Abstract
Phenotypic diversification of cells is crucial for developmental and regenerative processes in multicellular organisms. The diversification concept is described as the motion of marbles rolling down Waddington’s landscape, in which the number of stable states changes as development proceeds. In contrast to this simple concept, the complexity of natural biomolecular processes prevents comprehension of their design principles. We have constructed, in Escherichia coli, a synthetic circuit with just four genes, which programs cells to autonomously diversify as the motion on the landscape through cell–cell communication. The circuit design was based on the combination of a bistable toggle switch with an intercellular signaling system. The cells with the circuit diversified into two distinct cell states, “high” and “low,” in vivo and in silico, when all of the cells started from the low state. The synthetic diversification was affected by not only the shape of the landscape determined by the circuit design, which includes the synthesis rate of the signaling molecule, but also the number of cells in the experiments. This cell-number dependency is reminiscent of the “community effect”: The fates of developing cells are determined by their number. Our synthetic circuit could be a model system for studying diversification and differentiation in higher organisms. Prospectively, further integrations of our circuit with different cellular functions will provide unique tools for directing cell fates on the population level in tissue engineering.
Keywords: biological engineering, mathematical modeling, synthetic biology
Phenotypic diversification of cells in developmental and regenerative processes is conceptually modeled as the motion of marbles rolling down Waddington’s landscape (1). The simple concept of the landscape is helpful to interpret the dynamic phenotypic changes in natural phenotypic diversification from bacteria to mammalian cells (2). In contrast, the complex interactions of genes governing the natural diversification prevent the elucidation of its design principles, which are essential characteristics of phenomena, despite a wealth of knowledge on the individual genes governing the processes.
Because the landscape contains bifurcations, which are changes in the numbers of stable states, developing cells on the landscape differentiate into various cell states through developmental progression. Reprogramming of differentiated cells to pluripotent cells (3–5) revealed that the progression results from changes in the expression status of genes in individual cells, rather than simply the passage of time. For changes in the expression status, intercellular signaling cannot be ignored, because gene expression is regulated not only by genes in individual cells, but also by cell–cell communication. Such communication is also known to be important in cellular decision making in bacteria (2, 6).
To investigate the design principles for intricate natural phenomena, simple synthetic gene circuits that emulate these phenomena through the integration of theory and experiment (2, 7–15) can be efficient tools, regardless of whether the molecules used in the synthetic gene circuits are the same as those used in the natural circuits (15) or not (7–14, 16). The construction of a genetic toggle switch (8), a mutual inhibitory circuit that exhibited bistability in silico and in vivo, was an important milestone for the integration. The genetic toggle switch revealed that the circuits with balanced and imbalanced protein-synthesis rates resulted in bistability and monostability, respectively. Another example of the integration is the study of a population-control circuit (9), which includes intercellular signaling. This study demonstrated that population dynamics in vivo can be tuned by varying the stability of the intercellular signaling molecule, as predicted by numerical simulation. Other synthetic circuits with intercellular signaling systems have also been constructed to program cells to work as a population (9–12, 14, 17, 18). We expected that emulating the landscape with intercellular signaling would help to clarify the importance of cell–cell communication in the natural phenotypic diversification.
In this study, using an Escherichia coli population, we implemented a synthetic phenotypic diversification as the motion on the landscape, of which the bifurcation is mediated by the concentration of an intercellular signaling molecule autonomously produced by the cells (Fig. 1A). To implement the synthetic diversification, a diversity-generator circuit was designed with only four genes, based on the combination of a bistable toggle switch (8) with an intercellular signaling system (Fig. 1B). The Escherichia coli population diversified in silico and in vivo into two distinct cell states: high and low, when the cells were initialized to the low state. The phenotypic balance after the synthetic diversification primarily depended on the velocity of the signaling-molecule accumulation, which was mainly determined by the cell density and the rates of the signaling-molecule production in individual cells. This cell-number dependency with the landscape concept has implications for investigations and applications of developmental and regenerative processes in which the cell number is important. Thus, our simple circuit could be a model system to provide insights into fundamental processes of development and regeneration.
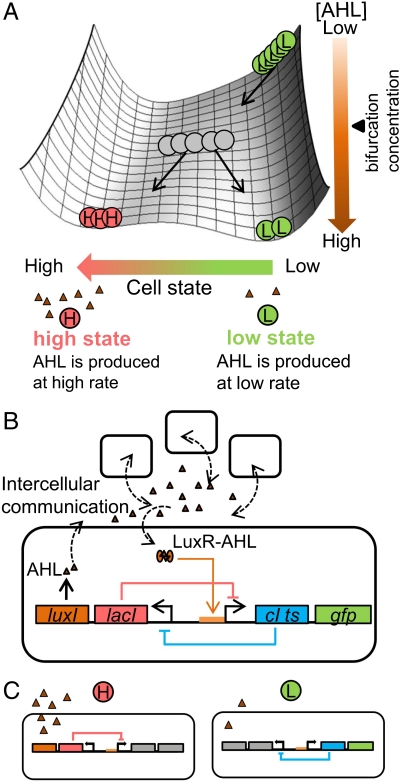
Fig. 1.
The landscape to conceptually describe the behavior of the synthetic diversification system and the network diagram of the synthetic circuit for the diversification. (A) Landscape. The horizontal axis to the landscape represents the cell state indicating the cellular concentration of LacI. The vertical axis to the landscape represents the AHL concentration. (B) Network diagram. CIts and LacI mutually inhibit each other. The Plux/lac promoter, which regulates CIts, can act as a Plac promoter with sufficient AHL. (C) The high-state cells (Left) produce AHL at a much higher rate than the low-state cells (Right).
Results
Design of the Diversity Generator to Implement the Synthetic Diversification.
The diversity-generator design includes an intercellular signaling mechanism and an intracellular bistability mechanism (Fig. 1B). The intercellular signaling mechanism was developed by employing the quorum sensing mechanism of Vibrio fischeri (19). The intracellular bistability mechanism was implemented by mutual inhibition in the genetic toggle switch (8) (Table S1). The diversity generator has two key promoters, the PL-1 con promoter and the Plux/lac promoter (the detail of the Plux/lac promoter is defined in Fig. S1); the PL-1 con promoter is repressed by the CI temperature-sensitive repressor protein (CIts) transcribed from the Plux/lac promoter, and the Plux/lac promoter is repressed by LacI transcribed from the PL-1 con promoter. In addition, the Plux/lac promoter does not work in the absence of acyl-homoserine lactone (AHL), which is enzymatically synthesized by LuxI. The LuxI and GFPmut3 coding sequences are arranged as the second cistrons downstream of PL-1 con and Plux/lac, respectively. We defined the high state as the state where LacI and LuxI are dominant, and the low state as that where CIts and GFPmut3 are dominant. Thus, the high-state cells produce AHL at a much higher rate than the low-state cells (Fig. 1C). The two cell states are both stable under conditions with sufficient AHL, but only the high state is stable in the absence of sufficient AHL. If all of the diversity-generator cells start from the low state in the absence of AHL, then as a consequence of the bifurcation with the increase in the AHL concentration, the cell population will finally exhibit a bimodal distribution with the low and high states (Fig. 1A).
In Silico Evaluation of the Diversity-Generator Design.
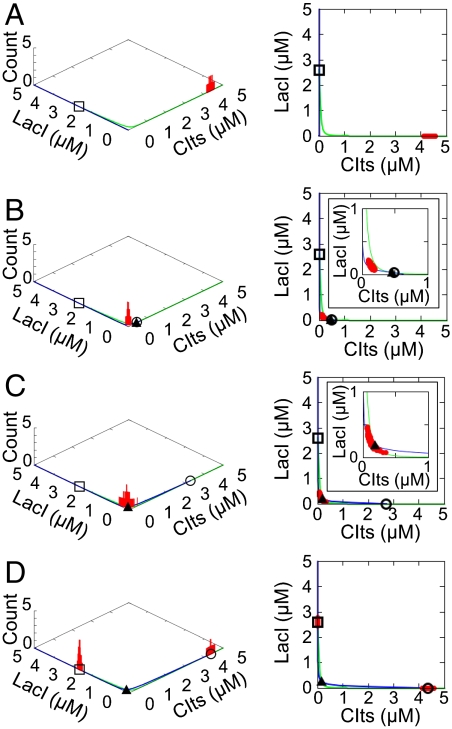
We built a stochastic model on the basis of simple biological reactions. Fig. 2 shows the dynamic behavior of the cell population with the nullclines in the two-dimensional protein concentration space. These figures also include unstable and stable steady states at each time point. All parameters used for Fig. 2 are listed in Table S2. When all of the diversity-generator cells were set in the low state and the AHL concentration in the medium was zero at the initial time (Fig. 2A), the cells started the transition to the high state. During the transition, the AHL production from the cells gradually increased. Sufficient accumulation of AHL caused the bifurcation, so that the high stable and unstable steady states appeared. The unstable steady state indicates a point on the separatrix between the basins of attraction for the high and low stable steady states (Fig. 2B). The position of the unstable steady state depends on the AHL concentration. With further AHL accumulation, the position asymptotically approached the point of the unstable steady state at the plateau concentration of AHL (Fig. 2C). Here, if the cells were distributed in both of the basins, then the population finally exhibited a bimodal distribution (Fig. 2D). On the other hand, the cell population distributed in only the low basin finally exhibited a monomodal distribution with the low state (Figs. S2A and S3A). Another monomodal distribution with the high state was observed from the cell population distributed in only the high basin (Figs. S2C and S3C). The velocity of AHL accumulation depends on two parameters: the initial cell density and the AHL-production rate of single cells. Therefore, appropriate combinations of these two parameters are required for the diversification.
Fig. 2.
Numerical predictions of the dynamic behavior in the diversity-generator cells. The cell-population histograms and the nullclines (Left), with their two-dimensional projections (Right). Blue and green lines represent nullclines of ordinary differential equations for [CIts] and [LacI], respectively. The open squares, open circles, and closed triangles indicate the high stable, low stable, and unstable steady states, respectively. Insets are enlarged views around the unstable steady state. (A) Cells are set in the low state, and the AHL concentration in the medium is zero. (B) Accumulation of AHL causes the bifurcation, so that the high stable and unstable steady states appear. (C) The position of the unstable steady state is in the cell population. (D) The cell population finally exhibits a bimodal distribution. All parameters used for the simulation are listed in Table S2.
Construction of the Diversity Generator.
To experimentally confirm the AHL-mediated bifurcation of the diversity generator, we constructed a pHT-toggle plasmid carrying a subcircuit of the diversity generator, by replacing the Ptrc-2 promoter on the genetic toggle switch pTAK132 (8) with the Plux/lac promoter (Fig. S1A). The HT-toggle cells can be initialized to either state by appropriate culture conditions. Both states were confirmed to be stable in the presence of AHL, by the fact that the addition of AHL just after the initializations resulted in minimal changes of the cell state from each initial state (Fig. S4). In addition to this bistability in the presence of AHL, monostability with the high state in the absence of AHL was confirmed by observations that the low-state cells transit to the high state, and the high-state cells maintain their state (Fig. S4). By changing the timing of the AHL addition, the pHT toggle provided additional experimental evidence that the diversification via the bifurcation with the increase in the AHL concentration could work (Fig. S5). During the above transition of the low-state cells in the absence of AHL, the addition of AHL made both states stable. Thus, part of the population returned to the low state and the rest transited to the high state when the timing of the AHL addition was appropriate (Fig. S5A), such as at 120 min in the experiments (Fig. S5B). As a result, the cell population finally exhibited a bimodal distribution. In contrast, early AHL addition, such as 0 min, resulted in a monomodal distribution with the low state. Another monomodal distribution with the high state was observed with later AHL addition, such as at 240 min.
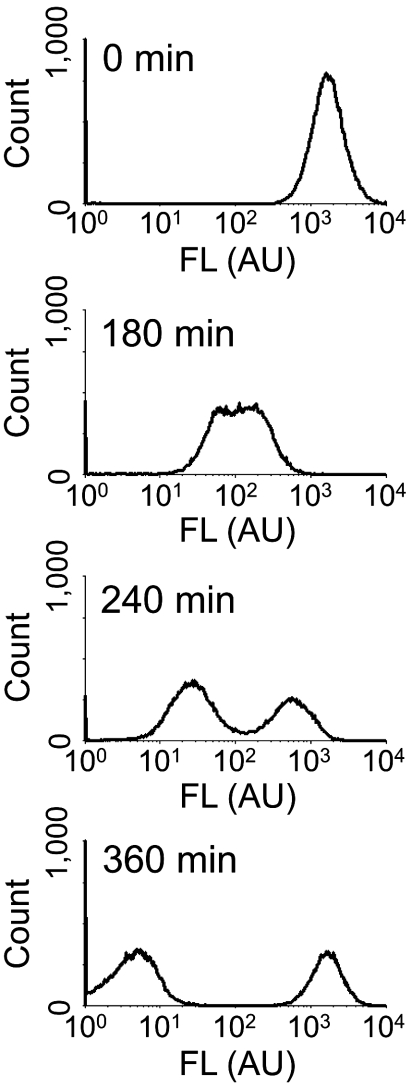
For autonomous signal production in our diversification, we placed a luxI variant, designed from known variants (20), downstream of the LacI-coding sequence on pHT toggle to obtain the diversity-generator plasmid, pHT_luxI1.5C (Fig. S1A). Similar to the HT-toggle cells, the HT_luxI1.5C cells can be initialized to either state. Because the HT_luxI1.5C cells can produce an appropriate amount of AHL by themselves, they autonomously diversified, as defined in Fig. 1A (Fig. 3), after the initialization to the low state, with high GFP fluorescence. Through the gradual decline of fluorescence, this monomodal distribution at 0 min turned into a monomodal distribution with midrange fluorescence at 180 min, at which the cell population started to divide. The cell population then exhibited a bimodal distribution at 240 min. After 360 min of incubation from the initialization, the cell population finally exhibited another bimodal distribution, in which the distance between the two fluorescent peaks was greater than that at 240 min.
Fig. 3.
Fluorescence histograms of HT_luxI1.5C cells at various time points. Cells with HT_luxI1.5C initialized to the low-state diversify into the high and low states. FL indicates the intensity of GFP fluorescence.
Dependency of the Synthetic Diversification on the Velocity of AHL Accumulation.
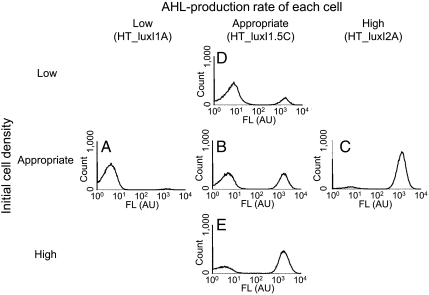
Based on the notion of the landscape (Fig. 1A) and the numerical simulations (Fig. 2, Figs. S2 and S3, and Tables S3 and S4), the velocity of AHL accumulation in the medium is important for the synthetic diversification. The velocity depends on the AHL-synthesis rate of single cells and the cell density. To experimentally confirm the importance of the velocity, we monitored the behavior after changing the AHL-synthesis rate of single cells. To change the rate, we constructed the pHT_luxI1A and pHT_luxI2A plasmids, which produce less and more AHL than the pHT_luxI1.5C plasmid, respectively (Table S3). The GFP expression histograms of the cells with each of the three plasmids are shown in Fig. 4 A–C. The low and high AHL-production rates led to monomodal cell distributions with the high state (Fig. 4A) and the low state (Fig. 4C), respectively. To confirm the importance of the velocity, we also monitored the behavior of the diversity-generator cells by preparing the initial cell cultures with a high, appropriate, or low density (Table S4). The fluorescence histograms of the HT_luxI1.5C cells, after the initialization and incubation under the three cell-density conditions, are shown in Fig. 4 D, B, and E. Similar to the AHL-synthesis rate of single cells, the difference in the cell-density conditions changed the cell distributions, in terms of the balance between the numbers of high- and low-state cells (Fig. 4 D and E).
Fig. 4.
Fluorescence histograms of HT_luxI-variant cells with different AHL-production rates of single cells and with different initial cell-density conditions. The AHL-production rate and the initial cell density play important roles in the diversification at 360 min. (A–C) The fluorescence histograms of the cells with pHT_luxI variants with low, appropriate, and high AHL-production rates, respectively. (D, B, and E) The fluorescence histograms of the HT_luxI1.5C cells under low, appropriate, and high initial cell-density conditions, respectively. FL indicates the intensity of GFP fluorescence.
Discussion
We demonstrated, in vivo and in silico, that the initial cell density plays an important role in the synthetic diversification (Fig. 4 D, B, and E), in which the phenotypic change of cells was described by the motion of marbles on Waddington’s landscape. The dependency on the cell number in our results is reminiscent of the community effect observed in developmental systems (21, 22). The dependency also resembles quite recent findings, such as the cell-number dependency of the embryonic cell population to direct cell fate (23–25). Our simple circuit design, consisting of just four genes, is a gene-network design candidate for natural diversifications depending on cell density, which is equivalent to cell number in a constant volume. Future investigations will include the comparison of our simple circuit to natural gene circuits governing natural diversification. The dynamic behavior in the synthetic diversification is also reminiscent of the maintenance of the phenotypic diversity of cells. According to a recent study, the utmost removal of pancreatic β-cells caused the transdifferentiation of α-cells into β-cells (26). Comparisons between the pancreatic circuits and ours, which involves mutual inhibition and intercellular signaling, may provide clues for investigations of pancreatic transdifferentiation.
In spite of frequent references to the concept of the landscape in various reviews about developmental and regenerative processes, the existence of genetic circuits that program cells to diversify as the motion on Waddington’s landscape had not been confirmed. In this study, we have realized, by using a synthetic genetic circuit, the concept of Waddington’s landscape in vivo. Constructions of synthetic genetic circuits that emulate natural phenomena have been used to understand their design principles (15, 16). Our synthetic circuit that emulates the natural diversification could be a model system to understand the design principles of the natural diversification, through the concept of the landscape.
To improve the behavior of engineered cells with a synthetic circuit, two approaches are usually taken: refinement of the circuit design and/or modification of the parameter values, such as promoter strengths. In this study, before our in vivo tests of the diversity generator including the CIts-regulated AHL-production mechanism, we also designed an AHL-constitutive circuit including a constant AHL-production mechanism. The AHL production in the diversity-generator manner gives rise to a negative feedback loop of AHL. In this circuit, the lower AHL concentration leads to the lower production rate of CIts and results in the higher production rate of AHL. Conversely, the higher AHL concentration results in the lower AHL-production rate. We expected that this negative feedback loop would relax the effect of the initial cell density on the AHL-accumulation velocity. Our numerical simulations actually showed that the diversity generator allowed a wider initial density range to realize a bimodal distribution, as compared to the AHL-constitutive circuit (Fig. S6). Thus, to implement the synthetic diversification, we selected the diversity generator design, rather than the AHL-constitutive circuit design. As previously reported, stochastic factors, such as Poissonian noise in protein synthesis/degradation, plasmid copy number, and cell-division timing, should be considered in the design of genetic circuits (27, 28). Indeed, the stochastic factors are important for the synthetic diversification, where the spreading of the cell distribution by the factors helps the distribution to cover the unstable steady state, when the position of the state has sufficiently approached that at the plateau concentration of AHL.
Our circuit design could provide a framework for a synthetic approach toward tissue engineering, because the community effect is important for the regulation of ES-cell differentiation (24). Mammalian synthetic circuits, such as a bistable toggle switch (29) and synthetic signaling systems (30, 31), have been developed. The integration of the above two systems by the construction of a multiresponsive promoter (32) could extend our design for synthetic phenotypic diversification to mammalian synthetic gene networks. Appropriate modifications of the signal-production rates may circumvent the extraordinary diversifications due to alterations in cell-population size. Moreover, a synthetic circuit based on our design for autonomous diversification would program the ratio of cell states in a population. The number of phenotypes in our synthetic diversifications can be increased, because the modularity of our synthetic circuit allows the integration of other mutual inhibitory and signaling machineries. Further integrations of our circuit with different cellular functions will provide unique tools for directing population fates.
Materials and Methods
Enzymes and Chemicals.
Restriction enzymes and polynucleotide kinase were purchased from New England Biolabs, TOYOBO, and TaKaRa. All oligonucleotides were from Operon Technologies, Inc. IPTG and standard chemicals were purchased from Nacalai, and N(β-ketocaproyl)-dl-homoserine lactone was obtained from Sigma-Aldrich Japan.
Strain Constructions.
E. coli strains used in this work are listed in Table S1. The host cell for all assays was E. coli strain JM2.300 [λ-, lacI22, rpsL135 (StrR), thi-1], which was used as the host cell for the genetic toggle switch (8). Because of the requirement of LuxR for the activation of the Plux/lac promoter, the pHT toggle and the diversity generator were used with a LuxR-producer plasmid, pLuxR (Fig. S1A), for the transformation of JM2.300. JM2.300-LuxR was transformed with pHT to create the pHT-toggle strain HT, and was then transformed with pHT_luxI1.5C to create the diversity-generator strain, HT_luxI1.5C.
Plasmid Constructions.
Plasmids used in this work are listed in Table S1. All genes and promoters were obtained by PCR amplification, using KOD-Plus (TOYOBO) polymerase and synthetic primers with overhanging ends containing the appropriate restriction sites. For the AHL-mediated bifurcation, the pHT-toggle was constructed by replacing the Ptrc-2 promoter on the genetic toggle switch pTAK132 (8) with a Plux/lac promoter, which has a LuxR-binding site and two LacI-binding sites (Fig. S1B). The LuxR-binding site is located upstream of the 10/-35 region of the promoter. The LacI-binding sites are located in the -10/-35 region and downstream of the -10/-35 region of the promoter (Fig. S1B). The LuxI-coding sequence with the BamHI and XmaI sites was amplified from the BBa_I751350 Biobrick plasmid by a forward primer, HT+luxI_fwd (GCGACCCGGGACGCGGCCGCCTAGAGAAAGAGGAGAAATACTAGATGAC), and a reverse primer, HT+luxI_rev (GCCTGGATCCGTTTATTAATTTAAGACTGCTTTTTTAAACTGTTC). The amplified LuxI-coding sequence with the restriction sites was cleaved by the restriction enzymes and inserted into the same sites of pHT-toggle to create the diversity-generator plasmid, pHT_luxIWT. The pHT_luxI variants were obtained by mutating the LuxI-coding sequence, using a QuickChange site-directed mutagenesis kit (Stratagene). We constructed several LuxI variants by combining mutations of three nucleotides in the LuxI-coding sequence, A101G, A117T, and A188G, as reported by Kambam et al. (20). The pHT_luxI1.5C plasmid was obtained by mutating A117T/A188G. LuxI1A and luxI2A contain the A101G and A101G/A117T/A188G mutations, respectively (20).
Diversity-Generator Assay Protocol.
An overnight culture of cells with an HT_luxI variant was diluted 100-fold into 3.0 mL of fresh LB medium with antibiotics (50 μg/mL carbenicillin and 30 μg/mL kanamycin), referred to as “basal medium.” The cells were grown at 37 °C until the cell population reached a sufficient density (OD590 = 0.35). For initialization of the cells, 1.0 mL of the culture with the sufficient density was washed with 1.0 mL of fresh basal medium by centrifugation. The washed cells were then inoculated into 100 mL of medium with a combination of 1.0 μM AHL and 2.0 mM IPTG, and incubated at 32 °C for 240 min for initialization to the low state. For the diversification, a 2.0 mL portion of the cell culture was washed three times with 1.0 mL of the basal medium by centrifugation, to remove the IPTG and AHL from the culture. The washed cells were then diluted to the target optical density, referred to as the “initial cell density,” into 100 mL of basal medium. After the dilution, the cells were grown at 32 °C for 360 min. We measured the GFP fluorescence of the cells at appropriate times after the dilution. The initial cell density for the assays, except for those shown in Fig. 4 D and E, was OD590 = 3.0 × 10-4. The initial cell densities in Fig. 4 D and E were OD590 = 1.0 × 10-5 and 2.0 × 10-3, respectively.
Fluorescence Measurement.
All fluorescence data were collected using a Becton-Dickinson FACSCalibur flow cytometer with a 488-nm laser and a 515–545-nm emission filter. Before measurement, the cells were washed with PBS by centrifugation.
Supplementary Material
Acknowledgments.
We thank J. Collins for providing pTAK132, and K. Aihara, T. Kobayashi, and T. Shibata for critical comments on the manuscript. This research was partially supported by the Precursory Research for Embryonic Science and Technology program of Japan Science and Technology (D.K.) and the Grant-in-Aid for Scientific Research (KAKENHI) programs from Ministry of Education, Culture, Sports, Science, and Technology (14085101 to M.H.; 14085203 to M.Y. and D.K.; 23119005 to D.K.; and 23119008 to M.Y.) and Japan Society for the Promotion of Science (21650018 to M.H. and D.K.; and 23680031 to D.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105901108/-/DCSupplemental.
References
- 1.Waddington CH. The Strategy of the Genes. London: Allen and Unwin; 1957. pp. 26–41. [Google Scholar]
- 2.Balázsi G, van Oudenaarden A, Collins James J. Cellular decision making and biological noise: From microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg A, Allis C, Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 5.Mohammad HP, Baylin SB. Linking cell signaling and the epigenetic machinery. Nat Biotechnol. 2010;28:1033–1038. doi: 10.1038/nbt1010-1033. [DOI] [PubMed] [Google Scholar]
- 6.Christopher M, Waters BLB. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 7.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 8.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 9.You L, Cox RS, Weiss R, Arnold FH. Programmed population control by cell-cell communication and regulated killing. Nature. 2004;428:868–871. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- 10.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 11.Brenner K, Karig DK, Weiss R, Arnold FH. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc Natl Acad Sci USA. 2007;104:17300–17304. doi: 10.1073/pnas.0704256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balagaddé FK, et al. A synthetic Escherichia coli predator-prey ecosystem. Mol Syst Biol. 2008;4(187):1–8. doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M. A tunable synthetic mammalian oscillator. Nature. 2009;457:309–312. doi: 10.1038/nature07616. [DOI] [PubMed] [Google Scholar]
- 14.Danino T, Mondragón-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ukai-Tadenuma M, et al. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Mukherji S, van Oudenaarden A. Synthetic biology: Understanding biological design from synthetic circuits. Nat Rev Genet. 2009;10:859–871. doi: 10.1038/nrg2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi H. Programmable cells: Interfacing natural and engineered gene networks. Proc Natl Acad Sci USA. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabor JJ, et al. A synthetic genetic edge detection program. Cell. 2009;137:1272–1281. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunlap PV, Greenberg EP. Control of Vibrio fischeri luminescence gene expression in Escherichia coli by cyclic AMP and cyclic AMP receptor protein. J Bacteriol. 1985;164:45–50. doi: 10.1128/jb.164.1.45-50.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kambam PKR, Sayut DJ, Niu Y, Eriksen DT, Sun L. Directed evolution of LuxI for enhanced OHHL production. Biotechnol Bioeng. 2008;101:263–272. doi: 10.1002/bit.21901. [DOI] [PubMed] [Google Scholar]
- 21.Gurdon JB. A community effect in animal development. Nature. 1988;336:772–774. doi: 10.1038/336772a0. [DOI] [PubMed] [Google Scholar]
- 22.Gurdon JB, Tiller E, Roberts J, Kato K. A community effect in muscle development. Curr Biol. 1993;3:1–11. doi: 10.1016/0960-9822(93)90139-f. [DOI] [PubMed] [Google Scholar]
- 23.Hwang YS, et al. Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11. Proc Natl Acad Sci USA. 2009;106:16978–16983. doi: 10.1073/pnas.0905550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balbach ST, et al. Governing cell lineage formation in cloned mouse embryos. Dev Biol. 2010;343:71–83. doi: 10.1016/j.ydbio.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Choi YY, et al. Controlled-size embryoid body formation in concave microwell arrays. Biomaterials. 2010;31:4296–4303. doi: 10.1016/j.biomaterials.2010.01.115. [DOI] [PubMed] [Google Scholar]
- 26.Thorel F, et al. Conversion of adult pancreatic α-cells to β-cells after extreme β-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenfeld N. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 28.Pedraza JM. Noise propagation in gene networks. Science. 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- 29.Kramer BP, et al. An engineered epigenetic transgene switch in mammalian cells. Nat Biotechnol. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- 30.Weber W, Daoud-El Baba M, Fussenegger M. Synthetic ecosystems based on airborne inter- and intrakingdom communication. Proc Natl Acad Sci USA. 2007;104:10435–10440. doi: 10.1073/pnas.0701382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnea G, et al. The genetic design of signaling cascades to record receptor activation. Proc Natl Acad Sci USA. 2008;105:64–69. doi: 10.1073/pnas.0710487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kramer BP, Fischer C, Fussenegger M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol Bioeng. 2004;87:478–484. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.