 3A). Also present was a predominant complex of (Hsp90)2(Hop)1 and minor charge state series for Hsp70, (Hsp90)1, and (Hsp90)2. Other minor complexes containing (Hsp90)2(Hop)2(Hsp70)1 and (Hsp90)2(Hop)2(Hsp70)2 were also formed (SI Appendix, Fig. S6). As observed in the binary experiments, the presence of ADP revealed no changes in the ratios of the complexes formed (SI Appendix, Fig. S5 C and D). These data show that Hsp70 binding to Hop is facilitated by formation of the (Hsp90)2(Hop)n complexes because Hop alone showed only a weak interaction with Hsp70 in the presence of ADP and was unable to bind directly to Hsp70 in the absence of nucleotide.
Fig. 3.
Hsp70 binds to (Hsp90)2 and Hop to form an intermediate complex. (A) MS of an intermediate complex (Hsp90)2(Hop)1(Hsp70)1 (pink) formed after addition of Hsp70 following Hop binding to the (Hsp90)2. B) Addition of FKBP52 to the intermediate complex to form (Hsp90)2(Hop)1(FKBP52)1(Hsp70)1 (olive green), (SI Appendix, Figs. S6 and S7 for additional quaternary complexes).
To examine if FKBP52 could bind to the intermediate complex, we first formed complexes of (Hsp90)2(Hop)n(Hsp70)n and subsequently added FKBP52 (Fig. 3B). Results showed unambiguously an additional complex that corresponds to (FKBP52)1(Hsp90)2(Hop)1(Hsp70)1 (SI Appendix, Fig. S7). The observation of Hsp70-containing complexes of (Hsp90)2(Hop)2(Hsp70)1, (Hsp90)2(Hop)2(Hsp70)2, and (FKBP52)1(Hsp90)2(Hop)1(Hsp70)1 allows us to conclude that the binding of Hsp70 is directly linked to the number of Hop subunits bound to (Hsp90)2.
A Kinetic Model for Hsp90 Complexes.
A model was constructed to show all feasible reaction pathways (SI Appendix, Fig. S8) and highlight all possible complexes observed in our experiments (Fig. 4). Starting with the Hsp90 dimer at micromolar concentrations (green oval) a population of monomeric Hsp90 is formed which is able to interact directly with FKBP52 but not Hsp70 or Hop. Dimeric Hsp90 is able to bind either FKBP52 or Hop to form binary complexes of (Hsp90)2(FKBP52)1 or (Hsp90)2(Hop)1. Having formed binary complexes containing the Hsp90 dimer with either FKBP52 or Hop both complexes can bind another copy of the same cochaperone to form (Hsp90)2(FKBP52)2 or (Hsp90)2(Hop)2 or bind the other cochaperone to form (Hsp90)2(FKBP52)1(Hop)1. The latter is favored statistically given equal protein concentrations of Hop and FKBP52. Our data shows a maximum of two copies of Hop or FKBP52 can bind per Hsp90 dimer. Interestingly, FKBP52 cannot bind complexes containing two Hop monomers and Hsp70 cannot bind to any complex that does not contain Hop and Hsp90.
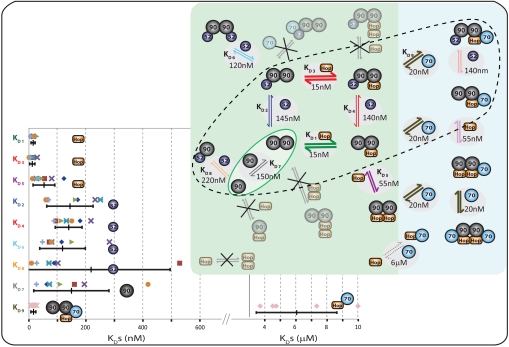
Fig. 4.
An interaction network for Hsp90, Hsp70, Hop, and FKBP52 KDS for Hop, (Hsp90)2 and FKBP52 determined from multiple spectra from eight different experimental datasets, including replicate experiments (green background). Favorable KDs are represented with thicker arrows. Addition of Hsp70 to the ternary (Hsp90)2(Hop)n(FKBP52)1 complexes leads to the formation of quaternary complexes with favorable KD values (blue background). Complexes likely to form, given the cellular concentrations of the four protein components, are circled (dotted line). KD values and associated errors are shown with values deduced form the same dataset represented by the same symbols.
To establish which complexes likely form under cellular conditions, we determined KD values using the intensity ratios of the 10 different species and monitored their population as a function of time (Fig. 2). We first established that the intensities of the peaks assigned to each Hsp90-containing complex are related to their concentrations in solution using an internal standard over appropriate concentration ranges (SI Appendix, Fig. S9). We assume that association and dissociation events follow second- and first-order kinetics respectively (SI Appendix). KD values are calculated by satisfying equations relating to the concentration of all components at every time point. We performed eight different sets of experiments including different concentrations, different order of addition as well as replicate experiments (SI Appendix, Table S2). This data yields a series of KD values that are reproducible within error between the different experimental conditions as well as replicate experiments (Fig. 4). All KD values that determine complex formations are in the range ∼10–250 nM.
These KD values reveal several interesting features. The binding of the first Hop to a binding site on the free (Hsp90)2 dimer to form (Hsp90)2(Hop)1 has a KD of 15 nM, whilst binding of the second Hop to form (Hsp90)2(Hop)2 has a KD of 55 nM. Binding of FKBP52 to the (Hsp90)2 to form either (Hsp90)2(FKBP52)1 or (Hsp90)2(FKBP52)2 is slightly less favorable than for Hop with corresponding KD values of 145 nM and 120 nM, respectively. Comparing the value obtained for Hop binding to (Hsp90)2(FKBP52)1 (KD = 15 nM), with that obtained for Hop binding to free (Hsp90)2 (KD = 15 nM) we deduce that prior binding of FKBP52 has no effect on subsequent binding of Hop to Hsp90. Likewise, adding FKBP52 to (Hsp90)2(Hop)1 or by addition of FKBP52 directly to (Hsp90)2, yields similar KD values of 140 nM and 145 nM respectively. This data not only shows that the subsequent binding of a TPR-containing cochaperone is independent of prior binding but also confirms the reproducibility of our approach. The KD values obtained for Hsp70 binding to the three Hsp90/Hop complexes yields low nanomolar values affirming our method and implying a favorable binding event for Hsp70 (KD = 20 nM) following the binding of Hop to Hsp90.
Discussion
The functionality of the Hsp90 machinery is known to depend on a number of chaperones and cochaperones to drive the cycle. The precise order of binding and the stoichiometries of the various complexes formed however have remained controversial. Using MS we have explored all possible complexes that can be formed by four proteins known to have critical roles in the Hsp90 chaperone cycle: Hsp90, Hsp70, Hop, and FKBP52. Initially, we found that Hsp90 was predominantly dimeric and FKBP52 monomeric, both were anticipated. Recently there has been some controversy over the oligomeric state of Hop reported as dimeric (6, 20, 34) or monomeric (22, 35) and Hsp70 can form high oligomeric species under some conditions (36). We have shown that under the conditions used here both proteins are monomeric.
To characterize the complexes formed between the components of the Hsp90 machinery we incubated different ratios of the four proteins, varying both their concentration and order of addition. The results enabled us to construct a network of possible complexes (Fig. 4). Despite the fact that all proteins are at micromolar concentration in solution, Hsp90 exists in monomeric and dimeric forms. Each Hsp90 monomer possesses a C-terminal MEEVD motif for TPR protein binding. We found that some complexes, which one might expect to form in theory, such as the (Hsp90)1(Hop)1 complex in practice did not form. For the 10 different species of Hsp90 and cochaperones complexes observed, we determined their binding stoichiometry as well as the likely assembly pathway. Starting with Hsp90 dimers (green oval Fig. 4), up to two Hop monomers can bind to form (Hsp90)2(Hop)1 and (Hsp90)2(Hop)2 complexes (Fig. 1
A and B). The absence of the (Hsp90)1(Hop)1 complex was in contrast to the results obtained for FKBP52, wherein the complex (Hsp90)1(FKBP52)1 was observed. Our observation implies that dimerization of Hsp90 is critical for Hop but not for FKBP52 binding, consistent with the 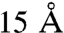 resolution EM structure of the Hsp90-Hop complex in which each Hop monomer interacts with both subunits of (Hsp90)2 (37).
resolution EM structure of the Hsp90-Hop complex in which each Hop monomer interacts with both subunits of (Hsp90)2 (37).
The binding affinities of FKBP52 and Hop for the two binding sites in the Hsp90 dimer, as well as additional binding of Hsp70, are all in the nanomolar range. However, there is a wide variability in the KD values reported for binary interactions between these proteins, both for human and yeast proteins (5, 6, 38) (SI Appendix, Table S3 and Fig. S10). This variability could arise from possible differences between the protein homologues, the varied solution conditions, and methods employed in their measurement. An apparent advantage of our method is that it gives a consistent set of KD values for a system of complexes, using the same method and solution conditions. Our data allows a direct comparison of the values determined for the different complexes formed in the cycle. Overall, our KD values are somewhat lower than those measured in an earlier study for C-terminal peptides of Hsp90 binding to isolated TPR domains (39) but are however very similar to some published values, where available, for the full-length human proteins (7, 40) (see SI Appendix, Table S3 and Fig. S10). Our KD values of 15 nM and 55 nM for the binding of the first and second Hop molecule are in accord with the proposal that binding of the first Hop results in a subtle decrease in affinity of the Hsp90 dimer for the second Hop (37).
The KD values for the binding of FKBP52 and Hop to either site in the (Hsp90)2 are similar. We speculate that the slight preference for Hop over FKBP52 binding may be due to the additional interactions Hop makes with the C-terminal and middle domain of Hsp90, as previously established using ITC (20) and more recently revealed by the Hsp90-Hop EM structure (37). Despite its slightly less favorable KD, FKBP52 is however able to form an asymmetric complex with Hop and Hsp90 (Fig. 1B). This finding is in accord with the recent observation that the ternary complex is preferentially formed (22). The asymmetric complex observed here forms regardless of the order of addition of either FKBP52 or Hop and is statistically favored and stable for at least 3 h under these conditions.
It has been proposed that Hop binds directly to Hsp70 with ADP present (7, 41). However, other studies have shown that Hop can bind to Hsp70 in the absence of nucleotide (42). Our results show evidence of a weak binding between Hsp70 and Hop but only when ADP is present (KD = 6 μM). Under the conditions used here, no binding was observed for Hsp70 and Hop in the absence of nucleotide. Favorable binding was however observed after prior binding of Hop to the Hsp90 dimer (KD = 20 nM). Our observation suggests a mechanism in which Hop and/or Hsp90 undergo a conformational change upon binding which renders the complex competent to bind Hsp70, and agrees with the recent EM structure where Hop binds a distinct conformation of Hsp90 which lies somewhere between the open conformation, observed for the apo state, and the N-terminally dimerized closed state (37). We find that the number of Hsp70 molecules that bind to the various Hsp90 assemblies is directly related to the number of Hop molecules within the complex. We also observe a complex with all four components (FKBP52)1(Hsp90)2(Hop)1(Hsp70)1 proposed previously (22) and discovered experimentally here. Interestingly, our KD values show that binding of Hsp70 is not affected by prior binding of FKBP52, Hop, or Hop and Hsp70 to the other binding site. Considering the cellular concentrations recently reported as 10 μM for Hsp70, 5 μM (Hsp90)2, and 3 μM for Hop (43), even though two Hop monomers can bind to an Hsp90 dimer the (Hsp90)2(Hop)1 complex is likely the physiologically relevant species. It has also been shown recently that a single Hop molecule is sufficient to completely inhibit the ATPase activity of Hsp90 (22). The (Hsp90)2(Hop)1 complex in turn can bind to Hsp70 to form the intermediate complex, that binds FKBP52 to form the functional chaperone complex (Fig. 4).
Given the ratios of (Hsp90)2 to Hop and FKBP52 (10∶1 and 15∶1 respectively) in the cell (44), it is perhaps surprising that the asymmetric complex is able to form by addition of an FKBP52 molecule. However, estimated concentrations do not take into account elevated protein levels in response to the stress environment, or the effects of crowding within the cell (45). The asymmetric complex is, however, statistically more likely to form than the (Hsp90)2(Hop)2 complex if concentrations of FKBP52 and Hop are comparable. Our results do show however that no additional FKBP52 molecule can bind to the (Hsp90)2(Hop)2 complex (Fig. 1), or those containing two Hop molecules with one or two Hsp70 molecules (SI Appendix, Fig. S7). Because FKBP52 is always present as a final component of the mature receptor complex (14), our data implies that binding of two Hop molecules renders the complex incompetent of further development into a functional chaperone complex. Our data is in contrast to earlier studies in which (Hsp90)2 binding to dimeric Hop was considered to be a prerequisite for chaperone function (4, 6, 7) but is consistent with the proposal that the asymmetric nature of Hsp90-Hop complexes are important in the Hsp90 cycle (22, 37).
The simultaneous binding of Hsp70 and Hsp90 to Hop has been proposed to bring the two proteins together into an active complex in which client proteins are transferred from Hsp70 to Hsp90 to complete their folding and maturation (15). Given the cellular concentration of Hsp70, which is higher than both (Hsp90)2 and Hop, the favorable KD for Hsp70 (20 nM) binding would ensure that the (Hsp90)2(Hop)1 complexes once formed can proceed to productive and competent folding complexes by binding FKBP52 to form (Hsp90)2(Hop)1(FKBP52)1(Hsp70)1. By contrast the complexes of (Hsp90)2(Hop)2(Hsp70)1 and (Hsp90)2(Hop)2(Hsp70)2, if formed, are incapable of subsequent FKBP52 binding and are therefore unlikely to play any further role in the chaperone cycle.
In summary, by incubating Hsp90 with different cochaperones, and exploring all possible reaction products, we have determined the dominant complexes formed during the Hsp90 cycle. We have also identified some key binding events that likely lead to the assembly of productive folding machinery in the Hsp90 chaperone cycle. The similarity of the KD values for the ten species studied here ensure that in the cellular environment the chaperone pathway will be populated with complexes that are both heterogeneous and dynamic and which can likely be finely tuned to respond to the stress conditions experienced. More generally, our results highlight the distinct advantages of MS in being able to determine simultaneously, and in real time, the composition and relative abundance of each complex in a highly heterogeneous system from many different datasets. This approach should have broad application to other systems as it not only gives insights into the interactions, but also leads to a self-consistent set of KD values as shown here for the different complexes formed in the Hsp90 chaperone cycle.
Methods
Protein Purification.
Hsp90 and cochaperones were human, expressed in Escherichia coli and purified as previously described (20, 29). Prior to MS analysis, 20 μL of the protein solutions were buffer exchanged into 50 mM ammonium acetate (AmAc), pH 7.5 at 4 °C using micro Bio-Spin® columns (Bio-Rad Laboratories).
Binding Studies.
Proteins were combined to give a range of concentrations from 1 to 5 μM in 50 mM AmAc pH 7.5 and incubated for 30 min on ice. For Hsp90 and Hop binary complexes were formed in 50 mM AmAc pH 7.5. As a control all complexes were formed in a standard buffer 50 mM Tris HCl pH 7.5. MS showed no difference in the ratios of the complexes formed. Competition experiments were performed by preforming binary complexes of Hsp90 with Hop or FKBP52 in 50 mM AmAc and adding FKBP52 or Hop immediately prior to MS. For nucleotide studies, ADP, AMPpNp, and ATP were prepared at 1 mM in 50 mM AmAc, pH 7.5 and added to give a final solution concentration of 100 μM and 300 μM nucleotide with an equivalent of Mg(OAc)2.
MS.
Spectra were obtained on a QToF II MS (Waters) modified for studying noncovalent interactions (27, 46). 2.5 μL aliquots were introduced into the MS from a gold-plated capillary needle (Harvard Apparatus). Capillary voltages were 1.5–1.8 kV, cone voltages from 80–130 V, and a collision voltages were from 50–150 V. Spectra were calibrated externally using caesium iodide.
Supplementary Material
Acknowledgments.
We thank funding from the European Union Prospects grant number (HEALTH-F4-2008-201648) (to N.M.) and Engineering and Physical Sciences Research Council for funding (to I.E.) Faculty of science and technology (FCT), Portugal for funding (to M.A.S.) and British Petroleum Centenary Murray Edwards College Fund and Cambridge Commonwealth Trust for funding (to S.D.). C.V.R. is a Royal Society Professor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106261108/-/DCSupplemental.
References
- 1.Pratt WB, Dittmar KD. Studies with purified chaperones advance the understanding of the mechanism of glucocorticoid receptor-hsp90 heterocomplex assembly. Trends Endocrin Met. 1998;9:244–252. doi: 10.1016/s1043-2760(98)00059-9. [DOI] [PubMed] [Google Scholar]
- 2.Richter K, Buchner J. Hsp90: chaperoning signal transduction. J Cell Physiol. 2001;188:281–290. doi: 10.1002/jcp.1131. [DOI] [PubMed] [Google Scholar]
- 3.Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siligardi G, et al. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50(cdc97) J Biol Chem. 2002;277:20151–20159. doi: 10.1074/jbc.M201287200. [DOI] [PubMed] [Google Scholar]
- 5.Siligardi G, et al. Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J Biol Chem. 2004;279:51989–51998. doi: 10.1074/jbc.M410562200. [DOI] [PubMed] [Google Scholar]
- 6.Prodromou C, et al. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez MP, Sullivan WP, Toft DO. The assembly and intermolecular properties of the hsp70-Hop-hsp90 molecular chaperone complex. J Biol Chem. 2002;277:38294–38304. doi: 10.1074/jbc.M206566200. [DOI] [PubMed] [Google Scholar]
- 8.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 9.Obermann WMJ, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grenert JP, Johnson BD, Toft DO. The importance of ATP binding and hydrolysis by hsp90 in formation and function of protein heterocomplexes. J Biol Chem. 1999;274:17525–17533. doi: 10.1074/jbc.274.25.17525. [DOI] [PubMed] [Google Scholar]
- 11.Krukenberg KA, Street TO, Lavery LA, Agard DA. Conformational dynamics of the molecular chaperone Hsp90. Q Rev Biophys. 2011;1:1–27. doi: 10.1017/S0033583510000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Southworth DR, Agard DA. Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol Cell. 2008;32:631–640. doi: 10.1016/j.molcel.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Owens-Grillo JK, et al. A model of protein targeting mediated by immunophilins and other proteins that bind to hsp90 via tetratricopeptide repeat domains. J Biol Chem. 1996;271:13468–13475. doi: 10.1074/jbc.271.23.13468. [DOI] [PubMed] [Google Scholar]
- 14.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 15.Chen SY, Smith DF. Hop as an adaptor in the heat shock protein 70 (Hsp70) and Hsp90 chaperone machinery. J Biol Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- 16.D’Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Scheufler C, et al. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 18.Morishima Y, et al. The Hsp organizer protein Hop enhances the rate of but is not essential for glucocorticoid receptor folding by the multiprotein Hsp90-based chaperone system. J Biol Chem. 2000;275:6894–6900. doi: 10.1074/jbc.275.10.6894. [DOI] [PubMed] [Google Scholar]
- 19.Richter K, Muschler P, Hainzl O, Reinstein J, Buchner J. Sti1 is a non-competitive inhibitor of the Hsp90 ATPase—binding prevents the N-terminal dimerization reaction during the ATPase cycle. J Biol Chem. 2003;278:10328–10333. doi: 10.1074/jbc.M213094200. [DOI] [PubMed] [Google Scholar]
- 20.Onuoha SC, Coulstock ET, Grossmann JG, Jackson SE. Structural studies on the co-chaperone Hop and its complexes with Hsp90. J Mol Biol. 2008;379:732–744. doi: 10.1016/j.jmb.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Hildenbrand ZL, et al. Hsp90 can accommodate the simultaneous binding of the FKBP52 and Hop proteins. Oncotarget. 2011;2:45–58. doi: 10.18632/oncotarget.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Richter K, Buchner J. Mixed Hsp90—cochaperone complexes are important for the progression of the reaction cycle. Nat Struct Mol Biol. 2011;18:61–66. doi: 10.1038/nsmb.1965. [DOI] [PubMed] [Google Scholar]
- 23.Young JC, Obermann WMJ, Hartl FU. Specific binding of tetratricopeptide repeat proteins to the C-terminal 12-kDa domain of hsp90. J Biol Chem. 1998;273:18007–18010. doi: 10.1074/jbc.273.29.18007. [DOI] [PubMed] [Google Scholar]
- 24.Radanyi C, Chambraud B, Baulieu EE. The ability of the immunophilin FKBP59-HBI to interact with the 90-kDa heat shock protein is encoded by its tetratricopeptide repeat domain. Proc Natl Acad Sci USA. 1994;91:11197–11201. doi: 10.1073/pnas.91.23.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharon M, Robinson CV. The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu Rev Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 26.Stengel F, et al. Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc Natl Acad Sci USA. 2010;107:2007–2012. doi: 10.1073/pnas.0910126107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 28.Karagoz GE, et al. N-terminal domain of human Hsp90 triggers binding to the cochaperone p23. Proc Natl Acad Sci USA. 2011;108:580–585. doi: 10.1073/pnas.1011867108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaughlin SH, Smith HW, Jackson SE. Stimulation of the weak ATPase activity of human Hsp90 by a client protein. J Mol Biol. 2002;315:787–798. doi: 10.1006/jmbi.2001.5245. [DOI] [PubMed] [Google Scholar]
- 30.McLaughlin SH, Ventouras LA, Lobbezoo B, Jackson SE. Independent ATPase activity of Hsp90 subunits creates a flexible assembly platform. J Mol Biol. 2004;344:813–826. doi: 10.1016/j.jmb.2004.09.055. [DOI] [PubMed] [Google Scholar]
- 31.Prodromou C, et al. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 32.Hernandez H, et al. Isoforms of U1-70k control subunit dynamics in the human spliceosomal U1 snRNP. PLoS One. 2009;4:e7202. doi: 10.1371/journal.pone.0007202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane LA, et al. Mass spectrometry reveals stable modules in holo and apo RNA polymerases I and III. Structure. 2011;19:90–100. doi: 10.1016/j.str.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Flom G, Behal RH, Rosen L, Cole DG, Johnson JL. Definition of the minimal fragments of Sti1 required for dimerization, interaction with Hsp70 and Hsp90 and in vivo functions. Biochem J. 2007;404:159–167. doi: 10.1042/BJ20070084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi F, Doudevski I, Regan L. HOP is a monomer: investigation of the oligomeric state of the co-chaperone Hop. Protein Sci. 2010;19:19–25. doi: 10.1002/pro.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benaroudj N, Batelier G, Triniolles F, Ladjimi MM. Self-association of the molecular chaperone HSC70. Biochemistry. 1995;34:15282–15290. doi: 10.1021/bi00046a037. [DOI] [PubMed] [Google Scholar]
- 37.Southworth DR, Agard DA. Client-loading conformation of the Hsp90 molecular chaperone revealed in the Cryo-EM structure of the human Hsp90∶Hop complex. Mol Cell. 2011;42:771–781. doi: 10.1016/j.molcel.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- 39.Cortajarena AL, Regan L. Ligand binding by TPR domains. Protein Sci. 2006;15:1193–1198. doi: 10.1110/ps.062092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pirkl F, Buchner J. Functional analysis of the Hsp90-associated human peptidyl prolyl Cis/Trans isomerases FKBP51, FKBP52 and cyp40. J Mol Biol. 2001;308:795–806. doi: 10.1006/jmbi.2001.4595. [DOI] [PubMed] [Google Scholar]
- 41.Johnson BD, Schumacher RJ, Ross ED, Toft DO. Hop modulates hsp70/hsp90 interactions in protein folding. J Biol Chem. 1998;273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- 42.Wegele H, Haslbeck M, Reinstein J, Buchner J. Sti1 is a novel activator of the Ssa proteins. J Biol Chem. 2003;278:25970–25976. doi: 10.1074/jbc.M301548200. [DOI] [PubMed] [Google Scholar]
- 43.Kundrat L, Regan L. Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry. 2010;49:7428–7438. doi: 10.1021/bi100386w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 45.Ellis RJ, Minton AP. Protein aggregation in crowded environments. Biol Chem. 2006;387:485–497. doi: 10.1515/BC.2006.064. [DOI] [PubMed] [Google Scholar]
- 46.Sobott F, Hernández H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.