Abstract
Toll-like receptors (TLRs) are expressed on innate immune cells and trigger inflammation upon detection of pathogens and host tissue injury. TLR-mediated proinflammatory-signaling pathways are counteracted by partially characterized anti-inflammatory mechanisms that prevent exaggerated inflammation and host tissue damage as manifested in inflammatory diseases. We biochemically identified a component of TLR-signaling pathways, A20-binding inhibitor of NF-κB (ABIN1), which recently has been linked by genome-wide association studies to the inflammatory diseases systemic lupus erythematosus and psoriasis. We generated ABIN1-deficient mice to study the function of ABIN1 in vivo and during TLR activation. Here we show that ABIN1-deficient mice develop a progressive, lupus-like inflammatory disease characterized by expansion of myeloid cells, leukocyte infiltrations in different parenchymatous organs, activated T and B lymphocytes, elevated serum Ig levels, and the appearance of autoreactive antibodies. Kidneys develop glomerulonephritis and proteinuria, reflecting tissue injury. Surprisingly, ABIN1-deficient macrophages exhibit normal regulation of major proinflammatory signaling pathways and mediators but show selective deregulation of the transcription factor CCAAT/enhancer binding protein β (C/EBPβ) and its target genes, such as colony-stimulating factor 3 (Csf3), nitric oxide synthase, inducible (Nos2), and S100 calcium-binding protein A8 (S100a8). Their gene products, which are intimately linked to innate immune cell expansion (granulocyte colony-stimulating factor), cytotoxicity (inducible nitric oxide synthase), and host factor-derived inflammation (S100A8), may explain, at least in part, the inflammatory phenotype observed. Together, our data reveal ABIN1 as an essential anti-inflammatory component of TLR-signaling pathways that controls C/EBPβ activity.
Inflammation is triggered by innate immune cells upon exposure to noxious stimuli, such as infectious agents and tissue injury, and is characterized by leukocyte infiltration, release of proinflammatory mediators, and plasma leakage into tissues (1). Different receptor systems are used to recognize such noxious stimuli, including the repertoire of Toll-like receptors (TLRs), which activate diverse signal-transduction pathways that control proinflammatory effector mechanisms, such as production of cytokines, growth factors, and cytotoxic mediators, including nitric oxide (NO). As part of a physiological immune response, inflammation is essential for immune defense; however, exaggerated or persistent inflammation leads to tissue injury and possibly organ failure.
Persistent, chronic inflammatory diseases, such as systemic lupus erythematosus (SLE) and psoriasis, usually entail involvement of the adaptive immune system, i.e., B and T cells, whose effector functions, such as antibody production and cytotoxicity, may contribute to innate immune cell activation and tissue injury. SLE can affect almost any organ, including skin, brain, joints, and kidney; psoriasis initially is limited to the skin but in as many as 10–30% of cases progresses toward arthritis (2, 3). Still, SLE and psoriasis seem to involve similar pathogenetic mechanisms, whereby tissue-derived nucleic acids, liberated upon cell death, form complexes with autoreactive antibodies (SLE) or antimicrobial peptides (psoriasis and SLE) that trigger specific TLRs (i.e., TLR9 and TLR7), thereby providing a possible link between tissue injury and perpetuation of inflammation (2–5). Although such observations support the idea that TLRs and their signaling pathways are involved in the promotion of inflammatory disease, the etiology of both SLE and psoriasis is largely unclear. Both diseases involve clear genetic components as revealed by familial aggregation studies (2), and a set of recent genome-wide association studies (GWAS) identified several susceptibility loci, including TNF-α–induced protein 3 (TNFAIP3) and TNFAIP3-interacting protein 1 (TNIP1), which encode the ubiquitin-modifying enzyme A20 and the ubiquitin-binding protein, A20-binding inhibitor of NF-κB (ABIN1), respectively (6–9). ABIN1 was cloned originally by yeast two-hybrid screening as an A20-interacting protein (10). Although no information about the function of ABIN1 in TLR-signaling pathways in primary immune cells is available, A20 has been characterized in detail as an essential anti-inflammatory protein acting in TNF receptor (TNFR) and TLR pathways (11, 12). Importantly, A20's function in TLR pathways is essential for protection from a lethal inflammatory disease, indicating a possible link between deregulation of TLR-signaling pathways and human disease (12).
TLR activation, e.g., through immunostimulatory DNA (CpG-DNA/TLR9) or LPS (TLR4), leads to recruitment of the intracellular adaptor molecule MyD88, whose receptor-mediated oligomerization translates receptor triggering into signal transduction (13–15). MyD88 is used by all TLRs except TLR3 and is recruited, either directly or via an additional adaptor protein, Toll-interleukin 1 receptor domain containing adaptor protein/MyD88-adapter-like (TIRAP/MAL), to the intracellular domain of TLR family members (16). Oligomerization of MyD88 initiates the formation of a signaling complex that contains several characterized proteins, including members of the IL-1 receptor–associated kinase (IRAK) family and the ubiquitin ligases TNF receptor-associated factor 3 (TRAF3) and TNF receptor-associated factor (TRAF6), which in turn activate different signaling pathways and transcription factors, such as NF-κB, MAPK, IFN regulatory factors (IRFs), and CCAAT/enhancer-binding protein β (C/EBPβ) (15, 16). TRAF-mediated nondegradative ubiquitination is essential for TLR-mediated signal transduction, at least in part because of the recruitment and activation of ubiquitin-binding proteins and their interaction partners (17) (18). Deubiquitination and degradation of ubiquitinated signaling proteins appears to be the key mechanism by which A20 limits TLR signaling as part of a negative feedback loop. Accordingly, A20 loss of function leads to prolonged activation of classic inflammatory signaling pathways, such as the NF-κB and MAPK pathways, and exaggerated production of proinflammatory cytokines, e.g., TNF-α and IL-6 (12, 19). Coexpression of ABIN1 together with A20 in HEK293 cells was shown to contribute to the negative regulatory function of A20 in TNF-α–mediated NF-κB activation (10). Although these data suggested a cooperative function of the two proteins in TNFR signaling, ABIN1 recently has been found to counteract TNF-α–mediated liver cell death during embryonic development, a function that is independent of A20 (ref. 20 and below). Therefore, ABIN1 and A20 may cooperate in some functions but not in others.
Here we describe the identification of ABIN1 as part of the activated TLR-signaling complex. We generated gene-deficient mice and studied ABIN1's function in vivo and in TLR-stimulated macrophages in vitro. We found that ABIN1-deficient mice develop an inflammatory disease postnatally that shares many characteristics of human SLE, consistent with ABIN1's function as an anti-inflammatory molecule. Surprisingly, in contrast to A20-deficient macrophages, ABIN1-deficient macrophages did not exhibit a deregulation of classic proinflammatory signaling pathways and effector functions upon TLR activation but displayed a selective up-regulation of the transcription factor C/EBPβ and C/EBPβ-target genes, such as colony-stimulating factor 3 (Csf3, G-CSF), Nos2 (inducible NOS, iNOS), and S100 calcium-binding protein A8 ( S100a8, S100A8).
Results
ABIN1 Is Part of the Activated TLR-Signaling Complex.
The molecular mechanisms that enable TLRs and their signaling pathways to maintain the required balance between pro- and anti-inflammatory functions are characterized only partially. To define the molecular composition of TLR-signaling pathways, we performed proteomic experiments using the well-characterized TLR-signaling protein MyD88 as bait. We previously demonstrated that coumermycin A1 (CM)-mediated dimerization of a MyD88-GyraseB fusion protein can be used to mimic physiological, TLR-mediated cell activation (15). Moreover, when coupled to affinity purification and mass spectrometry (MS), the composition of transiently assembled signaling complexes can be determined, as demonstrated by the identification of TRAF3 as a component in TLR-signaling complexes and regulator of type I interferons (15). Here we used a similar, more quantitative approach and purified MyD88-GyraseB from unstimulated and CM-treated cells, followed by separation by SDS/PAGE, and MS analysis (Fig. 1A). Degradation of inhibitor of κBα (IκBα) and recruitment of known MyD88-signaling components, such as TRAF6, TRAF3, and IRAK4 were confirmed by immunoblotting (Fig. 1B). In the MS analysis not only the known MyD88-signaling proteins were detected; ABIN1 also was identified unequivocally by 10 unique peptides in the activated MyD88 complex (Fig. 1C and Table S1). Complex formation between MyD88 and ABIN1 during physiological TLR activation was confirmed using RAW264.7 cells stably expressing epitope-tagged MyD88-GyrB and ABIN1 that were stimulated with CpG-DNA and LPS, which activate cells in an MyD88-dependent manner via TLR9 and TLR4, respectively (Fig. 1D) (13, 21–23). Interaction between endogenous MyD88 and endogenous ABIN1 was confirmed further in untransfected RAW264.7 cells during CpG-DNA stimulation (Fig. 1E). Together, the data show that ABIN1 is recruited into a MyD88-marshalled signaling complex upon TLR activation.
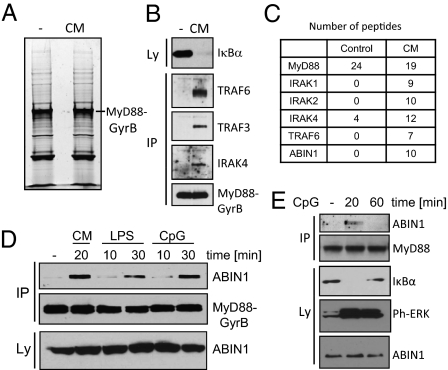
Fig. 1.
ABIN1 is recruited into the activated MyD88-signaling complex. (A and B) RAW264.7 cells expressing Flag-MyD88-GyrB-TAP were stimulated with CM for 6 min. Lysates were prepared, purified with antibodies against Flag, gel separated, and stained with SYPRO Ruby (A) or analyzed by immunoblotting (B). (C) MS analysis of lanes shown in A. The number of unique peptides of select proteins is shown. (D) RAW264.7 cells stably expressing Flag-MyD88-GyrB and HA-tagged ABIN1 were stimulated with CM, LPS, and CpG-DNA, followed by Flag immunoprecipitation and immunoblotting with antibodies against Flag (MyD88-GyrB) or HA (ABIN1). (E) RAW264.7 cells were stimulated with CpG-DNA, followed by immunoprecipitation with antibodies against MyD88 and immunoblotting with antibodies against ABIN1 and MyD88. IP, immunoprecipitation.
ABIN1 Is Essential for Protection from Embryonic Lethality and Postnatal Inflammatory Disease.
Given the genetic link between ABIN1 and inflammatory diseases and the identification of ABIN1 as part of the TLR-signaling complex, we hypothesized that ABIN1 may be involved in regulation of TLR-mediated inflammation. To investigate the function of ABIN1 in vivo, we established ABIN1-deficient mice which were based on ES cells carrying a gene trap cassette in the first intron of Tnip1, preceding the translation-initiating ATG located in the second exon (Fig. 2A). As expected, Tnip1 expression was abrogated completely both on the mRNA and protein level (Fig. 2 B and C). Consistent with a recent publication (20), the ratio of Tnip1−/− pups born alive from heterozygous crossings was far below Mendelian ratio (4.3% versus 25% expected), because of an essential antiapoptotic role of ABIN1 in TNF-α–mediated liver damage during embryogenesis (Tables S2 and S3). Embryonic lethality was influenced to some extent by the genetic background, because backcrossing from the 129S2 ES-cell background to C57BL/6 mice led to an increase of Tnip1−/− pups born alive at the F5 generation (10.3% vs. 25% expected) (Table S2). As reported, embryonic lethality could be prevented by simultaneous deletion of the genes encoding TNF-α (20) or TNFRI (Table S3). Interestingly, although live-born Tnip1−/− mice could not be differentiated macroscopically from WT littermates immediately after birth, they developed a cachectic disease and died prematurely within 4 mo after birth (Fig. 2 D and E). This disease was characterized by significant changes in the cellular composition of peripheral blood, lymph system, and different parenchymatous organs. Blood hemoglobin concentration was reduced, and the number of neutrophil granulocytes (neutrophils) in the peripheral blood was increased significantly (Fig. 2F). Also, as in patients with SLE and other chronic inflammatory conditions, peripheral blood and spleen contained a significant number of immature myeloid cells, characterized by low expression levels of Ly-6G (GR-1) and increased mRNA levels of neutrophil elastase (Ela2) and myeloperoxidase (Mpo) (Fig. 2 G and H) (24, 25). Lymph nodes and spleen were markedly increased in size and cellularity (Fig. 3A). More detailed analysis of spleen cells revealed an expansion of CD11b+ myeloid cells in Tnip1−/− mice, including GR-1high neutrophils, whereas B cell numbers were comparable to those in WT mice, and T cell numbers were reduced (Fig. 3B). T cells displayed an activated phenotype with increased surface levels of CD69 and CD44 and decreased levels of CD62L (Fig. 3C) accompanied by a general increase in Ig levels in the serum (Fig. 3D) and the spontaneous appearance of autoreactive antibodies against nuclear antigens and dsDNA (Fig. 3E). Several parenchymatous organs, such as liver, lung, and kidneys, were infiltrated with leukocytes, primarily F4/80+ myeloid cells and neutrophils (Fig. 3 F and G). Kidneys displayed significant glomerulonephritis, which correlated with proteinuria, reflecting tissue damage (Fig. 3H). Other organs, including joints, were not affected obviously by leukocyte infiltration (Fig. S1). Altogether, ABIN1-deficient mice develop an inflammatory disease with many characteristics of human SLE, including the appearance of immature granulocytes in the peripheral blood and development of autoreactive antibodies and glomerulonephritis.
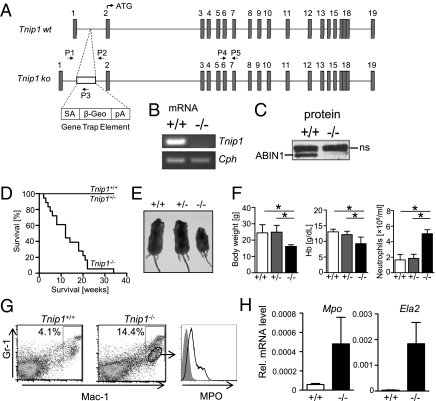
Fig. 2.
Generation of Tnip1−/− mice and development of runting disease and myeloid expansion. (A) Genomic WT and gene-trapped Tnip1 locus as generated in ES cells. Exons 1–19 are shown as gray squares; introns (drawn to scale) are shown as lines. Locations of primers used for genotyping PCR (P1–P3) and RT-PCR (P4, P5) are indicated. β-Geo, lacZ-neomycin fusion construct; pA, polyA signal; SA, splice acceptor site. Correct insertion of the gene-trap cassette was confirmed by PCR and DNA sequencing of the 5′ region. (B) mRNA from splenocytes of Tnip1+/+ and Tnip1−/− mice were analyzed by RT-PCR for Tnip1 and cyclophilin (Cph) expression using primers P4 and P5 (shown in A). (C) Protein extracts of splenocytes from Tnip1+/+ and Tnip1−/− mice were analyzed by immunoblotting for ABIN1 expression. ns, nonspecific. (D) Survival of live-born pups of indicated genotypes (n = 18 for each group). (E) Appearance of 6-wk-old mice (n = 7). (F) Body weight, hemoglobin (Hb) concentration, and neutrophil counts in blood of 6- to 8-wk-old mice (n = 9; *P < 0.01). (G) Splenocytes of Tnip1+/+ and Tnip1−/− mice were analyzed by flow cytometry using antibodies against the surface markers GR-1 and CD11b (Mac-1, macrophage-1 antigen) and intracellular MPO. MPO staining is shown for the CD11b+GR-1low subpopulation of a Tnip1−/− mouse as indicated. Data are representative of four experiments. (H) mRNA was prepared from peripheral blood leukocytes of Tnip1+/+ and Tnip1−/− mice, and expression levels of Mpo and Ela2 were determined by qPCR analysis. Error bars in F and H show SD.
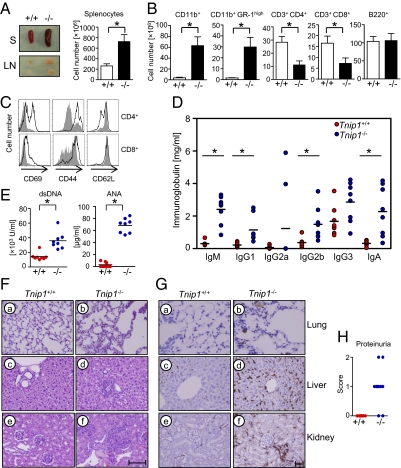
Fig. 3.
ABIN1 deficiency results in inflammatory disease. All experiments shown were done using 6- to 8-wk-old sex-matched WT and Tnip1−/− mice. (A) Appearance and total cell number of spleen (S) and lymph nodes (LN) of Tnip1+/+ and Tnip1−/− mice (n = 6). (B) Absolute numbers of splenic subpopulations as determined by flow cytometry (n = 6). CD11b+ depicts total of CD11b+ cells; CD11b+GR-1high depicts the GR-1high neutrophilic subpopulation of CD11b+ cells. (C) CD69, CD44, and CD62L expression on CD3+CD4+ and CD3+CD8+ splenocytes was analyzed by flow cytometry. Data shown are representative of the results of four independent experiments. (D) Serum immunoglobulin levels of Tnip1+/+ and Tnip1−/− mice as determined by ELISA (n = 8; *P < 0.001). (E) Serum levels of anti-DNA antibodies and anti-nuclear antibodies (ANA) of Tnip1+/+ mice (red circles) and Tnip1−/− mice (blue circles) as determined by ELISA (n = 8; *P < 0.01). (F) Microscopic image of H&E-stained tissue sections of lung (a and b), liver (c and d), and kidney (e and f). (Scale bar: 80 μM.) (G) Microscopic image of F4/80-stained tissue sections of lung (a and b), liver (c and d), and kidney (e and f). Brownish color identifies F4/80+ cells. (Scale bar: 80 μM). (H) Protein concentration in urine of Tnip1+/+ and Tnip1−/− mice (n = 9). For scoring, see Materials and Methods.
Deletion of TNFRI in Tnip1−/− Mice Protects from Embryonic Lethality but Not from Inflammation.
As mentioned, ABIN1 is required for protection from TNF-α–mediated liver apoptosis during embryonic development (ref. 20 and Table S2). However, Tnip1−/− TNF receptor superfamily member 1A (Tnfrsf1a)−/− double-knockout mice developed an inflammatory disease comparable to that in Tnip1−/− mice, as characterized by body weight loss, anemia, neutrophilia, leukocyte organ infiltrations, and glomerulonephritis, demonstrating that inflammatory disease measured by these parameters proceeds largely independently of ABIN1 function in the TNFRI pathway (Fig. S2 A–C). Still, the median survival of Tnip1−/− Tnfrsf1a−/− mice was extended significantly, by ∼2 mo, in comparison with Tnip1−/− Tnfrsf1a+/− or Tnip1−/− Tnfrsf1a+/+ mice, demonstrating some influence of TNFRI signaling for disease progression (Fig. S2D). Whether this effect results from the general inflammatory function of TNF-α or involves specific changes in TNF-α signaling because of the absence of ABIN1 remains to be established.
Bone Marrow-Derived Cells Drive Inflammation in ABIN1-Deficient Mice.
To determine whether cells of hematopoietic origin are responsible for the observed inflammatory disease, we established bone marrow-chimeric mice by transferring fetal liver cells from WT and Tnip1−/− embryos into lethally irradiated WT mice. Mice reconstituted with Tnip1−/− fetal liver cells developed an inflammatory disease comparable to that in Tnip1−/− mice, characterized by anemia, myeloid expansion, parenchymatous leukocyte infiltrations, and the activated phenotype of T and B cells (Fig. 4 A–C). These results indicate that bone marrow-derived cells trigger inflammatory disease in ABIN1-deficient mice.
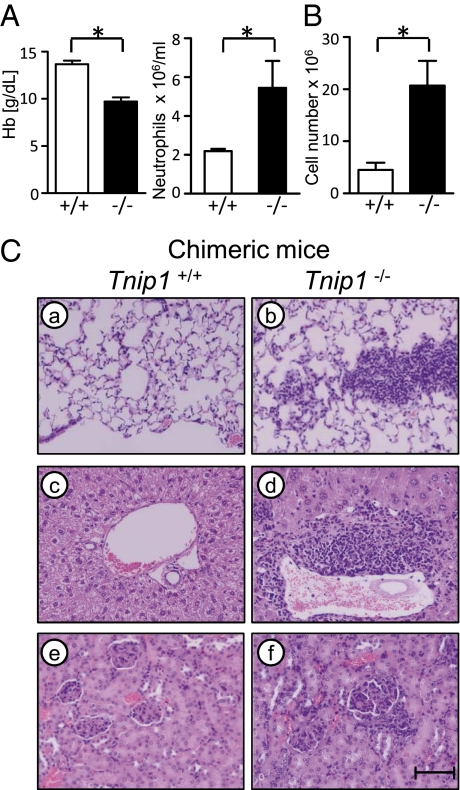
Fig. 4.
ABIN1-deficient bone marrow-derived cells promote inflammation. (A and B) Lethally irradiated WT mice were reconstituted with fetal liver cells of the indicated genotypes and analyzed 12 wk later for blood neutrophil counts and hemoglobin (Hb) concentration (A) and total splenic numbers of CD11b+ cells (B) (n = 5). (C) Chimeric mice were killed 12 wk post transplantation, and H&E-stained tissue sections of lung (a and b), liver (c and d), and kidney (e and f) were analyzed by microscopy. (Scale bar: 80 μM.)
ABIN1 Controls TLR-Induced C/EBPβ Activation.
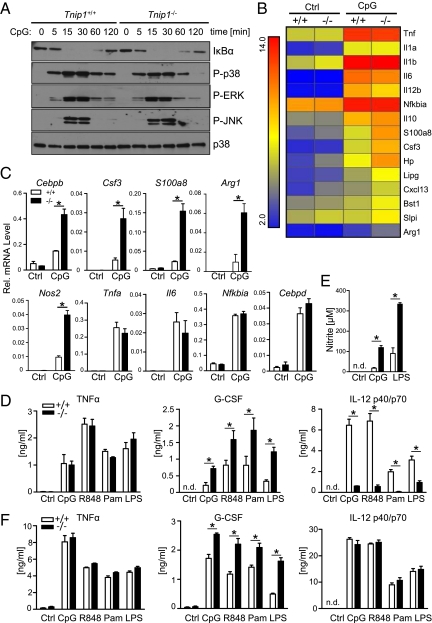
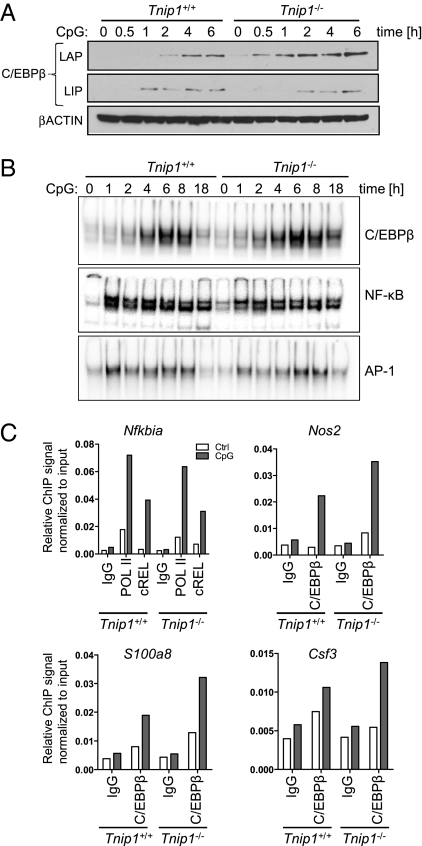
To identify TLR-signaling pathways and effector mechanisms controlled by ABIN1, we stimulated bone marrow-derived macrophages (BMM) from WT and Tnip1−/− mice with CpG-DNA and investigated classic TLR-signaling pathways, such as the NF-κB and MAPK pathways, by immunoblotting and gene regulation by microarray analysis. In stark contrast to A20-deficient cells (11), no differences were observed for IκBα degradation and resynthesis, reflecting NF-κB activation, or the phosphorylation of different MAPKs, including p38, ERK1/2, and JNK1/2 in WT and Tnip1−/− BMM upon CpG-DNA stimulation (Fig. 5A). Consistent with these data, detailed microarray analyses showed that genes encoding classic pro- and anti-inflammatory factors, such as TNF-α, IL-6, IL-1, IL-12, and IL-10, were up-regulated comparably in CpG-DNA-stimulated WT and Tnip1−/− BMM (Fig. 5B). In contrast, significant changes in mRNA expression levels were observed for a subset of genes that are established C/EBPβ-target genes, including Csf3, S100a8, and arginase, liver (Arg1). Differential regulation of these genes and of Nos2 and C/ebpb itself, which is up-regulated via a positive feed-forward loop, was confirmed by quantitative PCR (qPCR) analysis (Fig. 5C and Fig. S3) (26, 27). Notably, C/EBPβ as a potential ABIN1-regulated transcription factor also was identified by Cis-element overrepresentation (Clover) analysis based on the microarray dataset shown in Fig. 5B (Table S4). In this unbiased approach, 5-kb promoter regions 5′ upstream of genes deregulated (up-regulated) in Tnip1−/− BMM were extracted and analyzed for overrepresentation of transcription factor binding sites (28). C/EBPβ consensus binding sites were found to be significantly overrepresented in these promoters (P = 0.001), but NF-κB and activator protein 1 (AP-1) sites were not (significance threshold P < 0.01, for details see SI Materials and Methods). Increased levels of nitrite, reflecting iNOS activity, and G-CSF were found in the cell culture supernatants of CpG-DNA– and LPS-treated Tnip1−/− BMM (Fig. 5 D and E), whereas TNF-α was produced at levels comparable to those in WT BMM, and IL-12 levels were reduced (Fig. 5D). The reduction in IL-12 levels is consistent with data from C/EBPβ-mutant mice specifically lacking the major transcriptionally active form of C/EBPβ referred to as “liver-enriched transcriptional activator protein” (LAP) (29). Other TLR stimuli, i.e., the TLR2 agonist Pam3Cys and the TLR7 agonist resiquimod (R848) induced a similar pattern of cytokine release. Similar data also were obtained from bone marrow-derived dendritic cells (BMDC) [with the exception of IL-12 (Fig. 5F), whose secretion was not reduced in these cells, consistent with earlier observations demonstrating a cell type-specific regulation of this cytokine in BMM and BMDC (30)]. Also consistent with the up-regulation of C/EBPβ-target genes, the protein level of C/EBPβ LAP was increased and was accompanied by enhanced C/EBP DNA-binding activity in CpG-DNA–stimulated Tnip1−/− BMM (Fig. 6 A and B). Interestingly, a short form of C/EBPβ, referred to as “C/EBPβ liver-enriched inhibitory protein” (LIP), which lacks a transactivation domain, was found to be up-regulated to comparable levels in WT and Tnip1−/− BMM (Fig. 6A). Thus it appears that ABIN1 selectively controls the levels of the transcriptionally active LAP form of C/EBPβ. Consistent with the data obtained by immunoblotting and mRNA analysis, NF-κb and AP-1-DNA binding activity was induced comparably by CpG-DNA in WT and Tnip1−/− BMM (Fig. 6B). To determine whether increased expression of C/EBPβ and its target genes correlated with increased recruitment of C/EBPβ to relevant promoter regions, we performed ChIP assays from CpG-DNA–treated BMM. Indeed, C/EBPβ binding to promoter regions of Csf3, S100a8, and Nos2 was increased in Tnip1−/− BMM (Fig. 6C). In contrast, the NF-κB family member reticuloendotheliosis oncogene (c-REL) was recruited to the NF-κB polypeptide gene enhancer in B-cells inhibitor α (Nfkbia; IκBα) promoter comparably in WT and Tnip1−/− BMM (Fig. 6C). Taken together, the data show that ABIN1 specifically controls TLR-mediated C/EBPβ activity and expression of C/EBPβ-target genes.
Fig. 5.
ABIN1 controls the activation of TLR-mediated expression of C/EBPβ-target genes. (A) BMM from Tnip1+/+ or Tnip1−/− mice were stimulated with CpG-DNA, and total lysates were analyzed by immunoblotting for protein levels of IκBα and p38 and phosphorylation of p38, ERK1/2, and JNK1/2. (B) BMM of five 7- to 8-wk-old Tnip1+/+ and Tnip1−/− mice were stimulated for 4 h with CpG-DNA, and mRNA profiles were determined by microarray analysis. Classic inflammation regulators and genes up-regulated in Tnip1−/− over Tnip1+/+ BMM (fold change >2) are depicted. Gene arrays were done using mRNA from five independent BMM preparations (obtained from five independent mice) for each condition. Bst1, bone marrow stromal cell antigen 1; Cxcl13, chemokine (C-X-C motif) ligand 13; Hp, haptoglobin; Lipg, lipase, endothelial; Slpi, secretory leukocyte peptidase inhibitor. (C) mRNA levels determined by qPCR from BMM stimulated with CpG-DNA for 4 h [Cebpb, Csf3, S100a8, Tnfa, Il6, Nfkbia, and CCAAT/enhancer-binding protein δ (Cebpd)] or 24 h (Arg1, Nos2). (D and E) BMM were stimulated with indicated TLR agonists for 24 h, and TNF-α, G-CSF, and IL-12p40/p70 (D) and nitrite levels (E) were determined by ELISA and Griess reaction, respectively. (F) BMDC were stimulated with indicated TLR agonists for 24 h, and TNF-α, G-CSF, and IL-12p40/p70 levels were determined by ELISA. Data in C–F represent mean ± SD of three independent samples obtained from three separate BMM populations; data shown are representative of four (A), three (C–E), and two (F) independent experiments.
Fig. 6.
ABIN1 controls TLR-mediated C/EBPβ LAP activation. (A) BMM were stimulated with CpG-DNA, and C/EBPβ and βACTIN protein levels were determined by immunoblotting. LAP and LIP are visualized on the same blot by different exposure times. Note that the long form, LAP, contains the transactivation domain (which is missing in LIP) and therefore is transcriptionally active. (B) BMM were stimulated with CpG-DNA, and DNA-binding activity of C/EBP, NF-κB, and AP-1 was determined by EMSA. Representative data from three independent experiments are shown. (C) BMM from WT and Tnip1−/− mice were stimulated with CpG-DNA for 2 h, followed by ChIP analysis using antibodies against indicated factors or control IgG. Copurified chromosomal DNA of promoter regions of genes encoding G-CSF, S100A8, INOS, and IκBα were quantified using qPCR. qPCR values were normalized to qPCR values obtained from chromatin input sample and are displayed as relative ChIP signals. Representative data from three independent experiments are shown.
Discussion
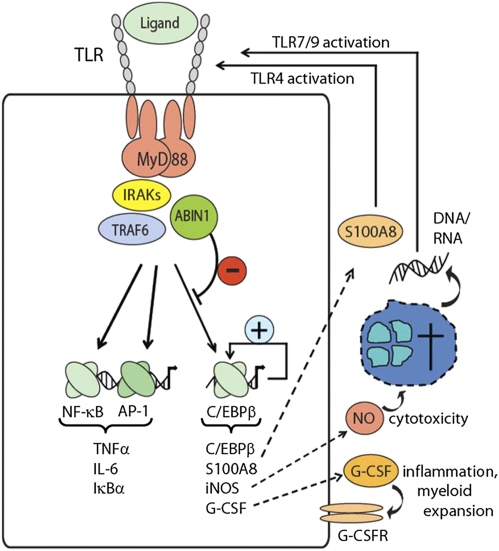
Our data show that ABIN1 is a negative regulator of TLR-induced C/EBPβ activity and that ABIN1 loss of function results in an inflammatory disease that reflects many characteristics of human SLE, an inflammatory disease of unclear etiology. A set of recent GWAS has revealed several disease-susceptibility loci for SLE, including TNIP1 and TNFAIP3, which encode ABIN1 and A20, respectively (6-8, 31, 32). Both genes also have been linked to psoriasis. In addition, TNFAIP3, but not TNIP1, has been linked to rheumatoid arthritis (8, 9, 33, 34). As mentioned, ABIN1 was cloned originally by a yeast two-hybrid system as an A20-interacting protein, and coexpression experiments in HEK293 cells demonstrated synergistic, negative regulatory activity of both proteins in the TNFR pathway (10). As such, it seemed possible that ABIN1 also contributes to A20-dependent activities in other pathways. Still, analysis of gene-deficient mice had demonstrated A20-independent activity of ABIN1, because only ABIN1-deficient mice, but not A20-deficient mice, die during embryogenesis as the result of TNF-α–mediated cell death (ref. 20 and Tables S2 and S3). Moreover, A20-deficient mice, but not ABIN1-deficient mice, develop significant joint inflammation, consistent with the above-mentioned link between A20 and rheumatoid arthritis (ref. 11 and Fig. S1). As shown here, TLR stimulation of ABIN1-deficient macrophages leads to increased C/EBPβ activation but does not provoke detectable deregulation of classic proinflammatory-signaling pathways, including NF-κB and MAPK pathways, a hallmark of A20-deficient macrophages (12). As a consequence, classic C/EBPβ-target genes encoding G-CSF, S100A8, iNOS, and ARG-1, but not NF-κB/AP-1-target genes encoding IκBα, TNF-α, IL-6, and IL-12, were up-regulated in ABIN1-deficient macrophages. Still, it currently is unclear whether ABIN1's function in the C/EBPβ pathway depends on A20 or whether the two proteins function entirely independently of each other. In fact, it is not clear at this point whether A20 has any role in C/EBPβ regulation.
As demonstrated, ABIN1 deficiency leads to deregulation of a rather select set of C/EBPβ-target genes, including Csf3, S100a8, and Nos2, whose characterized functions may explain several aspects of the observed phenotype. G-CSF is a major myeloid growth and survival factor (35), and myeloid expansion, including the appearance of increased numbers of macrophages and granulocytes, both in parenchymatous organs and peripheral blood, is a prominent phenotype of ABIN1-deficient mice. Notably, increased numbers of immature granulocytes also have been described in SLE patients (25, 24), and two recent reports suggested granulocytes as key players in human SLE through a cell death-related release of DNA, which potently activates innate immune cells via TLR9 when complexed with other host cell-derived factors such as antibacterial peptides or autoreactive antibodies (4., 5, 36, 37). INOS regulates the production of cytotoxic NO, which are essential for defense against intracellular pathogens but also can promote host tissue damage if produced at exaggerated levels (38, 39). S100A8, a protein whose serum levels correlate with SLE disease activity (40), has been demonstrated to trigger inflammation through direct binding of the TLR4/MD2 complex, a process shown to amplify other innate immune cell activation signals (e.g., LPS stimulation) (41). Based on these and other observations we propose a model for inflammatory disease development in which a low level of TLR stimulation (e.g., through TLR agonists released upon minor tissue injuries or through commensal bacteria) leads to increased production of described C/EBPβ-target genes. Their gene products promote amplification of inflammation (S1008A), cytotoxicity (iNOS), and liberation of cellular components such as DNA and RNA and increased myeloid expansion and cell turnover (G-CSF). Complex formation of liberated nucleic acids and antibacterial peptides or of nucleic acids and autoreactive antibodies then may trigger continued TLR activation and further promote a vicious circle of tissue injury, liberation of endogenous TLR agonists, and the perpetuating supply of myeloid cells. Notably, this model emphasizes innate immune cell deregulation as the primary culprit for the development of inflammatory disease, and it will be important to prove the contribution of identified factors in more detailed mouse models and possibly in human disease.
As demonstrated, ABIN1 is recruited into the MyD88-marshalled signaling complex and negatively regulates activation of C/EBPβ, a transcription factor whose mechanism of activation, particularly upon TLR stimulation, remains incompletely understood (42). Given that other MyD88-dependent signaling pathways, such as the NF-κB and MAPK pathways, proceed normally in ABIN1-deficient cells, it appears that ABIN1 contributes to the process of signal diversification downstream of MyD88. Still, the molecular mechanisms involved, with respect to both the proteins involved in ABIN1 recruitment and ABIN1's effector mechanism toward C/EBPβ, need to be established. A very recent report based on a knock-in approach using an ubiquitin-binding–deficient form of ABIN1 described the development of an autoimmune disease which also involved innate immune cells and lymphocyte activation (43). Embryos that were homozygous for this mutant form of ABIN1 did not die during embryogenesis; this result was unexpected given the earlier report demonstrating an essential role of ubiquitin binding in protection from TNF-α–mediated cell death (20). Also unexpected with respect to data obtained from ABIN1-deficient cells described in our work, TLR stimulation of innate immune cells and B cells from ABIN1-mutant mice showed deregulation of both NF-κB- and MAPK-signaling pathways, indicating additional effects of the mutant protein (43). It will be important to compare ABIN1-deficient cells and ABIN1-mutant cells side by side, possibly resolving the apparent discrepancies described.
TLR-mediated C/EBPβ activation in the absence of ABIN1 is reflected by increased C/EBPβ mRNA and protein levels and increased C/EBP DNA binding activity, all of which are consistent with the known regulation of C/EBPβ through a positive feed-forward loop that involves characterized C/EBPβ binding sites in the C/EBPβ promoter region (26, 27). Interestingly, we found that C/EBPβ LAP, but not C/EBPβ LIP, was increased selectively in ABIN1-deficient macrophages upon TLR stimulation. Both LAP and LIP forms are produced from the same Cebpb mRNA using alternative in-frame ATG codons for translation initiation because of a leaky ribosomal scanning mechanism (29). Although LAP contains DNA-binding and transactivation domains and therefore is able to drive transcription, LIP lacks the transactivation domain and is considered a negative regulatory protein (44). ABIN1 deficiency appears to shift the balance toward the transcriptionally active LAP form. The precise mechanism that regulates this process during TLR activation is still unclear; however, differential regulation of LAP vs. LIP forms is not unprecedented. A knock-in mutant of a catalytically inactive form of IRAK4 shows a defect in LIP production upon LPS stimulation, although LAP production is comparable to that in WT cells (45). Still, whether IRAK4 or any other additional protein is involved in ABIN1-dependent C/EBPβ regulation needs to be investigated. Notably, the pattern of gene deregulation observed in ABIN1-deficient macrophages matches very well those genes found to be affected in macrophages established from C/EBPβ-mutant mice that specifically lack the LAP form of C/EBPβ and strongly supports the interpretation that C/EBPβ LAP deregulation is responsible for the observed changes in gene expression in ABIN1-deficient mice (26).
Taking our results together, we identified a component of the TLR-signaling pathways, ABIN1, whose activity is essential for protection from inflammatory, lupus-like disease. As opposed to other, negative regulatory proteins, such as A20 or IRAK3, ABIN-1 selectively controls the TLR-activated C/EBPβ pathway but not NF-κB or MAPK pathways (12, 46). As a consequence, a select set of genes is deregulated, leading to increased production of the myeloid growth factor G-CSF, cytotoxic NOS, and inflammatory S100A8. Given the genetic link between ABIN1 and the human diseases SLE and psoriasis, the contribution of the C/EBPβ pathway and its individual target genes to the development of inflammatory disease needs to be investigated both in ABIN1-deficient mice and in human disease.
Materials and Methods
Mice and Cell Culture.
The gene (Tnip1)-trapped ES cell clone E059E05 was obtained from the German Gene Trap Consortium. Mice carrying the gene-trap mutation were generated by injection of ES cells into C57BL/6J blastocysts and crossing of germline chimeras with C57BL/6J mice. Tnfrsf1a−/− mice have been described (47) and were obtained from the Jackson Laboratory. For fetal liver transplantation, 2 × 106 fetal liver cells isolated from Tnip1+/+ or Tnip1−/− embryos on embryonic day 13.5 were injected into lethally irradiated (950 cGy) C57BL/6 mice. All mouse studies were carried out in accordance with protocols approved by the Institutional Animal Care and Use Committee at St. Jude Children's Research Hospital. BMM and BMDC were generated by cultivating bone marrow cells in Petri dishes for 6 d in the presence of 30% L-cell conditioned medium or medium containing GM-CSF, respectively. Stable RAW264.7 transfectants were generated and cultured as described (48).
Reagents and Plasmids.
Antibodies are described in SI Materials and Methods. ELISA kits were from eBiosciences (TNF-α), R&D Systems (G-CSF), Bethyl Laboratories (immunoglobulins), Alpha Diagnostic International (anti-nuclear antibodies and dsDNA). Griess assays to determine nitric oxide concentration were done as described (49). cDNA of mouse Tnip1 was amplified by RT-PCR using cDNA from BMM as template and cloned in-frame to an N-terminal triple-HA tag into a lentiviral, ubiquitin promoter-driven vector. CpG-DNA refers to the phosphothioate backbone containing oligonucleotide 1668 (1 μM, TCCATGACGTTCCTGATGCT) (TIB Molbiol). Other agonists used were LPS (10 ng/mL, Escherichia coli 0127:B8) (Sigma-Aldrich), coumermycin (0.1 μM) (Sigma-Aldrich), R848 (300 nM) (GLSynthesis), and tripalmitoyl cysteinyl lipopeptide (Pam3Cys; 100 ng/mL) (EMC Microcollections).
Protein Purification and Sample Preparation for MS Analysis.
Affinity purification of Flag-/TAP-tagged MyD88-GyrB has been described (15). In a variation of the original protocol, immobilized M2-Flag antibodies and a triple-Flag–tagged peptide (Sigma-Aldrich) were used for protein capture and elution, respectively. Following purification, samples were concentrated by trichloroacetic acid precipitation, separated on a 4–12% (wt/vol) Bis-Tris gel (Bio-Rad), and stained with SYPRO-Ruby (Invitrogen). Protein bands were subjected to in-gel trypsin digestion and analyzed by liquid chromatography-tandem MS using electrospray ionization coupled to a linear ion trap mass spectrometer (LTQ-XL; Thermo Scientific) as described in detail in SI Materials and Methods.
Histology, Complete Blood Cell Count, and Proteinuria Analysis.
Tnip1−/− mice and littermate controls (5- to 8-wk-old) were perfused with 10% (vol/vol) phosphate-buffered formalin and embedded in paraffin. Tissues were sectioned at 5 μm and stained with H&E. Complete blood cell counts were measured using the Forcyte Hematology System (Oxford Science). Protein concentration in urine was determined using Multistix 10 SG sticks (Siemens) and scored as 0 (negative or trace), 1 (30 mg/dL), or 2 (100 mg/dL).
Flow Cytometry Analysis.
Spleens were removed, and single-cell suspensions were prepared by straining through a 40-μm cell strainer. Cells were blocked with antibodies against CD16/CD32 (eBioscience) followed by staining for cell-surface markers. For intracellular staining, splenocytes that first were stained for the cell-surface markers were fixed with 2% (vol/vol) formaldehyde in PBS, followed by incubation with FITC-labeled antibodies against myeloperoxidase (or isotype control) in PBS containing 0.5% saponin. Flow cytometry analysis was done using a FACSCalibur instrument (Becton Dickinson).
Analysis of Gene Expression.
Total cellular RNA was prepared using TRIzol (Invitrogen) and analyzed by real-time qPCR or Affymetrix microarrays as described in SI Materials and Methods.
Electrophoretic Mobility Shift Assay.
Cells were lysed for 20 min in buffer A [20 mM Hepes/KCl (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.5% Nonidet P-40, Roche protease inhibitor mixture] on ice. Nuclei were pelleted, and the supernatant was saved as a cytoplasmic extract. Nuclear proteins were extracted in buffer A plus 400 mM NaCl for 20 min at 4 °C. For electrophoretic mobility shift assay (EMSA), 10 μg of nuclear protein was incubated for 30 min at room temperature with a 32P-labeled dsDNA probe in binding buffer [10 mM Tris/HCl (pH 7.6), 50 mM KCl, 1 mM EDTA (pH 8.0), 5% (vol/vol) glycerol, 1 μg/mL poly(dI:dc), 1 mM DTT]. Samples were separated on 5% (wt/vol) native polyacrylamide gels, and gels were dried, exposed to phosphor screens, and visualized by a Typhoon 9200 imaging system (GE Healthcare). Sequences of oligonucleotides used were NF-κB: AGTTGAGGGGACTTTCCCAGGC; AP-1: AGCTTACTCAGTACTAGTACGAATT; C/EBPβ: TGCAGATTGCGCAATCTGCA.
ChIP.
ChIP assays were performed according to standard protocols and are described in detail in SI Materials and Methods.
Statistical Analyses.
Data are expressed as mean ± SD. Statistically significant differences were assessed by two-tailed student's t test for two groups. Survival curves of ABIN-1–deficient mice at different Tnfrsf1a backgrounds were compared by log-rank test. P values less than 0.01 were considered significant in both tests.
Supplementary Material
Acknowledgments
We thank the staff of the Hartwell Center for Bioinformatics and Biotechnology, the animal resource center, and the Veterinary Pathology Core Laboratory and the Transgenic/Gene Knockout Facility at the St. Jude Children's Research Hospital for their excellent technical assistance. We also thank Paul Brindle and Lawryn Kasper for support related to ChIP assays, Peter Murray for technical advice related to ES cell culture, and Suraj Mukatira for the Clover analysis. This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Grant AI083443 (to H.H.), National Institutes of Health/National Cancer Institute Grant P30 CA2175, and the American Lebanese Syrian Associated Charities.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 17869.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106232108/-/DCSupplemental.
References
- 1.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 2.Flesher DL, Sun X, Behrens TW, Graham RR, Criswell LA. Recent advances in the genetics of systemic lupus erythematosus. Expert Rev Clin Immunol. 2010;6:461–479. doi: 10.1586/eci.10.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberson ED, Bowcock AM. Psoriasis genetics: Breaking the barrier. Trends Genet. 2010;26:415–423. doi: 10.1016/j.tig.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leadbetter EA, et al. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 5.Lande R, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 6.Gateva V, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han JW, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 8.Kawasaki A, et al. Association of TNFAIP3 interacting protein 1, TNIP1 with systemic lupus erythematosus in a Japanese population: A case-control association study. Arthritis Res Ther. 2010;12:R174–180. doi: 10.1186/ar3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nair RP, et al. Collaborative Association Study of Psoriasis. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heyninck K, et al. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J Cell Biol. 1999;145:1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee EG, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science Y. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boone DL, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 13.Häcker H, et al. Immune cell activation by bacterial CpG-DNA through myeloid differentiation marker 88 and tumor necrosis factor receptor-associated factor (TRAF)6. J Exp Med. 2000;192:595–600. doi: 10.1084/jem.192.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 15.Häcker H, et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 17.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 18.Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 20.Oshima S, et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–909. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 22.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 23.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science \ 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 24.Bennett L, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hacbarth E, Kajdacsy-Balla A. Low density neutrophils in patients with systemic lupus erythematosus, rheumatoid arthritis, and acute rheumatic fever. Arthritis Rheum. 1986;29:1334–1342. doi: 10.1002/art.1780291105. [DOI] [PubMed] [Google Scholar]
- 26.Uematsu S, et al. The C/EBP beta isoform 34-kDa LAP is responsible for NF-IL-6-mediated gene induction in activated macrophages, but is not essential for intracellular bacteria killing. J Immunol. 2007;179:5378–5386. doi: 10.4049/jimmunol.179.8.5378. [DOI] [PubMed] [Google Scholar]
- 27.Chang CJ, Shen BJ, Lee SC. Autoregulated induction of the acute-phase response transcription factor gene, agp/ebp. DNA Cell Biol. 1995;14:529–537. doi: 10.1089/dna.1995.14.529. [DOI] [PubMed] [Google Scholar]
- 28.Frith MC, et al. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 30.Häcker H, et al. CpG-DNA-specific activation of antigen-presenting cells requires stress kinase activity and is preceded by non-specific endocytosis and endosomal maturation. EMBO J. 1998;17:6230–6240. doi: 10.1093/emboj/17.21.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graham RR, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Musone SL, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plenge RM, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson W, et al. Wellcome Trust Case Control Consortium; YEAR Consortium. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39:1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Metcalf D. Hematopoietic cytokines. Blood. 2008;111:485–491. doi: 10.1182/blood-2007-03-079681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lande R, et al. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra19. doi: 10.1126/scitranslmed.3001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Romo GS, et al. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med. 2011;3:73ra20. doi: 10.1126/scitranslmed.3001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 39.Nagy G, et al. Central role of nitric oxide in the pathogenesis of rheumatoid arthritis and systemic lupus erythematosus. Arthritis Res Ther. 2010;12:210–215. doi: 10.1186/ar3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soyfoo MS, Roth J, Vogl T, Pochet R, Decaux G. Phagocyte-specific S100A8/A9 protein levels during disease exacerbations and infections in systemic lupus erythematosus. J Rheumatol. 2009;36:2190–2194. doi: 10.3899/jrheum.081302. [DOI] [PubMed] [Google Scholar]
- 41.Vogl T, et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med. 2007;13:1042–1049. doi: 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- 42.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: Structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nanda SK, et al. Polyubiquitin binding to ABIN1 is required to prevent autoimmunity. J Exp Med. 2011;208:1215–1228. doi: 10.1084/jem.20102177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci USA. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu YC, et al. Differential role for c-Rel and C/EBPbeta/delta in TLR-mediated induction of proinflammatory cytokines. J Immunol. 2009;182:7212–7221. doi: 10.4049/jimmunol.0802971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi K, et al. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 47.Pfeffer K, et al. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell. 1993;73:457–467. doi: 10.1016/0092-8674(93)90134-c. [DOI] [PubMed] [Google Scholar]
- 48.Häcker H, et al. Cell type-specific activation of mitogen-activated protein kinases by CpG-DNA controls interleukin-12 release from antigen-presenting cells. EMBO J. 1999;18:6973–6982. doi: 10.1093/emboj/18.24.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuehr DJ, Nathan CF. Nitric oxide. A macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J Exp Med. 1989;169:1543–1555. doi: 10.1084/jem.169.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]