Abstract
Basal transcription of human mitochondrial DNA (mtDNA) in vitro requires the single-subunit, bacteriophage-related RNA polymerase, POLRMT, and transcription factor h-mtTFB2. This two-component system is activated differentially at mtDNA promoters by human mitochondrial transcription factor A (h-mtTFA). Mitochondrial ribosomal protein L7/L12 (MRPL12) binds directly to POLRMT, but whether it does so in the context of the ribosome or as a “free” protein in the matrix is unknown. Furthermore, existing evidence that MRPL12 activates mitochondrial transcription derives from overexpression studies in cultured cells and transcription experiments using crude mitochondrial lysates, precluding direct effects of MRPL12 on transcription to be assigned. Here, we report that depletion of MRPL12 from HeLa cells by shRNA results in decreased steady-state levels of mitochondrial transcripts, which are not accounted for by changes in RNA stability. We also show that a significant “free” pool of MRPL12 exists in human mitochondria not associated with ribosomes. “Free” MRPL12 binds selectively to POLRMT in vivo in a complex distinct from those containing h-mtTFB2. Finally, using a fully recombinant mitochondrial transcription system, we demonstrate that MRPL12 stimulates promoter-dependent and promoter-independent transcription directly in vitro. Based on these results, we propose that, when not associated with ribosomes, MRPL12 has a second function in transcription, perhaps acting to facilitate the transition from initiation to elongation. We speculate that this is one mechanism to coordinate mitochondrial ribosome biogenesis and transcription in human mitochondria, where transcription of rRNAs from the mtDNA presumably needs to be adjusted in accordance with the rate of import and assembly of the nucleus-encoded MRPs into ribosomes.
Keywords: TFAM, TFB2M, translation, oxidative phosphorylation, gene expression
Mitochondria are dynamic cellular organelles that contain DNA (mtDNA). In mammals, mtDNA is maternally inherited and essential because it encodes genes required for oxidative phosphorylation. For example, human mtDNA is a 16,569 base-pair, double-stranded, circular molecule harboring thirty-seven genes: thirteen mRNAs, two rRNAs, and twenty-two tRNAs (1). The remaining ∼1,500 proteins in mitochondria (2) are nuclear gene products that are imported into the organelle, including many required for expression and replication of mtDNA (3). As one example, the dedicated set of mitochondrial ribosomes (entirely distinct from those in the cytoplasm) that translates the mtDNA-encoded mRNAs comprise ∼80 mitochondrial ribosomal proteins (MRPs) that are all products of nuclear genes and imported into the mitochondrial matrix to associate with the two mtDNA-encoded rRNAs (4–7). Thus, ribosome biogenesis is unique in mitochondria in that the protein and RNA subunits are encoded by two genomes. How transcription and processing of the rRNAs in the matrix is coordinated with nuclear gene expression and import of the MRPs for ribosome assembly is largely unknown, but may be a unique control point for mitochondrial gene expression and/or overall mitochondrial biogenesis (8).
Based on in vitro studies, transcription of human mtDNA requires a core two-component system (9): a single-subunit RNA polymerase, POLRMT (10), and human mitochondrial transcription factor B2, h-mtTFB2 (or TFB2M) (11, 12). POLRMT, like its yeast homolog Rpo41p, is related to the bacteriophage T-odd family of RNA polymerases (10, 13), but has a large N-terminal extension that contains two pentatricopeptide repeats of unknown function (14). Relevant to this study, h-mtTFB2 is homologous to the bacterial KsgA family of rRNA methyltransferases, an ortholog of which was inherited from the original bacterial endosymbiont ancestor of mitochondria (15, 16). This core two-component system is activated by the HMG-box transcription factor, h-mtTFA (or TFAM) (17–19), that binds at the mtDNA promoters LSP and HSP1 and interacts with h-mtTFB2 (20). The role of h-mtTFA as a transcriptional activator is well documented, but it also has proposed roles in packaging (21–23) and repair (24) of mtDNA. A paralog of h-mtTFB2, h-mtTFB1 (or TFB1M), that was originally characterized as a mitochondrial transcription factor in vitro (11, 25), is the functional ortholog of the bacterial KsgA rRNA methyltransferase that dimethylates a conserved stem-loop in the mitochondrial 12S rRNA (26) and, hence, has a conserved function in mitochondrial ribosome biogenesis (27) and translation (8, 12, 28) in vivo. That h-mtTFB1 and h-mtTFB2 are derived from an rRNA methyltransferase, suggests a long evolutionary linkage between mitochondrial proteins involved in transcription, translation, and ribosome biogenesis.
Transcription and translation are functionally linked in yeast mitochondria, where Nam1p and Sls1p bind to the N-terminal domain of mitochondrial RNA polymerase and couple transcription and translation activities (14, 29–31), presumably by delivering newly synthesized mRNAs to translation factors and ribosomes associated with the inner mitochondrial membrane (32, 33). Whether these events occur cotranscriptionally in yeast mitochondria is not known and, because no homologs of Nam1p and Sls1p have been identified in mammals, evidence for similar coupling mechanisms in mammalian mitochondria has not surfaced.
The mammalian 55S mitochondrial ribosomes comprise associated large (39S) and small (28S) ribosomal subunits (4, 5). In addition to their rRNA subunits, the large and small ribosomal subunits are each composed of multiple MRPs. We identified previously that MRPL7/L12 (referred to as MRPL12 from this point forward) binds directly to POLRMT and modulates mitochondrial gene expression when overexpressed in HeLa cells (34), providing a functional link between mitochondrial ribosomes/translation and transcription in human mitochondria. Here we have investigated this link further and conclude, based on both in vivo and in vitro evidence, that it is a “free” pool of MRPL12 (i.e., not associated with assembled ribosomes) that interacts with POLRMT to directly activate mitochondrial transcription. These results provide significant insight into the mechanism of human mitochondrial gene expression, as well as the proposed dual-function nature of several MRPs.
Results
Human MRPL12 Positively Regulates Mitochondrial Transcription and Translation In Vivo, without Grossly Affecting Ribosome Biogenesis.
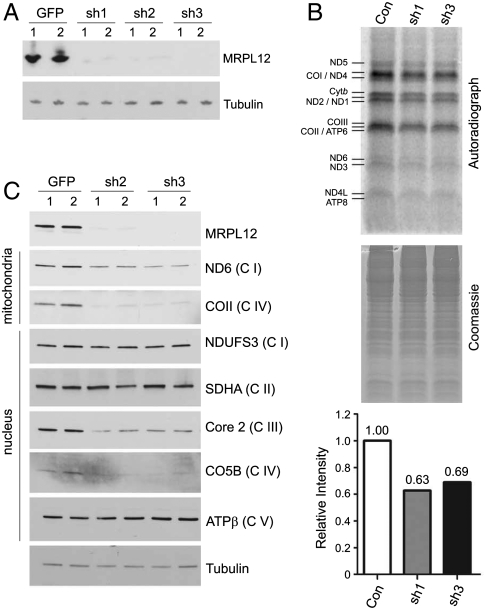
We showed previously that MRPL12 interacts with POLRMT in vitro and increases mitochondrial transcript levels when overexpressed in HeLa cells (34). To investigate further the role of MRPL12 in mitochondrial gene expression in vivo, we knocked it down using lentiviral-induced shRNA in HeLa and HEK293T cells. Western blot analysis revealed that three different shRNA constructs efficiently reduced MRPL12 compared to negative-control GFP shRNA-infected cells (Fig. 1A). As predicted from its essential role in ribosomes, the rate of mitochondrial translation was decreased globally in cells depleted of MRPL12 (Fig. 1B). Accordingly, the steady-state levels of mtDNA-encoded proteins (e.g., ND6 and COII) and some, but not all of the nuclear-encoded OXPHOS peptides were reduced (Fig. 1C). However, steady-state levels of several other MRPs analyzed were unchanged in MRPL12 knockdown cells 7 d after MRPL12 depletion (Fig. S1A). Moreover, sucrose density fractionation of mitochondrial ribosomes revealed that overall ribosome assembly was not significantly affected in MRPL12 knockdown cells compared to negative controls (Fig. S1B). These data, coupled with the known interaction with POLRMT, suggested to us that the observed effects of MRPL12 on mtDNA-encoded protein expression may involve not only its function in ribosomes and translation, but also a role for MRPL12 in transcription.
Fig. 1.
Analysis of mitochondrial translation rates and OXPHOS proteins in MRPL12-depleted cells. (A) Western blot analysis of MRPL12 protein in HeLa cells expressing MRPL12 shRNA (sh1-3) or GFP shRNA (GFP). (B) Labeling of mitochondrial translation products. Shown are an autoradiograph of the labeled mitochondrial peptides (top) and a Coomassie stained gel (middle). Radiolabeled mitochondrial proteins are indicated. Quantification results of combined band intensities in shMRPL12 lanes (sh1, 3) relative to control (Con) lane are shown at the bottom box. (C) Steady-state levels of indicated mtDNA-encoded (mitochondria) and nuclear-encoded (nucleus) OXPHOS subunits in shMRPL12 (sh2, 3) and shGFP cells. Corresponding OXPHOS complexes are labeled (C I–C V).
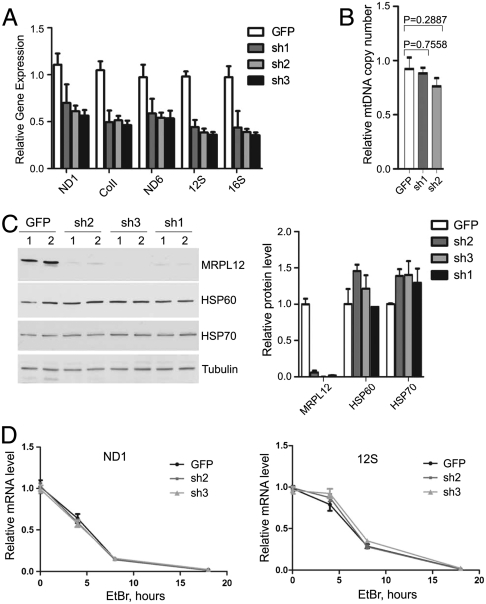
To examine this possibility, we next measured the steady-state level of mitochondrial transcripts in MRPL12 knockdown cell lines by quantitative real-time RT-PCR and observed a significant decrease in ND1, COII, and ND6 (representing mRNAs encoded on both strands of mtDNA) as well as 12S and 16S mitochondrial rRNAs (Fig. 2A). Similar results were obtained when MRPL12 was knocked down in a HeLa cell line that expresses HA-tagged POLRMT (Fig. S2 A and B). This effect was specific to depletion of MRPL12 because knockdown of another mitochondrial ribosomal protein, MRPS10, did not cause any significant changes in steady-state levels of mtDNA-encoded RNAs (Fig. S2C).
Fig. 2.
Analysis of mitochondrial gene expression, biogenesis, and mitochondrial transcript stability in MRPL12 knockdown cell lines. (A) Quantitative RT-PCR analysis of indicated mitochondrial transcripts in shMRPL12 (sh1-3) and shGFP cells. (B) Quantitative PCR measurement of mtDNA copy number in control (GFP) and shMRPL12 (sh1, 2) cells. P values from two-tailed unpaired student’s t-test are indicated. (C) Western blot analysis of MRPL12 knockdown (sh1-3) and shGFP control cells. Antibodies used for immunoblotting are indicated. Quantification of protein levels relative to tubulin loading control is shown. (D) Quantitative RT-PCR analysis of ND1 and 12S transcripts in control and shMRPL12 cells treated with ethidium bromide for 4, 8, or 16 h. For both transcripts, the level at time zero is set to one. In all boxes, results from three biological replicates are quantified and shown as the mean + /-standard deviation.
To address the basis for mtRNA reduction, mtDNA copy number, mitochondrial biogenesis, and transcript stability were analyzed in MRPL12 knockdown cells. Quantitative PCR analysis revealed that mtDNA copy number was not considerably changed between control and MRPL12 shRNA cells 7 d after viral infection (Fig. 2B, Fig. S2D). Moreover, levels of housekeeping mitochondrial proteins HSP60 and HSP70 relative to tubulin were unaffected in MRPL12-depleted cells (Fig. 2C, Fig. S2E), showing, in conjunction with the mtDNA copy number results, that overall mitochondrial biogenesis was not significantly affected. Finally, to assess whether MRPL12 affects RNA stability, mitochondrial transcription was inhibited with ethidium-bromide, and transcript decay rates were measured in control and MRPL12 knockdown cells. As expected, ethidium treatment inhibition led to a time-dependent decline in mtRNAs in all cell lines. However, despite the fact that MRPL12-knockdown cell lines start with less total mtRNA (Fig. 2A), the rates of transcript decay were identical in cells with or without MRPL12 knockdown (Fig. 2D, Fig. S3). These data demonstrate that MRPL12 does not affect transcript stability. Based on these results, we conclude that the ability of MRPL12 to regulate mitochondrial transcript abundance is most likely driven by a direct effect on mitochondrial transcription.
Human MRPL12 Stimulates Mitochondrial Transcription Directly In Vitro.
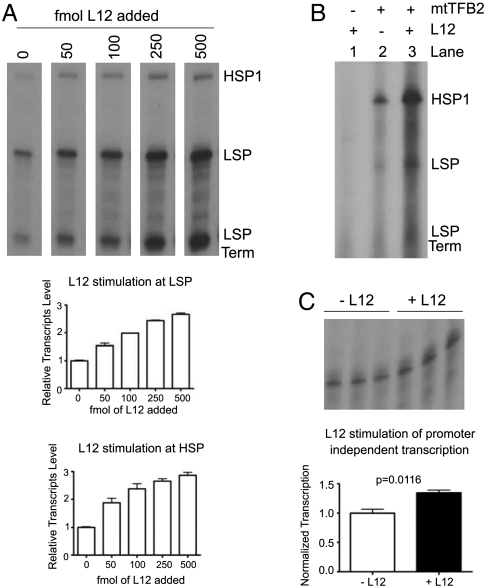
We have previously reported that MRPL12 stimulates mitochondrial transcription in vitro using partially purified, h-mtTFA-dependent mitochondrial extracts (34). To exclude potential caveats of other factors that may have been present in the crude lysates that mediated these effects of MRPL12 on mtDNA transcription, in vitro reactions were performed using a highly purified mitochondrial transcription system comprising recombinant POLRMT, h-mtTFB2, and h-mtTFA (35). As shown in Fig. 3A, MRPL12 activated transcription in this recombinant system in a dose-dependent manner, with transcription from both HSP1 and LSP being stimulated to a similar degree. Moreover, the h-mtTFA-independent transcription we described recently (9) (in the presence of POLRMT and h-mtTFB2 alone) was also stimulated by MRPL12 (Fig. 3B, compare lanes 2 and 3), while MRPL12 added to POLRMT alone yielded no activity (Fig. 3B, lane 1). We also tested the ability of MRPL12 to stimulate transcription in a promoter-independent fashion. Using a 3′-tailed DNA template that allows the RNA polymerase to bypass the initiation step of transcription, we observed modest, yet significant, stimulation in the presence of recombinant MRPL12 (Fig. 3C). Altogether, these data show that MRPL12 stimulates human mitochondrial transcription directly, possibly at multiple steps in the process, and its ability to do so does not require h-mtTFA.
Fig. 3.
Stimulation of promoter-dependent and promoter-independent mitochondrial transcription with recombinant MRPL12. (A) Representative in vitro transcription reaction in the presence of increasing amounts of recombinant MRPL12 (L12; 0-500 fmol). The HSP1, LSP, and LSP termination transcripts are indicated. (B) Promoter-specific h-mtTFA-independent in vitro transcription reaction. Shown is a representative reaction in the presence of POLRMT + /-MRPL12 (L12) and h-mtTFB2. (C) Promoter-independent in vitro transcription assay containing POLRMT alone (-L12) or POLRMT and MRPL12 (+L12). In (A and C), quantification of relative transcript level (mean + /-standard deviation of three independent experiments of the full-length, run-off product) is shown.
MRPL12 Associates with POLRMT in Complexes Distinct from Those Containing h-mtTFB2.
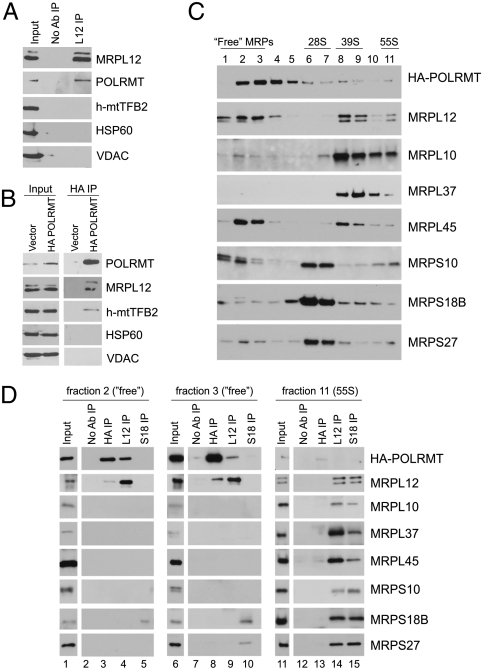
We have previously shown that POLRMT interacts with FLAG epitope-tagged MRPL12 in HeLa cells (34). To test whether MRPL12 interacts with h-mtTFB2, the other core component of the mitochondrial transcription initiation machinery, endogenous MRPL12 was immunoprecipitated (IP) from HeLa whole-cell extracts, and coprecipitated proteins were analyzed by Western blotting (Fig. 4A). Cells were crosslinked with DSP (dithiobis[succinimidylpropionate]) prior to lysis and IP to detect less stable or transient protein interactions. The specificity of the crosslinking and IP conditions was demonstrated by the absence of HSP60 and voltage-dependent anion channel (VDAC) (Fig. 4A), abundant mitochondrial matrix and outer membrane proteins, respectively. In agreement with previously published results, POLRMT was detected in MRPL12 IP (Fig. 4A). In contrast, h-mtTFB2 was not associated with MRPL12 (Fig. 4A), demonstrating that MRPL12 does not interact with the entire core transcription machinery, but rather POLRMT specifically.
Fig. 4.
Identification of a “free” pool of MRPL12 that selectively interacts with POLRMT. (A) Immunoblotting of proteins that co-IP with MRPL12 from HeLa whole-cell extracts. IP with no antibody added (No Ab IP) was used as a negative control. (B) Western blot analysis of proteins recovered in HA antibody IP (HA IP) from HeLA cell expressing HA-tagged POLRMT (HA-POLRMT) or empty vector as a negative control (vector). (C) Sedimentation of HA-POLRMT-expressing HeLa cell extracts through 10–30% sucrose gradients followed by Western blot analysis. Fraction numbers (1-lightest, 11-heaviest) are shown. Fractions enriched for “free” MRPs, small (28S) and large (39S) ribosomal subunits, and fully assembled ribosomes (55S) are underlined. (D) Western blot analysis of proteins that co-IP with HA-POLRMT, MRPL12 and MRPS18B antibodies. Fractions 2, 3, and 11 from (C) were used for pull-down (input). IP with no antibody added (No Ab IP) was used as a negative control. In all boxes, antibodies used for Western blot analysis are indicated.
Because available antibodies against endogenous POLRMT protein do not work in IP reactions in our hands, HeLa cells expressing HA epitope-tagged POLRMT were used for reciprocal POLRMT pull-down (Fig. 4B). Again, the specificity of the crosslinking and IP procedures was confirmed by the absence of the abundant mitochondrial markers, HSP60 and VDAC, in the pull-down fraction (Fig. 4B). Similar to h-mtTFB2, which is expected to co-IP with POLRMT as part of the transcription initiation complex, MRPL12 was found in HA-IP from cells expressing HA-POLRMT but not from negative-control cells expressing empty vector (Fig. 4B). Because only POLRMT is precipitated with MRPL12, while both MRPL12 and h-mtTFB2 are pulled-down with POLRMT, we conclude that MRPL12 and h-mtTFB2 bind POLRMT in distinct protein complexes.
A “Free” Pool of MRPL12 in the Matrix Interacts with POLRMT.
Given that MRPL12 is a known component of ribosomes, we next ascertained whether it activates transcription in this context or as a “free” (i.e., nonribosome associated) protein. Certainly the ability of recombinant MRPL12 alone to stimulate mitochondrial transcription in vitro implied that ribosome association is not absolutely required. To address this more directly in vivo, we determined whether there is a “free” pool of MRPL12 in the matrix to bind POLRMT. Extracts from HeLa cells expressing HA-tagged POLRMT were subjected to sedimentation through 10–30% continuous sucrose gradients. Western blot analysis of sucrose gradient fractions with antibodies against various mitochondrial ribosomal proteins revealed that small (28S) and large (39S) mitochondrial ribosomal subunits, as well as a fully assembled (55S: associated 28S and 39S) ribosomes were successfully separated (Fig. 4C). Notably, a substantial proportion of mitochondrial ribosomal proteins, including MRPL12, was found in the lowest density fractions (1–3), suggesting existence of a considerable “free” pool of these proteins (Fig. 4C).
To verify that “free” MRPs are present in lower sucrose density fractions, fractions 2, 3, and 11 were subjected to IP with MRPL12 and MRPS18B antibodies. As expected, interaction of both MRPL12 and MRPS18B with other MRPs from both the small and large ribosomal subunits was detected in fraction 11, which contains fully assembled mitochondrial ribosomes (Fig. 4D, lanes 14 and 15). MRPS18B IP from fractions 2 and 3 yielded none of the large subunit MRPs. However, another small ribosomal subunit protein MRPS27 was pulled-down in fraction 3, suggesting that these proteins cosediment in lower sucrose density fractions, likely as part of an intermediate in ribosome assembly or disassembly (Fig. 4D, lane 10). Importantly, none of the analyzed MRPs were detected in MRPL12 IP from fractions 2 or 3 (Fig. 4D, lanes 4 and 9), including MRPL10 protein, a direct binding partner of MRPL12 in the ribosome (36). In contrast, a strong HA-POLRMT signal was detected in Western blot analysis of proteins that co-IP with MRPL12 in both fractions 2 and 3 (Fig. 4D, lanes 4 and 9). Reciprocal IP of HA-POLRMT pulled-down MRPL12 from both fractions 2 and 3 (Fig. 4D, lanes 3 and 8). These data demonstrate clearly that there is a pool of “free” MRPL12 that interacts with POLRMT independently of ribosomes, consistent with a role in activating transcription as proposed.
Discussion
In this study, we have elucidated important mechanistic aspects of mitochondrial gene expression by demonstrating that MRPL12 has a second function outside of ribosomes in regulating mitochondrial transcription. The main conclusions we draw from our results are that (i) there is a significant pool of non-ribosome-associated (i.e., “free”) MRPL12 in human mitochondria, (ii) that it is “free” MRPL12 that interacts physically with POLRMT in vivo in a protein complex distinct from those containing h-mtTFB2, and (iii) that MRPL12 can promote mitochondrial transcription in vivo and activate it in vitro in a promoter-dependent and promoter-independent fashion. The basis for these conclusions is discussed below, as is the broader significance of the results with regard to control of human mitochondrial gene expression and function, and the dual-function nature of MRPL12 and other MRPs.
In addition to MRPL12 comigrating with large (39S) ribosomal subunit and fully assembled (55S) mitochondrial ribosomes, it is also reproducibly detected in a “free” pool, not associated with ribosomes (Fig. 4C) that interacts with POLRMT (Fig. 4D). That the POLRMT interacts with “free” MRPL12 selectively is supported by the fact that none of the other “free” MRPs in that fraction that we tested interacts with POLRMT (Fig. 4D), including MRPL10, a protein that is required for binding of the bacterial ortholog of MRPL12 to the ribosome (36). Based on both in vivo (Fig. 2, Fig. S2) and in vitro (Fig. 3) results presented herein, we conclude that “free” MRPL12 acts as a transcription factor that interacts directly with POLRMT to activate transcription. With regard to this function, MRPL12 stimulates both h-mtTFA-dependent and recently identified h-mtTFA–independent transcription initiation (9), demonstrating that h-mtTFA is not critical for the stimulatory activity of MRPL12. Because MRPL12 stimulates both promoter-specific and promoter-independent transcription in vitro (Fig. 3), it may have roles in both initiation and elongation or, perhaps most likely, aiding RNA polymerase transition between these two steps during the transcription cycle. With regard to the latter possibility, our finding that MRPL12 is not detected in POLRMT complexes containing the initiation factor h-mtTFB2 in vivo (Fig. 4 A and B) is consistent with it associating with RNA polymerase at a point in the initiation process after h-mtTFB2 has exited the complex (e.g., during the transition from initiation to elongation). This likely occurs soon after a few nucleotides are synthesized based on studies of the yeast mitochondrial transcription system (37). It is noteworthy that the only known transcriptional regulator of the related bacteriophage T7 RNA polymerase is T7 lysozyme, which is an allosteric inhibitor that impedes the transition of the initiation complex to the elongation complex (38). Given that POLRMT is a T7-related RNA polymerase (10, 13), it is tempting to speculate that MRPL12 may function similarly, but as an allosteric activator.
Accumulation of non-ribosome-associated L12 protein has been demonstrated in bacteria (39). Notably, bacterial “free” L12 controls ribosome biogenesis and coordinates the relative expression of ribosomal proteins and rRNA via binding to RNA sequences in its own operon and regulating protein expression by a negative feedback mechanism (40). We speculate that a somewhat analogous function of MRPL12 in mitochondrial ribosome biogenesis regulation is occurring in human mitochondria that involves the direct role of MRPL12 in mitochondrial transcription we have elucidated herein. Mitochondrial ribosomes are quite distinct from their bacterial and eukaryotic cytoplasmic counterparts, having smaller rRNA species and an apparently compensatory increase in protein content (5). The enhanced protein content is due, in part, to the presence of unique ribosomal proteins in mitochondrial ribosomes, as well as the acquisition of new domains in evolutionarily conserved proteins found in all ribosomes. From this evolutionary trend, other functions, and in some cases alternate cellular localizations, of certain MRPs have arisen. In addition to the proposed second function of free MRPL12 in transcriptional regulation discussed herein, additional activities of several MRPs have been described, including regulation of apoptosis and the cell cycle (3). It is tempting to speculate that the dynamics of mitochondrial ribosome biogenesis is being used as a critical cellular readout for overall mitochondrial or cell homeostasis. For example, inhibition of mitochondrial ribosome biogenesis, which could occur downstream of defective mtDNA expression of the rRNAs and/or defective nuclear expression or import of MRPs, could be signaled as an increase in the free pools of MRPs in the matrix. As free entities, these MRPs could, in principle, now be available to perform their alternative, nonribosomal functions. In the case of MRPL12, a new function is to bind POLRMT to increase transcription so that more rRNAs are produced to promote new ribosome biogenesis. While, in the case of MRPS29/DAP3 (41, 42) or MRPS30/PDCD9 (41), this alternative function could be to promote apoptosis should mitochondrial function decline to critical levels that inhibit normal cell function. These, of course, are speculative scenarios at this point, but worthy of consideration going forward. Our results provide an exciting platform for future studies aimed at understanding mitochondrial transcription, translation and ribosome biogenesis, and the interrelationships and relevance of these processes in vivo.
Materials and Methods
Generation of MRPL12 and MRPS10 Knockdown Cells and Cells Expressing HA-Tagged POLRMT.
Three different MRPL12 target sequences (sh1: 5′ GCCTCACTCTCTTGGAAAT 3′; sh2: 5′ TCAACGAGCTCCTGAAGAAA 3′; sh3: 5′ AACGTTGAAGATCCAGGATGT 3′) and an MRPS10 target sequence (5′ CCTGTGGTAACAATCTCTGAT 3′) were cloned into pLKO.1 vector, and lentiviral particles were packaged using 293FT cells (Invitrogen). Bulk infected HeLa cells were selected with 0.5 μg/mL of puromycin without clonal selection. All MRPL12 knockdown experiments described in the manuscript were performed with cells 7 d after viral infection. For HA-POLRMT cloning, an HA-epitope tag was inserted in POLRMT ORF between codons 41 and 42 downstream of mitochondrial localization sequence. HA-tagged POLRMT cDNA was ligated into pTRE2 vector (Invitrogen) and cotransfected with the plasmid pHyg (BD Biosciences) into HeLa Tet-On cells (Invitrogen). Hygromycin-resistant clones were selected and checked for doxycycline (dox)-inducible expression of HA-POLRMT that could be detected in mitochondrial fractions. Treatment of cells with dox (2 μg/mL) for 3 d was used for induction of HA-POLRMT expression.
Antibodies.
All antibodies used in this study are listed in Table S1.
Quantitative RT-PCR, mtDNA Copy Number Measurement, and EtBr-Mediated Transcription Inhibition.
For quantitative RT-PCR, 1.5–2 μg of total cellular RNA extracted with RNeasy Plus Mini kit (Qiagen) was used for reverse transcription with High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Gene expression was analyzed on a BioRad iCycler using iQ SYBR Green Supermix (BioRad), normalized to GAPDH cDNA levels, and calculated as a relative gene expression using 2-ΔΔCt method. Primers used for PCR are listed in Table S2. The copy number of mtDNA was measured as previously described (12). Unpaired two-tailed student’s t-test was used for statistical analysis of quantitative PCR results. For inhibition of mitochondrial transcription, cells were treated with 0.04 μg of EtBr per 1 mL of media.
DSP Crosslinking and Sucrose Density Fractionation.
DSP was purchased from Thermo Scientific and reconstituted with anhydrous DMSO (Sigma). Cells were washed with PBS and crosslinked with 1 mM DSP in PBS for 30 min at 4 °C. Crosslinking was quenched with 100 mM glycine for 15 min at 4 °C, and cells were washed again with PBS prior to cell lysis. For sucrose density fractionation, whole-cell extracts were prepared with WCE buffer [40 mM Tris HCl pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM EdTA, 1% NP40, 1× protease inhibitor cocktail (Roche), 1 mM PMSF]. Whole-cell extracts (3–10 mg) were loaded onto 10 mL of 10–30% linear sucrose gradient and centrifuged for 17.5 h at 80,000 × g. Fractions (1 mL) were collected and 15 μL aliquots were analyzed by immunoblotting. Gradients were prepared in 14 × 89 mm Beckman centrifuge tubes in the buffer described in (43).
Immunoprecipitation.
For IP, cell lysates (1–2 mg) in WCE buffer or 750 μL of sucrose gradient fractions diluted 1∶3 with WCE buffer were incubated with antibodies and protein G agarose (Pierce) overnight at 4 °C. Beads were then washed four times with WCE buffer, resuspended in SDS loading buffer (100 mM Tris HCl pH 6.8, 2% SDS, 15% glycerol, 5% β-mercaptoethanol, 0.1% bromophenol blue), boiled for 10 min, and analyzed by Western blotting.
Mitochondrial Run-Off Transcription Assays and Labeling of Mitochondrial Translation Products.
Recombinant MRPL12 was expressed in Escherichia coli and purified as described previously (34). Mitochondrial run-off transcription assays were performed as described (9). For promoter-independent transcription assay, linear DNA template was generated by annealing primers PITA-45 (5′ctagcctttctattagctcttagtaagattacacatgcaagcatc3′) and PITA-60 (5′gatgcttgcatgtgtaatcttactaagagctaatagaaaggctagccccccccccccccc 3′). Template, POLRMT, and MRPL12 were used in 3.4 nM, 16 nM, and 32 nM concentrations, respectively. The products were separated on 15%/7 M urea gels. Labeling of mitochondrial translation products was performed as described (44).
Supplementary Material
Acknowledgments.
The authors thank Vicki McCulloch, Bonnie Seidel-Rogol, and Hong Zhang for their contributions toward creating and characterizing the HA-POLRMT cell lines, Marcus Cooper for sharing ND1, ND6, and COII Q-PCR primer sequences, and Shadel lab members for helpful suggestions and for critically reading the manuscript. This work was supported by National Institutes of Health (NIH) grants R01 HL-059655 to G.S.S and National Research Service Award F32 GM093590-01 (to Y.V.S.) T.E.S. was supported by a postdoctoral fellowship from the United Mitochondrial Disease Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108852108/-/DCSupplemental.
References
- 1.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genom Hum G. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shutt TE, Shadel GS. A compendium of human mitochondrial gene expression machinery with links to disease. Environ Mol Mutagen. 2010;51:360–379. doi: 10.1002/em.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Brien TW. Properties of human mitochondrial ribosomes. IUBMB Life. 2003;55:505–513. doi: 10.1080/15216540310001626610. [DOI] [PubMed] [Google Scholar]
- 5.Sharma MR, et al. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 6.Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem. 2001;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- 7.Koc EC, et al. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem. 2001;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 8.Cotney J, McKay SE, Shadel GS. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum Mol Genet. 2009;18:2670–2682. doi: 10.1093/hmg/ddp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shutt TE, Lodeiro MF, Cotney J, Cameron CE, Shadel GS. Core human mitochondrial transcription apparatus is a regulated two-component system in vitro. Proc Natl Acad Sci USA. 2010;107:12133–12138. doi: 10.1073/pnas.0910581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiranti V, et al. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum Mol Genet. 1997;6:615–625. doi: 10.1093/hmg/6.4.615. [DOI] [PubMed] [Google Scholar]
- 11.Falkenberg M, et al. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 12.Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masters BS, Stohl LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 14.Rodeheffer MS, Boone BE, Bryan AC, Shadel GS. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J Biol Chem. 2001;276:8616–8622. doi: 10.1074/jbc.M009901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotney J, Shadel GS. Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J Mol Evol. 2006;63:707–717. doi: 10.1007/s00239-006-0075-1. [DOI] [PubMed] [Google Scholar]
- 16.Shutt TE, Gray MW. Homologs of mitochondrial transcription factor B, sparsely distributed within the eukaryotic radiation, are likely derived from the dimethyladenosine methyltransferase of the mitochondrial endosymbiont. Mol Biol Evol. 2006;23:1169–1179. doi: 10.1093/molbev/msk001. [DOI] [PubMed] [Google Scholar]
- 17.Fisher RP, Clayton DA. Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 19.Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J Mol Biol. 1995;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- 20.McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol Cell Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ekstrand MI, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 22.Kanki T, et al. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufman BA, et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canugovi C, et al. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair (Amst) 2010;9:1080–1089. doi: 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCulloch V, Seidel-Rogol BL, Shadel GS. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol Cell Biol. 2002;22:1116–1125. doi: 10.1128/MCB.22.4.1116-1125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- 27.Metodiev MD, et al. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Matsushima Y, Adan C, Garesse R, Kaguni LS. Drosophila mitochondrial transcription factor B1 modulates mitochondrial translation but not transcription or DNA copy number in Schneider cells. J Biol Chem. 2005;280:16815–16820. doi: 10.1074/jbc.M500569200. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Shadel GS. Stability of the mitochondrial genome requires an amino-terminal domain of yeast mitochondrial RNA polymerase. Proc Natl Acad Sci USA. 1999;96:8046–8051. doi: 10.1073/pnas.96.14.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryan AC, Rodeheffer MS, Wearn CM, Shadel GS. Sls1p is a membrane-bound regulator of transcription-coupled processes involved in Saccharomyces cerevisiae mitochondrial gene expression. Genetics. 2002;160:75–82. doi: 10.1093/genetics/160.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodeheffer MS, Shadel GS. Multiple interactions involving the amino-terminal domain of yeast mtRNA polymerase determine the efficiency of mitochondrial protein synthesis. J Biol Chem. 2003;278:18695–18701. doi: 10.1074/jbc.M301399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naithani S, Saracco SA, Butler CA, Fox TD. Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:324–333. doi: 10.1091/mbc.E02-08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shadel GS. Coupling the mitochondrial transcription machinery to human disease. Trends Genet. 2004;20:513–519. doi: 10.1016/j.tig.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J Biol Chem. 2007;282:12610–12618. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lodeiro MF, et al. Identification of multiple rate-limiting steps during the human mitochondrial transcription cycle in vitro. J Biol Chem. 2010;285:16387–16402. doi: 10.1074/jbc.M109.092676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Highland JH, Howard GA. Assembly of ribosomal proteins L7, L10, L11, and L12, on the 50 S subunit of Escherichia coli. J Biol Chem. 1975;250:813–814. [PubMed] [Google Scholar]
- 37.Mangus DA, Jang SH, Jaehning JA. Release of the yeast mitochondrial RNA polymerase specificity factor from transcription complexes. J Biol Chem. 1994;269:26568–26574. [PubMed] [Google Scholar]
- 38.Jeruzalmi D, Steitz TA. Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme. EMBO J. 1998;17:4101–4113. doi: 10.1093/emboj/17.14.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramagopal S. Accumulation of free ribosomal proteins S1, L7, and L12 in Escherichia coli. Eur J Biochem. 1976;69:289–297. doi: 10.1111/j.1432-1033.1976.tb10885.x. [DOI] [PubMed] [Google Scholar]
- 40.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 41.Cavdar Koc E, et al. A new face on apoptosis: death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Lett. 2001;492:166–170. doi: 10.1016/s0014-5793(01)02250-5. [DOI] [PubMed] [Google Scholar]
- 42.Hulkko SM, Zilliacus J. Functional interaction between the pro-apoptotic DAP3 and the glucocorticoid receptor. Biochem Biophys Res Commun. 2002;295:749–755. doi: 10.1016/s0006-291x(02)00713-1. [DOI] [PubMed] [Google Scholar]
- 43.Dennerlein S, Rozanska A, Wydro M, Chrzanowska-Lightowlers ZM, Lightowlers RN. Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem J. 2010;430:551–558. doi: 10.1042/BJ20100757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chomyn A. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 1996;264:197–211. doi: 10.1016/s0076-6879(96)64020-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.