Abstract
Rapid recognition of DNA target sites involves facilitated diffusion through which alternative sites are searched on genomic DNA. A key mechanism facilitating the localization of the target by a DNA-binding protein (DBP) is one-dimensional diffusion (sliding) in which electrostatic forces attract the protein to the DNA. As the protein reaches its target DNA site, it switches from purely electrostatic binding to a specific set of interactions with the DNA bases that also involves hydrogen bonding and van der Waals forces. High overlap between the DBP patches used for nonspecific and specific interactions with DNA may enable an immediate transition between the two binding modes following target site localization. By contrast, an imperfect overlap may result in greater frustration between the two potentially competing binding modes and consequently slower switching between them. A structural analysis of 125 DBPs indicates frustration between the two binding modes that results in a large difference between the orientations of the protein to the DNA when it slides compared to when it specifically interacts with DNA. Coarse-grained molecular dynamics simulations of in silico designed peptides comprising the full range of frustrations between the two interfaces show slower transition from nonspecific to specific DNA binding as the overlap between the patches involved in the two binding modes decreases. The complex search kinetics may regulate the search by eliminating trapping of the protein in semispecific sites while sliding.
Keywords: nonspecific binding, energy landscape, protein–DNA interactions
Many DNA-binding proteins (DBPs) recognize short DNA target sequences rapidly and specifically in an enormous genomic background with many sites of similar sequence to the specific target site. Various studies have suggested that proteins locate DNA target sites through a combination of multidimensional search mechanisms that allow a relatively few protein copies to activate or repress genes within reasonable response times (1, 2). DBPs bind with moderate affinity to nonspecific DNA sequences and randomly diffuse along the linear contour of the DNA (“sliding”). Sliding is interrupted by short-range dissociations to neighboring DNA segments (“hopping”) as well as dissociations to the bulk and transfers to distant DNA sequences that are spatially close (“intersegment transfer”).
The search kinetics for the target site may not be governed solely by the scanning efficiency of nonspecific sites along the genomic DNA: Its complexity can be increased and its pace slowed by other processes. For example, following the arrival of the protein at the target site, additional events may be required for specific DNA recognition to occur (3). This transition from the encounter complex that is stabilized by nonspecific interactions to the specific protein–DNA complex may involve conformational changes to one or both biomolecules. Most likely the transition is also coupled with expulsion of interfacial water molecules (4).
Probing the transition from nonspecific to specific protein–DNA binding is difficult because nonspecific interactions are transient in nature. Several structures of proteins bound to semispecific DNA sequences (i.e., with partial sequence similarity to the specific DNA target) have highlighted that nonspecific interactions are mostly dominated by electrostatic interactions between the positively charged protein side chains and the negatively charged DNA backbone (5–7). This notion is supported by a greater dependence of the nonspecific interactions on salt concentration in comparison with specific protein–DNA complexes (8–10). Generally, DBPs have substantial regions of positive electrostatic patches at their DNA-binding interface that complement the negatively charged DNA (11, 12). Negatively charged amino acids have a lower propensity in protein–DNA interfaces (13), though they might be observed and contribute to specificity by properly orienting the protein relative to the DNA (14) or interact with the DNA through a cation such as Mg2+, as observed in EcoRV endonuclease (15). Specificity in protein–DNA interactions is obtained mostly by a formation of hydrogen bonds between donors and acceptors from protein side chains and DNA bases (10, 16), stabilized by van der Waals and hydrophobic forces, electrostatics, and water-mediated interactions between polar groups (4, 17).
Recent studies indicated a high degree of similarity between specific and nonspecific protein–DNA binding. NMR studies of the HoxD9 homeodomain sliding dynamics and kinetics showed that the protein utilizes similar interfaces for both nonspecific and specific DNA binding and that the positive patch on the protein surface maintains a similar orientation with the DNA in the two binding modes (18). In addition, single molecule studies of DBPs diffusion along DNA indicated a rotation-coupled sliding along the DNA helical path, enabling a secondary structural element of the protein binding site to probe the DNA grooves (19). Molecular dynamic simulations also showed that the electrostatic potential of DBPs is sufficient to orient the protein during sliding at the binding mode of the specific interaction (20, 21).
Although the nonspecific and specific interactions of DBPs with DNA are often very similar, they are not identical. Differences between the two binding modes may originate from imperfect overlap between the patch of positive electrostatic potential that interacts with the DNA backbone and the patch that interacts with the base pairs. The need to switch from the nonspecific upon specific recognition reflects a frustration between the two modes because the protein has to exchange some electrostatic interactions with the DNA backbone with hydrogen bonding with the bases. Higher frustration between the two modes may involve varying degrees of conformational changes in the protein or the DNA (22). For example, the contents of lac repressor with DNA are dramatically altered in the transition from nonspecific DNA binding to the complex with its natural operator (7). In particular, base-pair interactions formed in the specific complex at the expense of a partial loss of nonspecific charge–charge interactions induce a tilt in the protein relative to the DNA upon the transition from the nonspecific to the specific complex, as well as distorting the canonical B-DNA form of the DNA (7, 10).
Here we investigated the existence of frustration between nonspecific and specific DNA binding by DBPs and its implications for DNA search mechanisms and kinetics. We asked to what extent the nonspecific binding modes of DBPs during sliding are similar to specific binding and in particular focused on whether the transition between the two introduces further kinetic complexity to the process of DNA target search by proteins.
Results and Discussion
Correspondence Between the Specific and Nonspecific Binding Patches on DBPs.
Under physiological conditions, DBPs have a reasonable affinity to any DNA sequence. This trait allows the protein to alternate between one-dimensional sliding to hopping and dissociations from the DNA. Positively charged patches are common in DBPs and have unique features compared with positively charged patches found in protein–protein interfaces (11, 12). For example, positively charged patches of DBPs are larger, have a higher α-helical content, greater hydrogen-bonding potency, and a higher degree of evolutionary conservation of positively charged residues than similar patches on non-DNA binding proteins (11).
Here, we analyzed a dataset of 125 DBPs for which the crystal structure with specific DNA sequence is available (12) (see SI Text) and explored the dual role the positively charged patch may have as a mediator of specific contact formation at the target DNA site and of an efficient protein translocation process along the DNA. For each DBP in the dataset, a subgroup of residues constitute the patch for specific binding to DNA was defined according to its structure. The positively charged patches of the DBPs were probed using the electrostatic potential of the protein obtained from Poisson–Boltzman calculations (11).
Fig. 1 (Upper) shows residues forming the specific DNA-binding patch together with the electrostatic potential of the protein surface. In the DNA-binding domain of Skn-1, both the specific DNA-binding residues and the positively charged region occupy the recognition helix that interact with the DNA major groove [Fig. 1A, Protein Data Bank (PDB) ID code 1skn]. The specific DNA binding residues of the chromosomal protein 7A are located in a β-sheet region, a less common DNA-binding motif, and are less localized in a region with positive electrostatic potential (Fig. 1B, PDB ID code 1c8c). In the monomeric lambda repressor, the positive potential at the protein surface is relatively nonuniform (Fig. 1C, PDB ID code 1lmb), with the specific DNA binding residues being dispersed among regions with differing electrostatic potentials. Variations in the electrostatic environment of each residue that specifically interacts with the DNA are also illustrated by highlighting neighboring positively (Arg and Lys) and negatively (Glu and Asp) charged residues in the vicinity of each residue of the specific patch (see Middle).
Fig. 1.
Electrostatic potential of the patch for specific DNA binding. Three DBPs with varying degrees of correspondence between the patches used for specific and nonspecific binding are shown for the Skn-1 DNA-binding domain 1sknA (A), chromosomal protein 7A 1c8cA (B), and the monomeric λ-repressor 1lmb3 (C). The residues that participate in specific DNA binding are shown as green spheres. (Upper) The electrostatic potential calculated using adaptive Poisson–Boltzmann solver (APBS) (35) is shown using a scale that ranges from red to blue. (Middle) The positively charged (Arg and Lys) and negatively charged (Asp and Glu) amino acids that surround each of the specific DNA binding residues (with a Cα–Cα distance of < 10 Å) are indicated with blue and red connecting lines, respectively. (Bottom) The values of χi (calculated according to Eq. 1), with green bars indicating the χi values for specific DNA binding residues (the mean of the green bars is denoted by χprot). The percentage overlap between the specific patch and the positively charged electrostatic patch [the largest blue patch (Upper)] is denoted by SI-PP.
The similarity between the specific DNA-binding patch on the DBP and the positively charged nonspecific binding patch, was quantified in two approaches. First, we calculated the overlap between the specific and nonspecific binding patches (denoted as SI-PP overlap), as the percentage of protein residues that specifically bind the DNA and also belong to the DBP largest positive electrostatic patch. Second, we evaluated the surrounding electrostatic environment for each individual residue i, with a measure χi that may range from −1 to 1 (for residues fully surrounded by negatively or positively charged residues) (see Methods, Eq. 1). For each protein, we then calculated χprot as the mean of the χi values for the residues involved in specific DNA binding. Clearly, a protein whose specific and nonspecific DNA patches overlap or even share a mutual common identity (i.e., fully correspond) will have a higher χprot value.
Fig. 1 (Lower) highlights the heterogeneous χi values observed in three different DNA-binding proteins as well as the localization of the specific DNA-binding residues relative to the positive patch region. In Skn-1 DNA-binding domain, with a χprot of 0.42, the specific DNA binding residues entirely overlap the positive patch region (whose residues are indicated by blue markers at the baseline). In the chromosomal protein 7A, only 63% of its specific DNA binding residues overlap with the positive patch and some are partially localized near negatively charged regions, giving rise to a χprot of 0.2. In the monomeric lambda repressor, many specific DNA binding residues are accommodated in the vicinity of negatively charged residues and are excluded from the positive patch region, resulting in a χprot of approximately 0 and less than 50% overlap with the positive patch.
Fig. 2 summarizes the two quantifications of the structural interplay between specific DNA binding residues and their surrounding electrostatic environment for the entire DBP dataset (divided into seven structural/functional groups). For reference purposes, we analyzed an additional dataset of 37 RNA-binding proteins (RBPs), a protein–protein dataset of 16 heterodimers in which the macromolecular interface is dominated by electrostatic interactions, and a dataset consisting of 129 homodimeric protein complexes in which the interfacial electrostatic observables discussed above are expected to be negligible (see SI Text for more details about the protein datasets).
Fig. 2.
Frustration between specific and nonspecific interfaces in DBPs, RBPs, and protein–protein complexes. The dataset of DBPs investigated includes 125 proteins (grouped into 7 categories based on their fold or function) and the dataset of RBPs includes 37 proteins. Protein–protein complexes are represented by 129 homodimeric and 16 heterodimeric proteins. The conflict between the specific and nonspecific interfaces is evaluated by (A) the overlap of the residues of the specific interface with the largest positive patch (SI-PP overlap) and (B) by the values of χprot (see Methods for additional details).
Fig. 2A shows that the positive patch overlap is higher for DBPs than for RBPs and protein–protein complexes. This finding is consistent with the observation that positive patches on DBPs tend to be larger than those on the surfaces of other proteins (11). However, for many DBPs, particularly enzymes, a significant fraction of the specific DNA binding residues is excluded from the positive patch, which results in a reduced overlap.
In Fig. 2B, higher χprot values are observed for DBPs than for RBPs and protein–protein complexes, as protein interfaces for DNA binding are, on average, more enriched with positive charges than are interfaces for RNA and protein binding. However, DBPs exhibit mostly moderate χprot values (ranging from approximately 0.05 to 0.25). We find that in many DBPs the observed χprot is below the maximal χprot available from an optimal rearrangement of the charges along the sequence (Fig. S1). This indicates a partial frustration between specific and nonspecific DNA binding, as many neutral and negatively charged residues surround residues that specifically bind DNA. Although examples exist for negatively charged residues in the interface (14), these surrounding residues may introduce local electrostatic repulsion from the DNA. DBPs with higher χprot values are mostly those recognizing DNA through α-helices (for example, leucine-zipper and zinc-coordinating proteins). By contrast, DBPs with low χprot values, such as enzymes, typically represent more complex DNA-binding sites with mixed positively and negatively charged residues. These observations suggest an implicit conflict within DBPs between their specific and nonspecific DNA-binding modes, which may induce structural differences between the specific binding and sliding conformations as well as raising a kinetic barrier to transition between the two.
Differences Between the Orientations Adopted by Proteins Relative to DNA During Sliding and Specific Binding.
During sliding, the protein is attracted to the DNA mostly by electrostatics and most of the interactions exist in specific DNA binding (hydrogen bonds, van der Waals) are absent (9, 10). We ask whether the different protein–DNA interactions observed for sequence specific binding compared with purely electrostatic binding, as quantified and described above, may dictate a different protein orientation relative to the DNA for nonspecific compared with specific binding.
To address this question, we studied the sliding dynamics of four proteins with varying χprot values. Sliding was studied along a nonspecific 100-bp dsDNA molecule at a low salt concentration. We used a coarse-grained model in which protein–DNA interactions were represented by electrostatic forces only and so mimicked nonspecific interactions (see Methods). The optimized protein configuration in sliding was captured by gradually decreasing the temperature. We then compared the orientation of the protein relative to the DNA during sliding with that seen in the experimental specific-sequence protein–DNA complex. Fig. 3 compares the relative orientations between the DBP and the DNA during specific and nonspecific DNA-binding modes. For the purpose of representation, we aligned each specific protein–DNA complex (shown in blue) with its corresponding sliding complex (protein shown in green, DNA in gray). The conformations of the DNA in the two complexes are quite similar, so discrepancies between the two binding modes make little contribution toward distorting the DNA from the canonical B-DNA form.
Fig. 3.
Comparison of nonspecific protein–DNA binding used during sliding, obtained from coarse-grained simulations (green) to specific binding modes of DBPs obtained from the crystal structure (blue). The DNA used in the simulation is shown in gray. Four proteins that span the whole range of χprot (0.42–0) found in native DBPs are shown: (A) Skn-1 DNA binding (1skn); (B) homeobox protein Hox-A9 (1puf); (C) Smad-MH1 domain (1ozj); and (D) BamHI endonuclease. The similarity between the two binding modes is measured by Δdsp-nsp, which is the average of the shortest distance of each residue of the specific interface to the DNA. For BamHI endonuclease 1bhm, additional crystal structure of the enzyme bound to semispecific DNA sequence (light blue, 1esg) is shown to compare the sliding conformation to both specific and semispecific DNA sequences.
The nonspecific and specific interactions of the Skn-1 DNA-binding domain (χprot = 0.42) with DNA are shown in Fig. 3A. The protein maintains a sliding orientation to the DNA that is very similar to the specific binding found in the crystal structure. The recognition helix can therefore efficiently sense the specific target sequence and readily form the network of hydrogen bonds that define the specific complex. We quantify the difference between the specific and nonspecific binding orientations by measuring the distance between the Cα atom of each residue that specifically bind DNA and the nearest DNA backbone phosphate atom in both the specific complex and the sliding conformation (each with its corresponding DNA). The mean difference between the distances in the specific complex and the sliding conformation (denoted as Δdsp-nsp) equals 1.3 Å for Skn-1, which supports the similarity between the two binding modes. Fig. 3 B and C show a comparison between the specific and sliding protein–DNA complexes of two other DBPs: homeobox protein Hox-A9 and Smad-MH1 protein. These proteins have lower χprot values of 0.35 and 0.18, respectively, which reflect a higher frustration (lower overlap) between their specific binding residues and charged residues. In these two proteins, the sliding orientation of the protein to the DNA differs more markedly from the orientation adopted in the specific complex than is observed for the Skn1 DNA-binding protein, which has a higher value of χprot. The increasing difference between the nonspecific and specific binding modes observed for these three proteins is reflected in increasing Δdsp-nsp values as χprot decreases.
A sliding conformation of a monomer from the restriction endonuclease BamHI is shown (Fig. 3D, green) together with the specific enzyme–DNA complex (blue) (23), as well as with a structure of the enzyme bound to a semispecific DNA sequence that differs by a single base pair from the specific sequence (light blue) (6). The low χprot value of 0.03 for this protein reflects a significant difference between the electrostatic qualities of the patch and the specific protein–DNA contact map. Indeed, the electrostatically favored orientation of the protein relative to the DNA captured in the simulation deviates significantly from both the specific and semispecific protein–DNA complexes. We note, however, that the sliding conformation is more similar to the semispecific complex ( ), than to the specific complex (
), than to the specific complex ( ). In agreement with an earlier study of the sliding of BamH1 on DNA using Poisson–Boltzmann calculations and Brownian dynamics (24), our observation indicates that the semispecific protein–DNA complex lies partway between protein sliding on entirely random sequences and a protein binding complex with its specific sequence.
). In agreement with an earlier study of the sliding of BamH1 on DNA using Poisson–Boltzmann calculations and Brownian dynamics (24), our observation indicates that the semispecific protein–DNA complex lies partway between protein sliding on entirely random sequences and a protein binding complex with its specific sequence.
Transition Kinetics from Sliding to Specific DNA Binding.
Structural differences between the nonspecific and specific interactions arising from the frustration between the patches used in each type of DNA binding may have kinetic effects on the overall search process for the target site. One can envision a time gap between localizing the target site and binding tightly to it during which the protein will have to switch its conformation. The time gap might be longer where the frustration between the two binding modes is larger, and this could reduce the likelihood of a successful switch from the nonspecific to specific binding mode.
The interplay between search kinetics and the degree of frustration between specific and nonspecific binding was studied by investigating the time scale for switching from nonspecific to specific binding using the coarse-grained model described above. We used the recognition helix of MAD (composed of 26 residues) from the heterodimer MAD-MAX that contains seven positively charged residues and three negatively charged residues. In addition to nonspecific interactions, the helix may bind a DNA target site (located at the center of the 100-bp DNA) defined based on the crystal structure, by forming specific interactions that are represented by the Lennard–Jones potential.
The helix translocation along the DNA (aligned along the Z axis) was monitored during the simulation, as were the formation of specific protein–DNA contacts with the target site (Fig. 4A). Fig. 4B exemplifies a typical sliding–binding trajectory for the wild-type recognition helix of MAD, for which χprot = 0.1, indicating a limited overlap of the positively charged electrostatic patch that dominates the sliding mode with the patch for specific protein–DNA interactions. Hence, although the protein locates the target (at Z = 0) relatively fast, its orientation is inappropriate for specific DNA binding, and it only forms a partial complex with the DNA (only up to 50% of the protein–DNA contacts are formed). Indeed, Fig. 4B shows the helix repeatedly undergoing several dissociations from the target, engaging in additional sliding, and then reassociating with the target again, until the protein–DNA interface is fully formed (defined as > 75% formation of the specific protein–DNA contacts).
Fig. 4.
Kinetics of the transition from nonspecific to specific DNA binding. (A) Illustration of a sliding–binding simulation for the recognition helix of MAD from the MAD-MAX heterodimer. The searching protein, shown in lucid blue, is initially positioned near the edge of the 100-bp dsDNA and starts sliding along the helical DNA path on nonspecific segments (I). It then locates the binding site, but the specific protein–DNA interface is not yet formed (II). The time required from the start of the search until the protein locates its binding site, τ1, is then recorded. The protein can then either switch from the trapped state to form high affinity interactions with the target site (IV) or continue to diffuse to other regions of the DNA (III) until it again locates the target and eventually fully binds to it (IV). The time elapsed from the start of the search until formation of the specific interface, τ2, is then recorded. Trajectories of three variant peptides with different χprot are shown: (A) wild-type peptide (χprot = 0.1), (B) peptide 33 (χprot = 0.4), and (C) peptide 19 (χprot = 0.6) (see Fig. S1). Each trajectory is analyzed in terms of the location of the protein along the DNA (blue) and the fraction of specific contacts formed at the binding site (gray). The times τ1 and τ2 are indicated by red dashed lines. The correlations between τ1 and τ2 from 100 simulations of each peptide system are shown and indicate greater correlation as χprot increases.
To study binding kinetics, we repeated the simulation 100 times and measured two time periods in each run: the time elapsed from the start of the search until target site localization (τ1) and the time until specific binding (i.e., formation of > 75% of the specific contacts) (τ2). Although in some of the simulations target localization was rapidly followed by target binding (i.e., τ2 ∼ τ1), in most cases, complete binding to the DNA target significantly lagged behind target localization (i.e., τ2 > τ1) and the overall correlation between the two time scales was weak (R2 = 0.2) (Fig. 4B, Right). A time gap between arrival at the specific site and binding to it implies the existence of a kinetic barrier in the switch from sliding to the specific binding. It further suggests that alternative protein–DNA nonspecific interfaces may have lower frustration with the specific DNA-binding residues and, thus, lower kinetic barrier that governs the transition from nonspecific to specific target binding.
To test this implication, we designed in silico a library of 150 variants of the recognition helix of MAD, in which we modified the composition and distribution of positively and negatively charged residues along the sequence while maintaining the protein–DNA specific contact map unmodified (Fig. S2). For each variant, we calculated its χprot and performed 100 sliding/binding simulations as described above. The effects of flanking nonspecific DNA on τ1 and τ2 were uniform for all peptides as all of the simulations started with the helix initially positioned near the DNA edge. Fig. 4C shows a typical sliding–binding trajectory for a variant of the recognition helix of MAD with χprot = 0.4 (peptide 33; see Fig. S2). In this variant, a relatively stable partial protein–DNA complex at the target sequence precedes the formation of the complete protein–DNA interface. Values for τ2 and τ1 are relatively similar with only a few outliers (R2 = 0.6). Fig. 4D demonstrates the binding kinetics a variant with χprot = 0.6 (peptide 19; see Fig. S2). The high χprot value of this peptide implies that it can bind specifically to the DNA while maintaining charge–charge interactions with the DNA similar to those it has in the sliding mode. Consequently, complete binding of this peptide variant to the target DNA sequence takes place rapidly upon localization of the target by the protein, with τ2 ∼ τ1 in all the simulations (R2 ∼ 1). Overall, these observations (Fig. 4) imply that lower frustration between the specific DNA-binding residues and the charged residues in the protein, i.e., increased χprot of the protein, may reduce the kinetic barrier to transition from the sliding mode to specific target binding.
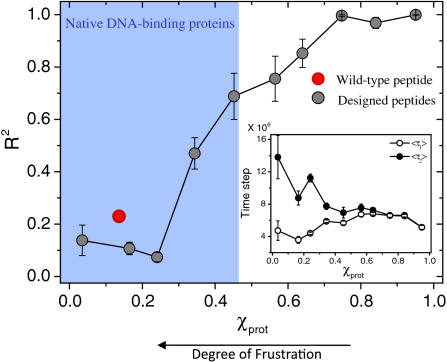
The histogram in Fig. 5 summarizes the interplay between search kinetics and the degree of frustration between sliding and specific binding to DNA. It shows the average values of the Pearson correlation coefficient between τ2 and τ1 against the χprot values of the simulated peptide library. Higher correlation between arrival and binding times is found for peptides with lower frustration between their two DNA binding modes (i.e., higher χprot values). The decrease of time gap between target localization and binding in peptides with high χprot values is reflected in Fig. 5, Inset. Returning to the main histogram, we note that the blue-shaded region indicates the χprot range for the natural DBP dataset described above, in which the value of the wild-type recognition helix resides (red symbol). It is therefore evident that most DBPs may experience a kinetic barrier in switching from nonspecific to specific DNA binding because of significant frustration in the DNA-binding interface between specific DNA binding residues and charged residues.
Fig. 5.
Interplay between frustration and the kinetics of target site search. Histogram of R2 (the correlation coefficient between τ1 and τ2; see Fig. 4) for the χprot values of the library of 150 MAD recognition helix variants is shown. Target localization and binding are significantly more correlated (R2 ∼ 1) in proteins with higher χprot values. The region shaded in blue marks the range of χprot values obtained from the dataset of natural DBPs, and the red circle corresponds to the wild-type peptide of the MAD recognition helix. Peptides with lower χprot values (i.e., with higher frustration between their specific and nonspecific binding modes) are characterized not only by a weak correlation between τ1 and τ2 but also by a gap between the two time scales (Inset) that disappear for less frustrated peptides.
Conclusions
The observation that the interface used by DBPs to specifically recognize DNA targets is often accommodated in a patch of positive electrostatic potential used for nonspecific interactions may suggest that both binding modes share similar structural properties. Although the degree of overlap between the positively charged patch and the patch that participates in hydrogen bonding is greater in DBPs comparing to RBPs or protein–protein complexes with a highly charged interface, it is nevertheless imperfect. The smaller the overlap is between the residues forming specific interactions and the positively charged patch (i.e., lower χprot), the larger the difference between the sliding and specific complexes and the greater the degree of frustration between the two binding modes to DNA.
Binding to the target site may therefore impose a free energy barrier in switching from the nonspecific orientation to the specific conformation. In support of this proposition, our comparison of BamHI endonuclease monomer bound to its specific, semispecific, and nonspecific DNA sequences indicates that the orientation of the protein to the DNA when bound to the semispecific sequence lies between the orientations adopted on completely random sequences and in specific complexes. A utilization of distinct DNA-binding modes for search and recognition has been suggested earlier to resolve the so-called “speed–stability” paradox as it allows the reduction of the ruggedness of the landscape for protein sliding (25, 29). Additional evidence for the existence of a multistate model of interactions with DNA can be found in several X-ray and NMR studies (7, 26, 27) and computer simulations (24, 28). The reorientation of the protein that is governed by the electrostatic interactions with the DNA is reminiscent of the electrostatic preorganization that is found in several cases in enzymatic catalysis in which the solvent reorients its dipoles to promote the reaction (30, 31).
To quantify the effect of the degree of overlap between the specific and nonspecific patches on search kinetics, we designed in silico a library of peptides that span a wide range of degrees of frustration between the specific and nonspecific DNA-binding interactions. We show that a time gap emerges between target localization and target binding due to a conflict between the two residue sets involved in each DNA-binding mode. A high degree of overlap results in almost immediate binding in what can be envisioned as a “lock and key” mechanism. However, limited overlap usually introduces additional kinetic steps for DNA binding during which the protein can become trapped in an intermediate complex with the target, and would switch to the specific orientation by forming hydrogen bonds with the DNA bases only at the expanse of several electrostatic interactions with the sugar-phosphate backbone. An example for such multistate kinetics of DNA sequence recognition has been described earlier for the C-terminal domain of the papillomavirus E2 protein with its target DNA sequence (3). The transition from nonspecific to specific binding might also be delayed by subsequent sliding periods following unproductive trapping. This scenario suggests that a selectivity principle operates whereby proteins avoid “wasteful” commitments to semispecific sites as they search the genome; thus, the ruggedness of the energy landscape for sliding is moderate and the overall search is rapid (25). Commitment to the specific site requires transition from the sliding to specific binding mode across a kinetic barrier in what could be considered an “induced fit” mechanism. This may involve at least reorientation of the protein relative to the DNA as observed for Dam methyltransferase substrate recognition (27) and, in some cases, also an intramolecular conformational change to the protein (22), the DNA (32), or both (7), although these were not addressed in the current study.
Our study serves as another example of the important role of frustration in biomolecular systems (33, 34). The prevalence of frustration in DBPs results in bimodal association with DNA and suggests an additional degree of kinetic complexity to the DNA target search problem. Clearly, additional factors can enrich the complexity of the facilitated diffusion that governs the search (e.g., the existence of roadblock proteins or packing of the DNA by the nucleosomes) that, on the one hand, may result in slower kinetics but, on the other hand, may introduce functional regulation. Zero frustration between specific and nonspecific binding would result in sliding that is interrupted by trapping, whereas a high degree of frustration would result in an extremely large barrier for switching. Accordingly, weak frustration is the optimal scenario for an efficient DNA search.
Methods
Calculation of χprot.
We first calculated χi for each residue i in the protein as follows:
 |
[1] |
where j′ denotes all the residues having their Cα atom closer than a cutoff distance of rc = 10 Å to the Cα atom of residue i. qj′ is a point charge of 1, –1, or 0, and rij′ is the Cα–Cα distance between residues i and j′. a = 2 is an exponential decay constant. A χi value approaching 1 is therefore expected for residues that are fully surrounded by positively charged residues. The value of χprot is the mean of the χi values for protein residues that belong to the interface used for specific binding to the DNA.
Simulation Model.
The dynamics of sliding along DNA was studied using a coarse-grained model used previously to explore various molecular aspects of the mechanism of DNA search (20) (see SI Text for more details). We studied the interaction of the recognition helix of MAD from the heterodimer MAD-MAX with DNA. To study how it makes the transition from its sliding to specific binding mode, we inserted the specific sequence of the DNA for MAD at the center of the 100-bp DNA molecule. One hundred simulations were performed for the wild-type and the 150 designed peptide variants (Fig. S2), starting at the exact same initial helix position to allow unified measurements for τ1 and τ2 in all of the simulations.
Supplementary Material
ACKNOWLEDGMENTS.
We are grateful to Shula Shazman for assistance with analyzing positively electrostatic patches of proteins. This work was supported by the Kimmelman Center for Macromolecular Assemblies and the Minerva Foundation with funding from the Federal German Ministry for Education and Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109594108/-/DCSupplemental.
References
- 1.von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 2.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez IE, Ferreiro DU, Dellarole M, de Prat-Gay G. Experimental snapshots of a protein-DNA binding landscape. Proc Natl Acad Sci USA. 2010;107:7751–7756. doi: 10.1073/pnas.0911734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lundback T, Hard T. Sequence-specific DNA-binding dominated by dehydration. Proc Natl Acad Sci USA. 1996;93:4754–4759. doi: 10.1073/pnas.93.10.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler FK, et al. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viadiu H, Aggarwal AK. Structure of BamHI bound to nonspecific DNA: A model for DNA sliding. Mol Cell. 2000;5:889–895. doi: 10.1016/s1097-2765(00)80329-9. [DOI] [PubMed] [Google Scholar]
- 7.Kalodimos CG, et al. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 8.Misra VK, Hecht JL, Yang AS, Honig B. Electrostatic contributions to the binding free energy of the lambdacI repressor to DNA. Biophys J. 1998;75:2262–2273. doi: 10.1016/S0006-3495(98)77671-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Record MT, Ha JH, Fisher MA. Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol. 1991;208:291–343. doi: 10.1016/0076-6879(91)08018-d. [DOI] [PubMed] [Google Scholar]
- 10.von Hippel PH. From “simple” DNA-protein interactions to the macromolecular machines of gene expression. Annu Rev Biophys Biomol Struct. 2007;36:79–105. doi: 10.1146/annurev.biophys.34.040204.144521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stawiski EW, Gregoret LM, Mandel-Gutfreund Y. Annotating nucleic acid-binding function based on protein structure. J Mol Biol. 2003;326:1065–1079. doi: 10.1016/s0022-2836(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 12.Szilagyi A, Skolnick J. Efficient prediction of nucleic acid binding function from low-resolution protein structures. J Mol Biol. 2006;358:922–933. doi: 10.1016/j.jmb.2006.02.053. [DOI] [PubMed] [Google Scholar]
- 13.Jones S, van Heyningen P, Berman HM, Thornton JM. Protein-DNA interactions: A structural analysis. J Mol Biol. 1999;287:877–896. doi: 10.1006/jmbi.1999.2659. [DOI] [PubMed] [Google Scholar]
- 14.Lamers MH, et al. The crystal structure of DNA mismatch repair protein MutS binding to a G·T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 15.Kostrewa D, Winkler F. Mg2+ binding to the active site of EcoRV endonuclease: A crystallographic study of complexes with substrate and product DNA at 2A resolution. Biochemistry. 1995;34:683–696. doi: 10.1021/bi00002a036. [DOI] [PubMed] [Google Scholar]
- 16.Rohs R, et al. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: A three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwahara J, Zweckstetter M, Clore GM. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc Natl Acad Sci USA. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blainey PC, et al. Nonspecifically bound proteins spin while diffusing along DNA. Nat Struct Mol Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Givaty O, Levy Y. Protein sliding along DNA: Dynamics and structural characterization. J Mol Biol. 2009;385:1087–1097. doi: 10.1016/j.jmb.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 21.Liu H, Shi Y, Chen X, Warshel A. Simulating the electrostatic guidance of the vectorial translocations in hexameric helicases and translocases. Proc Natl Acad Sci USA. 2009;106:7449–7454. doi: 10.1073/pnas.0900532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 23.Newman M, Strzelecka T, Dorner LF, Schildkraut I, Aggarwal AK. Structure of Bam HI endonuclease bound to DNA: Partial folding and unfolding on DNA binding. Science. 1995;269:656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- 24.Sun J, Viadiu H, Aggarwal AK, Weinstein H. Energetic and structural considerations for the mechanism of protein sliding along DNA in the nonspecific BamHI-DNA complex. Biophys J. 2003;84:3317–3325. doi: 10.1016/S0006-3495(03)70056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slutsky M, Mirny LA. Kinetics of protein-DNA interaction: Facilitated target location in sequence-dependent potential. Biophys J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman JI, Majumdar A, Stivers JT. Nontarget DNA binding shapes the dynamic landscape for enzymatic recognition of DNA damage. Nucleic Acids Res. 2009;37:3493–3500. doi: 10.1093/nar/gkp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horton JR, Liebert K, Hattman S, Jeltsch A, Cheng XD. Transition from nonspecific to specific DNA interactions along the substrate-recognition pathway of Dam methyltransferase. Cell. 2005;121:349–361. doi: 10.1016/j.cell.2005.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C, Pettitt M. The binding process of a nonspecific enzyme with DNA. Biophys J. 2011;101:1139–1147. doi: 10.1016/j.bpj.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou HX. Rapid search for specific sites on DNA through conformational switch of nonspecifically bound proteins. Proc Natl Acad Sci USA. 2011;108:8651–8656. doi: 10.1073/pnas.1101555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warshel A. Energetics of enzyme catalysis. Proc Natl Acad Sci USA. 1978;75:5250–5254. doi: 10.1073/pnas.75.11.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamerlin S, Sharma P, Chu Z, Warshel A. Ketosteroid isonearse provides further support for the idea that enzymes work by electrostatic preorganization. Proc Natl Acad Sci USA. 2010;107:4075–4080. doi: 10.1073/pnas.0914579107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouvier B, Zakrzewska K, Lavery R. Protein-DNA recognition triggered by a DNA conformational switch. Angew Chem Int Ed Engl. 2011 doi: 10.1002/anie.201101417. in press. [DOI] [PubMed] [Google Scholar]
- 33.Zhuravlev PI, Papoian GA. Protein functional landscapes, dynamics, allostery: A tortuous path towards a universal theoretical framework. Q Rev Biophys. 2010;43:295–332. doi: 10.1017/S0033583510000119. [DOI] [PubMed] [Google Scholar]
- 34.Ferreiro D, Hegler J, Komives E, Wolynes P. On the role of frustration in the energy landscapes of allosteric proteins. Proc Natl Acad Sci USA. 2011;108:3499–3503. doi: 10.1073/pnas.1018980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.