Abstract
Movement over and colonization of surfaces are important survival strategies for bacteria, and many find it advantageous to perform these activities as a group, using quorum sensing to sample population size and synchronize behavior. It is puzzling however, that swarming-proficient and virulent strains of Vibrio parahaemolyticus are silenced for the vibrio archetypal pathway of quorum sensing. Here we describe the S-signal, a pheromone that can be communicated between cells in coculture to regulate surface colonization. This signal was harvested in cell-free supernatants and demonstrated to stimulate swarming gene expression at low cell density. The S-signal was generated by the pyridoxal phosphate-dependent aminotransferase ScrA; signal reception required the periplasmic binding protein ScrB and the membrane-bound GGDEF-EAL domain-containing protein ScrC. ScrC is a bifunctional enzyme that has the ability to form and degrade the second messenger bis-(3′-5′) cyclic dimeric GMP (c-di-GMP). ScrA in neighboring cells was able to alter the activity of ScrC in a ScrB-dependent manner, transforming ScrC’s repressing ability to inducing activity with respect to swarming. Conversely, cell–cell signaling repressed capsule gene expression. In summary, we report that quorum sensing can stimulate swarming in V. parahaemolyticus; it does so via an alternative pathway capable of generating an autoinducing signal that influences c-di-GMP, thereby expanding the lexicon and language of cell–cell communication.
Keywords: swarming motility, cyclic diguanylate, autoinducer signaling, bacterial communication
Bacteria use chemical signals to communicate with themselves and with others. These signals are diverse and include linear and cyclic peptides, long- and short-chain N-acyl-homoserine lactones, γ-quinolones, and unsaturated fatty acids (reviewed in refs. 1 and 2). Some of these signals are easily diffusible small molecules, whereas others are more hydrophobic and can be membrane- or vesicle-associated (3). Many of these communication signals are highly genus or species specific, although one, the furanosyl borate diester product of LuxS, seems a more universally synthesized and recognized molecule (reviewed in ref. 4). These molecules are collectively called “autoinducers.” It is thought that bacteria produce and use such signaling molecules to detect not only their neighbors and cell-population density, but also as a gauge to sample aspects of their environment, such as diffusion and confinement (reviewed in refs. 5 and 6). In a broad sense, quorum sensing allows coordination of population-wide activities, including swarming motility on surfaces, bioluminescence, virulence factor production, and biofilm dynamics (reviewed in refs. 7–9).
In this context, the personality of Vibrio parahaemolyticus is perplexing because this bacterium clearly participates in behaviors that are social in nature: it is an excellent swarmer, a robust biofilm former, and a pathogen (reviewed in ref. 10). However, many of these group activities occur in a seemingly asocial cell type (11, 12). Specifically, V. parahaemolyticus displays On/Off phase switching with respect to the archetypal pathway of quorum sensing in the Vibrionaceae (13). Curiously, and in contrast to what is found for many bacteria, it is the cell type with disruptions in this pathway that is the most proficient in swarming and expression of some virulence traits.
The second messenger bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) also regulates many of these same group activities of V. parahaemolyticus (14), doing so in the quorum-sensing–disrupted cell type (11), suggesting the intriguing hypothesis that this second messenger might be co-opted for novel quorum-sensing–mediated control mechanisms. Given the ubiquitous nature and diversity of c-di-GMP systems, it seems remarkable that there is only a single example of such a scenario. In this case, the diffusible signaling factor of the plant pathogen Xanthomonas campestris pathovar campestris is linked to c-di-GMP degradation via a HD-GYP domain-containing response regulator (15).
In V. parahaemolyticus, the lateral flagellar (laf) regulon and capsular polysaccharide (cps) gene expression are inversely tuned to the level of this second messenger. In fact, laf genes can be released from their normal, surface-dependent mode of gene control and induced in liquid by expressing scr genes, which modulate the cellular c-di-GMP concentration (16, 17). The scrABC operon is up-regulated in response to growth on a surface, and ScrC, a GGDEF-EAL domain-containing protein, acts as a phosphodiesterase to decrease the c-di-GMP concentration of the cell during swarming (18, 19). Here we provide evidence that swarming in V. parahaemolyticus is a social activity: It is influenced by a distinct cell–cell signaling pathway that controls the activity of a membrane-bound GGDEF-EAL protein participating in setting the cellular c-di-GMP concentration.
Results
ScrB Modulates the Activity of ScrC Independently of ScrA.
The GGDEF-EAL domain-containing protein ScrC is a cytoplasmic membrane-bound enzyme that also contains a potential periplasmic sensory domain of ∼200 aa flanked by two transmembrane regions. This protein is encoded as part of a three-gene operon, scrABC (VPA1513-11). BLAST searches indicate that scrA encodes a predicted Class V pyridoxal-phosphate dependent aminotransferase (pfam00266; E-value = 2.0e-35). Although a ScrA-TnlacZ fusion protein partitions to the cytoplasm (16), ScrA contains a predicted twin arginine translocation signal and the native protein may be transported to the periplasm. ScrB is a predicted family-3 bacterial solute-binding protein (pfam00947; E-value = 2.2 e-17); it was localized in the periplasm by using a ScrB-TnphoA fusion construct (16). ScrC is a bifunctional enzyme: it can act as a phosphodiesterase (PDE) to decrease the cellular c-di-GMP pool when it is coproduced with ScrA and ScrB; conversely, ScrC produced in the absence ScrA and ScrB works as a diguanylate cyclase (DC) to generate c-di-GMP (18). During swarming, ScrC functions as a PDE and cellular levels of c-di-GMP are low (18, 19).
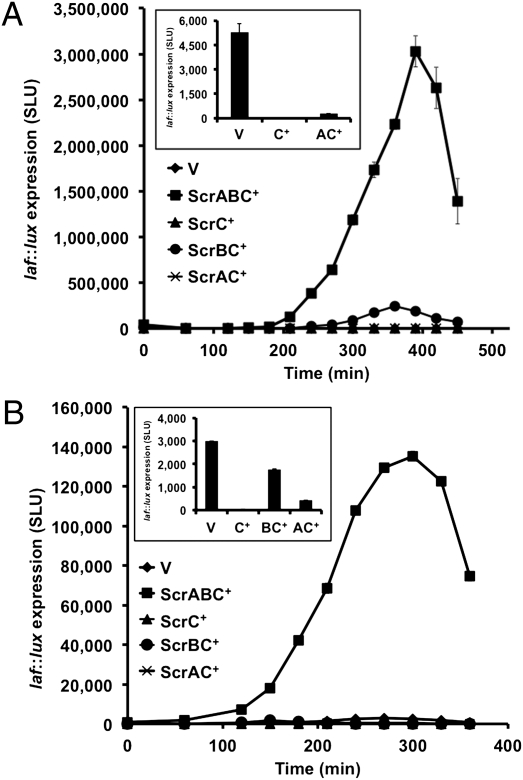
To dissect the flow of information through the pathway, additional combinations of the scr genes were expressed using an isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible plasmid; specifically, plasmids were designed to coproduce ScrBC and ScrAC. Ectopic expression scrA+ or scrB+ was previously shown to have little effect on lateral flagellar gene expression (16). The plasmids were examined in the ΔscrABC strain LM6565 that contains a luminescence reporter for laf gene expression and an allele of the regulator luxO, which locks the canonical quorum-sensing pathway in the low cell-density state (13). Consistent with prior findings, maximal activation of laf::lux by scrABC+ was 3,000,00 SLU (specific light units, normalized to cell density) during growth in heart infusion (HI) medium, whereas ScrC production severely prevented laf::lux induction, resulting in maximal light of ∼10 SLU (Fig. 1A). In comparison, the maximal light produced by the vector control strain was ∼5,300 SLU. Coproduction of ScrA with ScrC also resulted in repression of luminescence (∼300 maximal SLU); however, ScrB and ScrC together significantly reversed the repressing activity of ScrC. In fact, coexpression of scrBC+ resulted in the stimulation of laf::lux to ∼243,000 maximal SLU. Thus, under these conditions the presence of ScrB was found to profoundly alter the activity of ScrC with respect to its ability to influence laf expression.
Fig. 1.
ScrB neutralizes the repressing activity of ScrC independently of ScrA, but ScrA is required for high-level activation of laf gene expression. The effects of ectopic expression of IPTG-inducible scr genes were examined using laf::lux ΔscrABC reporter strains. Strains: LM9429 (vector), LM9430 (ScrABC+), LM9431 (ScrC+), LM9432 (ScrBC+), and LM10149 (ScrAC+). Strains were grown in complex HI medium (A) and minimal Mops medium (B). (Insets) Maximal expression for the samples below the scale. Luminescence was normalized to OD600 and is expressed as SLU. Error bars represent the SD of triplicate light measurements. P values, calculated using maximal SLUs, were < 0.0001 for all comparisons.
The regulation of lux activity by the Scr proteins was also examined in minimal medium (Fig. 1B). As in HI medium, expression of scrABC+ stimulated luminescence in this growth condition, although activation was lower in minimal than complex medium (i.e., ∼135,000 vs. 3,000,00 maximal SLU, respectively). The vector-containing control strain produced ∼3,000 SLU in minimal medium. ScrC-mediated repression of laf::lux was severe (∼8 SLU) and equivalent to the repression observed in HI medium. The effect of coproduction of ScrA and ScrC on laf::lux was also similar in minimal (∼440 SLU) compared with HI medium (∼300 SLU). Coproduction of ScrB with ScrC elicited ∼1,800 SLU, restoring luminescence to a level not too different from the vector-control strain (0.6-fold similar). Thus, ScrB neutralized ScrC-mediated repression of laf::lux in minimal medium more than 200-fold; however, unlike the effect in HI medium, ScrB failed to stimulate ScrC’s ability to cause activation of laf::lux. Taken together, the data indicate that ScrB modulates the activity of ScrC independently of ScrA, the ScrB-mediated effect on ScrC activity is influenced by environmental conditions (i.e., complex vs. minimal media), and the system requires ScrA for full activation of laf expression.
ScrA Produces an Extracellular Signal.
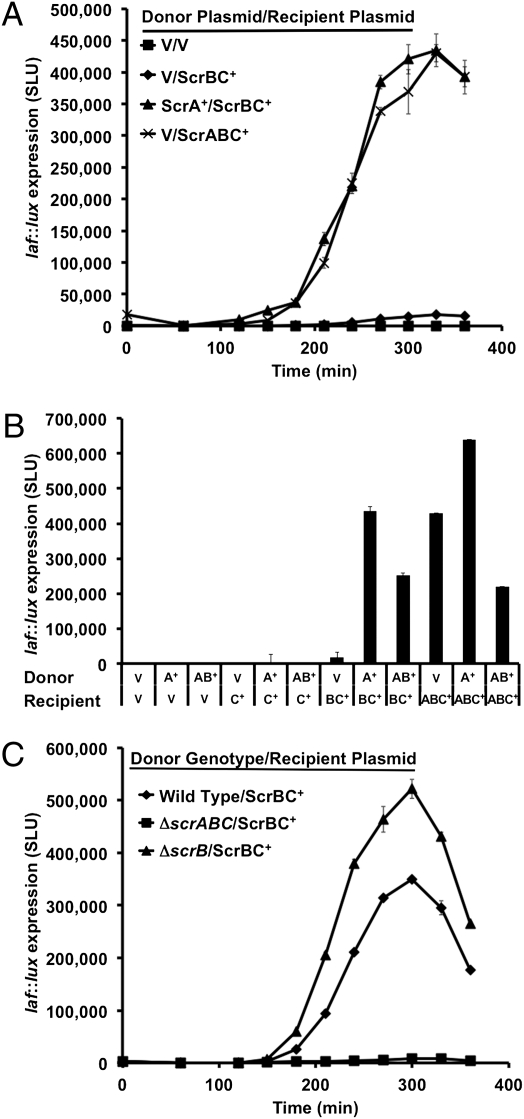
In keeping with the requirement of ScrA for full stimulation of the system, as well as the predicted enzymatic function of ScrA, we hypothesized that ScrA produced a signaling molecule capable of interacting with ScrB. Given that ScrB is a periplasmic solute-binding protein, we wondered if the product of ScrA needed to be within the cell or whether it could be supplied by neighboring cells. To test this theory, an extracellular complementation assay was designed. Nonluminous ΔscrABC donor strains expressing combinations of the scrABC operon from an IPTG-inducible plasmid or the vector control were cocultured at a 3:1 ratio in HI medium, with luminous ΔscrABC recipient strains containing other combinations of the scrABC operon expressed from IPTG-inducible plasmids. During coculture, the ΔscrABC/scrA+ donor strain activated laf::lux expression in the ΔscrABC/scrBC+ recipient strain to the levels observed for a strain with the resident signaling pathway intact; that is, with the neutral ΔscrABC/vector donor and the ΔscrABC/scrABC+ recipient strain (Fig. 2A). (Note that the maximal light is ∼fourfold lower in this experiment than in Fig. 1A, consistent with only one-quarter of the population in coculture being luminous.)
Fig. 2.
In coculture, ScrA makes an extracellular signal that activates laf::lux expression through ScrBC. (A and B) Cell–cell complementation and (C) physiological level of signal production. (A) Nonluminous ΔscrABC donor strains (with expression plasmids) were grown with luminous laf::lux ΔscrABC recipient strains (with plasmids) in HI. Donors: LM8038 (V) or LM8040 (ScrA+); Recipients: LM9429 (V), LM9430 (ScrABC+), or LM9432 (ScrBC+). Light production was monitored throughout growth. The ScrA+ donor significantly stimulated luminescence compared with the vector donor (P < 0.0001). (B) A similar experiment as in A was performed with an expanded complement of strains including the nonluminous donor strain LM8044 (ScrAB+) and the luminous recipient strain LM9431 (ScrC+). The bars represent the maximum SLU during the time course. (C) A similar coculture but performed with donor strains LM5674 (wild-type), LM6567 (ΔscrABC), and LM4897 (ΔscrB) and recipient LM9432 (laf::lux ΔscrABC/ScrBC+). The wild-type and ΔscrB donors significantly stimulated lux compared with the ΔscrABC donor (P values < 0.0001).
To further examine the requirements for signal transmission, the ΔscrABC/scrA+ donor strain was tested with recipient reporter strains that contained only ScrC or no Scr proteins at all. The ScrA+ donor failed to stimulate expression in the ΔscrABC/scrC+ recipient or the ΔscrABC/vector recipient (Fig. 2B). Furthermore, a donor strain coproducing ScrA and ScrB (ΔscrABC/scrAB+) could not activate laf::lux in the ScrC+ recipient strain. Thus, and consistent with a model in which ScrB and ScrC interact physically, these two Scr proteins must be present in the same cell to up-regulate laf::lux expression. The ScrA+ donor strain also stimulated laf::lux expression in the ScrABC+ recipient ∼50% more than the vector donor strain, suggesting dose-dependent activation of laf::lux. Interestingly, laf::lux expression was decreased by ∼twofold or greater when ScrB was coproduced in the donor with ScrA (P values < 0.0003). Coproduction of ScrB with ScrA may sequester some of the signal produced by ScrA, resulting in decreased yield in the supernatant.
The complementation experiments in Fig. 2 A and B were performed using ectopic expression plasmids in ΔscrABC backgrounds. We wondered whether ScrA signal production and extracellular complementation were physiologically relevant; that is, if a wild-type strain was capable of donating sufficient signal to elicit a response in a recipient rather than a donor with an IPTG-controlled scrA gene. To test this theory, a similar coculture experiment was conducted, but using wild-type or mutant donor strains. The wild-type donor was able to stimulate laf::lux expression in the ΔscrABC/scrBC+ recipient to a level almost as high as that found when the scrA+ plasmid-borne strain was used (350,000 vs. 440,000 SLU, in Fig. 2 C and A, respectively). The negative control donor ΔscrABC mutant failed to induce luminescence, whereas the ΔscrB mutant donor stimulated light production to a degree even higher than that of which the wild-type was capable, a result consistent with the posited ability of ScrB to bind and sequester the signal.
Extracellular Signal Can Be Harvested from Cell-Free Supernatant.
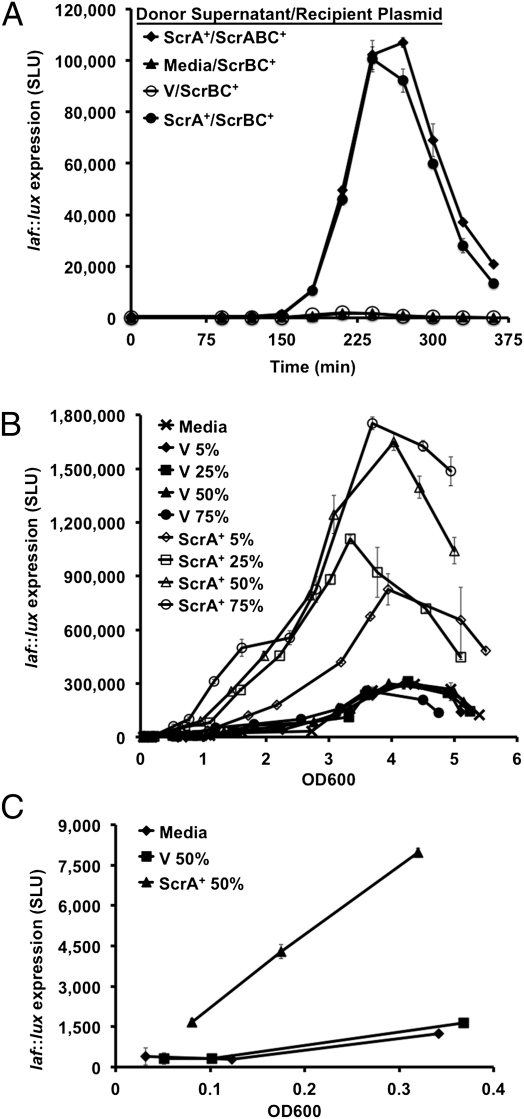
The previous coculture experiments demonstrated that the ScrA-generated signal could pass between cells. To determine if the signal might be isolated in cell-free supernatants, we harvested spent supernatants. For these experiments, the cultures were grown in minimal medium, in part to avoid the basal activation of ScrB in HI medium and also with an aim toward eventual purification of the signal. The ΔscrABC strains carrying the plasmid-borne scrA+ or the vector were induced with IPTG and grown to OD600 = 1. Cultures were centrifuged and the supernatant passed through a 0.22-μm filter to remove the residual cells. The cell-free supernatant from the ScrA+ strain stimulated laf::lux expression in the ΔscrABC/ scrBC+ recipient strain to 100,000 maximal SLU (Fig. 3A). Recipient ScrBC+ cultures spiked with supernatant prepared from the donor carrying the vector or with fresh medium were not bright (2,100 and 1,670 maximal SLU, respectively). Thus, the ScrA+ supernatant was capable of stimulating light production ∼50-fold. The light elicited in the ScrBC+ strain on provision of ScrA+ supernatant was similar to the level observed in the ScrABC+ recipient grown with ScrA+ supernatant (107,000 maximal SLU).
Fig. 3.
Cell-free supernatant stimulates laf::lux expression. (A) ScrA+ supernatant stimulates ScrBC+-mediated laf::lux expression; (B) ScrA+ supernatant shifts the timing of laf::lux induction in the ΔscrA strain; and (C) cell-free supernatant stimulates precocious swarming gene expression during growth on plates. (A) Cell-free supernatants were harvested from cultures of ΔscrABC strains carrying expression plasmids that were exponentially growing in minimal medium: LM8047 (V) and LM8050 (ScrA+). Luminous receptor strains (ΔscrABC laf::lux) with plasmids were inoculated into minimal medium that was amended with supernatant (or plain medium) at 50% final concentration: LM9430 (ScrABC+) and LM9432 (ScrBC+). Light was monitored throughout growth. Maximal SLU elicited by the ScrA+ donor was significantly different from that produced using the vector donor or media control (P values < 0.0001). (B) Cell-free supernatant was harvested from cultures of LM8047 (ΔscrABC/V) and LM8050 (ΔscrABC/ScrA+) that were exponentially growing in HI medium. LM9432 (ΔscrABC laf::lux/ScrBC+) was inoculated into HI medium that was amended with supernatant (or plain medium) at 0%, 5%, 25%, 50%, and 75% final concentration. (C) HI plates containing 50% ScrA+ supernatant, vector supernatant, or plain medium control were spread with LM1017 (laf::lux). During growth, individual plates were harvested by suspending cells in a fixed volume of medium to measure total OD600 and light. Throughout growth, the ScrA+ supernatant elicited a response that was significantly different from that produced by the vector supernatant or medium control (P values < 0.0001).
ScrA-Generated S-Signal Stimulates the Timing and Degree of laf::lux Induction.
The previous experiments demonstrated that ScrA produces a signal that stimulates laf::lux expression in ΔscrABC/scrBC+ recipient strains and that this signal, which we name the “S-signal” to indicate its effects on swarming, can occur extracellularly. To examine whether the concentration of signal could act to induce and coordinate induction of laf gene expression in a population, we supplied various amounts of supernatant to the ScrBC+ recipient strain (Fig. 3B). Growth rates were similar among the conditions. Increasing concentrations of ScrA+ supernatant incrementally increased the time of onset and level of induction of laf::lux expression. All concentrations of control supernatant failed to stimulate laf::lux expression above the levels of the blank control (i.e., the strain receiving fresh HI medium not supernatant). Thus, this experiment revealed that the timing of laf gene induction could be altered and suggested that S-signal accumulation could be a determining parameter of the induction of swarming.
To test this hypothesis directly, the effect on timing of activation by this extracellular signal was examined in another way. Lateral flagellar gene expression was monitored during growth on HI agar plates in strain LM1017, which contained only the laf::lux allele (i.e., it was not the ΔscrABC laf::lux reporter strain used in Fig. 3 A and B). HI plates were prepared containing 50% cell-free supernatant harvested from the ΔscrABC/scrA+ donor, the ΔscrABC/vector control strain, or with fresh medium. LM1017 was spread on multiple plates and after a 2.5 h incubation at 30 °C, individual plates were harvested every 30 min and examined for OD600 and luminescence measurements. Growth rates were similar on the various kinds of plates. Plates amended with ScrA+ supernatant induced laf::lux expression (normalized to OD600 and reported as SLU) at a lower OD600 compared with plates containing the vector supernatant or plain medium (Fig. 3C). For example, the ScrA+ supernatant elicited 1,660 SLU at OD600 = 0.08, but the control supernatant induced 325 SLU (OD600 = 0.10) and the media control induced 279 SLU (OD600 = 0.12). Thus, cell-free supernatant can trump cell density: provision of S-signal in the supernatant elicited activation of gene expression at low cell density to provoke precocious laf gene expression.
Cell–Cell Signaling Occurs During Swarming.
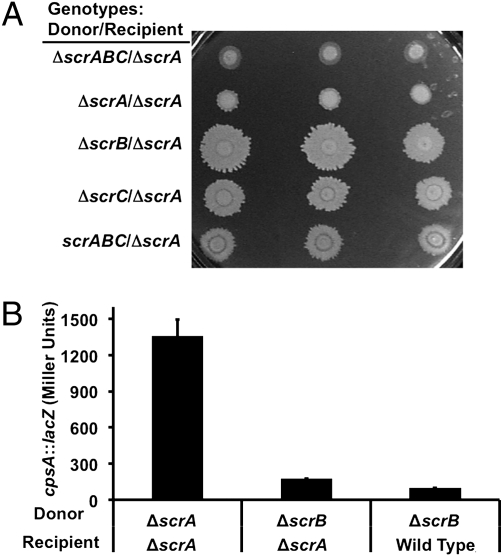
To dissect the roles of the individual scr genes, the prior experiments examined the consequence of various combinations of IPTG-controlled scr gene expression on laf::lux expression (either configured as donor strains or recipient strains). Although these combinations of strains provided insight into the function and information flow through the signal transduction pathway, we wanted to examine the importance of signaling during swarming on a surface. To do this assessment, wild-type and mutant strains were mixed and spotted on plates. The donor strains were swarm-defective because of a lux insertion in the laf structural gene encoding the flagellar hook; they also contained various scr alleles. The recipient strain, which was disrupted in canonical quorum sensing because of a deletion in the output regulator opaR (13), was also swarm impaired, but solely because of the ΔscrA allele. All of the mixtures containing donors with the wild-type scrA gene showed increased swarming compared with the ΔscrA or ΔscrABC donors (Fig. 4A, third to fifth rows compared with the top two rows). Thus, the signal produced by strains with an intact scrA allele was sufficiently received by ΔscrA neighboring cells to rescue their swarming defect.
Fig. 4.
Cell–cell signaling occurs during swarming. (A) Swarm-defective donors were mixed at equal OD600 with the signal-deficient recipient LM10150 (ΔscrA). Donors: LM6565 (ΔscrABC laf::lux), LM5545 (ΔscrA laf::lux), LM5547 (ΔscrB laf::lux), LM5549 (ΔscrC laf::lux), and LM1017 (laf::lux). Mixtures were spotted in triplicate in rows onto HI plates and incubated overnight. (B) Capsular polysaccharide gene expression was monitored over time by introducing a cpsA::lacZ reporter on cosmid pLM3122. The recipient strains LM10153 (ΔscrA/pLM3122) and LM10191 (wild type/pLM3122) were grown on HI tetracycline plates with the donors LM10151 (ΔscrA/vector) and LM10152 (scrB1::Tn5/vector). β-Galactosidase activity was measured at 18 h. The differences between ΔscrA and scrB1 donors were significant (P values < 0.0001).
The Scr circuit regulates surface colonization and controls more than just the laf regulon (18). In addition to being swarm-defective, scr mutants show enhanced biofilm formation in part because of excessive production of capsular polysaccharide. To examine cell–cell communication and regulation of cps gene expression within a colony, a similar experiment to that shown in Fig. 4A was conducted but with the introduction of a plasmid carrying a cpsA::lacZ reporter into the ΔscrA recipient strain, which was then mixed with either the ΔscrA or ΔscrB donor. The ΔscrA/cpsA::lacZ recipient grown in mixed colony with ΔscrA donor produced ∼eightfold more β-galactosidase than when it was grown mixed with the ΔscrB donor (Fig. 4B). The amount of reporter enzyme produced when the signaling pathway was reconstituted via cell–cell interactions was similar to that observed for a wild-type strain grown in similar configuration (i.e., an scrABC+ strain carrying the LacZ reporter cosmid cocultured with the ΔscrB strain). Thus, Scr-mediated cell–cell signaling reciprocally controls laf and cps gene expression during growth in a colony on a surface.
Discussion
Our findings support a model (diagrammed in Fig. 5) of an alternative cell–cell signaling pathway in V. parahaemolyticus that is distinct from the archetypal vibrio quorum sensing cascade. This pathway involves quorum-modulated control of the activity of a GGDEF-EAL protein that affects c-di-GMP in the cell. We have identified the genes responsible for signal generation (scrA) and detection (scrBC). In coculture, the signal can be produced by one cell type and detected in another; furthermore, cell-free supernatants can elicit prematurely responsive gene expression when presented to cells at low cell density.
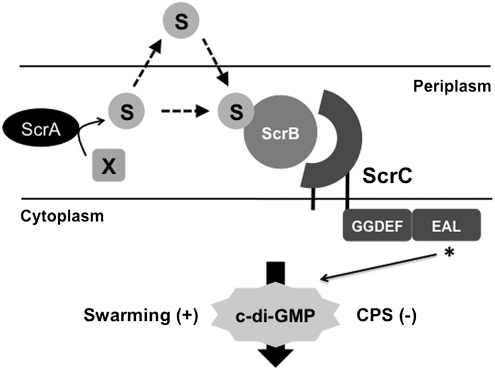
Fig. 5.
Quorum sensing and c-di-GMP. ScrC is a membrane-bound protein, with a periplasmic domain and cytoplasmic GGDEF and EAL domains. Prior work has shown that ScrC (in the absence of ScrA and ScrB) acts as a DC, whereas in the presence of ScrA and ScrB, ScrC displays PDE activity. Activation of the PDE reduces the amount of c-di-GMP in the cell, resulting in an increase in swarming and a decrease in CPS production. Experimental evidence in this work demonstrates that the presence of the periplasmic solute-binding protein ScrB is sufficient to neutralize the DC activity of ScrC as indicated by using a swarming reporter (laf::lux); however, full potentiation of ScrC’s PDE activity requires ScrA. ScrA is a predicted pyridoxal phosphate-dependent aminotransferase; it may localize to the periplasm because of a twin arginine translocation signal sequence. Although the substrate of this enzyme is not known (“x”), ScrA produces the autoinducer S-signal, which can exit the cell to communicate with ScrB in neighboring cells. Cell-free supernatants prepared from a ScrA-overproducing strain or the wild-type activate swarming and repress capsule gene expression in an ScrBC-dependent manner. The S-signal can promote precocious laf expression in a dose-dependent manner. Thus, the ScrABC circuit participates in directing quorum control of c-di-GMP-modulated swarming and sticking in V. parahaemolyticus.
This is a unique demonstration that swarming in V. parahaemolyticus is controlled by a quorum. It is a satisfying observation because swarming seems clearly a multicellular behavior and is known to be a group activity in many bacteria [e.g., Serratia species, Pseudomonas aeruginosa, and Rhizobium etli (reviewed in ref. 9)]; however, until now evidence suggested that swarming proficiency was associated with a quorum-disrupted cell type. Although in this work we have focused mainly on the regulation of the laf gene system, S-signaling is pertinent to global gene control. S-signaling represses expression of the capsular polysaccharide (CPS) biosynthetic locus. Importantly, the laf gene system is part of a larger regulon of genes that are specifically induced by growth on a surface (19). In addition to the surface motility system, the surface-sensing regulon includes other genes pertinent to surface colonization and virulence, including a type-three secretion system.
In many vibrios it is thought that virulence gene expression is repressed at high cell density, a scenario quite opposite from other bacterial species (reviewed in ref. 1). Taken together, the implications of our findings are that a second quorum system provides a unique and alternate mode of communication in V. parahaemolyticus, a system that has the potential to coordinate swarming, sticking, and virulence gene expression at high cell density. We note with interest that similar pathways may exist in other bacteria: there are ScrA homologs linked to genes encoding periplasmic binding proteins as well as GGDEF- and EAL-type proteins in some (but not all) Vibrio species and other proteobacteria.
Intracellular c-di-GMP seems an almost universal messenger in bacteria and it serves to potentiate diverse decision making events; for example, controlling the switch between sessile cells contained in biofilms and motile cells, influencing bacterial virulence, and determining cell-cycle progression (reviewed in refs. 20 and 21). Linking population density control with second messenger signaling expands the potential for an important means of orchestrating gene control. The decision to swarm or stick is a complex one, and by using the currency of c-di-GMP diverse input signals—environmental and social—can be factored into that decision.
This work also provides some insight into the dual nature of ScrC. Previously we concluded that although ScrC is a bifunctional diguanylate cyclase and phosphodiesterase, it most likely functions in its c-di-GMP–degrading capacity in the cell because it is always expressed in an operon with scrA and scrB (18). Our current findings now suggest a potential active role for both domains: at low cell density (in the absence of the S-signal) the GGDEF domain elevates c-di-GMP and represses swarming, but at high cell density the accumulation of the S-signal activates the phosphodiesterase domain. Differentiation to the swarmer cell is a large commitment of cellular resources; there are ∼70 genes in the regulon and many of these are highly expressed as hundreds of lateral flagella are produced by the cell (18, 19). Securing a tight Off/On switch response to cell density could be an advantageous survival strategy for such a profound event as swarmer cell differentiation.
Although we do not yet know the molecular identity of the signal, based on the predicted activities of ScrA and ScrB, it seems likely to be derived from an amino acid. ScrA is classified as a fold-type I aminotransferase (22); ScrB belongs to family 3 of the extracellular solute-binding proteins, which are involved in binding polar amino acids (23). Furthermore, the activity of ScrB seems partially responsive to something in complex compared with minimal medium. Many of the characterized autoinducer molecules produced by proteobacteria are amino acid-based. The N-acyl-homoserine lactones and AI-2 (the product of LuxS) are derived from methionine and S-adenosyl methionine; similarly are the products of the CsqA synthases derived, which are aminotransferases (reviewed in ref. 2). CsqA was originally discovered in Vibrio cholerae; however, orthologs exist in diverse proteobacteria and recent work suggests that many may produce distinct signaling molecules (24). Although both CsqA and ScrA are aminotransferases, they have little identity beyond being PLP-dependent enzymes that share a common fold. ScrA is not CsqA: V. parahaemolyticus has its own CsqA ortholog, which is 82% identical to that of Vibrio harveyi, and also possesses the cognate sensor CsqS. Importantly, the S-signal functions in cells that are disrupted in the classic quorum signaling pathway of the Vibrionaceae, by either mutation in the terminal output regulator of the pathway opaR, or using an allele locking LuxO in the low cell-density state (13). ScrA represents a distinct class of enzymes capable of generating the S-signal autoinducer and this signal works independently of the LuxO pathway; thus, our findings expand the potential vocabulary and syntax of bacterial communication.
Materials and Methods
Bacterial Strains and Growth Media.
The strains and plasmids used in this study are described in Table S1 Strains were grown at 30 °C in HI medium (2.5% Bacto Heart Infusion broth, 1.5% NaCl) or modified Mops minimal medium (40 mM Mops pH 7.2, 4 mM Tricine, 50 mM KCl, 10 mM NH4Cl, 5 mM MgSO4, 10 mM KH2PO4, 428 mM NaCl, 0.4% galactose) (25). Media were supplemented when appropriate with 25 μg/mL gentamicin, 10 μg/mL tetracycline, or 0.5 mM IPTG.
The markerless ΔscrA2 strain LM10150 was derived from LM5793 (ΔscrA2::Kanr) by using the FLP recombinase to eliminate the drug resistance cassette (26). The IPTG-inducible scrAC+ plasmid pLM3604 was derived similarly from pLM3603. Plasmid pLM3603 was created by introducing the ΔscrB::Camr allele into pLM2796 (scrABC+), and then the drug-resistance cassette eliminated to create pLM3604. This deletion removed 844 of the 963-bp scrB coding sequence. The other scr expression plasmids have been described previously (16).
Reporter Assays.
For luminescence, exponentially growing cultures in HI or overnight minimal-grown cultures were diluted into growth medium with IPTG to an OD600 of 0.05 and grown with shaking at 30 °C. At intervals, samples were removed to measure OD600 and relative light units (RLU). RLU were determined in triplicate by measuring 100 μL samples for 15 s in a TD20/22 luminometer (Turner Designs). Specific light units (SLUs) are RLU per minute per milliliter per unit of optical density at OD600. Coculture experiments in liquid used a donor to recipient ratio of 3:1.
For β-galactosidase, exponentially growing cultures propagated in HI with tetracycline to maintain the reporter or a control cosmid were diluted to OD600 of 0.2, mixed at equal concentrations, and 50 μL spread onto HI tetracycline plates (1.5% agar). At 18 h, cells were harvested from these plates by suspension in 5 mL medium at times suited to bracket maximal reporter expression. OD600 was recorded, the cells were lysed (27), and LacZ activity was determined in triplicate for each sample (28).
Supernatant Preparation.
Exponentially growing cultures were harvested at OD600 of 1.0 for minimal-grown cells by centrifuging at 10,000 × g. The supernatant was passed through a 0.22-μm filter (MILLEX-GS; Millipore), aliquoted, and stored at −80 °C. Supernatants were prepared using strains containing a mutation in the capsular polysaccharide locus to eliminate the confounding effects of CPS during preparation.
Coculture to Examine Swarming.
Cells growing exponentially in HI were diluted to an OD600 of 0.05, mixed in a 1:1 ratio of donor and recipient, and 1 μL of the mixture was spotted onto HI plates (1.4% Bacto agar). The plates were incubated overnight at 30 °C and photographed.
Statistical Significance.
All experiments presented herein have been performed at least three times, and a representative experiment is shown. Error bars represent the SD of the mean of triplicate measurements determined for samples in the experiment shown. Statistical significance was assessed by using Student t test (two-tailed distribution with two-sample, equal-variance calculations).
Supplementary Material
Acknowledgments
We thank Daniel Chodur and Cindy Gode-Potratz for their input, and Hank Kimbrough for initial experiments examining the ScrA-generated signal. This work was supported by National Science Foundation Grant 0817593 and National Institutes of Health Training Grant 2T32AI007511-16 (to M.J.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113790108/-/DCSupplemental.
References
- 1.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winans SC. A new family of quorum sensing pheromones synthesized using S-adenosylmethionine and Acyl-CoAs. Mol Microbiol. 2011;79:1403–1406. doi: 10.1111/j.1365-2958.2011.07551.x. [DOI] [PubMed] [Google Scholar]
- 3.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 4.Xavier KB, Bassler BL. LuxS quorum sensing: More than just a numbers game. Curr Opin Microbiol. 2003;6:191–197. doi: 10.1016/s1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 5.Fuqua C, Winans SC, Greenberg EP. Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 6.Boyer M, Wisniewski-Dyé F. Cell-cell signalling in bacteria: Not simply a matter of quorum. FEMS Microbiol Ecol. 2009;70:1–19. doi: 10.1111/j.1574-6941.2009.00745.x. [DOI] [PubMed] [Google Scholar]
- 7.Parsek MR, Greenberg EP. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005;13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Antunes LC, Ferreira RB, Buckner MM, Finlay BB. Quorum sensing in bacterial virulence. Microbiology. 2010;156:2271–2282. doi: 10.1099/mic.0.038794-0. [DOI] [PubMed] [Google Scholar]
- 9.Daniels R, Vanderleyden J, Michiels J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol Rev. 2004;28:261–289. doi: 10.1016/j.femsre.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 10.McCarter L. The multiple identities of Vibrio parahaemolyticus. J Mol Microbiol Biotechnol. 1999;1:51–57. [PubMed] [Google Scholar]
- 11.McCarter LL. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J Bacteriol. 1998;180:3166–3173. doi: 10.1128/jb.180.12.3166-3173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaques S, McCarter LL. Three new regulators of swarming in Vibrio parahaemolyticus. J Bacteriol. 2006;188:2625–2635. doi: 10.1128/JB.188.7.2625-2635.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gode-Potratz CJ, McCarter LL. Quorum sensing and silencing in Vibrio parahaemolyticus. J Bacteriol. 2011;193:4224–4237. doi: 10.1128/JB.00432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yildiz FH. Cyclic dimeric GMP signaling and regulation of surface-associated developmental programs. J Bacteriol. 2008;190:781–783. doi: 10.1128/JB.01852-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan RP, et al. Cell-cell signal-dependent dynamic interactions between HD-GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris. Proc Natl Acad Sci USA. 2010;107:5989–5994. doi: 10.1073/pnas.0912839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boles BR, McCarter LL. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J Bacteriol. 2002;184:5946–5954. doi: 10.1128/JB.184.21.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim YK, McCarter LL. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J Bacteriol. 2007;189:4094–4107. doi: 10.1128/JB.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira RB, Antunes LC, Greenberg EP, McCarter LL. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J Bacteriol. 2008;190:851–860. doi: 10.1128/JB.01462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gode-Potratz CJ, Kustusch RJ, Breheny PJ, Weiss DS, McCarter LL. Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol Microbiol. 2011;79:240–263. doi: 10.1111/j.1365-2958.2010.07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Römling U, Simm R. Prevailing concepts of c-di-GMP signaling. Contrib Microbiol. 2009;16:161–181. doi: 10.1159/000219379. [DOI] [PubMed] [Google Scholar]
- 21.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 22.Percudani R, Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003;4:850–854. doi: 10.1038/sj.embor.embor914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tam R, Saier MH., Jr Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng WL, et al. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol Microbiol. 2011;79:1407–1417. doi: 10.1111/j.1365-2958.2011.07548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Putnam SL, Koch AL. Complications in the simplest cellular enzyme assay: Lysis of Escherichia coli for the assay of beta-galactosidase. Anal Biochem. 1975;63:350–360. doi: 10.1016/0003-2697(75)90357-7. [DOI] [PubMed] [Google Scholar]
- 28.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1972. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.