Abstract
Most solid tumors are aneuploid, and many missegregate chromosomes at high rates in a phenomenon called chromosomal instability (CIN). CIN reflects the erosion of mitotic fidelity, and it correlates with poor patient prognosis and drug resistance. The most common mechanism causing CIN is the persistence of improper kinetochore–microtubule attachments called merotely. Chromosomes with merotelic kinetochores often manifest as lagging chromosomes in anaphase, suggesting that lagging chromosomes fail to segregate properly. However, it remains unknown whether the lagging chromosomes observed in anaphase segregate to the correct or incorrect daughter cell. To address this question, we tracked the segregation of a single human chromosome during cell division by using LacI-GFP to target an integrated LacO array. By scoring the distribution of each sister chromatid during mitosis, we show that a majority of lagging chromosomes in anaphase segregate to the correct daughter cell. Instead, sister chromatids that segregate erroneously frequently do so without obvious evidence of lagging during anaphase. This outcome is expected if sister kinetochores on a chromosome bind microtubules oriented toward the same spindle pole, and we find evidence for syntelic kinetochore attachments in cells after treatments that increase missegregation rates. Thus, lagging chromosomes in anaphase are symptomatic of defects in kinetochore–microtubule attachment dynamics that cause chromosome missegregation associated with CIN, but the laggards rarely missegregate.
Keywords: aneuploidy, syntely, MCAK, micronuclei, genome instability
Solid tumors are frequently aneuploid and many missegregate chromosomes at high rates in a phenomenon called chromosomal instability (CIN; refs. 1 and 2). CIN is associated with poor patient prognosis, and various studies have shown that it correlates with advanced tumor stage including acquisition of metastatic potential and drug resistance (3–5). It has been proposed that by frequently changing the karyotype of tumor cells, that CIN provides an agent of change that drives the evolution of tumor cell phenotypes (3–8). The treatment difficulties encountered in advanced stage tumors underscores the importance of determining the mechanisms of CIN and how they contribute to tumor growth.
Various mechanisms have been proposed to cause CIN including dysfunction of the spindle assembly checkpoint, defects in sister chromatid cohesion, and defects in the attachment of chromosomes to spindle microtubules (2). Recently, live cell imaging demonstrated that the most common cause of CIN is the persistence of errors in the attachment of spindle microtubules to chromosomes (9, 10). Microtubules bind to chromosomes at specialized structures called kinetochores. Each chromosome has a pair of kinetochores, and faithful chromosome segregation arises when single kinetochores bind microtubules oriented toward only one spindle pole resulting in the biorientation of chromosomes on the spindle. However, errors in the orientation of kinetochore–microtubule (k-MT) attachments frequently occur, particularly in early phases of mitosis, as a consequence of the stochastic interactions between microtubules and kinetochores (11). A prominent error is when a single kinetochore binds microtubules oriented toward both spindle poles. This error is called merotely (12, 13). The persistence of merotely undermines chromosome segregation because merotelic kinetochores experience poleward force toward both spindle poles. As a consequence, merotely often results in the appearance of lagging chromosomes in anaphase, and tumor cells with CIN have elevated rates of lagging chromosomes and merotelic attachments (9). Moreover, it was shown that increasing the correction rate of merotely by stimulating the dynamics of k-MT attachment suppressed CIN, providing a causative relationship between the persistence of improper k-MT attachments and CIN (10).
Using quantitative measurements, there is a strong correlation between the frequency of lagging chromosomes and the rate of chromosome missegregation (9, 10, 14), suggesting that lagging chromosomes frequently missegregate to cause CIN. In contrast, live imaging of marsupial cells in mitosis with merotelic kinetochores failed to identify missegregation of chromatids with a merotelic kinetochore (13, 15). Thus, it remains controversial if lagging chromosomes that are observed in anaphase ultimately segregate to the incorrect daughter to generate missegregation that is observed in human tumor cells with CIN. To resolve this controversy, we tracked the fate of each sister chromatid of a single human chromosome during segregation by using an integrated LacO array illuminated by LacI-GFP.
Results
To track the fate of lagging chromosomes during mitosis in human cells, we used live imaging of cells expressing GFP-tagged histone H2B (9). Human U2OS osteosarcoma cells are aneuploid and chromosomally unstable and frequently show lagging chromosomes in anaphase (10). Live imaging of chromosome segregation in mitosis in these cells revealed that 78% (n = 23) of anaphase cells with lagging chromosomes resulted in at least one daughter cell with a distinct micronucleus (Fig. S1). We also examined fixed cells for the presence of lagging chromosomes in anaphase and for micronuclei in pairs of daughter cells. Lagging chromosomes occurred in 21.7% (±1.7%, SEM) of anaphases, and micronuclei were present in at least one daughter cell in 21.4% (±1.7%, SEM) of daughter cell pairs, indicating a strong correlation between the appearance of laggards and micronuclei in these cells. HCT116 cells behaved similarly when the levels of merotelic kinetochore attachments were increased by recovery from monastrol- or nocodazole-induced mitotic delay. Eighty percent (n = 31) of HCT116 cells with lagging chromosomes in anaphase resulted in daughter cells with distinct micronuclei. These data demonstrate that most lagging chromosomes in anaphase result in micronuclei in a daughter cell at the completion of mitosis.
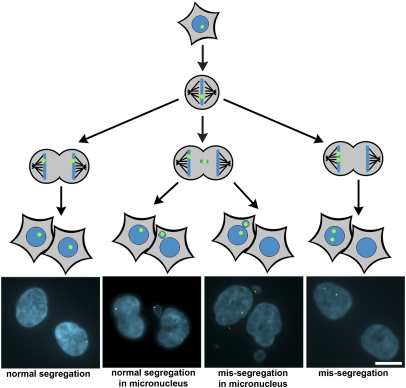
We next followed the fate of each chromatid of a single human chromosome during mitosis in HCT116 cells by using a LacO array integrated into a single chromosomal locus and detected by expression of LacI-GFP (16–18). The probability of witnessing a lagging chromatid from this specific chromosome using live cell imaging is expected to be quite rare (1/45 chromosomes tagged × 1 lagging chromosome observed in every 3 HCT116 cells recovering from monastrol- or nocodazole-induced mitotic delay = 1/135 cells would display an error in this specific chromosome; ref. 9). Therefore, we relied on fixed cell analyses for our experiments. Mitotic cells were harvested and allowed to complete mitosis at very low cell density so that the fate of this marked chromosome could be unequivocally determined in the two daughter cells. In principle, there are four possible fates for the two chromatids (each segregates to major nuclei of opposite daughter cells, micronuclei in the correct daughter cell, micronuclei in the incorrect daughter cell, and both segregate to the major nucleus of the same daughter cell), and we observed each fate to different degrees depending on the conditions (Fig. 1).
Fig. 1.
Fates of sister chromatids of a single chromosome during mitosis. Corresponding daughter HCT116 cells expressing lacIGFP with LacO arrays integrated into a single chromosome were fixed and imaged after inducing high levels of merotely with MCAK siRNA or monastrol washout. Schematic of possible segregation fates of the marked chromosome (Upper) and representative images of each fate (Lower) are shown. (Scale bar: 10 μm.)
HCT116 cells are not chromosomally unstable and faithfully segregate chromosomes with an error detected in the segregation of this marked chromosome in only 0.16% of daughter cells (Table 1). The most common error in untreated HCT116 cells is when the marked chromosome is in a micronucleus. However, the micronucleus with the marked chromosome was nearly 10 times more likely to reside in the correct daughter cell than the incorrect daughter cell, and that would not alter the karyotype of either daughter cell. Thus, the frequency of missegregation (i.e., the two sister chromatids residing in the same daughter cell) of this chromosome is quite rare in untreated HCT116 cells.
Table 1.
Chromosome segregation errors in HCT116 cells
| Normal segregation (%) | Normal segregation in micronucleus (%) | Missegregation in micronucleus (%) | Missegregation (%) | |
| Control (n = 7,531) | 7,519 (99.84) | 9 (0.12) | 1 (0.01) | 2 (0.03) |
| Monastrol washout (n = 9,262) | 9,032 (97.52) | 114 (1.23) | 32 (0.35) | 84 (0.91) |
| MCAK knockdown (n = 7,572) | 7,490 (98.92) | 30 (0.40) | 8 (0.11) | 44 (0.58) |
| Monastrol washout + MCAK knockdown (n = 6,410) | 6,201 (96.72) | 65 (1.02) | 55 (0.86) | 89 (1.39) |
In contrast, there is a dramatic increase in the rate of chromosome segregation errors when the prevalence of merotelic attachments is increased, by increasing their rate of formation by recovery from monastrol treatment, reducing their rate of correction by knockdown of the kinesin-13 protein MCAK, or both (Table 1). We have performed MCAK knockdown by using siRNA (16) and used the significant increase in lagging chromosomes in anaphase to confirm efficient knockdown in these experiments (Fig. S2). Using either the drug recovery strategy or the loss of MCAK activity to increase levels of merotely, we found that when the marked chromosome was in a micronucleus, it was approximately four times more likely to segregate to the correct daughter cell than to the incorrect daughter cell. This ratio was reduced slightly when the two treatments were combined, but the trend was the same with the chromosome segregating to the correct daughter cell more frequently than to the incorrect daughter cell. Given that the fate of most lagging chromosomes is to become a micronucleus in one of the daughter cells (Fig. S1), these data indicate that lagging chromosomes that form micronuclei tend to segregate to the correct daughter cell even when the frequency of merotelic attachments is substantially increased.
The most striking change in chromosome segregation observed when the prevalence of merotelic attachments is increased is where one daughter cell has both sister chromatids in the major nucleus with no observable micronucleus (Fig. 1 and Table 1). This event is rare in untreated cells, but is quite common in cells that recovered from monastrol treatment, lack MCAK activity, or both. Indeed, in all circumstances, the frequency of one daughter cell with both sister chromatids in the major nucleus is higher than the frequency of cells with the marked chromosome in a micronucleus in the incorrect daughter cell. This finding is most extreme in cells depleted of MCAK where it is five times more likely that a missegregated chromosome will be in the major nucleus compared with a micronucleus. Assuming all chromosomes behave similarly in cells lacking MCAK activity, ≈26% of mitoses (0.58% for one chromosome × 45 chromosomes) would have both sister chromatids in the major nucleus of one daughter cell, compared with only ≈5% of mitoses (0.11% for one chromosome × 45 chromosomes) yielding a chromatid in a micronucleus of the incorrect daughter cell. Because the fraction of lagging chromosomes that do not result in micronuclei is too small to account for the frequency of this event, these data suggest that there is a substantial population of cells under these conditions where both sister chromatids segregate to the same daughter cell without overtly lagging.
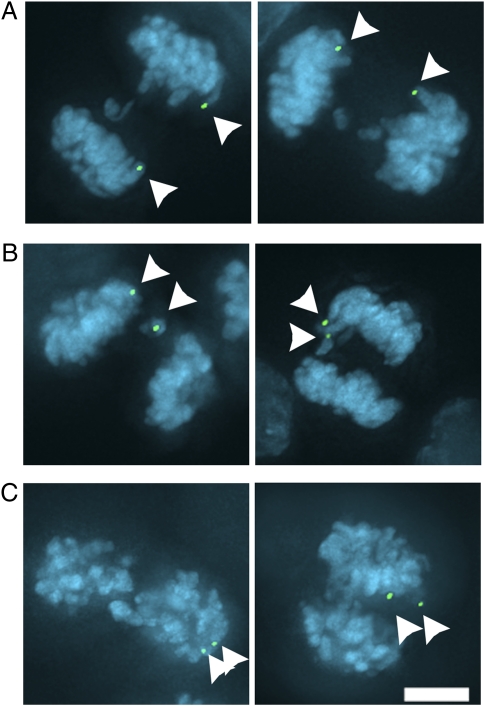
To verify that sister chromatids missegregate without lagging, we followed the fate of each sister chromatid of the marked chromosome during anaphase (Fig. 2 and Table 2). There are three possible fates for the two chromatids in anaphase (segregation to opposite poles, lagging, and segregation to the same pole), and we observed each fate to different degrees depending on the conditions (Fig. 2). As expected, chromosomes rarely missegregate in anaphase in untreated control cells (Table 2). In contrast, increasing the prevalence of merotely either by recovery from monastrol or through depletion of MCAK activity significantly increases the error rate (Table 2). As expected, the frequency with which both chromatids segregate to the same pole without overtly lagging in anaphase (Table 2) is near the frequency of both chromatids being in the major nucleus of the same daughter cell (Table 1). Importantly, the chromosomal locus marked by LacI-GFP on each chromatid in these anaphase cells is well-separated, indicating that chromosome missegregation is not caused by the failure of sister chromatids to disjoin in anaphase.
Fig. 2.
Fates of sister chromatids of a single chromosome during anaphase. HCT116 cells in anaphase expressing lacIGFP with LacO arrays integrated into a single chromosome were fixed and imaged after inducing high levels of merotely with MCAK siRNA or monastrol washout. Representative images show the marked chromosome segregating correctly, with one chromosome in each anaphase plate (A), lagging in the spindle midzone (B), or missegregating with no evidence of lagging (C). Arrowheads point to the LacIGFP labeled chromosome. (Scale bar: 5 μm.)
Table 2.
Chromosome segregation errors in anaphase in HCT116 cells
| Normal segregation (%) | Lagging (%) | Missegregation without lagging (%) | |
| Control (n = 300) | 299 (99.7) | 0 (0) | 1 (0.3) |
| Monastrol washout (n = 1,391) | 1,373 (98.7) | 8 (0.6) | 10 (0.7) |
| MCAK knockdown (n = 508) | 497 (97.8) | 8 (1.6) | 3 (0.6) |
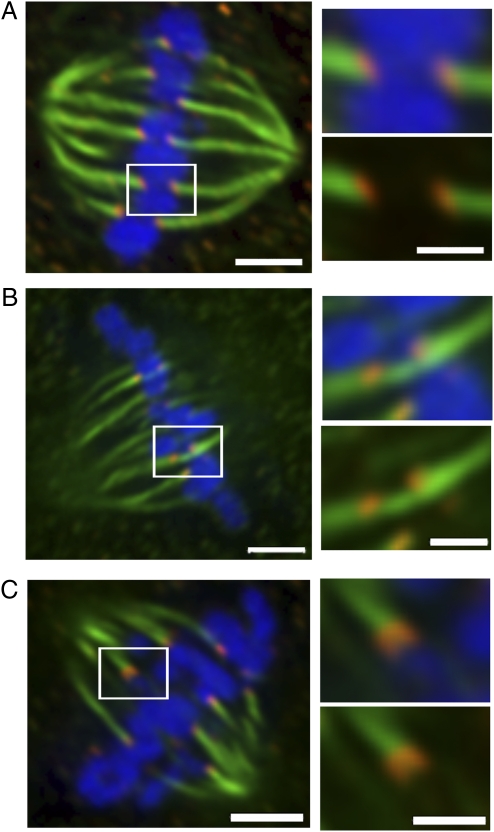
The most likely mechanism causing both chromatids to segregate to the same pole after they dissociate at anaphase onset is through attachment of both kinetochores of that chromosome to microtubules oriented toward the same spindle pole (e.g., syntely). Here, we sought evidence for the presence of syntelic chromosomes in human cells. For these experiments, we used the diploid, chromosomally stable RPE1 cells and induced the formation of k-MT attachment errors by using recovery from monastrol treatment. To enhance the contrast of k-MTs during mitosis, non–k-MTs were depolymerized with a calcium-containing buffer before fixation and stained for microtubules and the kinetochore protein Hec1 (Fig. 3 and Fig. S3). Most chromosomes are amphitelic as expected (Fig. 3A), and we could easily identify merotelic attachments in metaphase (Fig. 3B). We also identified chromosomes that appeared to be predominantly syntelic in early anaphase as judged by the broadened and bilobed structure of the kinetochore on one side of a chromosome by using staining for Hec1 (Fig. 3C and Fig. S4). The large number of kinetochore pairs around the metaphase plate precluded us from using imaging to directly quantify the frequency of these syntelic-like chromosomes that were most readily identifiable near the periphery of the metaphase plate. Nevertheless, these results provide evidence that segregation of both sister chromatids to the same daughter cell without overtly lagging is possible through a syntelic-like k-MT attachment.
Fig. 3.
Various kinetochore–microtubule attachments. RPE1 cells recovering from monastrol treatment to induce k-MT attachment errors were extracted in the presence of calcium; fixed; stained for kinetochores (Hec1) in red, microtubules (green), and DNA (blue); and imaged by using confocal microscopy to identify k-MT attachments. Single focal planes showing amphitely (A), merotely (B), and syntely (C) are shown in Insets. (Scale bars: Left, 2.5 μm; Right, 1 μm.)
Discussion
Human cancer cells with CIN display lagging chromosomes in a substantial fraction of cells during anaphase because of the persistence of k-MT attachment errors. By tracking sister chromatids of a single human chromosome under conditions that promote these errors, we demonstrate here that the fate of most lagging chromosomes in anaphase is to form a micronucleus in the correct daughter cell. Chromosomes in micronuclei with an intact centromere undergo typical condensation in the subsequent mitosis and reassociate with the other chromosomes (Fig. S5), resulting in no karyotypic change in either daughter cell. Thus, chromosomes that lag in anaphase tend not to missegregate (i.e., cause nondisjunction) in human cells. These results are in agreement with data from nonhuman model cell systems showing that lagging chromosomes rarely missegregate (19, 20).
Instead, our evidence shows that chromosome missegregation in human cells with elevated rates of k-MT attachment errors is rooted in sister chromatids that separate, but segregate to the same spindle pole without obviously lagging in anaphase. The most likely cause of this mode of chromosome missegregation is that sister kinetochores attach to spindle microtubules oriented toward the same spindle pole. We observe chromosomes with the hallmarks of syntely in human cells and note that syntelic chromosomes have been observed to missegregate during meiosis in insect cells (19). Syntelic chromosomes are unlikely to satisfy the checkpoint (21), and we have never observed anaphase onset before alignment of all chromosomes in numerous CIN and stable human cell lines (9). Thus, syntelic k-MT attachments most likely become converted into merotelic attachments to promote chromosome alignment and checkpoint satisfaction. Because the correction of both syntely and merotely relies on the dynamic attachment of microtubules to kinetochores, the contribution of syntelic precursors to chromosome missegregation is fully consistent with the fact that faithful chromosome segregation can be restored in human cancer cells with CIN by overexpression of enzymes that promote error correction by decreasing the stability of k-MT attachments (10). Thus, lagging chromosomes in anaphase rarely missegregate, yet their presence is symptomatic of an underlying defect in k-MT attachment dynamics that causes chromosome missegregation that can lead to CIN.
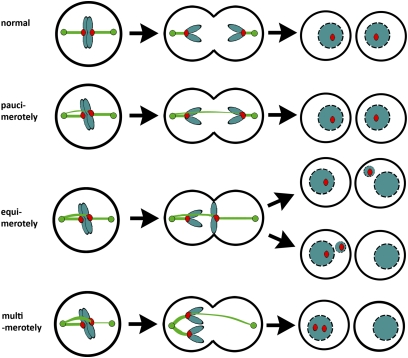
These results indicate that only specific types of k-MT attachment errors render chromosomes prone to missegregation. The spindle assembly checkpoint prevents anaphase onset in the presence of chromosomes with monotelic or syntelic but not merotelic kinetochores. However, not all merotelic kinetochores are equivalent because they differ in the quantity of microtubules oriented toward the wrong spindle pole (Fig. 4). Thus, we propose a nomenclature for discriminating these different versions of merotely based on the Latin root terms for few (pauci), equal (equi), and many (multi). We would expect chromosomes with pauci-merotelic kinetochores (those with few microtubules oriented toward the wrong pole) to segregate properly, perhaps without even lagging in anaphase. If so, then the prevalence of paucimerotely may be unrecognized. Lagging chromosomes in anaphase are most likely caused by equimerotelic kinetochores (those with approximately equal numbers of microtubules oriented toward the correct and wrong poles). Roughly equal poleward force experienced by an equimerotelic kinetochore toward both spindle poles causes the chromosome to lag in anaphase. However, our data and data from others (15) show that most lagging chromosomes end up in the correct daughter cell as micronuclei. That bias is most likely provided by the back-to-back geometry of sister kinetochores that would encourage a preponderance of k-MT attachments to be oriented toward the correct spindle pole even within an equimerotelic configuration. Finally, our data indicate that chromosomes with multimerotelic kinetochores (those with many microtubules oriented toward the wrong pole) make the largest contribution to missegregation without obvious lagging in anaphase (Fig. 4). Multimerotely most likely arise as a direct consequence of attachment of syntelic kinetochores to microtubules oriented toward the opposite spindle pole. The contribution of multimerotely to chromosome missegregation fits with predictions by Salmon and colleagues (13, 15) and has been visualized during meiosis in insect cells (19). Based on the frequencies of chromosome missegregation in cells depleted of a key element of the machinery responsible for correction of k-MT attachments errors (e.g., MCAK), we estimate that multimerotely arises in ∼25% of cells. This percentage underestimates the frequency of these errors, because other parts of the correction machinery remain intact in MCAK-deficient cells (e.g., Aurora B and Kif2b). Thus, multimerotely as a consequence of syntely may be relatively common during unperturbed mitosis in human cells. This finding would provide a strong selective pressure for the development of robust error correction machinery.
Fig. 4.
Kinetochore microtubule attachments and chromosome fate. Schematic shows numerical microtubule differences in merotelic attachments (pauci, few; equi, equal; multi, many) and the likely fate of these attachment errors if they persist into anaphase.
Materials and Methods
Cell Culture.
Cells were maintained at 37 °C with 5% CO2 in McCoy’s 5a (HCT116) or Dulbecco’s Modified Eagle Medium (RPE1, U2OS) supplemented with 10% FBS, 50 IU/mL penicillin, and 50 μg/mL streptomycin. HCT116 cells expressing LacIGFP with LacO integrated on a single chromosome (16) were grown as above, maintaining selection for the chromosome mark by alternating treatment with blasticidin (2 μg/mL) and hygromycin (300 μg/mL) approximately every 7 d. HCT116 H2B cells were grown as described above under 2 μg/mL blasticidin selection.
Live Cell Imaging.
HCT116 H2BGFP cells were imaged for micronuclei reincorporation by acquiring a single focal plane image every minute in the GFP channel by using a 0.45 N.A. Plan Fluor 20× ELWD objective on a Nikon Eclipse Ti microscope at 37 °C equipped with a cooled charge-coupled device camera (Clara; Andor Technology). HCT116 and U2OS H2BGFP cells were imaged for micronuclei formation as described above with a Plan Fluor 40× N.A. 0.6 ELW air objective on a Nikon TE 2000-E. Autocontrast was applied to images using Photoshop CS2 (Adobe).
RNAi.
Published sequences were used to deplete MCAK (22) (5′-GAUCCAACGCAGUAAAUGGUtt-3′); dsRNA (200 nM; Ambion) was transfected into cells using oligofectamine as described (23). Cells were analyzed or used for subsequent experiments 72 h later.
Segregation Assays.
For analyzing segregation in anaphases, cells were treated with siRNA for 72 h or with drugs (100 μM monastrol or 100 ng/mL nocodazole) for 8 h. Drug washouts were performed by washing twice with PBS and then incubated for 50 min with fresh medium. Cells were fixed with 3.5% paraformaldehyde and stained with DAPI. For analyzing segregation in interphase cells, mitotic cells treated with siRNA for 72 h and/or treated with drugs for 8 h were isolated by shakeoff, plated at low density on coverslips, and were fixed 14 h later. Cells were fixed to coverslips by placing coverslip in 50 mL conical tube full of polyacrylamide (to form a flat platform), spinning briefly until reaching ∼1000 × g, fixing in 1% gluteraldehyde in PBS for 5 min, washing twice with sodium borohydride (5 min each wash), and staining with DAPI in TBS-BSA for 5–10 min before placing on slide with mounting medium. Spinning coverslips ensured that the chromosome mark would be on the same z plane in every cell. Cells were imaged by using the Nikon microscope with 60 × 1.4 N.A. oil immersion lens. Optical slices in the z axis were acquired at 0.25-μm steps for anaphase images, and iterative restoration was performed by using Phylum Live software. Autocontrast was applied to images by using Photoshop CS2.
Indirect Immunofluorescence Confocal Imaging.
RPE1 cells growing on coverslips were treated with 100 μM monastrol for 16 h. Drug was washed out as described above for 30–45 min. Cells were fixed to visualize merotely by first extracting in calcium buffer (100 mM pipes, 1 mM MgCl2, 1 mM CaCl2, and 0.5% Triton X-100 at pH 6.8) at room temperature for 3–5 min, then fixing in 1% gluteraldehyde in PBS for 10 min followed by two 10-min washes in sodium borohydride. Fixed cells were washed with TBS-BSA (10 mM Tris, 150 mM NaCl, 10% BSA, and 0.1% sodium azide), then stained sequentially with Hec1 antibody (Novus Biologicals) at 1:250 for 2 h, Texas red anti-mouse secondary at 1:500 + DAPI for 2 h, DM1α (Sigma) at 1:1,000 for 25 min, and fluorescein anti-mouse secondary at 1:500 for 20–25 min. Coverslips were mounted on slides with Prolong Gold Antifade (Invitrogen).
Images were acquired by using a 1.4 N.A. 100× PlanApo VC oil immersion lens on an Eclipse Ti Nikon microscope equipped with a Quorum Wave FX-X1 spinning disk confocal system with 403-, 491-, and 561-nm lasers with an EM-CCD digital camera (Hamamatsu) with Metamorph software (Molecular Devices). To capture full spindle volume, 101 0.1-μm slices in the z axis were obtained per cell. Huygens software (Scientific Volume Imaging) was used to deconvolve images and Imaris software (Bitplane AG) was used to analyze images and obtain snapshots of single planes. Autocontrast was applied to images by using Photoshop CS2.
Lagging Chromosome Rates.
Anaphase chromosomes were counted as “lagging” if they contained CREST (ACA) kinetochore staining and were located in the spindle midzone, separated from kinetochores at the anaphase plate. Rates for each condition were measured in at least three independent experiments.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM008704 (to S.L.T.) and GM51542 (to D.A.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109720108/-/DCSupplemental.
References
- 1.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 2.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–R295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heilig CE, et al. Chromosomal instability correlates with poor outcome in patients with myelodysplastic syndromes irrespectively of the cytogenetic risk group. J Cell Mol Med. 2010;14:895–902. doi: 10.1111/j.1582-4934.2009.00905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi CM, et al. Chromosomal instability is a risk factor for poor prognosis of adenocarcinoma of the lung: Fluorescence in situ hybridization analysis of paraffin-embedded tissue from Korean patients. Lung Cancer. 2009;64:66–70. doi: 10.1016/j.lungcan.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 5.Lee AJ, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71:1858–1870. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao C, et al. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci USA. 2007;104:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuukasjärvi T, et al. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res. 1997;57:1597–1604. [PubMed] [Google Scholar]
- 8.McClelland SE, Burrell RA, Swanton C. Chromosomal instability: A composite phenotype that influences sensitivity to chemotherapy. Cell Cycle. 2009;8:3262–3266. doi: 10.4161/cc.8.20.9690. [DOI] [PubMed] [Google Scholar]
- 9.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimini D, Moree B, Canman JC, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci. 2003;116:4213–4225. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- 12.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salmon ED, Cimini D, Cameron LA, DeLuca JG. Merotelic kinetochores in mammalian tissue cells. Philos Trans R Soc Lond B Biol Sci. 2005;360:553–568. doi: 10.1098/rstb.2004.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol. 2009;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cimini D, Cameron LA, Salmon ED. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 16.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinett CC, et al. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 19.Janicke MA, Lasko L, Oldenbourg R, LaFountain JR., Jr Chromosome malorientations after meiosis II arrest cause nondisjunction. Mol Biol Cell. 2007;18:1645–1656. doi: 10.1091/mbc.E06-10-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torosantucci L, De Santis Puzzonia M, Cenciarelli C, Rens W, Degrassi F. Aneuploidy in mitosis of PtK1 cells is generated by random loss and nondisjunction of individual chromosomes. J Cell Sci. 2009;122:3455–3461. doi: 10.1242/jcs.047944. [DOI] [PubMed] [Google Scholar]
- 21.Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cassimeris L, Morabito J. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol Biol Cell. 2004;15:1580–1590. doi: 10.1091/mbc.E03-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganem NJ, Compton DA. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J Cell Biol. 2004;166:473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.