Abstract
Since the sensational discovery of a living coelacanth off the east coast of South Africa, the geographic distribution of viable coelacanth populations has been a subject of debate. In the past, the coelacanths off the African mainland were thought to be strays from the Comoros because most coelacanths captured were caught in the waters surrounding the Comoros archipelagos. However, in recent years, a large number of coelacanths were captured off the coast of Tanzania, including nine living specimens observed in a remotely operated vehicles survey. Thus, it is possible that there is a reproducing population inhabiting waters off the Tanzania coast. We have sequenced the complete mitochondrial genomes of 21 Tanzanian and 2 Comoran coelacanths and analyzed these sequences together with two additional full mitochondrial genomes and 47 d-loop sequences from the literature. We found that the coelacanth population off the northern Tanzanian coast is genetically differentiated from those of the southern Tanzania coast and the Comoros, whereas no significant genetic differentiation occurs between the latter two localities. The differentiation between the northern and southern Tanzanian coast populations is consistent with the hypothesis that the existence of northward-flowing ocean current along the Tanzanian coast may reduce or prevent gene flow from the northern to the southern population. Finally, we estimated that the population localized to the southern Tanzanian coast and the Comoros diverged from other coelacanths at least 200,000 y ago. These results indicate that the coelacanths off the northern Tanzania coast are not strays but a genetically distinct group. Our study provides important information for the conservation of this threatened “living fossil.”
At present, Latimeria chalumnae (1) in the western Indian Ocean (WIO) and Latimeria menadoensis (2) in the Pacific Ocean are described as the only two extant representatives of the subclass Coelacanthimorpha (coelacanths). Since the sensational discovery of the first living coelacanth off the estuary of the Chalumna River of South Africa in 1938, this group has been regarded as one of the most important animals in evolutionary biology because they are the only survivors of an ancient Devonian lineage of crossopterygian fish close to the root of tetrapods (3). After the discovery of a second specimen, which was found in the Comoros archipelagos (4), the existence of a viable coelacanth population in this region was confirmed (5). In addition to the Comoros archipelagos, several coelacanths had been captured off the coasts of Mozambique (6), Madagascar (7), and Kenya (8), indicating that coelacanths are widely distributed throughout the WIO. However, because the majority of the coelacanths caught were from the Comoros archipelagos, the coelacanths captured or observed off African coastal regions were thought to be strays or dead-end drifters from the main population in Comoros (6, 9), making it unclear whether any viable populations existed outside of the Comoros archipelagos. Indeed, J. L. B. Smith, the discoverer of the living coelacanths, had also suggested that the first South African coelacanth individual may be a stray (4).
To address the above question, several molecular population genetic studies have been conducted. First, Schliewen et al. (6) investigated the genetic differentiation between coelacanths from Mozambique and Comoros by comparing partial sequences of the mtDNA d-loop region and band-sharing frequencies of multilocus DNA fingerprints. This study revealed extremely low genetic differentiation, implying that Mozambique coelacanths originated from the Comoran population. Next, Schartl et al. (9) examined the sequences of mtDNA cytb gene and the d-loop region and microsatellites in nuclear DNA among 47 individuals from Comoros, South Africa, Madagascar, Mozambique, and Kenya. No significant genetic differences between the populations were found. Although these results implied that coelacanths in the WIO form a large panmictic population, the strong South Equatorial Current might prevent gene flow from the south or north of the east African coast to the Comoros (9). Accordingly, it was suggested that all coelacanths living outside the Comoros were strays because of passive transport by the strong current from Comoros to coastal regions. Alternatively, the non-Comoran coelacanths could be founders of young subpopulations. Therefore, the biogeographic history of coelacanth populations in the WIO remains unresolved.
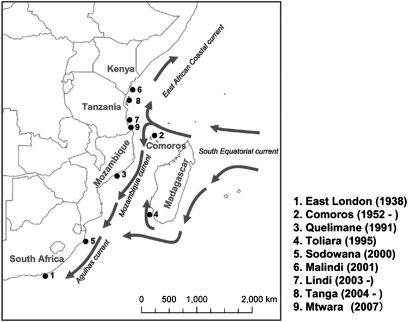
In recent years, a large number of coelacanth individuals were captured off the Tanga region in north Tanzania (Fig. 1 and Table S1). Although several coelacanths have been captured in south Tanzania (Lindi and Mtwara), most of the captures were in the north. Furthermore, in 2007 an international coelacanth research team (Aquamarine Fukushima, African Coelacanth Ecosystem Program, Sustainable Seas Trust, Tanzania Fisheries Research Institute) conducted underwater surveys off Tanga using remotely operated vehicles and encountered nine live individuals in their natural habitat (10) (Movie S1). These observations raised the possibility that a reproducing population exists in this area. To examine the genetic background of the Tanzanian coelacanths, Sasaki et al. (11) determined the complete mtDNA sequences of two individuals collected from northern and southern coasts of Tanzania and analyzed these data and the 47 (36 Comoro, 6 South Africa, 3 Madagascar, 1 Mozambique and Kenya) mtDNA d-loop region sequences reported by Schartl et al. (9). Although two additional sequences were not enough to provide a definitive conclusion, they showed that haplotype 5, defined by Schartl et al. (9), was observed only in Kenya and northern Tanzania but not in the Comoros, implying that this haplotype may distinguish the population of Tanzania from that of Comoros (11). In the present study, we determined the complete mtDNA genomes of 21 additional Tanzanian and 2 Comoran coelacanth individuals and analyzed them together with the published sequences to examine whether a genetically distinct, reproducing coelacanth population exists off the Tanzanian coast. The recent submersible research on coelacanths suggests that the survival of the coelacanth is severely threatened with extinction by the accidental catches of local fishermen. This study may provide important information in developing an effective plan to manage and conserve the severely threatened coelacanths.
Fig. 1.
The locations of the coelacanth individuals captured or observed in the WIO. The location numbers indicate the order of the captures. The location names and the dates are summarized at the right bottom of the map. Ocean currents in the WIO are indicated by gray arrows. The map is drawn according to Schartl et al. (9).
Results and Discussion
Comparison of Entire mtDNA Sequences.
We determined the entire mtDNA sequences from 21 Tanzanian and 2 Comoran coelacanths (GenBank accession nos. AP012177 to AP012199) (Table S1). The lengths and gene organization of the mtDNA determined in the present study were concordant with those previously reported by Sasaki et al. (11). The length of full mtDNA genomes of coelacanths in the WIO is 16,445 bp for all samples.
We first performed multiple alignments of the full mtDNA genomes determined in this study and those determined by Sasaki et al. (11) to estimate the nucleotide variation among individuals. In this alignment, a Comoran individual determined previously (12) was not included because of several critical sequence errors, such as a 21-nt deletion in ATPase6 and a 3-nt insertion in COI (see ref. 11 for more details). In total, 23 Tanzanian and 2 Comoran sequences were aligned. Table S2 shows the variable sites identified in 25 coelacanths in the WIO. Only 14 segregating sites were found in 16,445 bp, which amounted to only 0.018% nucleotide diversity. Such extremely low genetic variation among WIO coelacanths may be because of a very recent population subdivision or a very slow evolutionary rate in their mitochondrial genomes (see Are Coelacanths off African Mainland Strays, Recent Founders, or Distinct Populations?). Based on the 14 segregating sites, the sequences were divided into 12 haplotypes. These haplotypes were named according to Schartl et al. (9), in which only d-loop sequences were used. For example, Haplotype 1 based on the segregating sites in d-loop sequences was further subdivided into four haplotypes (1-1 to 1-4) by four independent segregating sites at nt 5370, 5437, 11712, and 12125. Similarly, Haplotype 3 was further subdivided into three haplotypes (3-1 to 3-3). These haplotype names (1-Xs and 3-Xs) were used only for the analyses of full mtDNA.
Fig. S1 shows the genealogical relationships of the 25 mtDNA sequences, including additional haplotypes in the WIO, deduced from the segregating sites found in the full mtDNA genomes. The coelacanth individuals represented by blue, red, and white characters were captured in northern Tanzania, southern Tanzania, and Comoros, respectively. The 12 haplotypes were separated into two large groups: Group A, consisting of Hap_3-Xs, 5, 7, 8, 9; and Group B, consisting of Hap_1-Xs and 10. These two groups were separated by three nucleotide substitutions: nt 16176 G to A, nt 3950 G to A, and nt 15772 G to A. We did not find the intermediate haplotypes, which connect groups A and B. This trend is similar in d-loop sequences, as shown later in Fig. 2. This haplotype network tree shows that identical haplotypes are not shared between Tanzania and Comoros. This result is quite unexpected because previous studies (6, 9) showed little genetic differentiation among coelacanths in the WIO.
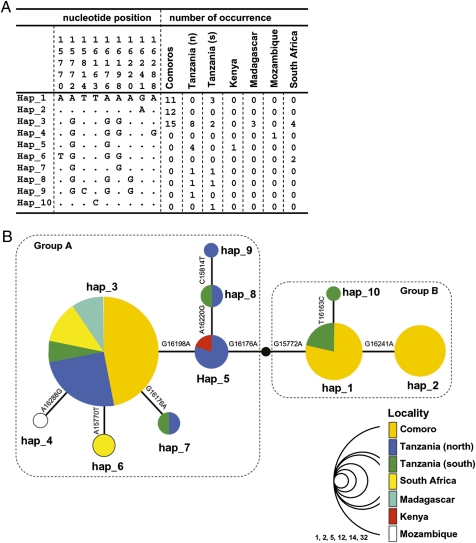
Fig. 2.
The observed mitochondrial haplotypes of coelacanths. (A) Alignment of 10 d-loop haplotypes of L. chalumnae in the WIO and the number of occurrences in each locality. The nucleotide positions are according to Sasaki et al. (11). The (n) and (s) of Tanzania indicate the northern and southern coastal regions, respectively. A Comoran sequence determined previously was not included because of several critical sequence errors. (B) Haplotype network of the d-loop haplotypes of L. chalumnae in the WIO. The size of the circle corresponds to the number of occurrences. Nucleotide substitutions between haplotypes are shown above each branch. Haplotypes of each locality are represented by different colors. The hypothetical haplotype that was not observed in the present study is denoted by a black circle.
Comparison of Coelacanth d-Loop Sequences in the WIO.
We analyzed the d-loop sequences from the present study and from Schartl et al. (9) to investigate the biogeographic relationships of coelacanth populations in the WIO. Although d-loop sequences might be less informative than entire mtDNA sequences, the 47 d-loop sequences published by Schartl et al. (9) allow us to include the individuals in different areas of the WIO into the analysis. The 73 d-loop sequences analyzed were divided into 10 haplotypes, four of which were newly characterized in the present study. Fig. 2A shows the nucleotide sequence alignment of the 10 d-loop haplotypes and the number of occurrences in each locality. Fig. 2B shows the genealogical relationships of these haplotypes. Consistent with the description of Schartl et al. (9) and Sasaki et al. (11), haplotype_3 was shared by most of the coelacanths caught in Comoros, Tanzania, South Africa, and Madagascar. The sharing of a haplotype among broad areas of the WIO may support the idea that the coelacanths captured off African mainland were drifters or very recent founders (4, 6, 9). However, we also found distinct differences in the haplotype frequencies among populations of Tanzania and Comoros. For example, previously unrecorded haplotypes_7, _8, _9, and _10 were found off the northern Tanzania coast but not in the Comoros, suggesting that the two populations have differentiated.
We then investigated the frequencies of d-loop haplotypes in each population more in detail, focusing on populations of Tanzania and Comoros. As in the analysis of entire mtDNA, d-loop haplotypes were subdivided into groups A and B, which are separated by two substitutions. Group A consists of haplotypes 3 to 9, and Group B consists of haplotypes 1, 2, and 10. This grouping was supported by the phylogenetic analysis of these 10 d-loop haplotypes by using Indonesian coelacanth, L. menadoensis, as an outgroup (Fig. S2). Other methods of clustering, such as hierarchical clustering (Fig. S3), also identified Groups A and B. It is important to note that the coelacanths captured off northern Tanzania did not include any haplotype in Group B, whereas coelacanths off southern Tanzania appeared in both groups A and B (Table 1). This finding may point to the possibility that populations of northern and southern Tanzania are dissimilar in their genetic compositions. Furthermore, in view of the observation that the coelacanths in Comoros also included both groups A and B, the genetic composition of southern Tanzania is likely to be more similar to Comoros than to northern Tanzania.
Table 1.
Differentiation (Fst values) between Tanzania and Comoros coelacanth populations
| Comoro (n = 38) | Northern Tanzania (n = 15) | |
| Tanzania (all n = 23) | 0.176†,** | — |
| Northern Tanzania (n = 15) | 0.341*** | — |
| Southern Tanzania (n = 8) | 0.000 (ns) | 0.226* |
*P < 0.05, **P < 0.01, ***P < 0.001, (ns) not significant.
†P value of Z* by permutation test (11).
To examine the degree of genetic differentiation between coelacanth populations of northern Tanzania, southern Tanzania, and Comoros, we calculated the Fst (estimation of genetic diversity and population differentiation) values between them (Table 1). We first calculated the Fst value between Comoros and Tanzania (Fst = 0.176, P < 0.01, permutation test of Z*) (13). This result indicates that the coelacanth populations of Tanzania and Comoros are distinct in their haplotype frequencies, although the genetic differences are relatively small. We then calculated the Fst values between northern Tanzania and Comoros and between southern Tanzania and Comoros, separately. Interestingly, the Fst value between northern Tanzania and Comoros (0.341) became much higher and statistically more significant (P < 0.001), whereas the Fst value between southern Tanzania and Comoros approached zero (almost no differentiation). Furthermore, the Fst value between northern and southern Tanzania was relatively high (0.226) and statistically significant (P < 0.05). Taken together, the population off the northern coast of Tanzania is genetically differentiated from the Comoros population, whereas the population off the coast of southern Tanzania is genetically close to the Comoros population. The present results reinforce the expectation by Sasaki et al. (11) that coelacanth captured off southern area of Tanzania (Songa Mnara) might be a stray from Comoros.
Divergence Time Between Northern Tanzania and Comoros Populations.
Finally, we estimated the divergence time between the populations of northern Tanzania and Comoros using a molecular clock based on the d-loop region. We first conducted a relative rates test between groups A and B, and found that the data were compatible with a molecular clock (P > 0.5 for all tests for both full mtDNA as well as d-loop alone) (see Methods). Because the Group B is a monophyletic group of coelacanths found only in Comoros (and possible strays in southern Tanzania) and Group A is group B’s monophyletic sister group, consisting of all localities, the age of the sharing of the last common ancestor between Comoros and northern Tanzania can be estimated by the divergence time between Groups A and B. The relative rate test does not reject the molecular clock between groups A and B (Table S3), indicating that we can interpolate the divergence times based on other estimates from the literature. The divergence time between the two coelacanth species was estimated in three studies (14–16). Holder et al. (15) used mtDNA sequences of 4,823 bp and estimated the divergence time at 4.7 to 6.3 Mya. On the other hand, using the entire mtDNA sequences (except for the d-loop) and Bayesian methods, Inoue et al. (16) estimated the divergence time at 30 to 40 Mya, much older than the estimate of Holder et al. (15). Sudarto et al. (14) also used Bayesian analysis and proposed the divergence time to be 28 Mya, similar to the estimate of Inoue et al. (16). However, the authors were cautious about this older dating because they also found that evolutionary distance and the pattern of nucleotide substitutions between L. chalumnae and L. menadoensis were very similar to those observed between Pan troglodytes and Pan paniscus, divergence of which may not be older than 10 Mya. In the present analysis, we used all three estimated divergence times to determine how long ago coelacanths of northern Tanzania and Comoros last shared a common ancestor (Table 2). We compared the rates of pair-wise nucleotide substitutions between individuals in populations A and B to those between L. chalumnae and L. menadoensis (Table 2 and Table S4). Because our data fail to reject a molecular clock (Table S3), we estimated the divergence times using the estimates of the substitution rates from sources in the literature (14–16). Our results based on Holder et al.’s estimate of 4.7 to 6.3 Mya for the divergence between L. chalumnae and L. menadoensis (15) indicate a mean divergence time between populations A and B of 350 to 470 kya (range 200–700 kya) (Table S4). Estimates based on older divergence times of 28 Mya (14) and 30 to 40 Mya (16) lead to even older mean divergence estimates of 2.1 Mya (range: 1.2–3.1 Mya) or 2.2 to 3.0 Mya (range: 1.29–4.4 Mya), respectively. Although the estimated divergence times differed greatly, the minimum estimate was at least 200,000 y ago. This result supports the view that the coelacanths in the coastal area of northern Tanzania are a reproducing population distinct from those near Comoros.
Table 2.
Estimated age of the sharing of the last common ancestor between L. chalumnae populations of northern Tanzania and Comoros
|
L. chalumnae – L. menadoensis divergence time (years ago) |
||||
| 4,700,000 (15) | 28,000,000 (14) | 30,000,000 (16) | ||
| Mean divergence time between Groups A and B (years ago) | 348,000 | 2,070,000 | 2,220,000 | |
Mean divergence times between mtDNA haplotype Groups A and B was estimated according to the mean value for the rate of pair-wise nucleotide substitutions between individuals in populations A and B to those between L. chalumnae and L. menadoensis of 0.074 (see Table S4 for more details).
Are Coelacanths off African Mainland Strays, Recent Founders, or Distinct Populations?
Previously, Schliewen et al. (6) and J. L. B. Smith (4) proposed that the coelacanth individuals caught or observed off the African mainland were strays. The argument for the “strays hypothesis” is as follows. Coelacanths in the surrounding waters of the Comoros are bottom-drift hunters inhabiting the relatively calm waters of the rock slopes. In contrast, the South African and Mozambique locations are exposed to the current and have flat, sandy, shallow bottoms, which might be unsuitable for coelacanths, making it unlikely that viable populations inhabit such regions. This hypothesis was further supported by several molecular studies (6, 9), none of which showed genetic differentiation between Comoros and African coastal regions. However, recent submersible expeditions in Sodowana, South Africa (17), and in Tanga, Tanzania (10) (Movie S1) discovered living coelacanth populations in their natural habitat and also several gravid females. Indeed, recent bathymetric study in the WIO (18) raised the potential coelacanth habitats in northern Mozambique and South Africa and suggested that Tanzania and Madagascar should not be ignored as potential habitats of coelacanths. Furthermore, our molecular clock analysis indicated that the divergence time between northern Tanzania and Comoros is likely to be more than 200,000 y ago. Thus, it is likely that the coelacanths in Tanga, northern Tanzania, are neither strays nor recent founders but rather are a genetically distinct reproducing group. Given that coelacanths caught off South Africa, Mozambique, and Madagascar possess distinct haplotypes (Fig. 2A), they likely also form viable populations. Given the similarity of coelacanths to sharks in their ecology, large body size, and long generation times, it is plausible that coelacanths exhibit a slow nucleotide substitution rate in their mitochondrial genomes similar to that observed in sharks (19). Thus, it is possible that the rate of substitution in the coelacanth mitochondrial genome is unusually slow, limiting the rate of genetic differentiation between populations. Concerning the population off south Tanzania, present analysis did not detect genetic differentiation from that of Comoros, implying the possibility of gene flow from Comoros, perhaps by passive transport through the South Equatorial current. However, comparison of entire mtDNA haplotypes showed that even southern Tanzania and Comoros did not share identical haplotypes (Fig. S1). Accordingly, entire mtDNA sequences and especially nuclear DNA markers of a sufficiently large number of coelacanth individuals in Comoros will help detect genetic differentiation between populations of southern Tanzania and Comoros.
Origin, Biogeography, and Conservation of Coelacanths in the WIO.
The phylogenetic and network tree of d-loop haplotypes suggest that the coelacanths in the WIO are divided into two large groups. Probably, in the common ancestor of coelacanths in the WIO, haplotypes were first diverged into these two groups. The coelacanths in Comoros possess both haplotype groups A and B (Fig. 2), whereas the others possess only group A. Therefore, the ancestral coelacanth population in the WIO might have originated in Comoros. Then, coelacanths possessing haplotype group A (probably haplotype_3) dispersed to different areas of the WIO by passive transport through the South Equatorial, Mozambique, and East African current. Although the date of this dispersal from Comoros to other localities is difficult to estimate at present, our analysis suggests that the northern Tanzania population diverged more than 200,000 y ago, and perhaps considerably earlier (Table 2 and Table S4).
According to the recent submersible observations, the living coelacanths are now severely threatened because of accidental catches by local fishermen (21–25). It is noteworthy that the number of haplotypes in the Comoros population was just 3 in the 38 individuals studied, whereas the number in the northern Tanzania population was 5 in the 15 individuals studied (Fig. 2A). Furthermore, the Tajima’s D statistic was significantly positive for only the Comoros population (Table S5). The demographic processes, such as a population reduction or bottleneck, population subdivision, or migration can produce significantly positive Tajima’s D in a neutral DNA marker (26). Because immigration is not likely for coelacanths in the Comoros, a recent population reduction or bottleneck may be more likely. As a genetically differentiated population, the coelacanths off the northern Tanzania coast should be important as a conservation unit. In the near future, we will obtain more information from larger datasets, such as whole genome sequences, which will lead to further understanding of the population structure of coelacanths. Recently, the Tanzanian government has begun promoting a large national marine park (The Coelacanth Marine Park) along the northern coastal region of Tanzania. We strongly support the goal of this project to conserve the coelacanths, a priceless heritage from the past (27).
Methods
Source of Tanzanian and Comoran Coelacanths.
In the present study, we determined the complete mitochondrial genomes of 21 Tanzanian and 2 Comoran coelacanths. The capture dates, localities, and ID numbers of the specimens are summarized in Table S1. The ethanol preserved tissues (muscles, fins, or gills) were transferred from Tanzania Fisheries Research Institute to Tokyo Institute of Technology in accordance with international regulations under the Convention on International Trade in Endangered Species of Wild Fauna and Flora. Total genomic DNA was extracted from ethanol preserved tissue samples using DNeasy Tissue kit (Qiagen) and stored at 4 °C in TE buffer until use.
PCR and Direct Sequencing of Entire mtDNA.
We determined the sequences of complete mtDNA for 23 coelacanths according to Sasaki et al. (11). Briefly, we first amplified the entire mtDNA sequence of each individual in two fragments. The long PCR amplification was performed with LA PCR kit (TaKaRa) according to the manufacturer’s instructions. By the second amplification, the entire mtDNA sequence was divided into seven fragments, which overlap each other, allowing us to obtain reliable sequences. The sequencing of each fragment was performed with BigDye terminator v3.1 cycle sequencing kit (Applied Biosystems) according to the manufacturer’s instructions. A total of 93 primers were used for sequence determination of both L and H strands of the mt genome.
Population Genetic Analyses.
The sequences of fragmented PCR products were assembled and edited by GENETYX-Windows version 10.1. The MEGA 4.0 software (28) was used for aligning the sequences, neighbor-joining tree construction, calculation of genetic distance, and divergence time estimation. The haplotype network was constructed using TCS (29). DnaSP 4.5 (30) was used for the estimation of genetic diversity and population differentiation (Fst). The hierarchical clustering analysis was conducted by calculating the distance matrix using the R function dist and then processing it using the hclust function (31). The test of the molecular clock was conducted using Phylemon 2.0 (32–34). The samples were grouped as (Group A, Group B, Outgroup: L. chalumnae group A, Group B, and L. menadoensis, respectively). Each clade comprising group A and B was treated as a star phylogeny (i.e., no topology was supplied for these individuals).
Supplementary Material
Acknowledgments
We thank the Tanzania Commission for Science and Technology for research permission and the Tanzania Fisheries Research Institute for permission to use their facilities. This work was supported by research grants from the Japan Society for the Promotion of Science AA Science Platform Program and the Ministry of Education, Culture, Sports, Science and Technology of Japan (to N.O.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AP012177–AP012199).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115675108/-/DCSupplemental.
References
- 1.Smith JLB. A living fish of Mesozonic type. Nature. 1939;143:455–456. [Google Scholar]
- 2.Pouyaud L, et al. [A new species of coelacanth. Genetic and morphologic proof] (Translated from French) C R Acad Sci III. 1999;322:261–267. doi: 10.1016/s0764-4469(99)80061-4. [DOI] [PubMed] [Google Scholar]
- 3.Gorr T, Kleinschmidt T, Fricke H. Close tetrapod relationships of the coelacanth Latimeria indicated by haemoglobin sequences. Nature. 1991;351:394–397. doi: 10.1038/351394a0. [DOI] [PubMed] [Google Scholar]
- 4.Smith JLB. The second coelacanth. Nature. 1953;171:99–101. doi: 10.1038/171099a0. [DOI] [PubMed] [Google Scholar]
- 5.Fricke H, et al. Habitat and population size of the living coelacanth Latimeria chalumnae. Environ Biol Fishes. 1991;32:287–300. [Google Scholar]
- 6.Schliewen U, Fricke H, Schartl M, Epplen JT, Pääbo S. Which home for coelacanth? Nature. 1993;363:405. [Google Scholar]
- 7.Heemstra PC, Freeman AL, Wong HY, Hensley DA, Rabesandratana HD. First authentic capture of a coelacanth, Latimeria chalumnae (Pisces: Latimeriidae), off Madagascar. S Afr J Sci. 1996;92:150–151. [Google Scholar]
- 8.De Vos L, Oyugi D. First capture of a coelacanth, Latimeria chalumnae Smith, 1939 (Pisces: Latimeriidae), off Kenya. S Afr J Sci. 2002;98:345–347. [Google Scholar]
- 9.Schartl M, Hornung U, Hissmann K, Schauer J, Fricke H. Genetics: Relatedness among east African coelacanths. Nature. 2005;435:901. doi: 10.1038/435901a. [DOI] [PubMed] [Google Scholar]
- 10.Mahika CG, Ngatunga BP, Mwakosya C, Kalombo H. Proceedings of the International Symposium on “The Coelacanth. Fathom the Mystery,”. Iwaki Meisei University; 2007. [Nov. 24, 2007]. ‘Living fossil’ found off the coast of Tanzania delights conservationists; pp. 12–15. available at http://www.marine.fks.ed.jp/coelacanth/symposium2007.html. [Google Scholar]
- 11.Sasaki T, et al. Mitogenomic analysis for coelacanths (Latimeria chalumnae) caught in Tanzania. Gene. 2007;389:73–79. doi: 10.1016/j.gene.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 12.Zardoya R, Meyer A. The complete DNA sequence of the mitochondrial genome of a “living fossil,” the coelacanth (Latimeria chalumnae) Genetics. 1997;146:995–1010. doi: 10.1093/genetics/146.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Mol Biol Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- 14.Sudarto , et al. Mitochondrial genomic divergence in coelacanths (Latimeria): Slow rate of evolution or recent speciation? Mar Biol. 2010;157:2253–2262. [Google Scholar]
- 15.Holder MT, Erdmann MV, Wilcox TP, Caldwell RL, Hillis DM. Two living species of coelacanths? Proc Natl Acad Sci USA. 1999;96:12616–12620. doi: 10.1073/pnas.96.22.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue JG, Miya M, Venkatesh B, Nishida M. The mitochondrial genome of Indonesian coelacanth Latimeria menadoensis (Sarcopterygii: Coelacanthiformes) and divergence time estimation between the two coelacanths. Gene. 2005;349:227–235. doi: 10.1016/j.gene.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Venter P, et al. Discovery of a viable population of coelacanths (Latimeria chalumnae Smith, 1939) at Sodwana Bay, South Africa. S Afr J Sci. 2000;96:567–568. [Google Scholar]
- 18.Green A, Uken R, Ramsay P, Leuci R, Perritt S. Potential sites for suitable coelacanth habitat using bathymetric data from the western Indian Ocean. S Afr J Sci. 2009;105:151–154. [Google Scholar]
- 19.Martin AP, Naylor GJ, Palumbi SR. Rates of mitochondrial DNA evolution in sharks are slow compared with mammals. Nature. 1992;357:153–155. doi: 10.1038/357153a0. [DOI] [PubMed] [Google Scholar]
- 21.Fricke H, Hissman K. Natural habitat of coelacanths. Nature. 1990;346:323–324. [Google Scholar]
- 22.Fricke H. Coelacanth tissue bank. Nature. 1992;357:105. [Google Scholar]
- 23.Fricke H, Hissmann K. Home range and migrations of the living coelacanth Latimeria chalumnae. Mar Biol. 1994;120:171–180. [Google Scholar]
- 24.Fricke H, Hissmann K, Schauer J, Plante R. Yet more danger for coelacanths. Nature. 1995;374:314. [Google Scholar]
- 25.Hissmann K, Fricke H, Schauer J. Population monitoring of the coelacanth (Latimeria chalumnae) Conserv Biol. 1998;12:759–765. [Google Scholar]
- 26.Maruyama T, Fuerst PA. Population bottlenecks and nonequilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics. 1985;111:675–689. doi: 10.1093/genetics/111.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith JLB. The atomic bomb and the coelacanth. The Daily Dispatch. 1963 10 December 1963. [Google Scholar]
- 28.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 29.Clement M, Posada D, Crandall KA. TCS: A computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 30.Rozas J, Rozas R. DnaSP, DNA sequence polymorphism: An interactive program for estimating population genetics parameters from DNA sequence data. Comput Appl Biosci. 1995;11:621–625. doi: 10.1093/bioinformatics/11.6.621. [DOI] [PubMed] [Google Scholar]
- 31.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 32.Sánchez R, et al. Phylemon 2.0: A suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Res. 2011;39:W470–W474. doi: 10.1093/nar/gkr408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson-Rechavi M, Huchon D. RRTree: Relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics. 2000;16:296–297. doi: 10.1093/bioinformatics/16.3.296. [DOI] [PubMed] [Google Scholar]
- 34.Robinson M, Gouy M, Gautier C, Mouchiroud D. Sensitivity of the relative-rate test to taxonomic sampling. Mol Biol Evol. 1998;15:1091–1098. doi: 10.1093/oxfordjournals.molbev.a026016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.