Abstract
The causative agent of Legionnaires disease, Legionella pneumophila, injects several hundred proteins into the cell it infects, many of which interfere with or exploit vesicular transport processes. One of these proteins, LidA, has been described as a Rab effector (i.e., a molecule that interacts preferentially with the GTP-bound form of Rab). We describe here the structure and biochemistry of a complex between the Rab-binding domain of LidA and active Rab8a. LidA displays structural peculiarities in binding to Rab8a, forming a considerably extended interface in comparison to known mammalian Rab effectors, and involving regions of the GTPase that are not seen in other Rab:effector complexes. In keeping with this extended binding interface, which involves four α-helices and two pillar-like structures of LidA, the stability of LidA-Rab interactions is dramatically greater than for other such complexes. For Rab1b and Rab8a, these affinities are extraordinarily high, but for the more weakly bound Rab6a, Kd values of 4 nM for the inactive and 30 pM for the active form were found. Rab1b and Rab8a appear to bind LidA with Kd values in the low picomolar range, making LidA a Rab supereffector.
Keywords: adenylylation, Legionella containing vacuole, vesicular trafficking
Vesicular trafficking is a fundamentally important process in eukaryotic cells allowing material exchange between different intracellular organelles. A well-known example for an externally triggered process involving vesicle transport is the uptake of pathogens by cells of the immune system and their subsequent degradation in the lysosomal pathway. Such processes need tight spatial and temporal control and coordination, and a class of regulatory factors called Rab proteins has evolved to orchestrate intracellular vesicular trafficking (1, 2). Rabs are small GDP/GTP binding proteins that make up the largest subfamily branch of the small Ras-like G proteins. When in the GDP state, Rabs are resting and inactive, whereas they are active when loaded with GTP. In the active state, each Rab can interact with Rab-effector proteins that promote a specific trafficking step. The transition between the active and inactive state is mediated by accessory proteins, the guanine nucleotide exchange factors (GEFs), which catalyze the replacement of GDP on Rabs by cytosolic GTP, whereas GTPase activating proteins (GAPs) assist in switching off the active (GTP-bound) Rabs by stimulating the intrinsically low GTPase activity of Rab proteins.
Immune cells have developed elaborate mechanisms to take up and destroy potentially harmful bacteria. Professional phagocytes engulf bacteria by phagocytosis and degrade the intruder by subjecting it to the lysosomal degradative pathway. This pathway involves the transition of phagosomes to lysosomes, which is realized by subsequent fusion of the phagosome with early and late endocytotic vesicles. In the acidic milieu of the lysosome, the bacterium is eventually lysed and cleared from the eukaryotic cell. Pathogens that are to survive this process (e.g., by macrophages) have to develop a means to evade the phagolytic pathway and to reprogram the engulfing cell to promote bacterial survival. The gram-negative bacterium Legionella pneumophila (termed Legionella from here on) has evolved intricate ways to ensure its survival and replication in macrophages (3). In the course of infection, the bacterium releases 200–300 Legionella proteins directly into the host cytosol by injection via a type-IV secretion system (Dot/Icm), and these proteins modify cellular host processes in various ways. Because vesicular trafficking is an important process in defending eukaryotic cells against bacterial invasion, it is not surprising that many of the secreted Legionella proteins affect components of the vesicle transport machinery. Rab1 in particular appears to be an important target of Legionella proteins. Rab1 regulates trafficking from the endoplasmatic reticulum (ER) to the Golgi apparatus, interacting with several GTPase effector proteins to fulfill its function (4–10). Rab1 is activated by multiprotein GEFs, the transport protein particle I or II (TRAPP-I or TRAPP-II) complexes, and is deactivated by the GAP TBC1D20. Legionella releases its own set of Rab1 regulatory molecules. First the protein DrrA, also known as SidM, is translocated into the eukaryotic cytosol and binds to the cytosolic surface of the Legionella containing vacuole (LCV) by means of a C-terminal phosphatidylinositol-4-phosphate (PI-4-P) binding domain (P4M). A central GEF domain of DrrA then recruits and activates Rab1 on the LCV. Subsequently, the N-terminal adenylylation domain of DrrA catalyzes the transfer of an adenosine monophosphate (AMP) moiety from ATP to a specific tyrosine residue (Y77) of Rab1 (= Rab1-AMP). The consequences of this covalent modification on the interaction with Rab1-binding partners are diverse. Although in the GTP state, Rab1-AMP is rendered incapable of interacting with MICAL-3, and its deactivation by GAP proteins (TBC1D20, LepB) is impaired. However, binding to the Legionella protein LidA still persists, despite the presence of the AMP group in an important effector-interaction interface (11).
LidA is translocated by Legionella into the host cytosol at the beginning of infection, and it localized to the LCV at the cytosolic surface, which is probably mediated by its PI-4-P and PI-3-P (phosphatidylinositol-3-phosphate) binding activity (12, 13). The protein consists of 729 amino acids and shows no obvious sequence similarity to any known protein. LidA was shown to interfere with the early steps in the secretory pathway when expressed heterologously (in the absence of an infection) in yeast cells or mammalian COS1 cells (14). Because LidA can bind to active or inactive Rab1 (15), which regulates transport from the ER-Golgi intermediate compartment to the Golgi, the Rab-binding activity of LidA could be responsible for the observed interference with early secretory vesicular trafficking. Interestingly, LidA binds not only to Rab1, but also at least to Rab6 and Rab8 (15). In addition, we could show that adenylylation of Rab1 by DrrA on Y77 still allows interaction with LidA, whereas the binding with the human effector proteins MICAL-3 is abrogated (11).
The mode of interaction between Rabs and a number of their effectors is very similar at the structural level. In the majority of cases, effectors interact with the so-called switch I and switch II regions (i.e., the regions that undergo the most characteristic conformational changes between the GDP and GTP states). The adenylylated Tyr77 (Tyr77-AMP) is located centrally in the switch II region, thus explaining the impairment of MICAL-3 binding to Rab1GTP-AMP. However, why LidA is still able to bind Rab1-AMP is not understandable from known Rab-effector interactions. Here we show that LidA has exceptional Rab-binding properties, making it a “supereffector.” The crystal structure of a Rab8a∶LidA complex gives valuable insight into the recognition of Rab proteins by LidA. Furthermore, the affinity of LidA toward Rab proteins is exceptionally high even in the GDP form, but even higher for active Rabs. The high affinity explains why Rab1-AMP is still able to interact with LidA, albeit with reduced affinity.
Results
Structure of the Complex Between Active Rab8a6–176 and LidA201–503.
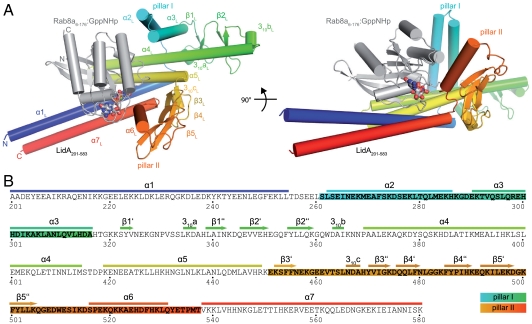
The Legionella protein LidA has been reported to bind to human Rab1, Rab6, and Rab8, and surprisingly to bind Rabs—in contrast to other Rab effectors—also in the inactive, GDP state (15). In order to analyze the structural basis for Rab binding, we embarked on the determination of the structure of numerous Rab∶LidA complexes by crystallography. We succeeded in solving the crystal structure of a complex between the truncated forms of LidA (LidA amino acids 201–583: LidA201–583) and active Rab8 (lacking the hypervariable region at the C terminus; Rab8 amino acids 6–176: Rab86–176) bound to the nonhydrolyzable GTP analogue GppNHp to a resolution of 2.5 Å (for data collection and refinement statistics, see Table S1). The central domain of LidA (201–583) represents a unique fold, consisting of seven α-helices, five antiparallel β-sheets, and three 310-helices (Fig. 1 and Movie S1). The base of the protein is formed by two antiparallel coiled-coil structures (α1L + α7L and α4L + α5L). Two parallel pillar-like structures extend almost perpendicularly from the center of this helical platform, giving the protein a sledge-like shape (most easily seen in Fig. 2A). The N-terminal pillar (amino acids 261–317, termed pillar I) and the C-terminal pillar (amino acids 450–537, termed pillar II) consist of helices α2L - α3L, and β-sheets β3L - β5L and helix α6L, respectively. Interestingly, the N and C termini are adjacent in the structure and hence suggest that the as yet uncharacterized terminal domains (aa 1–200 and aa 584–729) are in close spatial proximity in the wild-type protein.
Fig. 1.
The Rab8a6–176∶LidA201–583 complex crystal structure. (A) Cartoon representation of the Rab8a6–176∶LidA201–583 complex structure with Rab8a in gray and LidA colored from blue (N terminus) to red (C terminus), shown in two different orientations rotated by 90°. GppNHp is shown in stick representation and a magnesium ion as a green sphere. (B) Amino acid sequence of LidA201–583, indicating the secondary structure elements and the pillar-like structures (highlighted sequences).
Fig. 2.
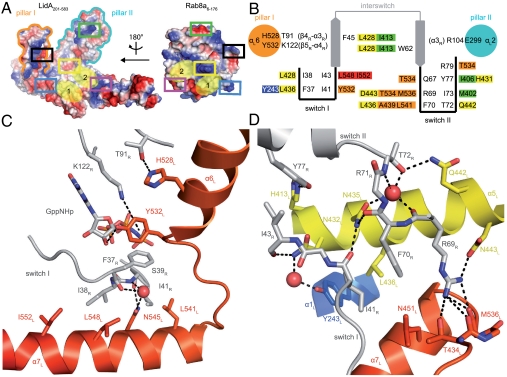
Intermolecular interactions in the Rab8a8–176∶LidA201–583 complex. (A) Surface representation of LidA (Left) and Rab8a (Right) colored by electrostatic potential represented in an open view to show the complex interface (boxes: interacting regions between Rab8a and LidA indicated by color coding). (B) Schematic representation of the amino acid interactions between Rab8a and LidA. (C) Interface of the hydrophobic patch II formed by α-helices α6L + α7L of LidA (I552L, L548L, L541L, and Y532L) and switch I of Rab8a. Polar interactions between α6L + α7L of LidA and Rab86–176 are depicted with dashes. (Red sphere: water molecule.) (D) Selected residues of LidA and Rab8a that contribute to contacts in hydrophobic patch I. Polar interactions between Rab86–176 and α-helices α1L + α5L + α7L of LidA are depicted with dashes. (Red spheres: water molecules.) (Coloring of B and C according to Fig. 1.)
Rab8a6–176 shows the typical GTPase fold, in which a central six-stranded β-sheet is surrounded by five α-helices (1) (Fig. 1A). The switch I (α1R - β2R-loop) and switch II (β3R - α2R-loop plus α2R-helix) regions are important structural elements that undergo the largest conformational changes between the GDP and GTP states, thus allowing Rab effectors to discriminate between active and inactive Rabs. In keeping with this, switch I and switch II face LidA and position the Rab8a∶GppNHp complex in a perpendicular groove of LidA, which comprises pillar I, pillar II, and the basal helices α1R, α4R, α5R, and α7R. The interswitch region of Rab8a (β2R + β3R) is in contact with helices α4R and α5R, whereas the GDP/GTP-binding pocket is oriented toward pillars I and II. Thus, LidA binds to active Rab8a by interacting with β1R and β2R and structural elements involved in GDP/GTP binding.
LidA Interacts with Rabs in a Unique Manner.
Most of the known eukaryotic Rab effectors bind to their cognate Rab via a binding platform that consists of α-helical coiled-coil structures. In such complexes, the site of interaction is centered on a conserved hydrophobic triad of the Rab protein (F45R, W62R, Y77R of human Rab8a) and contacts are established by two parallel α-helices of the effector (1), although variations on this scheme exist (10). Specificity for a Rab or a subset of Rabs and for their activation states (either GDP or GTP) is achieved by sensing specific amino acids and nucleotide-dependent conformations of the hydrophobic triad and the switch regions. The rigid rod-like structure of coiled coils commonly allows only the covering of a relatively small surface area on the approximately spherical Rab.
LidA provides a platform consisting of four helices, α1L, α4L, α5L, and α7L, to bind Rab8a (Fig. 1A and Fig. S1). Mainly hydrophobic interactions are formed in this interface and two hydrophobic patches on LidA are apparent: Patch I is formed by amino acid residues Y243L, M402L, I413L, L428L, L436L, and A439L of helices α1L, α4L, and α5L, and patch II by Y532L, L541L, L548L, and I552L of loop α6L - α7L (pillar II) and helix α7L (Fig. S1 and Fig. 2A). The hydrophobic triad of Rab8a (F45R, Y77R, W62R) is positioned on the edge of the Rab8∶LidA-complex interface and interacts exclusively with LidA hydrophobic patch I (Fig. 2B). Residues from both switch regions of Rab8a (I41R, I43R, F70R, and I73R) contribute to the binding of patch I, whereas patch II is almost exclusively contacted by switch I residues (F37R, I38R, and I41R) with F70R from switch II as an exception (Fig. 2 B and C). Thus, I41R and F70R are the only hydrophobic Rab8a residues that are in contact with both hydrophobic patches of LidA.
Most of the polar interactions are formed by switch II (Q67R, R69R, T72R, Y77R, and R79R), connecting Rab8a to pillar II (Y532L, T534L, and M536L) and to amino acid residues in patch I (E403L, H431L, Q442L, and D443L) (Fig. 2C). Additionally, the side chains of N432L and N435L (centrally located in patch I) establish hydrogen bonds to the main chain atoms of switch I I41R and I43R, thereby possibly assisting in correctly positioning these important Rab residues.
Despite the expected observation that LidA binds to the hydrophobic triad and to the switch regions in order to discriminate between the GDP/GTP states, unique interactions are seen that have not been observed between a Rab protein and any other effector or regulatory molecule. Pillars I and II of LidA embrace switch II and the nucleotide binding pocket of Rab8a, respectively, hence blocking access to the GDP/GTP-binding site to a great extent. Additional contacts are formed by amino acids of pillar I and pillar II between amino acids of Rab8a that are outside the switch and interswitch regions: Pillar I contributes E299L (α3L) for binding to R104R (α3R) of Rab8a; pillar II binds with H528L and Y532L to T91R and K122R, respectively, the latter being part of the guanine base recognition sequence (G4-motif: NKxD) of Rab8a.
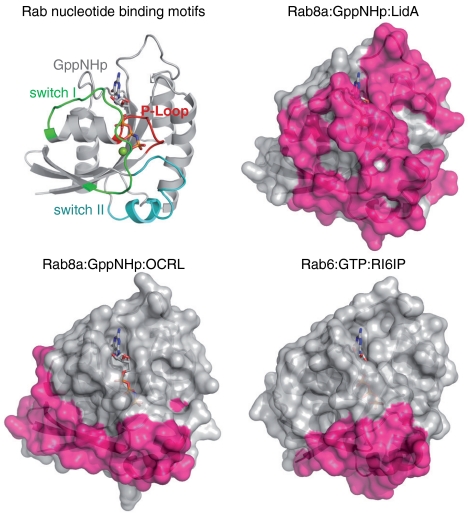
LidA displays a unique and more complex binding mode than other known Rab effectors. Instead of using only one or two α-helices for Rab binding (1), LidA creates a Rab-binding platform consisting of four α-helices. Additionally, LidA interacts with amino acids in regions of the Rab that were not previously observed to be involved in effector binding. As a consequence, LidA covers considerably more surface area on Rab8a than other Rab effectors [Rab8a6–176∶LidA201–583, 1,567.7 Å2; Rab86–176∶OCRL1540–678∶921.2 Å2 (10); Rab68–195∶RI6IP707–1145∶797.8 Å2 (16)] (Fig. 3).
Fig. 3.
LidA covers an unusually large surface area on Rab8a. The area (magenta) covered on Rab8a by LidA (Top Right) or OCRL1 (Bottom Left) (10) and by RI6IP on Rab6 (Bottom Right) (16) are compared. Rab8a from the Rab86–176∶LidA201–583 structure indicating the positions of the nucleotide binding region is shown in cartoon representation for comparison (Top Left).
The Affinity Between LidA and Rab Proteins Is Exceptionally High.
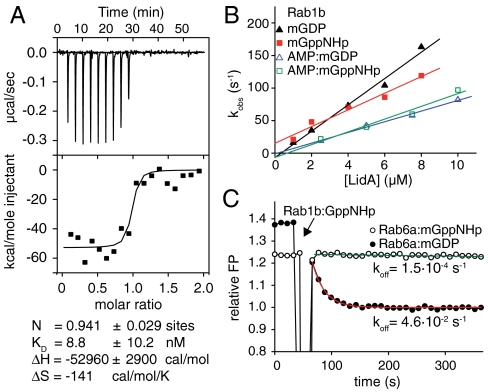
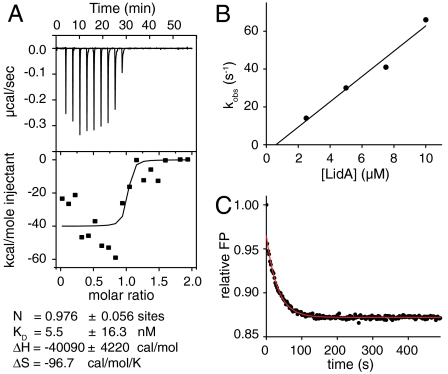
The structural peculiarities of the Rab8a-LidA complex prompted us to test whether the extensive molecular interactions result in particular biochemical properties. We first confirmed the published binding of Rab1b, Rab6a, and Rab8a with LidA (11, 15) using highly purified proteins by analytical size exclusion chromatography (Fig. S2). Subsequently, we aimed at determining the affinity between LidA201–583 and inactive and active Rab1b by isothermal titration calorimetry (ITC) (Fig. 4A). Surprisingly, the complex affinity was too high to be determined by ITC, with a dissociation equilibrium constant (Kd) significantly lower than 10 nM, in contrast with other effectors that usually bind to their Rabs micromolar affinity (10, 17). As an alternative we tried to independently quantify the association and dissociation rate constants (kon and koff) for the calculation of the Kd (Kd = koff/kon). To determine kon, the association rates of Rab proteins loaded with fluorescent 2′3′-(N-methyl-anthraniloyl) (mant) GDP/GppNHp analogues were measured in dependence on the LidA201–583 concentration by stopped-flow kinetics (Fig. 4B and Fig. S3). The kon values for the mantGDP and mantGppNHp forms of Rab1b, Rab6a, and Rab8a are in the range of 4.9·106 to 43.6·106 M-1 s-1 (Table 1). However, the determination of koff for Rab1b and Rab8a failed because no displacement of the fluorescent Rabs from LidA with nonfluorescent proteins was observed (Fig. S4A). Only Rab6∶mGDP could be displaced from LidA by an excess of Rab1∶GppNHp (Fig. 4C), resulting in a koff value of 0.048 s-1 and combined with kon the Kd was calculated to Kd = 4.9 nM. Complete displacement and thus accurate koff determination of the Rab6∶mantGppNHp∶LidA complex was not possible, but the koff could be estimated from a slow initial decrease in the signal to be of the order of koff = 1·10-4 s-1 (Fig. 4C). Hence, the Kd for the Rab6∶mantGppNHp∶LidA complex is approximately 30 pM. Thus, the affinity of LidA for active and inactive Rab6 differs by approximately two orders of magnitude. Because Rab1∶mGDP and Rab8∶mGDP, in contrast to Rab6∶mGDP, could not be displaced from LidA to a measurable extent (Fig. S4A), their koff values appear to be smaller than 10-4 s-1. Assuming a factor of 100 for the tighter binding of the GTP form means that their Kd values must be in the picomolar range. Interestingly, the affinity of Rab6∶mGDP for full-length LidA was comparable to that of LidA201–583 (Fig. S4 B and C), indicating that the N- and C-terminal regions of LidA do not contribute to Rab binding.
Fig. 4.
Kinetics of Rab-LidA interactions. (A) ITC of 4 μM Rab1b∶GDP with 40 μM LidA201–583. (B) Determination of the association rate constants (kon) for Rab1b-LidA201–583 interaction. The slope of a linear fit of the observed rate constants (kobs) in dependence on the LidA201–583 concentration gives kon (see Table 1). The kobs values have been obtained from fitting the change in fluorescence polarization upon shooting 1 μM Rab1b against increasing concentrations of LidA201–583 to a single exponential. (C) Displacement of 0.2 μM Rab6a∶mGDP or Rab6a∶mGppNHp from LidA201–583 by nonfluorescent Rab1b∶GppNHp (2 μM) monitored by fluorescence polarization (FP).
Table 1.
Kinetic constant of Rab-LidA interactions
| Rab | kon, 106 M-1 s-1 | koff, s-1 | Kd, nM | Signal |
| Rab1∶GDP | — | — | < 10* | ITC |
| Rab1-AMP∶GDP | — | — | < 10* | ITC |
| Rab1∶mGDP | 20.2 | < 10-4* | < 0.01* | Fl |
| Rab1-AMP∶mGDP | 8.2 | < 10-4* | < 0.01* | FP |
| Rab1∶mGppNHp | 12.8 | Fl | ||
| Rab1-AMP∶mGppNHp | 9.8 | FP | ||
| Rab6∶mGDP | 9.8 | 0.048 | 4.9 | FP |
| Rab6∶mGDP† | 3.5 | 0.018 | 5.1 | FP |
| Rab6∶mGppNHp | 4.6 | 1.4·10-4 | 0.03 | FP |
| Rab6-AMP∶mGppNHp | 6.7 | 0.032 | 4.8 | FP |
| Rab8∶mGDP | 39.3 | < 10-4* | < 0.01* | Fl |
| Rab8-AMP∶mGDP | 16 | FP | ||
| Rab8∶mGppNHp | 43.6 | < 10-4* | < 0.01* | Fl |
| Rab8-AMP∶mGppNHp | 12.8 | FP |
*Values are only estimates; FP: fluorescence polarization; Fl: fluorescence.
†Determined with full-length LidA.
Adenylylation of Rab Proteins Reduces Their Affinity to LidA.
Rab1b and Rab8a are subject to adenylylation by Legionella DrrA/SidM on Y77R (11). Superimposition of adenylylated Rab1b (Rab1b-AMP) with the Rab8a∶LidA complex structure reveals steric clashes of the AMP group with several Rab8a binding residues (I413L, L428L, and H431L) and helices α4L and α5L of LidA (Fig. S5), demonstrating that the AMP moiety cannot be accommodated conveniently in the conformation suggested by the determined Rab1-AMP structure. Like the GEF domain of DrrA (11), LidA is still able to interact with adenylylated Rab1b and Rab8a (Fig. S6 and ref. 11), although the AMP group is located centrally in the LidA and DrrA binding interface of the Rab protein (Fig. S5). We therefore speculated that adenylylation of Rabs should weaken the affinity for LidA, which would thus enable the determination of the Kd between LidA and adenylylated Rabs. We were unable to obtain ITC data from an interaction between LidA with adenylylated or nonadenylylated Rab8a due to a very low enthalpy of the reaction. However, an ITC experiment of Rab1b-AMP∶GDP with LidA201–583 gave identical results to those with nonadenylylated Rab1b∶GDP (Kd < 10 nM) (Fig. 5A). Binding of Rab1b-AMP∶mGDP, Rab1b-AMP∶mGppNHp, and Rab6a-AMP∶mGppNHp to LidA could be confirmed by determining the kon rates (Figs. 4B and 5B and Table 1), but the displacement of fluorescent Rab1-AMP∶mGDP from a complex with LidA was not possible on a reasonable time scale (Fig. S4A). However, the presence of the AMP group on Rab6a significantly accelerated the dissociation rate of Rab6a-AMP∶mGppNHp from LidA (Fig. 5C), leading to a koff value of 0.032 s-1 and a Kd of 4.8 nM. Thus, adenylylation of Rab6a decreased the affinity for LidA by roughly two orders of magnitude, and it seems reasonable to assume that a similar factor will apply to other Rabs that can be adenylylated.
Fig. 5.
Interaction of adenylylated Rabs with LidA. (A) ITC of 4 μM Rab1b-AMP∶GDP with 40 μM LidA201–583. (B) Determination of the association rate constant (kon) for the Rab6a-AMP∶mGppNHp interaction with LidA201–583 (for experimental details, see Fig. 4B). (C) Dissociation rate constant (koff) determination for Rab6a-AMP∶mGppNHp (1 μM) displaced from a complex with LidA201–583 displaced by 10 μM nonfluorescent Rab1b∶GppNHp. (For determined constants, see Table 1.)
Discussion
The structural and biochemical analysis of the interaction between the Legionella protein LidA and several Rab proteins revealed surprising previously undescribed aspects of Rab effector interaction. At the structural level, an extended binding interface between LidA and Rab8a is seen, and in contrast to other known effectors the intermolecular interactions comprise four α-helices of LidA instead of one or two as commonly observed. Quite unusually, the LidA binding extends significantly beyond the switch regions and the hydrophobic triad. This extensive binding interface results in extraordinarily high affinities of LidA for Rab proteins. The affinities for both the GDP and GTP-bound forms of Rab1b and Rab8a are immeasurably high using established methods. At least in the case of Rab6a, LidA fulfills the formal requirement for a Rab effector; i.e., it binds more strongly to the GTP (Kd = 30 pM) than the GDP form (Kd = 4.9 nM) of Rab. The displacement of Rab1b∶mGDP or Rab8a∶mGDP from LidA appeared to be much slower than for Rab6a∶mGppNHp (koff,Rab6∶mGppNHp∶LidA = 10-4 s-1; KD,Rab6∶mGppNHp∶LidA = 30 pM), thus allowing the conclusion that the koff and Kd values for inactive forms of Rab1b and Rab8a are koff < 10-4 s-1 and Kd < 30 pM, respectively. Assuming that LidA can discriminate between inactive and active Rab1b/Rab8a by a factor similar to that for Rab6a∶mGDP and Rab6a∶mGppNHp, a Kd in the picomolar or even lower range can be expected for the Rab1b/Rab8a∶GTP/GppNHp∶LidA interaction. The affinity of Rab proteins in the GTP form for their effector is usually in the micromolar Kd regime (10, 17), although moderate nanomolar affinities have also been reported (18–20). Thus, LidA is unusual because it binds to active Rabs at least three orders of magnitude more strongly than any Rab effector reported to date, so that LidA might be regarded as a Rab supereffector. However, the extremely high affinities, even to the GDP form, suggest that it might not play the role of a classical effector. In their classical role, effectors are in labile equilibrium with their cognate Rabs, which is a necessary requirement for the inactivation of Rabs by GTP hydrolysis under the influence of GAPs. Because GAPs cannot act directly on Rab-effector complexes, spontaneous dissociation has to occur to allow access of GAPs to the active site of the GTPase. The spontaneous dissociation of LidA-Rab-effector complexes appears to be orders of magnitude too slow for this scenario, so that other roles must be considered. Possible roles could include blockage of Rab activity or sequestering of transport vesicles with cognate Rabs on their surface.
The modification of Rab6a∶mantGppNHp by Legionella DrrA decreased the affinity for LidA by two to three orders of magnitude and suggests that adenylylation of Rab1b and Rab8a affects LidA binding to a similar extent. Therefore, LidA might have evolved to interact with Rab proteins with a remarkable high affinity in comparison to other Rab effectors in order to tolerate the weakening presence of the AMP group, the latter being necessary to block GAP-stimulated GTP hydrolysis (11).
The high Rab-LidA affinity appears to result from the large number of molecular interactions seen in the Rab8a∶LidA complex crystal structure: In contrast to other effectors that bind Rabs with one or two α-helices (1), LidA instead uses four α-helices as a basic interaction scaffold and additionally contacts areas outside of the interswitch and switch regions via pillars I and II. It is therefore tempting to speculate that interference with any of these regions, potentially by an allosteric mechanism, could modulate the affinity of LidA for Rab proteins and thereby lower the Rab-LidA complex affinity. Such an interference with Rab-LidA binding appears to be necessary considering several observations from the recent literature. First, deadenylylation of Rab1-AMP can be catalyzed by the deadenylylase SidD that is released by Legionella at later stages of infection (21, 22). However, the enormous affinity of Rab1 for LidA is not decreased dramatically by the AMP group, and this raises the question of how SidD could access the adenylylated Y77R in Rab1b to hydrolytically liberate the AMP moiety. Second, at later stages of infection, Rab1 disappears from the LCV (23). This process requires Rab1 to return to the GDP-bound state, because only inactive but not active Rabs can interact with and be extracted by the protein GDP dissociation inhibitor (GDI) (24–26). To return Rab1∶GTP to the GDP state, the Rab1-GAPs LepB (from Legionella) (23) or the human GAP TBC1D20 (27) needs to access Rab1 and stimulate GTP hydrolysis. The extensive interactions of LidA with the Rab switch I and II regions, which are important for GAP binding (28), suggest that access of GAPs to Rab1 is blocked as long as LidA is bound. The affinity of GAPs for GTP-bound Rab proteins is low [Kd: 20–200 μM (28, 29)] so that an efficient competition with the at least six orders of magnitude stronger binding LidA appears impossible. Thus, decreasing the Rab-LidA affinity by some unknown factor in order to displace LidA might be required.
The structure of LidA allows speculations on how a potential LidA displacement factor could act to influence the interaction with Rab proteins. From the Rab-binding site of LidA three regions protrude that could easily be contacted by additional proteins (Fig. S7). Two possible regions (1 and 2) are provided by LidA pillars I and II because one side of each pillar faces the bound Rab, whereas the opposite sides are freely accessible. In particular, pillar II could be an attractive displacement factor target because a number of interactions are formed with the GTPase. The third potential area comprises the region opposite to the Rab-binding sites of the two pillars. Here, the N-terminal half of helix α4L and the β-sheets β1L - β5L could be bound and influence the conformation of the pillars and therefore liberate Rab1/Rab1-AMP for interactions with SidD, LepB, and GDI.
In the course of Legionella infection, Rab1, Rab6, and Rab8 appear on the LCV membrane. However, a Legionella targeting factor (DrrA/SidM) has been identified only for Rab1 (15, 30), but not for Rab6 and Rab8. Factors that target Rab proteins to intracellular membranes need to compete with GDI, which keeps the prenylated Rab soluble in the cytosol. It has been demonstrated that GEF-catalyzed GDP/GTP exchange can be utilized to abrogate GDI binding due to the low affinity of GTP-bound Rab for GDI (24, 25). But displacement of the strongly bound GDI could be also mediated in a competitive manner by proteins that have a higher affinity for Rab∶GDP than GDI. The LidA and GDI-binding sites overlap on the Rab protein (Fig. S8), but the high affinity of LidA for the GDP state of Rabs (picomolar to nanomolar Kd) can efficiently compete with prenylated Rabs binding to GDI (nanomolar Kd) (24, 25). Thus, thermodynamically, LidA has the potential to displace GDI competitively and hence to recruit Rab proteins to the LCV. Therefore, it will be of interest to investigate the relevance of the enormous Rab-LidA affinity for Legionella infection and for the recruitment of Rabs and ER-derived vesicles by creating amino acid substitutions on LidA that systematically lower the affinity for Rab proteins. However, if LidA is not sufficient for depositing Rab6 and/or Rab8 on the LCV, the existence of other targeting factors is probable.
Another possible role of LidA was identified in earlier work that suggested that DrrA and LidA cooperate in the recruitment of ER-derived vesicles to the LCV (15). This would be similar to the role played by classical tethering factors for vesicle recruitment via Rabs. However, LidA does not conform to the two basic types of tethering factors seen (i.e., multicomponent complexes or extended coiled-coil structures).
The enormous affinity of LidA for RabGTPases allows use of the protein for various displacement approaches and as a biochemical tool. So far, Rab-binding partners are known that have high affinity for only the GDP [GDI, Rab escort protein (24, 25)] or the GTP state [GAPs, Rab effectors (18–20, 28, 29)], but not for both. LidA is a remarkable exception in this respect because it can bind to Rabs even in the inactive, GDP-bound form with at least nanomolar affinity.
Methods
Protein Preparation.
Rab8a, Rab8a6–176, Rab1b, and Rab6a were prepared as described (10, 24, 31). Preparative adenylylation of Rab1b and Rab6a were achieved using DrrA/SidM (11). Full-length LidA was produced as GST-fusion protein as reported (15). LidA201–583 was cloned by the Dortmund Protein Facility (http://www.mpi-dortmund.mpg.de/misc/dpf/) into a pOPINF vector (N-terminal His6-tag followed by a PreScission protease cleavage sequence) by the infusion cloning method (32). The protein was expressed in Escherichia coli BL21 (DE3) by induction with 0.3 mM isopropyl-β-dithiogalactopyranoside at 20 °C for 12 h. LidA201–583 was purified from the bacterial lysate by a combination of Ni-NTA affinity and size exclusion chromatography in 20 mM Hepes pH 8.0, 50 mM NaCl. The His6-tag was cleaved by PreScission protease. Selenomethionine LidA201–583 was expressed using the methionine biosynthesis inhibition method (33). Preparative loading of Rabs with nucleotide analogous was carried out as described previously (10).
Crystallization and Structure Determination of Rab86–176∶LidA201–583.
Native and selenomethionine-labeled LidA201–583 in complex with Rab86–176 were crystallized by the hanging-drop vapor diffusion method at 20 °C by mixing 1 μL protein (45 mg/mL) in buffer (20 mM Hepes pH 8.0, 50 mM NaCl, 1 mM MgCl2, 10 μM GppNHp, 2 mM dithioerythritol) with 1 μL of a reservoir solution containing 50% (vol/vol) 2-methyl-2,4-pentandiol, 10 mM spermidine, and 0.1 M sodium cacodylate pH 5.6. Diffraction data were collected on beamline X10SA of the Swiss Light Source and processed with X-ray Detector Software (34). Selenium atoms were located and a model was obtained with phenix.autosol and structure refinement was completed by using phenix.refine (35) and followed by manual building in Coot (36). The protein crystallizes in space group P43212 with one molecule of Rab86–176∶LidA201–583 in the asymmetric unit. Molecular presentations were prepared with PyMOL (37).
Rab1-LidA ITC.
Interaction studies by ITC were performed using an iTC200 microcalorimeter (MicroCal). Measurements were carried out in 50 mM Hepes buffer pH 7.5, 50 mM NaCl, 1 mM MgCl2, 1 mM tris(2-carboxyethyl)phosphine (TCEP), 1 μM GDP at 25 °C. LidA201–583 (40 μM) titrated into cell containing the Rab protein (Rab1b∶GDP, Rab1b∶GppNHp, or Rab1b-AMP∶GDP) at 4 μM. Data were analyzed using the Software (Microcal, LLC ITC) provided by the manufacturer.
Rab-LidA Kinetics.
Kinetic measurements were performed with a stopped-flow apparatus (Applied Photophysics) and a fluorescence spectrometer (Fluoromax-3, Horiba Jobin Yvon). All measurements were carried out in 20 mM Hepes buffer pH 7.5, 50 mM NaCl, 1 mM MgCl2, and 1 mM TCEP at 25 °C. LidA201–583 was used for the experiments unless stated otherwise. Association and dissociation kinetics were monitored by changes in fluorescence polarization [excitation: 366 nm, emission: 440 nm (Fluoromax-3) or 420 nm cutoff filter].
Analytical Gel Filtration.
Complex formation of Rab proteins with LidA was confirmed with analytical gel filtration. An amount of 400 μg LidA201–583 was mixed with 150 μg Rab in a final volume of 100 μL and complex elution was analyzed by monitoring the elution from an analytical gel filtration column (Superdex 200 10/30, GE Healthcare) by UV absorption at 280 nm.
Supplementary Material
Acknowledgments.
We thank N. Bleimling for invaluable technical assistance, the staff of Beamline X10SA at the Paul Scherrer Institute, and the X-ray communities at the Max-Planck-Institute (MPI) Dortmund and the MPI Heidelberg. Matthias Machner (National Institutes of Health, Bethesda, MD) is acknowledged for providing the LidA full-length expression construct. The Dortmund Protein Facility is acknowledged for assistance in cloning and protein expression/purification. This work was supported by a grant from the Deutsche Forschungsgemeinschaft SFB642, project A4.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3TNF).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113133108/-/DCSupplemental.
References
- 1.Itzen A, Goody RS. GTPases involved in vesicular trafficking: Structures and mechanisms. Semin Cell Dev Biol. 2011;22:48–56. doi: 10.1016/j.semcdb.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Novick PJ, Hutagalung AH. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: Making a cosy niche inside host cells. Nat Rev Microbiol. 2008;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer J, Weide T, Barnekow A. The MICAL proteins and rab1: A possible link to the cytoskeleton? Biochem Biophys Res Commun. 2005;328:415–423. doi: 10.1016/j.bbrc.2004.12.182. [DOI] [PubMed] [Google Scholar]
- 5.Weide T, Teuber J, Bayer M, Barnekow A. MICAL-1 isoforms, novel rab1 interacting proteins. Biochem Biophys Res Commun. 2003;306:79–86. doi: 10.1016/s0006-291x(03)00918-5. [DOI] [PubMed] [Google Scholar]
- 6.Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: Programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- 7.Moyer BD, Allan BB, Balch WE. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic. 2001;2:268–276. doi: 10.1034/j.1600-0854.2001.1o007.x. [DOI] [PubMed] [Google Scholar]
- 8.Diao A, Rahman D, Pappin DJC, Lucocq J, Lowe M. The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J Cell Biol. 2003;160:201–212. doi: 10.1083/jcb.200207045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satoh A, Wang Y, Malsam J, Beard MB, Warren G. Golgin-84 is a Rab1 binding partner involved in Golgi structure. Traffic. 2003;4:153–161. doi: 10.1034/j.1600-0854.2003.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou XM, et al. A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1. EMBO J. 2011;30:1659–1670. doi: 10.1038/emboj.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller MP, et al. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 12.Conover GM, Derre I, Vogel JP, Isberg RR. The Legionella pneumophila LidA protein: A translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol. 2003;48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- 13.Brombacher E, et al. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem. 2009;284:4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derre I, Isberg RR. LidA, a translocated substrate of the Legionella pneumophila type IV secretion system, interferes with the early secretory pathway. Infect Immun. 2005;73:4370–4380. doi: 10.1128/IAI.73.7.4370-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell. 2006;11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Recacha R, et al. Structural basis for recruitment of Rab6-interacting protein 1 to Golgi via a RUN domain. Structure. 2009;17:21–30. doi: 10.1016/j.str.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Hu B, Zimmermann B, Kilimann MW. Rim1 and rabphilin-3 bind Rab3-GTP by composite determinants partially related through N-terminal alpha -helix motifs. J Biol Chem. 2001;276:32480–32488. doi: 10.1074/jbc.M103337200. [DOI] [PubMed] [Google Scholar]
- 18.Chung SH, Joberty G, Gelino EA, Macara IG, Holz RW. Comparison of the effects on secretion in chromaffin and PC12 cells of Rab3 family members and mutants. Evidence that inhibitory effects are independent of direct interaction with Rabphilin3. J Biol Chem. 1999;274:18113–18120. doi: 10.1074/jbc.274.25.18113. [DOI] [PubMed] [Google Scholar]
- 19.McKiernan CJ, Stabila PF, Macara IG. Role of the Rab3A-binding domain in targeting of Rabphilin-3A to vesicle membranes of PC12 cells. Mol Cell Biol. 1996;16:4985–4995. doi: 10.1128/mcb.16.9.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuda M. Distinct Rab27A binding affinities of Slp2-a and Slac2-a/melanophilin: Hierarchy of Rab27A effectors. Biochem Biophys Res Commun. 2006;343:666–674. doi: 10.1016/j.bbrc.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Neunuebel MR, et al. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science. 2011;333:453–456. doi: 10.1126/science.1207193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan YH, Luo ZQ. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature. 2011;475:506–509. doi: 10.1038/nature10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 24.Schoebel S, Oesterlin LK, Blankenfeldt W, Goody RS, Itzen A. RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol Cell. 2009;36:1060–1072. doi: 10.1016/j.molcel.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 25.Wu YW, et al. Membrane targeting mechanism of Rab GTPases elucidated by semisynthetic protein probes. Nat Chem Biol. 2010;6:534–540. doi: 10.1038/nchembio.386. [DOI] [PubMed] [Google Scholar]
- 26.Alory C, Balch WE. Organization of the Rab-GDI/CHM superfamily: The functional basis for choroideremia disease. Traffic. 2001;2:532–543. doi: 10.1034/j.1600-0854.2001.20803.x. [DOI] [PubMed] [Google Scholar]
- 27.Haas AK, et al. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci. 2007;120:2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- 28.Pan X, Eathiraj S, Munson M, Lambright DG. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- 29.Albert S, Will E, Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 1999;18:5216–5225. doi: 10.1093/emboj/18.19.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murata T, et al. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 31.Bergbrede T, Pylypenko O, Rak A, Alexandrov K. Structure of the extremely slow GTPase Rab6A in the GTP bound form at 1.8 A resolution. J Struct Biol. 2005;152:235–238. doi: 10.1016/j.jsb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Berrow NS, et al. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35:e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 34.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 35.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.New York: Schrödinger, LLC; The PyMOL Molecular Graphics System, Version 1.3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.