Abstract
New protein functions can require complex sequence changes, but the minimal path is not well understood. The guanylate kinase enzyme (GKenz), which catalyzes phosphotransfer from ATP to GMP, evolved into the GK domain (GKdom), a protein-binding domain found in membrane associate guanylate kinases that function in mitotic spindle orientation and cell adhesion. Using an induced polarity assay for GKdom function, we show that a single serine to proline mutation is sufficient to switch extant GKenz into a functional GKdom. The mutation blocks catalysis (GKenz function) but allows protein binding and spindle orientation (GKdom function). Furthermore, whereas the GKenz undergoes a large closing motion upon GMP binding, fluorescence quenching and NMR demonstrate that the S → P mutation inhibits GMP-induced GK movements. Disrupting GK closing with a mutation at a different position also leads to GKdom function, suggesting that blocking the GKenz closing motion is sufficient for functional conversion of GKenz to GKdom. Although subtle changes in protein function can require complex sequence paths, our work shows that entirely new functions can arise from single mutations that alter protein dynamics.
Keywords: protein interactions, neofunctionalization, protein engineering, molecular evolution
Proteins perform remarkably diverse functions including catalysis, small molecule binding, and protein recognition. Molecular evolution seeks to understand how the diversity of known protein functions arose, whereas the goal of protein engineering is to create new ones (1). An evolutionary process for creating new protein functions is the divergent evolution of duplicated genes, in which gene duplication is followed by functional evolution [neofunctionalization; (2)]. However, loss of function (pseudogene formation or nonfunctionalization) competes with functional evolution and may be much more likely (3). The degree to which neofunctionalization can “win out” over pseudogene formation depends on the number of mutations required to attain the new function and the possible paths in sequence space to the new function. Thus, neofunctionalization is a competitive process that is dependent on the number of required mutations and how protein function is achieved. Insight into the mechanisms of neofunctionalization should improve our ability to develop previously undescribed protein functions.
We examined neofunctionalization in the context of a dramatic functional change: creation of a mitotic spindle-orienting protein from a nucleotide kinase within the membrane associated guanylate kinase (MAGUK) family of proteins. MAGUKs are proteins that mediate intricate multicellular functions such as cell junction formation and mitotic spindle orientation and have been found only in metazoans and choanoflagellates (4, 5). A defining MAGUK feature is the guanylate kinase (GK) domain (GKdom) that diverged from the guanylate kinase enzyme (GKenz), which is broadly distributed throughout evolution [(4, 6); Fig. 1A]. GKenz catalyzes phosphoryl transfer from ATP to GMP (Fig. 1B). The MAGUK GKdom functions as a protein interaction domain and is involved in more sophisticated cellular processes such as stabilizing cell–cell adhesions and orientation of the mitotic spindle (5, 7, 8). Although GKenz and GKdom have high sequence and structural similarity (9–11), GKenz has enzymatic activity but no known peptide ligands, whereas GKdom has multiple peptide ligands but no known enzymatic activity (5, 12). An understanding of the sequence differences between GKenz and GKdom that support their distinct functions may illuminate the molecular processes that lead to the creation of new protein functions.
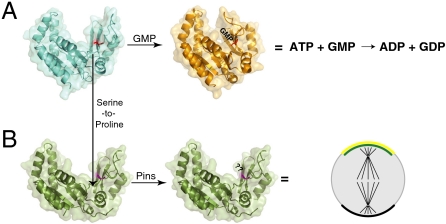
Fig. 1.
An induced polarity spindle orientation rescue assay for measuring GKdom function. (A) GK enzymes are widely distributed, whereas GK domains are restricted to animals and choanoflagellates (4). (B) Comparing GK enzyme and domain activity (18). The reaction catalyzed by the GK enzyme is shown. (Bottom) Schematic of discs-large (Dlg) interaction with Pins is also shown. The linker domain from Pins (PinsLINKER) is phosphorylated by the mitotic kinase Aurora A (7). Phosphorylated PinsLINKER interacts with the Dlg GK domain (TPR: tetratricopeptide repeat; L: linker domain; 1–3: GoLoco domains; PDZ: PSD-95, Dlg, ZO-1 domains; SH3: Src Homology 3 domain; GK: guanylate kinase domain). (C) GK phylogenetic tree. The Drosophila GK enzyme has diverged relatively little from the common ancestor (circle). Calculated from ref. 4. (Dm: Drospohila melanogaster; Sc: Saccharomyces cerevisiae; Ec: Escherichia coli; At: Arabidopsis thaliana; “0.5” indicates average substitutions per site) (D) Induced polarity spindle orientation assay. Adherence of S2 cells occurs through homophilic, intercellular interactions of membrane-bound echinoid molecules. Cell clusters redistribute this cortical Ed:Pins (shown in green using intrinsic GFP fluorescence) specifically to sites of cell–cell contact. The orientation of the mitotic spinde (shown in red with anti-α-tubulin stain) is measured (white hash marks) with respect to the center of the Ed:Pins crescent (white arrowhead). Anti-phosphohistone-H3 (blue) marks the chromosomes as an additional mitotic reference point. (E) Spindle orientation rescue assay. Dlg is inactivated by RNAi and proteins such as the GK domain are expressed to determine if they rescue Dlg activity downstream of Ed:Pins. (F) The GK domain, but not the GK enzyme, rescues spindle orientation. The cumulative percentage of cells with a spindle angle less than the indicated value is shown. (+) indicates both Ed:Pins expression and dlg RNAi. (++) indicates Ed:Pins expression, dlg RNAi and expression of indicated rescue protein. Dm, Drosophila melanogaster. Asterisks denote statistical significance (** represents P < 0.01; Dunnett’s post hoc test) from +dlg RNAi.
One example of GKdom protein interaction is binding of the MAGUK Dlg to phosphorylated partner of inscuteable [Pins; Fig. 1B; (7, 13)]. This interaction is important for mitotic spindle positioning during oriented divisions such as in Drosophila neuroblasts, which polarize during cell division to segregate distinct fate determinants into the daughter cells (14–16). Dlg orients the mitotic spindle in response to polarity cues, and this activity can be reconstituted in cultured S2 cells (7). Dlg is recruited to the cell cortex through interactions between its GKdom and the Pins linker domain (PinsLINKER), which is phosphorylated on S436 by the mitotic kinase Aurora A (7). In addition to spindle orientation, the GKdom functions in a wide range of physiological processes such as the formation of epithelial cell adhesions and scaffolding of the postsynaptic density at neuronal synapses (5). These activities are mediated by a diverse set of binding partners including GKAP (17), MAP1A (18, 19), GukH (20), and calmodulin (21).
Although the GKenz to GKdom transition led to the creation of a protein recognition domain, the mechanism of nucleotide kinase to protein-binding domain conversion has been unclear. The critical sequence differences between GKenz and GKdom that are responsible for their functional differences have not been identified, nor has the mechanism by which these differences disrupt catalytic activity and give rise to protein binding. We have used a combined biochemical and cell biological approach to answer these fundamental questions.
Results
A Single Mutation Imparts Spindle-Orienting Activity to the GK Enzyme.
How could a nucleotide kinase be converted into a phosphoprotein recognition domain that regulates spindle orientation? To answer this question, we asked which of the sequence differences between GKenz and GKdom are sufficient to convert the extant Drosophila GK enzyme into a functional spindle orientation protein. The Drosophila GK enzyme has diverged relatively little from the common ancestor of GK enzymes and domains [Fig. 1C; (4)]. As the protein-binding surface is expected to be significantly larger than the nucleotide-binding interface, we expected that numerous mutations would be required to convert GKenz into GKdom. To test for GKdom activity, we used a spindle orientation assay we recently developed (7). In this assay, Pins crescents are induced in cultured Drosophila S2 cells by fusing Pins to the adhesion protein echinoid (Ed). Clusters of adhered cells restrict Ed and the attached Pins protein to the area of cell–cell contact, and during mitosis the spindle becomes aligned with the center of the Ed–Pins crescent (Fig. 1D). As Ed–Pins requires Dlg for spindle orientation activity (7), we attempted to rescue its Dlg-dependent spindle orientation activity as an assay for GKdom function (Fig. 1E). In this assay, Dlg inactivation by RNAi results in complete loss of spindle orientation by Ed–Pins, but alignment can be nearly fully restored by Dlg GKdom expression (Fig. 1F). Expression of GKenz does not rescue spindle alignment, however, indicating that the extant enzyme does not have GKdom activity (Fig. 1F).
Although we expected that numerous mutations would be required to introduce GKdom activity to GKenz, we first examined a number of single mutations. We strategically chose single mutations guided by multiple sequence alignments of several GKenz and GKdom sequences (Fig. S1). All GKenz proteins with single point mutations that we examined failed to rescue spindle orientation, except for one (Fig. 2A). Surprisingly, when serine 68, a residue near the GMP-binding subdomain (GBD), is changed to the corresponding residue from GKdom, which is a proline (S68P; Drosophila enzyme numbering), the spindle is oriented to a level indistinguishable from Dlg GKdom (Fig. 2A). As the proline at this position is highly conserved in GKdom sequences (Fig. S1), we tested other substitutions to examine the necessity of proline. Mutation of the conserved serine to either alanine or threonine (S68A and S68T) had no effect on GKenz function in spindle orientation, suggesting a unique requirement for proline in GK functional evolution. We confirmed that the mutation induces spindle orientation activity in an extant fungal GKenz (S35P; Saccharomyces enzyme numbering), a lineage that lacks MAGUKs [Fig. 2A; (4, 6)]. Thus, a single mutation is sufficient for the functional evolution of a nucleotide kinase to a spindle-orienting domain within the GK protein fold.
Fig. 2.
A serine to proline mutation converts the guanylate kinase enzyme into a phosphoprotein recognition domain that orients the spindle. (A) The GKenz serine to proline mutation rescues spindle-orienting ability. The cumulative percentage of cells with a spindle angle less than the indicated value is shown. Symbols are as in Fig. 1D. The S → P mutation (Drosophila: S68P; yeast: S35P) causes gain of spindle-orienting activity. Other tested single mutations do not impart spindle-orienting function to GKenz. Two letter codes indicate species (Dm, Drosophila melanogaster; Sc, Saccharomyces cerevisiae). (B) The Sc GKenz S35P binds phosphorylated PinsLINKER. A GST fusion with PinsLINKER can pull down Sc GKenz S35P but not wild-type GKenz. An anti-FLAG immunoblot was used to detect bound GKenz. (C) The serine to proline mutation causes loss of enzyme activity. The dependence of initial rate of GMP hydrolysis as a function of initial GMP concentration is plotted for both Sc GKenz wild-type and S35P. (D) The serine to proline mutation does not affect ATP binding. The amount of ATP bound to equivalent amounts of the Dlg GKdom, and both Sc GKenz wild-type and S35P, as determined by filter binding with radioactive ATP is shown.
The Serine (S) to Proline (P) Mutation Converts a Nucleotide Kinase into a Phosphoprotein Binding Domain.
We next tested if the GKenz S → P mutant has the expected attributes of a GKdom. Consistent with the gain of spindle-orienting activity, the S → P mutation leads to PinsLINKER binding in an in vitro pull-down assay (Fig. 2B). Interestingly, although the mutation leads to binding of the phosphorylated form, it also binds to the unphosphorylated form and is therefore not as specific as the extant GKdom is thought to be (7), indicating that other sequence differences between GKenz and GKdom must impart specificity. In terms of GKenz, serine is conserved at this position (Fig. S1), and we find that the proline mutation abrogates catalytic activity of the fungal GK [Fig. 2C; (12)]. Although the proline mutant lacks nucleotide kinase activity, it retains the ability to bind ATP unlike the extant GKdom (Fig. 2D). Thus, with respect to the GKenz sequence, nucleotide kinase and spindle orientation functions are mutually exclusive, but a single mutation can reversibly switch between them.
The Serine to Proline Mutation Inhibits GMP-Induced Closing of GK.
The proline mutation could alter GK function by changing its conformation. We tested this possibility by determining the GK S35P structure by X-ray crystallography at 2.4 Å (Fig. 3A; Table S1). The S → P mutant GKenz contains a large cleft between the ATP-binding LID and GBD subdomains (Fig. 3 A and B) like the wild-type protein crystallized without bound ATP or GMP. The two proteins are very similar (rmsd for backbone atoms of 1.2 Å) save for a small twist in the GBD relative to the CORE domain that opens the cleft slightly more than in the wild-type protein. Thus, the GKenz proline mutant maintains an “open” conformation like the apo wild-type enzyme.
Fig. 3.
Structure of the yeast guanylate kinase enzyme serine to proline mutant. (A) Structure of yeast GKenz with the serine to proline mutation that confers spindle-orienting function. LID (blue), CORE (green), and GBD (red) subdomains are shown. The mutated residue is marked by its sequence number (“35”). “Hinge” denotes residues that undergo large dihedral angle changes upon GMP-induced closing (9). The arrow shows the extent of cleft opening between LID and GBD subdomains. (B) Structure of wild-type apo GKenz [PDB ID code 1EX6; (9)]. (C) Structure of GMP-bound GKenz [PDB ID code 1EX7; (9)]. Note the proximity of the LID (blue) and GBD (red) domains compared with B, demonstrating the large GMP-induced conformational change in the enzyme GK fold.
Several additional lines of evidence led us to hypothesize that protein dynamics may play an important role in switching between catalysis and spindle orientation functions. First, only a single mutation is required to switch functions, and this S → P mutation is unlikely to fabricate a protein interaction interface within the GKenz fold. Second, nucleotide binding to GKenz causes a large conformational change that closes the binding cleft to position donor and acceptor phosphates next to one another [Fig. 3 B and C; (9)]. Third, the critical serine residue is part of a “hinge” that mediates this nucleotide-induced closing motion (Fig. 3A), and proline has relatively few allowable backbone conformations (22, 23), which could affect GK closing motions. Fourth, as PinsLINKER is much larger than GMP, closing could introduce significant steric overlap between the GKdom and PinsLINKER. Finally, the closed form of the related adenlyate kinase is significantly populated, even in the absence of nucleotide (24, 25). Based on these observations, we hypothesized that the S → P mutation prevents the GK closing conformational change that is normally required for catalysis but detrimental to protein binding and spindle orientation.
We used fluorescence quenching and NMR to directly test if the S → P mutation prevents GK closing motions. When fluorescein is attached to cysteines introduced in the LID and GBD subdomains of GKenz (see Fig. 3A), addition of GMP caused a dose-dependent decrease in steady-state fluorescence that is specific to GMP (Fig. 4A and Fig. S2). Quenching could result from the direct effect of GMP on fluorescein quantum yield or from the GMP-induced conformational change. To distinguish between these possibilities we measured the effect of GMP binding on GK enzymes with only a single fluorescein and observed no quenching when the dye was attached to either the LID or GBD (Fig. S2), indicating that both dyes must be present for quenching to occur. The requirement for dyes at both LID and GBD positions to observe GMP-dependent quenching is consistent with quenching arising from GK closing motions.
Fig. 4.
Loss of GMP-induced GK closing movements accompanies functional evolution. (A) Fluorescence quenching upon ligand (GMP or Pins) binding to the GK enzyme. The fluorescence at 520 nm (F520) was used to monitor emission of Sc GK with fluorescein attached to cysteines at residues 43 and 138 (in the LID and GBD, respectively; a C96S mutation was also made to prevent labeling at that position; see Materials and Methods). Fluorescence was normalized to the fluorescence at 520 nm in the absence of ligand (F520 at 0 μM GMP). GMP binding induces significant quenching of the wild-type protein, and this effect requires dyes at both LID and GBD positions (Fig. S1), indicating that it is not due to direct effects of GMP binding. In the S35P mutation, quenching by GMP is not observed even at concentrations approximately 10 times the Kd (12) nor by binding of the Pins linker domain (“Pins”). The binding curve shown represents a single-site binding isotherm with a Kd of 17 μM. (B) Effect of GMP binding on NMR 1H-15N HSQC chemical shifts of the wild-type and S35P yeast GK enzymes. A portion of the spectra in the absence (gray) and presence of increasing GMP (orange) is shown. Residue assignments are shown for the protein without GMP. The full HSQC spectra are shown in Fig. S3. (C) GMP-induced 1H-15N HSQC chemical shift changes for wild-type and S35P GK enzyme mapped onto the apo structure. Saturating concentrations of GMP to the WT GK enzyme (9 mM GMP; see Fig. S3E) causes significant chemical shifts (> 2.0 ppm) to 56 residues, including those that directly contact GMP and those that are influenced by GK closing motions. In contrast, only residues that directly contact GMP have chemical shifts that are perturbed by saturating GMP in the S35P GK enzyme (42.5 mM GMP; see Fig. S3E), indicating that it is binding but not inducing GK closing.
Conformational changes can also be observed by NMR, which has the advantage of many position-dependent signals. We analyzed GMP binding to GKenz by NMR using 1H-15N heteronuclear single quantum coherence (HSQC) experiments. Here, each hydrogen that is covalently bound to nitrogen gives rise to a signal that is dependent on the environment of both atoms. Addition of saturating concentrations of GMP to wild-type GKenz leads to shifts in a large number of signals, including those from residues that directly contact GMP and others that are influenced by GMP-induced GK closing (Fig. 4 B and C and Fig. S3). This effect is specific to GMP, as it does not occur with cGMP (Fig. S3). We conclude that both fluorescence quenching and NMR can accurately detect the GMP-induced GK conformational change.
We analyzed the effect of the S → P mutation on GMP-induced closing of GKenz. In the fluorescence quenching assay, we found that a GKenz containing this mutation is not quenched by GMP, consistent with a loss of the GK conformational change (Fig. 4A and Fig. S2). Furthermore, the Pins linker domain has no effect on the fluorescence signal suggesting that it does not induce GK closing (Fig. 4A). We confirmed the effect of the mutation in the NMR assay. Whereas GMP induces shifts in a large number of residues in wild-type GKenz (due to direct binding and GK closing), GMP only caused a much smaller subset of residues to shift in S → P GKenz (Fig. 4 B and C and Fig. S3). This subset of residues whose chemical shifts are perturbed by GMP correspond to those that directly contact GMP in the GBD. The lack of movement of broadly distributed signals from residues that did shift in wild-type GKenz indicate that GMP-induced closing no longer occurs in S → P GKenz, whereas the shift of residues that directly contact GMP demonstrates that the mutation does not prevent GMP binding. We conclude that the S → P mutation inhibits GMP-induced GK closing motions.
Inhibition of GK Closing Is Sufficient for Functional Conversion.
These results support an alternate view for functional evolution that places an emphasis on protein dynamics. Rather than gaining function through a series of mutations that each has a small contribution to overall activity, functional evolution may occur by a single mutation that alters protein conformational movements. We sought to determine if inhibition of the GK conformational change is sufficient for functional evolution by introducing a mutation at a hinge site distinct from the naturally occurring one (yeast: S33P; Drosophila: S66P; Fig. 3A). This mutation leads to loss of GMP-induced closing as assessed by lack of GMP-induced fluorescence quenching (Fig. 5A and Fig. S2), as is seen with other hinge mutation. Furthermore, mutation at this site also results in a gain of spindle-orienting activity (Fig. 5B), indicating that there are multiple sequence paths to inhibiting the GMP-induced conformational change and functional evolution.
Fig. 5.
Multiple paths to GK functional evolution. (A) Fluorescence quenching upon GMP binding to the GK enzyme. Data are displayed as in Fig. 4A. See Fig. S2 for full emission spectra. In the S33P mutation, quenching by GMP is not observed even at high concentrations. (B) Mutation of Dm S66 to proline causes gain of spindle-orienting function. Cumulative percentage graph and symbols are shown as in Fig. 1F. Asterisks denote statistical significance (** represents P < 0.01, * represents P < 0.05; Dunnett’s post hoc test) from +dlg RNAi.
Discussion
We have investigated the sequence differences that support nucleotide kinase versus protein recognition functions in the GK fold and found that the sequence path between the two can be remarkably small. In other examples of neofunctionalization that alter protein-binding activity, numerous sequence changes can be required. In the case of steroid hormone receptors, mutations that alter the specificity for particular hormones can be remarkably complex, requiring mutations in the binding site and other “permissive” mutations of residues distant from the ligand binding pocket (1, 26). However, other neofunctionalization events have been identified that also can occur on a short sequence path. For example, a single amino acid substitution in a carboxylesterase causes insecticide resistance in blowflies because it converts the enzyme to an organophosphorus hydrolase (27). It is likely that a diversity of sequence paths will be found as more examples of neofunctionalization are studied, with the constraint that longer sequence paths may be less likely to occur.
Our findings suggest a role for protein dynamics in neofunctionalization. We have shown that conversion of a nucleotide kinase into a protein-binding domain alters the protein’s conformational response to ligand binding such that catalytic activity is lost but protein binding and spindle orientation activities are gained (28). Dramatic changes in function caused by altering protein dynamics could occur relatively rapidly because there are potentially many sites at which mutations could modulate conformational movements.
Another instance of evolution of a protein interaction domain from an enzyme is the cholinesterase-like (ChEL) domain found in the adhesion protein neuroligin-2 (29) and the precursor of thyroid hormone synthesis, thyroglobulin (30). This domain is derived from the enzyme cholinesterase, which catalyzes the hydrolysis of acetylcholine, but has gained protein-binding function to mediate oligomerization. The use of enzymes as “raw material” for neofunctionalization, as has happened with the GK and ChEL domain, raises the possibility that currently labeled pseudogenes may instead be neofunctionalized active proteins. The degree to which genes that encode enzymes have been harnessed for previously undescribed protein activities will require further study. Interestingly, kinases are one of the largest families of proteins in the mammalian “psuedogenome” (3), although apparently dead enzymes have been found to be active (31). Modulating conformational changes within this pool of proteins, as has happened in the MAGUK GK domain, may represent a particularly efficient method of functional evolution.
Materials and Methods
Plasmid Construction and Protein Purification.
Pins constructs were designed from the Drosophila melanogaster sequence (GenBank accession no. AAF64499.1). Dlg constructs were designed from the Drosophila melanogaster isoform-G sequence (GenBank accession no. AAS65312.1). Guanylate kinase enzyme constructs were designed from either the Drosophila melanogaster isoform (NCBI Reference Sequence: NP_648408.1) or the Saccharomyces cerevisiae isoform (GenBank accession no. AAA34657.1).
Plasmids were transformed into BL21(DE3) competent Escherichia coli and grown as an overnight starter culture under ampicillin (100 μg/mL) selection. Six liters of LB were inoculated with starter culture, allowed to grow to and OD600 = 0.7, and induced with the addition of 200 μM IPTG overnight at 20 °C. Protein purification was carried out using sequential NiNTA affinity, anion exchange, and size-exclusion chromatographies. All proteins eluted as predicted monomers from the size-exclusion column, and their purities were assessed to be > 95% by Coomassie staining of an SDS-PAGE gel. Proteins were concentrated using Vivaspin concentrators (Sigma-Aldrich), flash frozen in liquid nitrogen, and stored at -80 °C in buffer (20 mM Tris, pH 7.5, 50 mM NaCl, 5% glycerol, 2 mM DTT).
Cell Culture and Echinoid Cell-Adhesion Assays.
Maintenance of S2 cells, construction of expression plasmids (including echinoid fusion sequences), and cell-adhesion assays have recently been detailed elsewhere (7). Briefly, S2 cells were transfected using the Effectene reagent (Qiagen) with 0.4–1 μg total DNA for 24–48 h. For Dlg RNAi treatment, transfected cells were incubated for 1 h in serum-free media containing approximately 1 μg RNAi followed by 72 h in normal growth media. Protein expression was then induced by the addition of 500 μM CuSO4 for 24 h. Cell-adhesion clustering was induced by constant rotation at approximately 175 rpm for 1–3 h.
For immunostaining, clustered cells were fixed in 4% paraformaldehyde for 20 min, washed, and incubated with primary antibodies overnight at 4 °C in buffer (0.01% saponin plus 0.1% albumin diluted in phosphate-buffered saline). Slides were subsequently washed and fluorescently linked secondary antibodies were added for 2 h at room temperature. Finally, slides were again washed and mounted using Vectashield Hardset medium (Vector Laboratories). All images were collected using a Leica SP2 confocal microscope with a 60 × 1.4 NA lens. The spindle angles for at least 20 cells were measured for each condition (each point in the cumulative percentage plots represents a unique measurement).
Antibodies used were as follows: rat anti-α-tubulin (Abcam; 1∶500), rabbit antiphosphohistone-3 (Upstate; 1∶1,000), and mouse anti-FLAG (Sigma; 1∶500).
Fluorescence Quenching.
To fluorescently label GKenz constructs, the naturally occurring cysteine at residue 96 was changed to a serine, and cysteines were introduced at the solvent-exposed GBD and LID residues, A43C and T138C, respectively (versions of the protein with single mutations were also constructed and analyzed to confirm that the observed effects required the presence of dyes at both positions; Fig. S1). To label the resulting single and double cysteine-containing proteins, 10 mg protein was diluted to 100 μM in labeling buffer (10 mM Tris, pH 7.5 and 50 mM NaCl). A 5-fold molar excess of fluorescein maleimide was incubated with protein for 2 h at room temperature. Reactions were quenched by addition of 2-mercaptoethanol, and unreacted dye was removed using size-exclusion chromatography followed by extensive dialysis. Labeled proteins were concentrated and stored in labeling buffer.
For fluorescence-quenching measurements, proteins were diluted to 1 μM in labeling buffer and emission scans (500–600 nm) were taken from a 490 excitation wavelength using an ISS PC1 fluorometer with sample maintained at 20 °C using a circulating water bath. GMP or other indicated ligands were subsequently titrated in sequentially under constant stirring, and individual emission scans were taken.
GST Pulldowns.
GST-tagged PinsLINKER (residues 399–466) was absorbed to glutathione agarose for 30 min at 4 °C and subsequently washed 3 times with PBS. GST-Pins was then incubated in the presence or absence of Aurora-A (Millipore) in kinase buffer (20 mM Tris, pH 8, 50 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, and 100 μM ATP) for 30 min at room temperature. Following phosphorylation of S436, 50 μg of GKdom or GKenz proteins were added for 1 h at 4 °C. Reactions were washed (20 mM Tris, pH 7.5, 100 mM NaCl, 5 mM MgCl2, and 0.5% NP-40), and samples were analyzed using anti-FLAG Western blot.
NMR Spectroscopy.
NMR experiments were carried out on a Bruker Avance II 600-MHz spectrometer equipped with a triple resonance z-axis gradient CryoProbe®. All NMR samples contained 0.9 mM (WT yeast GKenz) or 0.85 mM (S35P) 15N- or 15N/13C-labeled protein and were prepared in 90% H2O/10% D2O containing 20 mM NaHPO4, 100 mM NaCl, and 0.05% sodium azide (pH = 7.4). All NMR experiments were at 25 °C. Resonance assignments of GKenz WT and GKenz S35P were obtained from the following experiments: CCONH, HNCA, HNCACB, HNCO, HNCOCA, and HNCACO. NMR data were processed with the NMRPipe software package (32). Chemical shifts were referenced to the methyl signal of internal 2,2-dimethylsilapentane-5-sulfonic acid directly for 1H and indirectly for 13C and 15N. The XEASY program was used for resonance assignments (33). GMP titrations utilized > 99% pure GMP dissolved in 20 mM NaHPO4 100 mM NaCl buffer at pH = 7.4. GKenz WT was measured by 15N-HSQC at 0, 0.5, 1.0, 2.0, 5.0, and 10.0 M equivalent additions of GMP. GKenz S35P was measured by 15N-HSQC at 0, 0.5, 1.0, 2.0, 5.0, 10.0, 30.0, and 50.0 M equivalent additions of GMP.
Guanylate Kinase Activity Assay.
To assess the catalytic function of wild-type and S35P guanylate kinases, we used a coupled-enzyme assay as described previously (34). Briefly, phosphate release is detected by oxidation of NADH by lactate dehydrogenase following the decrease in absorbance at 340 nm. Guaunylate kinase enzyme was at 250 nM in assay buffer (100 mM Tris, pH 7.5, 100 mM KCl, 10 mM MgCl2, 1.5 mM sodium phosphoenolpyruvate, 300 mM NADH, 4 mM ATP, and pyruvate kinase and lactate dehydrogenase). Initial GMP concentrations ranged from 500 nM to 10 mM. The reaction was initiated by addition of GMP and brief mixing, followed by absorption measurement using a Tecan Safire fluorometer. Reactions were carried out at 30 °C and measured at 15-s intervals for 30 cycles. Data were analyzed and plotted using GraphPad Prism. Reaction rates are plotted as initial velocity (change in absorption at 340 nm/s) as a function of GMP concentration.
ATP-Binding Assay.
We tested for ATP-binding activity of wild-type and S35P guanylate kinase enzymes and the GK domain from Dlg using a filter binding assay. GK proteins at a concentration of 1 μM in assay buffer (20 mM Tris, pH 7.5, 100 mM NaCl) were allowed to equilibrate with 10 μM ATP at 30 °C for 10 min. The ATP contained a small amount of [32γP]-ATP at approximately 2 × 105 cpm/μL. The reaction was spotted on individual filter discs and washed 3 times with ice cold wash buffer (20 mM Tris, pH 7.5 and 100 mM NaCl) for 5 min each. Bound [32γP]-ATP was quantified using liquid scintillation counting.
Crystallography and Structure Determination.
The 6x-His-tagged S35P mutant enzyme was purified to homogeneity using sequential Ni2+-NTA affinity, anion exchange, and size-exclusion chromatography (35, 36). The final preparation was then concentrated to 43 mg/mL in storage buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 1 mM DTT). For crystallization, S35P protein was mixed 1∶1 with crystallization buffer (1.6M LiSO4, 0.1M Hepes, pH 7.5) using the sitting-drop vapor diffusion method at 15 °C. Crystals appeared within 3–5 d and grew to maximum dimensions within 2 wk. Crystals were transferred and cryoprotected using a quick (approximately 30 s) soak in crystallization buffer supplemented with 20% glycerol. Crystals were then flash cooled and stored in liquid nitrogen prior to data collection.
Remote-access data collection was performed using beamline 8.2.1 at the Berkeley National Laboratory Advanced Light Source synchrotron. A complete dataset (0–150° at 1°/frame) was collected under constant cryostream using 1 min /frame exposures. Data were indexed and scaled using HKL2000 software (37). Crystals belong to the P43212 space group with two molecules in the asymmetric unit (see Table S1).
The S35P structure was determined using the Phaser molecular replacement program within the CCP4 software suite (38). The structure of wild-type guanylate kinase from Saccharomyces cerevisiae (PDB ID code 1EX6; ref. 9) was used as a search model. Automated search functions revealed a clear solution with a high Phaser confidence score of 24.5 and initial Rcryst/Rfree factors of 34/40.
Iterative rounds of model adjustment and restrained refinement were performed using Coot (39) and REFMAC. The S35P mutant was introduced in clearly defined electron density. As the S35P mutant enzyme assumes a slightly more open conformation (see Fig. 3), much of the GMP-binding subdomain (residues 54–80) required remodeling of the initial solution structure. Because of insufficient electron density, a short loop (residues 132–145) in molecule B is missing in the final structural model. In later refinement stages, four Translation/Libration/Screw groups were used for each of the two GKs in the asymmetric unit.
Supplementary Material
Acknowledgments.
The authors thank J. Thornton and B. Nolen for helpful comments on the manuscript, D. Jensen and C. Platt for NMR sample preparation and screening, and F. Peterson for help with NMR data collection and analysis. C.A.J. is a Damon Runyon Cancer Research Foundation postdoctoral fellow. C.Q.D. is an Investigator of the Howard Hughes Medical Institute. K.E.P. acknowledges financial support from the American Heart Association (Grant-in-aid) and the National Institutes of Health (Grants GM068032 and GM087457). The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 17863.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3SQK).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104365108/-/DCSupplemental.
References
- 1.Harms MJ, Thornton JW. Analyzing protein structure and function using ancestral gene reconstruction. Curr Opin Struct Biol. 2010;20:360–366. doi: 10.1016/j.sbi.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prince VE, Pickett FB. Splitting pairs: The diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 3.Torrents D, Suyama M, Zdobnov E, Bork P. A genome-wide survey of human pseudogenes. Genome Res. 2003;13:2559–2567. doi: 10.1101/gr.1455503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Mendoza A, Suga H, Ruiz-Trillo I. Evolution of the MAGUK protein gene family in premetazoan lineages. BMC Evol Biol. 2010;10:93. doi: 10.1186/1471-2148-10-93. 10.1186/1471-2148-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 6.te Velthuis AJ, Admiraal JF, Bagowski CP. Molecular evolution of the MAGUK family in metazoan genomes. BMC Evol Biol. 2007;7:129. doi: 10.1186/1471-2148-7-129. 10.1186/1471-2148-7-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston CA, Hirono K, Prehoda KE, Doe CQ. Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell. 2009;138:1150–1163. doi: 10.1016/j.cell.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegrist SE, Doe CQ. Microtubule-induced cortical cell polarity. Genes Dev. 2007;21:483–496. doi: 10.1101/gad.1511207. [DOI] [PubMed] [Google Scholar]
- 9.Blaszczyk J, Li Y, Yan H, Ji X. Crystal structure of unligated guanylate kinase from yeast reveals GMP-induced conformational changes. J Mol Biol. 2001;307:247–257. doi: 10.1006/jmbi.2000.4427. [DOI] [PubMed] [Google Scholar]
- 10.McGee AW, et al. Structure of the SH3-guanylate kinase module from PSD-95 suggests a mechanism for regulated assembly of MAGUK scaffolding proteins. Mol Cell. 2001;8:1291–1301. doi: 10.1016/s1097-2765(01)00411-7. [DOI] [PubMed] [Google Scholar]
- 11.Tavares GA, Panepucci EH, Brunger AT. Structural characterization of the intramolecular interaction between the SH3 and guanylate kinase domains of PSD-95. Mol Cell. 2001;8:1313–1325. doi: 10.1016/s1097-2765(01)00416-6. [DOI] [PubMed] [Google Scholar]
- 12.Olsen O, Bredt DS. Functional analysis of the nucleotide binding domain of membrane-associated guanylate kinases. J Biol Chem. 2002;278:6873–6878. doi: 10.1074/jbc.M210165200. [DOI] [PubMed] [Google Scholar]
- 13.Sans N, et al. mPins modulates PSD-95 and SAP102 trafficking and influences NMDA receptor surface expression. Nat Cell Biol. 2005;7:1179–1190. doi: 10.1038/ncb1325. [DOI] [PubMed] [Google Scholar]
- 14.Neumuller RA, Knoblich JA. Dividing cellular asymmetry: Asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23:2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prehoda KE. Polarization of Drosophila neuroblasts during asymmetric division. Cold Spring Harb Perspect Biol. 2009;1:a001388. doi: 10.1101/cshperspect.a001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 17.Naisbitt S, et al. Characterization of guanylate kinase-associated protein, a postsynaptic density protein at excitatory synapses that interacts directly with postsynaptic density-95/synapse-associated protein 90. J Neurosci. 1997;17:5687–5696. doi: 10.1523/JNEUROSCI.17-15-05687.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenman JE, et al. Localization of postsynaptic density-93 to dendritic microtubules and interaction with microtubule-associated protein 1A. J Neurosci. 1998;18:8805–8813. doi: 10.1523/JNEUROSCI.18-21-08805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reese ML, Dakoji S, Bredt DS, Dotsch V. The guanylate kinase domain of the MAGUK PSD-95 binds dynamically to a conserved motif in MAP1a. Nat Struct Mol Biol. 2007;14:155–163. doi: 10.1038/nsmb1195. [DOI] [PubMed] [Google Scholar]
- 20.Mathew D, et al. Recruitment of scribble to the synaptic scaffolding complex requires GUK-holder, a novel DLG binding protein. Curr Biol. 2002;12:531–539. doi: 10.1016/s0960-9822(02)00758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuko N, et al. Interaction of NE-dlg/SAP102, a neuronal and endocrine tissue-specific membrane-associated guanylate kinase protein, with calmodulin and PSD-95/SAP90. A possible regulatory role in molecular clustering at synaptic sites. J Biol Chem. 1999;274:5782–5790. doi: 10.1074/jbc.274.9.5782. [DOI] [PubMed] [Google Scholar]
- 22.Ramachandran GN, Sasisekharan V. Conformation of polypeptides and proteins. Adv Protein Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 23.Ting D, et al. Neighbor-dependent Ramachandran probability distributions of amino acids developed from a hierarchical Dirichlet process model. PLoS Comput Biol. 2010;6:e1000763. doi: 10.1371/journal.pcbi.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanson JA, et al. Illuminating the mechanistic roles of enzyme conformational dynamics. Proc Natl Acad Sci USA. 2007;104:18055–18060. doi: 10.1073/pnas.0708600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henzler-Wildman KA, et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 26.Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461:515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newcomb RD, et al. A single amino acid substitution converts a carboxylesterase to an organophosphorus hydrolase and confers insecticide resistance on a blowfly. Proc Natl Acad Sci USA. 1997;94:7464–7468. doi: 10.1073/pnas.94.14.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 29.Ichtchenko K, et al. Neuroligin 1: A splice site-specific ligand for beta-neurexins. Cell. 1995;81:435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- 30.Swillens S, Ludgate M, Mercken L, Dumont JE, Vassart G. Analysis of sequence and structure homologies between thyroglobulin and acetylcholinesterase: Possible functional and clinical significance. Biochem Biophys Res Commun. 1986;137:142–148. doi: 10.1016/0006-291x(86)91187-3. [DOI] [PubMed] [Google Scholar]
- 31.Mukherjee K, et al. CASK functions as a Mg2+-independent neurexin kinase. Cell. 2008;133:328–339. doi: 10.1016/j.cell.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Bartels C, Xia TH, Billeter M, Guntert P, Wuthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 34.Agarwal KC, Miech RP, Parks RE., Jr Guanylate kinases from human erythrocytes, hog brain, and rat liver. Methods Enzymol. 1978;51:483–490. doi: 10.1016/s0076-6879(78)51066-5. [DOI] [PubMed] [Google Scholar]
- 35.Qian Y, Prehoda KE. Interdomain interactions in the tumor suppressor discs large regulate binding to the synaptic protein GukHolder. J Biol Chem. 2006;281:35757–35763. doi: 10.1074/jbc.M607057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ricketson D, Johnston CA, Prehoda KE. Multiple tail domain interactions stabilize nonmuscle myosin II bipolar filaments. Proc Natl Acad Sci USA. 2010;107:20964–20969. doi: 10.1073/pnas.1007025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otwinowsk Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]