Abstract
Environmental stresses and nutrition availability critically affect animal development. Numerous animal species across multiple phyla enter developmental arrest for long-term survival in unfavorable environments and resume development upon stress removal. Here we show that compromising overall microRNA (miRNA) functions or mutating certain individual miRNAs impairs the long-term survival of nematodes during starvation-induced L1 diapause. We provide evidence that miRNA miR-71 is not required for the animals’ entry into L1 diapause, but plays a critical role in long-term survival by repressing the expression of insulin receptor/PI3K pathway genes and genes acting downstream or in parallel to the pathway. Furthermore, miR-71 plays a prominent role in developmental recovery from L1 diapause partly through repressing the expression of certain heterochronic genes. The presented results indicate that interactions between multiple miRNAs and likely a large number of their mRNA targets in multiple pathways regulate the response to starvation-induced L1 diapause.
Keywords: age-1, developmental timing, GW182, unc-31, ain-1
Food deprivation is a life-threatening challenge that animals frequently face as individuals and as species. Different organisms have developed versatile growth arrest strategies to overcome starvation-induced metabolic and developmental problems. The coordinated entrance into developmental arrest, long-term survival, and the reinitiation of development upon food availability are important biological processes to investigate.
The nematode Caenorhabditis elegans responds to starvation by entering developmental arrest at multiple stages of its life cycle (1). When late, first larval stage (L1) worms sense unfavorable conditions, they enter an alternative and long-lived larval stage called dauer larvae (or dauer diapause). However, when newly hatched L1 worms encounter an environment with no food, developmental programs arrest and the worm enters L1 diapause. Unlike dauer diapause, L1 diapause is not accompanied by life cycle changes and has not been shown to require certain signaling pathways that control the formation of dauer diapause [such as TGF-β signaling (daf-1, daf-7) and nuclear hormone receptor (daf-12)] (2, 3). Furthermore, worms that are long-lived due to dietary restriction or decreased mitochondrial respiratory rates are short-lived during L1 diapause, suggesting that the mechanisms controlling L1 starvation survival are different at least in some aspects from those controlling aging (3).
Previous studies showed that the release of postdocking calcium-regulated dense-core vesicles, the insulin receptor (InsR) pathway, the AMPK pathway, and protein chaperones are required for the long-term survival of starved L1 worms (2–4). The roles of InsRs have also been implicated in arresting the cell cycle in germ cells and a portion of somatic cells during L1 diapause (2, 4). Upon entering L1 diapause, RNA polymerase II quickly accumulates and pauses at promoter regions, and this accumulation was speculated to stop transcription and facilitate the immediate reinitiation of gene expression when food becomes available (2). However, the mechanisms that coordinate the long-term survival, overall developmental arrest, and reinitiation remain to be investigated.
microRNAs (miRNAs) are well known for their functions in controlling developmental timing in the nematode (5, 6). Surprisingly, the majority of the C. elegans miRNAs and miRNA families are not essential for development or viability under normal culture conditions (7, 8). The key properties of miRNA-mediated gene silencing, such as one miRNA targeting multiple mRNAs and one target gene being regulated by multiple miRNAs, suggest that (i) miRNAs are excellent candidate regulators for coordinating multiple aspects of molecular and cellular activities for specific physiological functions and (ii) miRNAs likely function in a synergistic or additive manner to regulate sets of genes under specific physiological conditions that are not limited to those related to development (9). Consistent with these ideas, several recent lines of evidence suggest that miRNA let-7 and the heterochronic genes lin-42 and hbl-1 are required to regulate the starvation-induced dauer diapause (10–12) and that a number of miRNAs including lin-4 and mir-71 are involved in regulating life span (13, 14). Furthermore, a recent study suggests that the expression of certain miRNAs is differentially regulated by starvation-induced dauer diapause (15). However, it remains unclear how, and to what extent, miRNAs coordinate animal survival and development in response to stresses. In this study, we addressed the questions of whether and how miRNAs impact developmental arrest and the long-term survival of early L1 stage worms in response to food starvation.
Results
Intestinal miRNAs Play Critical Roles in L1 Starvation Survival.
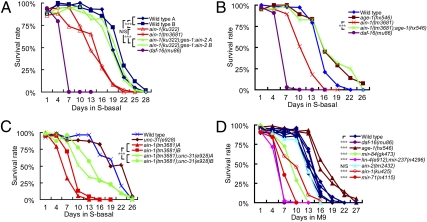
AIN-1 and AIN-2 are GW182 family proteins that are essential and partially redundant components of miRNA-induced silencing complexes (miRISCs) in C. elegans (16, 17). To investigate the roles of miRNAs in animal survival during starvation-induced L1 diapause, we impaired the overall miRISC function with loss-of-function (lf) mutants of ain-1 (ku322, ku425, and tm3681) and ain-2(tm2432) and examined their L1 starvation survival rate (Materials and Methods). We found that ain-1 but not ain-2 mutants displayed a significant reduction in L1 starvation survival rate compared with that of wild type (Fig. 1 A and D). We further found that this survival rate reduction of ain-1 mutants was overcome by ectopic expression of the AIN-2 protein in the intestine but not in the muscle (Fig. 1A and Fig. S1A). This is consistent with the previous reports that AIN-1 and AIN-2 are functional homologs with overlapping biochemical roles (16, 17). These results suggest that miRNAs act in the intestine, and possibly in other tissues, to promote L1 starvation survival. The overall effect of miRNAs on L1 starvation survival is expected to be significantly stronger than that reflected by the data in Fig. 1A because the ain-1 mutations reduce, but do not eliminate, miRISC functions.
Fig. 1.
Compromising overall miRNA function dramatically reduces the survival rate of L1 worms in starvation-induced diapause, and the effect can be significantly suppressed by an age-1/PI3K mutation. (A) Survival rate curves of wild-type and mutant strains, as indicated. The two ain-1 loss-of-function alleles displayed significant reductions in L1 starvation survival rate. The reduced survival of ain-1(ku322, lf) worms was suppressed by an intestine-expressed ges-1promoter:ain-2::GFP transgene (A and B represent two replicates). The result of a negative control using a myo-3promoter:ain-2::GFP transgene is shown in Fig. S1. Wild-type strains A and B are an N2 strain recently obtained from the C. elegans Genetic Center (reference 257) and an N2 strain from the laboratory stock, respectively. (B) Survival rate of single and double mutants to indicate the functional relationship between ain-1 and age-1. The low survival rate associated with ain-1(tm3681, lf) was largely overcome by the addition of the age-1(rf) allele. (C) The reduced L1 starvation survival rate of ain-1(lf) mutants was significantly suppressed by a null allele of unc-31. A and B represent replicated experiments of the indicated genotypes. (D) A representative chart of the L1 starvation survival rates of different miRNA mutants.
Previous studies indicate that the InsR pathway plays a dominant role in regulating L1 starvation survival and that reducing the activity of the insulin receptor daf-2, the PI3Kinase age-1, or the upstream regulator unc-31 results in increased L1 starvation survival rate (2, 3). We found that the reduced survival rate of ain-1 was suppressed by either reduction of age-1 function or loss of unc-31 function (Fig. 1 B and C), suggesting that a significant portion of the overall miRNA functions in L1 diapause is upstream of, or in parallel to, the InsR pathway.
To identify individual miRNAs that play prominent roles in L1 diapause, we screened 72 available mutant strains of individual miRNAs and miRNA families (87 miRNAs in total) using the L1 starvation assay. We identified 10 miRNA mutants that showed reduced survival rates with a stringent standard, as well as a few miRNA mutants with slightly increased survival rates (Table S1, Fig. 1D, and Fig. S1B). The effect observed in ain-1(lf) mutants is likely the consequence of the combined effects of attenuating functions of these individual miRNAs.
mir-71 Deletion Drastically Reduces L1 Starvation Survival and the Defect Is Partially Suppressed by Mutations in age-1/PI3K and unc-31.
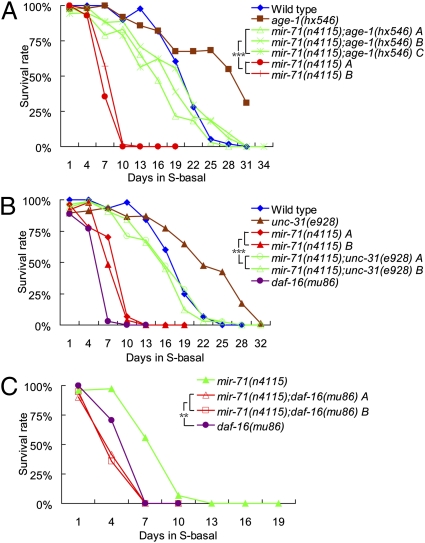
Among short-lived miRNA mutants, a mir-71 deletion mutant, mir-71(n4115) (referred to as mir-71(lf) hereafter), displayed a severe reduction in L1 starvation survival rate (Table S1 and Fig. 2A). Because the InsR pathway was previously shown to play a prominent role in L1 diapause (2, 3), we examined genetic interactions between miR-71 and different components of the InsR pathway. Reduction-of-function mutation (rf) in the age-1/PI3 kinase gene, age-1(hx546), made worms long-lived in the L1 starvation assay and was able to suppress the reduced L1 survival rate of mir-71(lf); the rate of the double mutants was comparable to that of wild type (Fig. 2A). We next examined the relationship between miR-71 and UNC-31, which functions upstream of AGE-1 during L1 diapause by regulating calcium-regulated dense-core vesicle fusion and the release of an insulin-like ligand (3). We found that the unc-31(e928 null) mutation was also able to partially, but significantly, suppress the reduced survival rate associated with mir-71(lf) (Fig. 2B). These results suggest that a significant portion of the miR-71 activities in L1 diapause survival may be devoted to regulating the activities of UNC-31–mediated InsR/PI3K signaling and that the rest of miR-71 activity may regulate UNC-31–independent pathways.
Fig. 2.
Mutating miR-71 drastically reduces the survival rate of animals in L1 diapause, and the effect can be suppressed by mutations of insulin receptor pathway genes age-1 and unc-31. (A) The mir-71(n4115, lf) mutant displayed severe reduction in L1 starvation survival rate, and the reduced survival rate of mir-71(lf) was suppressed by a reduction-of-function allele of age-1(hx546). (B) The severely reduced survival rate of the mir-71(lf) mutant was suppressed by a null allele of unc-31(e928). (C) The poor survival rate of daf-16(mu86, null) was enhanced by mir-71(lf). A, B, and C represent replicated experiments of the indicated genotypes.
We further examined the functional relationship between miR-71 and DAF-16, a FOXO transcription factor acting critically and negatively downstream of AGE-1/PI3K in the InsR pathway. We found that the poor survival rate of daf-16(mu86)(lf) was further decreased by mir-71(lf) (Fig. 2C), consistent with the notion that a portion of miR-71 activities regulate genes that act in parallel to UNC-31–mediated InsR/PI3K signaling for long-term survival during L1 diapause.
miR-71 Likely Directly Represses the Expression of age-1 and unc-31 by Acting on Their 3′ Untranslated Regions.
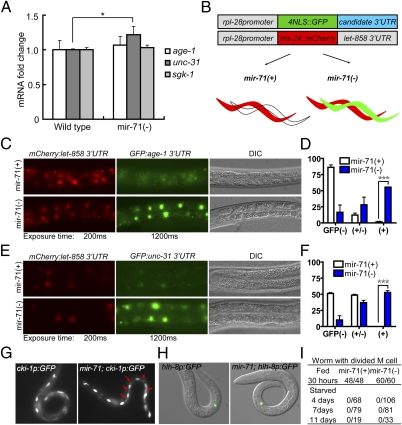
To test whether the activity of the InsR pathway was down-regulated by miR-71, we first examined the endogenous expression of components of the InsR pathway in mir-71(lf). We found that the mRNA level of UNC-31 was up-regulated by about 20% in mir-71(lf) (Fig. 3A). Because miRNA-mediated gene silencing may cause translational inhibition or mRNA degradation or both (19), the relatively small increase of UNC-31 in mir-71(lf) animals was still consistent with unc-31 being a target of miR-71.
Fig. 3.
miR-71 represses the expression of age-1 and unc-31 through the actions on their 3′UTR, but miR-71 is not required for arresting M cell division during L1 diapause. (A) Results of quantitative RT-PCR analysis of mRNA levels of indicated genes in 1-d–starved wild type and mir-71(lf) [labeled in Figs. 3 and 4 as mir-71(−)] L1 worms. The transcript level of unc-31 was increased in mir-71(lf) worms, compared with that of wild-type controls that were normalized to the value of 1. *P < 0.05. (B) A cartoon drawing illustrating the design of the dual-color 3′UTR reporter system (details in Materials and Methods) (18). (C) Fluorescence and differential interference contrast (DIC) images showing that the age-1 3′UTR reporter was repressed in mir-71(+) worms (3/4 transgenic lines) but not in mir-71(lf) worms (4/4 transgenic lines). (D) Fractions of worms that carry 3′UTR reporter transgene and show no GFP expression [GFP(−)], weak GFP expression [GFP(+/−)], and comparable GFP expression to mCherry [GFP(+)]. The proportions of GFP(+) worms were compared between mir-71(+) and mir-71(−) mutants; n = 128 and 71, respectively. (E) Fluorescence and DIC images showing that the unc-31 3′UTR reporter was repressed in mir-71(+)worms (2/2 transgenic lines) but not in mir-71(lf) worms (4/4 transgenic lines). (F) Two transgenic lines from each genotype in E were analyzed in the same ways as those in D. n = 26 for mir-71(+) and n = 33 for mir-71(−). (G) Fluorescence images showing the lateral views of 7-d–starved L1 worms expressing a cki-1promoter:GFP(cki-1p:GFP). The reporter is strongly expressed in H and V cells in both wild-type and mir-71(lf) worms. Note that there are extra GFP-positive cells (red arrows) in mir-71(lf) mutants. See Fig. S3 for the top view images. (H and I) Fluorescence images (H) and statistical data (I) showing that the M cell diveded in fed animals but remained undivided in 4-, 7-, or 11-d–starved L1 wild-type and mir-71(lf) worms.
We found that the 3′UTRs of several genes of the InsR pathway, including unc-31, age-1, pdk-1, akt-2, and sgk-1, contain predicted miR-71 targeting sites (as predicted by TargetScan and mirWIP). The computation-based prediction that age-1 and pdk-1 are potential targets of miR-71 was also reported in a recent study focusing on miRNA functions in aging where the mRNA level of pdk-1 was shown to be up-regulated in mir-71 worms (14). Among these potential miRNA targets, the predicted miR-71–targeting sites in the 3′UTRs of age-1 and unc-31 are conserved between C. elegans and Caenorhabditis briggsae, leading us to focus further analyses on these two genes.
We used a dual-color 3′UTR reporter system (18) to test the computational, prediction-based hypothesis that the 3′UTRs of age-1 and unc-31 are directly regulated by miR-71 (Fig. 3B and Materials and Methods). Specifically, an rpl-28:histone-24::mCherry:let-858 3′UTR construct that drives constitutive and ubiquitous mCherry expression was used as an internal control, and a 4×NLS::GFP construct driven by the same rpl-28 promoter and containing the 3′UTR of age-1 or unc-31 was used as the reporter (Fig. 3B). The reporter construct, the control plasmid, and a transformation marker plasmid were coinjected into worms to generate the extrachromosomal arrays for analysis. If the 3′UTR of age-1 or unc-31 is repressed by miR-71, the GFP expression will be repressed in tissues where miR-71 is expressed in wild-type worms, but derepressed in the same tissues of mir-71(lf) worms. We focused our analyses on intestine cells because (i) mir-71promoter:GFP was ubiquitously expressed in the pharynx, neurons, intestine cells, and other tissues throughout development (20) (Fig. S2A); (ii) we found strong expression of rpl-28 promoter-driven reporters in the intestine nuclei; and (iii) more importantly, our result presented in Fig. 1A and Fig. S1A indicated a dominant role of intestinal miRNAs in regulating L1 starvation survival. We observed constitutive expression of histone-24::mCherry in both the control and mir-71(lf) worms (Fig. 3 C and E). In contrast, the nuclear-localized GFP expression under the control of the 3′UTR of age-1(Fig. 3 C and D) or unc-31 (Fig. 3 E and F) was strongly repressed in the control worms, but prominently derepressed in mir-71(lf) mutant worms. These results suggest that miR-71 regulates the expression of unc-31 and age-1 through their 3′UTRs.
miR-71 Is Not Required for Arresting Seam Cell or M-Cell Divisions During L1 Diapause.
DAF-16 (the FOXO homolog in C. elegans) has been shown to play an important role in cell cycle arrest and developmental progression partly by promoting cki-1 expression in some somatic cells during L1 arrest (2). To determine whether miR-71 also plays a prominent developmental role in L1 diapause, we first examined the expression of a cki-1promoter:GFP (21) in V cells (epithelial seam cells dividing in early and mid-L1 stages). Interestingly, we found that the cki-1promoter:GFP was highly expressed in mir-71(lf) worms, and there were even additional cells that expressed the GFP in 7-d–starved mir-71(lf) L1 worms (Fig. 3G and Fig. S3A). Furthermore, the divisions of M cells and seam cells, as well as the expression of lin-4promoter:YFP, were suppressed in starved mir-71(lf) L1 worms similar to starved wild-type L1 worms (Fig. 3 H and I and Fig. S3 B and C). These results indicate that miR-71 is not essential for arresting seam cell or M-cell divisions during L1 diapause, suggesting that miR-71 function is distinct from DAF-16 function.
miR-71 Regulates the Timing of Vulval Cell Division in Animals Recovering from L1 Diapause.
Consistent with reported analyses (7, 14), we did not observe any obvious developmental defects associated with well-fed mir-71(lf) mutant animals. We thus asked whether miR-71 was required for the reinitiation of developmental programs during the recovery phase after L1 starvation. We noted that even though >90% of the mir-71(lf) worms were able to recover from 4 d of L1 starvation, most of them displayed defects in vulval development (protruding vulva or vulvaless) and a severe reduction in brood size (Fig. S4A).
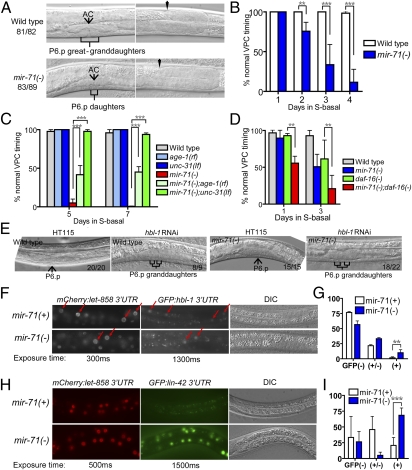
We further examined worms recovering from 4 d of L1 starvation and found that around 90% of the mir-71(lf) mutants displayed retarded vulval precursor cell (VPC) division, compared with less than 5% in wild type (Fig. 4A). Furthermore, mir-71promoter:GFP is highly expressed in the VPCs at L3 stage (Fig. S2B), and the severity of the VPC timing defect of the mir-71(lf) mutants depended on the length of time that mir-71(lf) L1 mutants were starved (Fig. 4B). In worms that recovered from 4 d of L1 starvation, we also found that a significant portion of the mir-71(lf) mutants displayed egg-laying defects and overproliferating or precociously reflexed gonads.
Fig. 4.
miR-71 regulates vulval cell division during recovery of starved L1 worms. (A) Differential interference contrast (DIC) images showing L4 worms recovered from 4-d–starved L1 worms. Whereas the vulva of wild-type worms developed into the pyramidal stage (81 of 82 worms), the P6.p of mir-71(n4115, lf) mutant worms divided only once (83 of 89 worms). (Right panels) The gonad of the same animals in the Left panels to indicate the similar developmental stage. AC, anchor cell. (B) Bar graph showing the correlation between the severity of the retarded vulval precursor cell (VPC) timing defect of mir-71(lf) mutants and the duration of L1 starvation. n > 40 for each genotype at every time point. (C) Bar graph showing that the delayed VPC timing defects of mir-71(lf) worms was suppressed by an unc-31(lf) mutation and partially suppressed by an age-1(rf) mutation. n > 40 for every genotype at each time point. (D) Bar graph showing that the delayed VPC timing defect of mir-71(lf) worms was enhanced by daf-16(lf) after 1 or 3 d of L1 starvation. Note that the daf-16(lf) worms recovering from 3 d of L1 starvation displayed a ∼12-h delay in overall development and that the mir-71(lf); daf-16(lf) double mutants displayed an ∼24-h delay. n > 30 for every genotype at each time point. (E) DIC images showing that hbl-1(RNAi) caused precocious VPC divisions in late L2/early L3 in both wild-type and mir-71(lf) worms recovered from 4 d of L1 starvation. The numbers on each image indicate how many worms of the examined ones displayed the indicated phenotype. (F) Fluorescence and DIC images showing that an hbl-1 3′UTR reporter was repressed in mir-71(+) worms and slightly derepressed in mir-71(lf) mutants. Red arrows point to representative intestine nuclei. (G) Statistical analysis of data in F; n = 60 for each. (H) Fluorescence and DIC images showing that a lin-42 3′UTR reporter was repressed in mir-71(+) worms (2/2 transgenic lines) and prominently derepressed in mir-71(−) worms (2/2 transgenic lines). (I) Statistical analysis of data in H; n = 30 for each.
miR-71 May Regulate Developmental Timing During Recovery from L1 Diapause by Repressing the Expression of Genes in both UNC-31/AGE-1/DAF-16–Dependent and –Independent Pathways.
We found that the vulval defects of recovering mir-71(lf) worms were strongly suppressed by an unc-31(null) mutation and partially suppressed by the age-1(rf) mutation (Fig. 4C and Fig. S4B), indicating that UNC-31 and AGE-1 have negative effects on proper vulval development during L1 diapause recovery and that miR-71 may antagonize such effects by repressing their expression.
Because daf-16(mu86 lf) worms displayed slow growth after 3 d of L1 starvation (delayed on average ∼12 h, n = 23), and because the insulin receptor pathway negatively regulates the activity of DAF-16, we speculated that the VPC timing defect of starved mir-71(lf) worms may be due mainly to the reduced activity of daf-16. If this were true, the starved mir-71(lf); daf-16(lf) double-mutant worms should show a slow growth phenotype similar to that of daf-16(lf) worms, but no specific VPC timing defect. However, we found that the VPC timing defect of mir-71(lf) was dramatically enhanced in mir-71(lf); daf-16(lf) double mutants recovered from either 1 or 3 d of L1 starvation and that the mir-71(lf); daf-16(lf) double mutants displayed an even slower rate (∼24 h delay) of overall growth than that of daf-16(lf) worms (∼12 h delay) when recovered from 3 d of L1 starvation (Fig. 4D). These results indicate that miR-71 plays a significant role in larval development of animals recovering from L1 diapause and likely does so by regulating the expression of components of the insulin receptor/DAF-16 pathway, as well as factors acting downstream, or in parallel to, DAF-16.
hbl-1 and lin-42 Are Likely Two Additional miR-71 Targets with Roles in Timing Regulation of VPC Division in Animals Recovering from L1 Diapause.
To understand how miR-71 affects VPC division, we searched its predicted targets for potential genes involved in regulating developmental timing. We found that the known developmental timing genes, hbl-1, lin-42, and lit-1, were at the top of the list (TargetScan). All three genes play roles in specifying the second larval stage (L2) developmental programs; loss-of-function mutations in these genes are associated with precocious expression of the L3 cell fates in hypodermal cell lineage (skipping the L2 program) (22–25). Moreover, the expression of hbl-1 is repressed by let-7 family miRNAs at L3 during normal development, and the hyperactivity of hbl-1 caused by failure of miRNA regulation leads to retarded development (26). Therefore, the retarded vulval development phenotype associated with mir-71(lf) mutants could potentially be caused by a collective effect of hyperactivities in these known timing regulators.
To test the hypothesis that these developmental timing genes mediate the regulatory role of miR-71 in larval development during recovery from starvation-induced L1 diapause, we examined whether knocking down HBL-1 function can suppress the retarded VPC timing defect of mir-71(lf). Consistent with the observation described above, the 4-d–starved mir-71(lf) mutants recovering on the RNAi control plates displayed the highly penetrant retarded defect in VPC division. In contrast, the mir-71(lf) mutant worms recovering on hbl-1(RNAi) displayed precocious VPC divisions similar to that seen in wild type (Fig. 4E). This result suggests that miR-71 likely functions upstream of, or in parallel to, HBL-1 in regulating VPC timing. In starved L1 worms, we detected only a slight increase in the mRNA level of hbl-1 in mir-71 mutants compared with that in wild type (∼10%), which may not be biologically significant. We then compared the expression of a hbl-1 3′UTR reporter (18) in the mir-71(lf) mutants with that in wild type and found that the expression of this reporter was slightly derepressed at L3 in the mir-71 mutant (Fig. 4 F and G). This is consistent with hbl-1 being one of the downstream targets of miR-71, although this modest effect alone is not expected to account for the vulval developmental phenotype in mir-71 mutant. This result is also consistent with the prediction from a miRISC immunoprecipitation analysis that hbl-1 is likely a target of one or more miRNAs, in addition to the let-7 family miRNAs, during early development (18). The strong suppression of the mir-71(lf) defect by hbl-1(RNAi), and the relatively weak effect of miR-71 on hbl-1 expression, are consistent with the idea that miR-71 exerts its role by modulating activities of multiple genes related to hbl-1 function in developmental timing.
To determine the functional relationship of miR-71 with LIN-42 and LIT-1, mir-71(lf); lin-42(lf) L1 worms were starved for 4 d and recovered on lit-1(RNAi) plates. Knocking down lit-1 by RNAi in mir-71(lf); lin-42(lf) double mutants caused no significant suppression of the VPC timing defects of mir-71(lf) worms. However, we found that the reporter transgene with the lin-42 3′UTR was significantly repressed in wild-type worms, but derepressed in the mir-71(lf) worms (Fig. 4 H and I). These results indicate that lin-42 is likely one of the targets regulated by miR-71 during the L1 starvation recovery phase, but the major developmental defects of mir-71(lf) are due to the collective effect of changes in expression of many target genes, including those acting downstream of, or in parallel to, lin-42 to regulate VPC divisions.
Discussion
Although the complete removal of miRNA functions causes embryonic lethality or infertility in worms, a partial disruption of overall miRNA functions by mutating either ain-1 or ain-2 provides an effective way to investigate miRNA functions (16, 17). Our analysis of L1 diapause using the ain-1(lf) mutants has generated valuable information regarding miRNA functions for long-term L1 survival, suggesting that miRNAs expressed in the intestine play critical roles during L1 diapause and that a significant portion of these miRNA activities are involved in modulating the UNC-31–InsR pathway. These results compelled us to examine specific interactions between individual miRNAs and their targets to gain mechanistic insights. Although the L1 diapause defect associated with ain-1(lf) mutations was quite robust, it reflected only a part of miRNA functions because the ain-1(lf) alleles themselves reduce but do not eliminate miRISC functions. The effects of eliminating all miRNA functions are expected to be much stronger, which is perhaps consistent with the additive effect of the defects associated with mutations of many individual miRNAs shown in Fig. 1D.
Following the studies by Horvitz and colleagues (7, 8) that showed that single mutations of the vast majority of miRNAs did not reveal obvious developmental or growth defects, our study reinforces the idea that most miRNAs do not regulate specific physiological functions through a robust regulatory interaction between one miRNA and one target. Instead, many specific physiological functions, such as the starvation-induced stress response, are regulated by a miRNA-target network, often involving multiple miRNAs and a large number of their targets. On one hand, we showed that deletions of a good number of miRNAs have varying impacts on the L1 diapause survival rate, although they may effect the rate through different mechanisms. On the other hand, the role of a particular miRNA (miR-71) is executed by repressing the expression of many genes in multiple pathways. Our genetic analysis indicated that for both L1 diapause survival and developmental recovery functions, miR-71 regulates expressions of genes in both the insulin receptor-dependent and -independent pathways. Furthermore, the observed derepression of individual genes by mir-71(lf) seemed too weak to account for the phenotype, consistent with the idea that a prominent phenotype of an miRNA mutation is caused by the collective effect of changing expression in many genes, an important property of miRNA-mediated gene regulation. To gain a thorough understanding of the miRNA–target interactions for starvation-induced L1 diapuase, biochemical and genomic approaches (17, 18, 27) may be helpful in identifying other targets of miR-71 and targets of other miRNAs involved in the process, given the limitations of the existing target prediction programs (28).
The InsR pathway has been repeatedly shown to be critical in stress responses (2, 3). Components of the InsR pathway, including age-1, have recently been predicted to be targets of miR-71 in its role in aging (14). Our data provide the experimental evidence that two components of the InsR pathway are likely direct targets of miR-71 in its role in a specific physiological process, L1 diapause (see a model in Fig. S5). It is also worth mentioning that multiple components of the InsR pathway, including age-1, pdk-1, akt-2, and daf-16, are predicted to be targets of the let-7 family miRNAs. It seems plausible that miRNAs that control developmental timing are also involved in regulating the metabolic rate through repressing the InsR pathway activity. As pointed out above, multiple miRNAs in addition to miR-71 and the let-7 family miRNAs have roles in L1 diapause, and they may regulate the expression of many diverse targets that may include, but are not limited to, factors involved in UNC-31–InsR-signaling activities.
Unlike classical heterochronic miRNAs such as lin-4 and let-7, the role of miR-71 in vulval cell division is essential in animals recovering from starvation-induced L1 diapause, but not in animals hatched on plates with food. We speculate that the expression of heterochronic genes controlling the L2/L3 programs, including that of hbl-1 and lin-42, are increased during L1 diapause to arrest the developmental progression, and miR-71 is probably required to suppress these “excess” signals during the recovery phase (Fig. S5). However, miR-71 does not appear to regulate all postembryonic development during L1 diapause recovery. For example, we observed a robust retarded mutant phenotype in the vulval lineage but did not see obvious defects in seam cell differentiation or alae formation. The latter results are consistent with the observation that miR-71 is not expressed in the seam cells. It is possible that other miRNAs, including those in the let-7 family, control developmental timing in other tissues during the recovery phase after L1 starvation. We would also like to point out that, similar to its role in starvation survival, miR-71 likely regulates the expression of multiple targets in addition to the tested genes (unc-31, hbl-1, and lin-42) to influence developmental recovery; the developmental defects may not necessarily be caused mainly by changed expressions of the genes tested in our study.
A recent study showed that the expression of miR-71 was significantly increased relative to other miRNAs in starved L1 worms (15). This result is consistent with the observation that miR-71 is specifically required for the starvation-induced stress response (Fig. S5). Using an mir-71promoter:GFP reporter (20), we did not observe obvious difference between the levels of GFP expression in 1-d–starved and nonstarved L1 animals, raising a possibility that miR-71 is maintained at a high level during L1 diapause when the expression of many other genes is reduced. We asked whether the expression of miR-71 was regulated by DAF-16, which is required during L1 diapause for long-term survival (2). However, we found that the expression of mir-71promoter:GFP was not decreased in daf-16(lf) L1 mutants that were starved for 24 h (Fig. S6). This result suggests that the high expression of miR-71 during L1 diapause is induced or maintained by other signaling pathways.
Materials and Methods
Nematode Strains and Methods.
Worms strains were grown and maintained at 20 °C as described (29). The following strains were used: N2 (wild type), age-1(hx546), daf-16(mu86), MH2385 ain-1(ku322), ain-1(ku425), ain-1(tm3681), ain-2(tm2432), DA509 unc-31(e928), MT2257 lin-42(n1089), VT2084 unc-119(ed3) III; maIs352[unc-119(+) + Pmir-71:GFP], RG733 wIs78 IV, JR667 unc-119(e2498::Tc1) III; wIs51, NH263 hlh-8::GFP, VT825 dpy-20(e1282) IV; maIs113, PS4997 unc-119(e2498) III; syIs179, MH3489 hbl-1 3′UTR reporter. MT12993 mir-71(n4115) worms were outcrossed with N2 for four generations before any test except the initial screen. hbl-1– and lit-1–feeding RNAi experiments were carried out following standard protocols (30, 31).
L1 Starvation Survival Assay and Statistical Analysis.
L1 starvation assay was adapted from a previously described protocol (3). Briefly, worms were well fed for at least two generations, and gravid adults were bleached with hypochlorite and sodium hydroxide. The eggs were transferred to plates seeded with HB101 and bleached again 3 d later. The resulting eggs were hatched in 4–6 mL S-basal without cholesterol in 15-mL tubes (Greiner), which were placed on an end-over-end rocker (VWR) at 20 °C. A total of 16–24 h later, the density of newly hatched L1 worms was adjusted to three to five worms per microliter S-basal. To determine viability, 20-μL aliquots (60–100 worms) were placed every 3 d onto two 6-cm nematode growth medium (NGM) plates seeded with OP50, and the numbers of L1 worms were recorded as number of plated worms (Np). Three days later, the number of worms that were L2 or older was recorded as number of survived worms (Ns), and the survival rate was calculated as Ns/Np, which is an estimation of survived worms in the whole population. Survival curves were drawn in Excel. To compare the survival rates between strains, we simulated the survival rate of each genotype to 100 arbitrary “individual worms” and performed the log-rank test in Graphpad Prism 4. Also with Prism 4, the median life span was calculated with a nonlinear regression analysis using the following equation: Y = bottom + (top − bottom)/{1+10^[(LogEC50-X) × HillSlope]}. In this equation, bottom and top were set as 0 and 100, respectively. The median life span is equal to EC50. The data for 3′UTR expression and for VPC timing were analyzed using χ2 test. In Figs. 1–4, “*”, “**” and “***”represent a P value equal or less than 0.05, 0.005, and 0.001, respectively. Bar diagrams represent mean and SD in Figs. 1–4.
3′UTR Reporters and Microscopy.
PCR and molecular cloning were carried out following standard protocols. 3′UTRs of genes of interest were cloned into the modified pPD129.57 vector as described previously (18). The primers that were used to amplify the 3′UTR of candidate genes are available upon request. Individual GFP reporter constructs for candidate genes (4 ng/μL) and the mCherry internal control plasmid (4 ng/μL) were mixed with unc-119 rescuing plasmid (20 ng/μL) and pBluescript KS+ (72 ng/μL) and coinjected into unc-119(ed3) and mir-71(n4115); unc-119(ed3) worms following standard protocols (32). Non-Unc stable transgenic lines were maintained, and the expression of GFP and mCherry were observed under a Zeiss Axiovision II microscope. For examining the age-1 3′UTR reporter, the rol-6(d) marker (100 ng/μl pRF4) was used instead of the unc-119(+) plasmid. Images were pseudocolored in Photoshop CS3 (Adobe) and assembled in Illustrator CS3 (Adobe).
Supplementary Material
Acknowledgments
We thank the laboratories of R. Horvitz, V. Ambros, S. Mitani, and the C. elegans Genetic Center for many mutants of miRNA and other genes. We also thank B. Kudlow, M. Kniazeva, C. Chi, H. Zhu, R. Wang, M. Cohen, and Q. Zhou for reagents and technical assistance; A. Sewell, M. Cohen, and B. Weaver for comments on the manuscript; and B. Kudlow, B. Wood, T. Blumenthal, Y. Rui, and members of the M.H. laboratory for valuable discussions during the course of this study. This study is supported by National Institutes of Health Grant GM47869 (to M.H.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105982108/-/DCSupplemental.
References
- 1.Ogawa A, Sommer RJ. Developmental biology. Strategies to get arrested. Science. 2009;326:944–945. doi: 10.1126/science.1183272. [DOI] [PubMed] [Google Scholar]
- 2.Baugh LR, Sternberg PW. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr Biol. 2006;16:780–785. doi: 10.1016/j.cub.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 3.Lee BH, Ashrafi K. A TRPV channel modulates C. elegans neurosecretion, larval starvation survival, and adult lifespan. PLoS Genet. 2008;4:e1000213. doi: 10.1371/journal.pgen.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuyama M, Rougvie AE, Rothman JH. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr Biol. 2006;16:773–779. doi: 10.1016/j.cub.2006.02.073. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 6.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 7.Miska EA, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibáñez-Ventoso C, Driscoll M. MicroRNAs in C. elegans aging: Molecular insurance for robustness? Curr Genomics. 2009;10:144–153. doi: 10.2174/138920209788185243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammell CM, Karp X, Ambros V. A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2009;106:18668–18673. doi: 10.1073/pnas.0908131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tennessen JM, Opperman KJ, Rougvie AE. The C. elegans developmental timing protein LIN-42 regulates diapause in response to environmental cues. Development. 2010;137:3501–3511. doi: 10.1242/dev.048850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karp X, Ambros V. The developmental timing regulator HBL-1 modulates the dauer formation decision in Caenorhabditis elegans. Genetics. 2011;187:345–353. doi: 10.1534/genetics.110.123992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 14.de Lencastre A, et al. MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol. 2010;20:2159–2168. doi: 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karp X, Hammell M, Ow MC, Ambros V. Effect of life history on microRNA expression during C. elegans development. RNA. 2011;17(4):639–651. doi: 10.1261/rna.2310111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding L, Spencer A, Morita K, Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, et al. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Hammell M, Kudlow BA, Ambros V, Han M. Systematic analysis of dynamic miRNA-target interactions during C. elegans development. Development. 2009;136:3043–3055. doi: 10.1242/dev.039008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez NJ, et al. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 2008;18:2005–2015. doi: 10.1101/gr.083055.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong Y, Roy R, Ambros V. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development. 1998;125:3585–3597. doi: 10.1242/dev.125.18.3585. [DOI] [PubMed] [Google Scholar]
- 22.Abrahante JE, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 23.Lin SY, et al. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 24.Ren H, Zhang H. Wnt signaling controls temporal identities of seam cells in Caenorhabditis elegans. Dev Biol. 2010;345:144–155. doi: 10.1016/j.ydbio.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Tennessen JM, Gardner HF, Volk ML, Rougvie AE. Novel heterochronic functions of the Caenorhabditis elegans period-related protein LIN-42. Dev Biol. 2006;289:30–43. doi: 10.1016/j.ydbio.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 26.Abbott AL, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammell M, et al. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008;5:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood WB. The Nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 30.Fraser AG, et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 31.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 32.Mello CC, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.