Abstract
The NFX1-LIKE1 (NFXL1) and NFXL2 genes were identified as regulators of salt stress responses. The NFXL1 protein is a nuclear factor that positively affects adaptation to salt stress. The nfxl1-1 loss-of-function mutant displayed reduced survival rates under salt and high light stress. In contrast, the nfxl2-1 mutant, defective in the NFXL2 gene, and NFXL2-antisense plants exhibited enhanced survival under these conditions. We show here that the loss of NFXL2 function results in abscisic acid (ABA) overaccumulation, reduced stomatal conductance, and enhanced survival under drought stress. The nfxl2-1 mutant displayed reduced stomatal aperture under all conditions tested. Fusicoccin treatment, exposition to increasing light intensities, and supply of decreasing CO2 concentrations demonstrated full opening capacity of nfxl2-1 stomata. Reduced stomatal opening presumably is a consequence of elevated ABA levels. Furthermore, seedling growth, root growth, and stomatal closure were hypersensitive to exogenous ABA. The enhanced ABA responses may contribute to the improved drought stress resistance of the mutant. Three NFXL2 splice variants were cloned and named NFXL2-78, NFXL2-97, and NFXL2-100 according to the molecular weight of the putative proteins. Translational fusions to the green fluorescent protein suggest nuclear localisation of the NFXL2 proteins. Stable expression of the NFXL2-78 splice variant in nfxl2-1 plants largely complemented the mutant phenotype. Our data show that NFXL2 controls ABA levels and suppresses ABA responses. NFXL2 may prevent unnecessary and costly stress adaptation under favourable conditions.
Introduction
ABA plays a major role in seed maturation, control of germination, and other phases of plant development, but perhaps the most important function of ABA is to regulate plant water balance and osmotic stress tolerance. ABA-deficient mutants and ABA-insensitive mutants readily wilt under drought stress. The role of ABA under drought stress is at least twofold: ABA maintains the water balance mainly through guard cell regulation, and ABA induces genes that confer dehydration tolerance [1].
Significant progress has been made in understanding the signal transduction processes linking the hormone to target responses [2], [3]. The PYRABACTIN RESISTANCE1 (PYR1) and PYR1-LIKE (PYL) receptor family was recently shown to bind ABA and to inhibit activity of protein Ser/Thr phosphatases type 2C (PP2Cs) functioning as negative regulators [4]–[7]. Inhibition of the PP2Cs allows phosphorylation and activation of SnRK2 protein kinases [8]–[12], which in turn may activate transcription factors [13] and ion channels [14]–[17]. This central signalling module represents the earliest events of ABA signal transduction. Further physiological responses to ABA under water stress conditions are largely brought about by changes in gene expression. Transcription factors of various families (e.g., bZIP, B3, HD-ZIP, NAC, WRKY, bHLH, and Zn-finger classes) control genomic ABA responses [18]. The AREB/ABF (ABA-responsive element-binding protein or ABA-responsive element-binding factor) bZIP transcription factors such as AREB1/ABF2, AREB2/ABF4, and ABF3 are key regulators of ABA signalling in response to osmotic stress [13], [19]–[23]. The AREB/ABF proteins regulate ABRE-dependent gene expression during osmotic stress conditions. Transcription factors of the DREB/CBF (dehydration-response element-binding or C-repeat binding factor) family physically interact with AREB/ABF family transcription factors and control ABA sensitivity [24]. Members of other transcription factor classes have been reported to be both positive and negative regulators of ABA-responsive gene expression [18].
Under drought and salt stress, the endogenous ABA level increases. ABA biosynthesis is regulated at several steps and coordinated with the upstream metabolism [1], [25]. When dehydrated plants are rehydrated, the ABA level decreases rapidly.
The changes in gene expression patterns [18], metabolism [26], [27], and physiology [28], [29] in response to ABA, osmotic and salt stress have been intensely analyzed. Little information is available concerning the upstream sensing and signalling that determines ABA accumulation under abiotic stress [30]. The reduction of turgor may be sensed as a mechanical change that stimulates ABA accumulation. Protein kinases (e.g. histidine sensor kinases, mitogen-activated protein kinases [MAPKs], calcium-dependent protein kinases [CDPKs], and SnRK2s) may play important roles.
In this study, we characterize the Arabidopsis gene NFX1-LIKE2 (NFXL2). NFXL2 was identified as homologue of the NFXL1 gene [31]. NFXL proteins are found in animals, fungi, plants, and protists, and may be ubiquitous in eukaryotes [32]. The human NFX1 protein constituted the protein family [33]–[35]. Unique structural features of the NFX1 and NFXL proteins are the Cys-rich region and the specific RING finger motif. The Cys-rich region frequently comprises more than 500 amino acids and harbors several NFX1-type zinc finger domains [32]. The pattern C–X(1–6)-H-X-C-X3-C(H/C)-X(3–4)-(H/C)-X(1–10)-C describes the NFX1-type zinc finger (X can be any amino acids; two positions can be either His or Cys). The Cys-rich region is furthermore characterized by additional highly conserved residues (predominantly Cys residues). The Cys-rich region of the human NFX1 protein is required for binding to specific promoter elements [33], [34]. The N-termini of the Cys-rich regions usually are identical between isoforms, but the C-termini vary. The structure and complexity of the Cys-rich region may represent a major determinant of protein function. The whole Cys-rich region rather than individual NFX1-type zinc finger motifs may specify binding properties.
The second unique feature of the NFX1 and NFXL proteins is a specific RING finger motif. The RING finger is characterized by a C4HC3 Zn ligand signature and additional conserved amino acids [32]. E3 ubiquitin ligase activity is an intrinsic function of many RING finger proteins, and NFX1-like proteins may also have E3 activity [36]. A number of E3s have been identified that control ABA-biosynthesis and ABA responses [37]. For example, the RING-type E3 ligase SDIR1 is involved in ABA-related stress signal transduction [38], and the RING-H2 domain-containing XERICO protein controls ABA biosynthesis [39]. RING domain-containing proteins also function as part of multi-subunit E3 complexes such as SKP-Cullin-F-box (SCF) complexes. For example, the F-box protein DOR serves as negative regulator of ABA-induced stomatal closure under drought stress [40].
Plants use the ubiquitin proteasome system to alter their proteome to mediate changes required for responses to abiotic stress [41], [42]. However, ubiquitination is a versatile post-translational modification that is not only used for targeting to the proteasome. For example, monoubiquitination can control nuclear localization and transcriptional activity of transcription factors [43]. Further nuclear proteins such as histones and RNA polymerase II are subject to ubiquitination [44]. The functional relevance of the putative E3 ligase activity of the NFX1 and NFXL proteins is unknown.
In this study, we functionally characterize the Arabidopsis NFXL2 gene. The loss of NFXL2 resulted in elevated ABA-levels, reduced stomatal aperture, and enhanced survival of water stress. NFXL2 may prevent unnecessary and costly stress adaptation under favourable conditions. Deactivation of NFXL2 action may be inevitable for proper stress responses.
Results
Structure and subcellular localisation of NFXL2 proteins
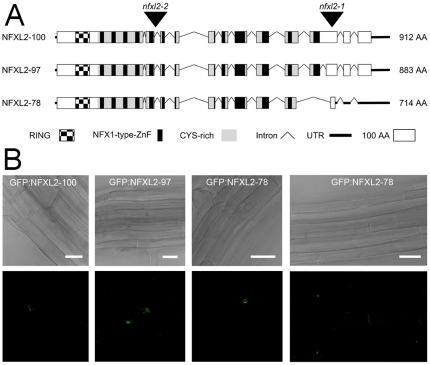
Screening of RACE-libraries resulted in the identification of three NFXL2 splice variants termed NFXL2-78, NFXL2-97, and NFXL2-100 according to the putative molecular weight of the encoded proteins. The NFXL2-97 isoform differs only slightly in the C-terminus from the previously identified NFXL2-100 isoform. An 87 bp intron corresponding to amino acids 732 to 760 of the NFXL2-100 protein is removed in the NFXL2-97 isoform (Figure 1A). The N-terminal primary structure of the NFXL2-78 isoform (comprising 697 amino acids) is identical to the NFXL2-97 and NFXL2-100 isoforms, but alternative donor and acceptor sites account for sequence divergence of the C-termini (Figure 1A). The three putative NFXL2 proteins show the characteristic features of NFXL proteins (Figure 1A), namely the N-terminal C4HC3 RING-finger motif and the Cys-rich region. The length of the Cys-rich region of the NFXL2-78 isoform is reduced in comparison to the NFXL2-97 and NFXL2-100 isoforms (Figure 1A). The NFXL2-78 protein comprises 10 NFX1-type zinc finger motifs, the NFXL2-97 and NFXL2-100 isoforms comprise 11 NFX1-type zinc finger motifs.
Figure 1. Structure and subcellular localization of NFXL2 proteins.
A. Exon-intron structure and domain organisation of NFXL2 proteins. Exons are represented by boxes, untranslated regions by thick lines, and introns by thin lines. T-DNA insertions in the nfxl2-1 (SALK_140301) and nfxl2-2 (GABI-Kat line 668B09) mutants are indicated. B. Subcellular localisation of GFP:NFXL2-100, GFP:NFXL2-97, and GFP:NFXL2-78 proteins. GFP-fluorescent images and bright field images of roots of stably transformed Arabidopsis plants. The scale bar represents 25 µm.
To determine whether NFXL2-78 is imported into the nucleus, a GFP (green fluorescent protein)-tagged NFXL2-78 protein was stably expressed in Arabidopsis plants. Green fluorescence was detectable in the nuclei of root cells (Figure 1B). Thus, the NFXL2-78 protein directs GFP to the nucleus. Analysis of plants carrying the 35S::GFP:NFXL2-97 and 35S::GFP:NFXL2-100 constructs suggests that the NFXL2-97 and NFXL2-100 isoforms presumably are also nuclear proteins.
NFXL2 expression pattern
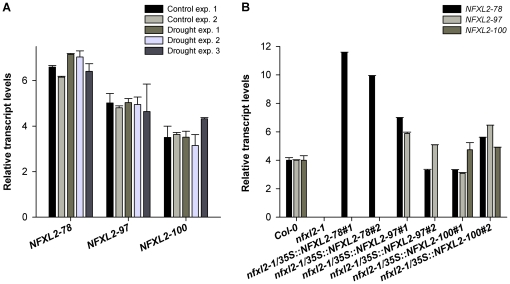
Web-based platforms such as Genevestigator [45] and the Arabidopsis eFP Browser [46] indicate the presence of relatively steady levels of NFXL2 transcript in most tissues and cell types. Transcript levels barely change under biotic and abiotic stress conditions, upon treatment with elicitors and other chemicals, and under different light and nutrient conditions. The NFXL2 gene is comparably expressed in mesophyll cells and guard cells [47], and is not controlled by ABA and other phytohormones [45]–[47]. Since common expression profiling platforms (i.e., the Affymetrix ATH1 and Affymetrix Tiling arrays) do not discriminate between the NFXL2 splice variants, NFXL2-78, NFXL2-97, and NFXL2-100 transcript levels were determined in shoot material of well-watered and drought-stressed plants by means of quantitative RT-PCR. Transcript levels were not significantly different under both conditions (Figure 2A).
Figure 2. NFXL2 transcript levels.
A. Real-time RT-PCR analysis of NFXL2 transcript levels in shoots of wild-type (Col-0) plants. For drought treatment, four-week-old soil-grown plants were withheld from water for 3 d. Higher bars indicate higher transcript levels. A difference of one indicates a fold change of two. Error bars: SE of three technical replicates. B. Real-time RT-PCR analysis of NFXL2 transcript levels in shoots of four-week-old nfxl2-1/35S::NFXL2-78, nfxl2-1/35S::NFXL2-97, and nfxl2-1/35S::NFXL2-100 plants in comparison to the wild type and nfxl2-1 mutant. Wild-type transcript levels were arbitrarily adjusted to a relative value (i.e. 4). The nfxl2-1 mutant is not able to produce the NFXL2-78/97/100 mRNAs. Transcript levels of the transgenic lines were given relative to the wild type. Higher bars indicate higher transcript levels. A difference of one indicates a fold change of two. Error bars: SE of three technical replicates.
Loss of NFXL2 prevents wilting under drought stress
In a previous study, the nfxl2-1 loss-of-function mutant and NFXL2-antisense lines showed higher survival rates under salt stress/high light conditions in comparison to the wild type [31]. Higher survival rates were accompanied by higher Fv/Fm ratios, indicating less damage to the photosynthetic apparatus. These findings suggested that NFXL2 suppresses salt stress responses.
The nfxl2-1 mutant showed only minor phenotypic changes under well-watered conditions. The mutant had a tendency to early flowering (data not shown), the dry weight content was slightly increased (Table S1), but the mutant did not display obvious morphological alterations. An additional T-DNA insertion mutant was identified and termed nfxl2-2. The nfxl2-1 and nfxl2-2 mutants exhibited similar phenotypes (see below).
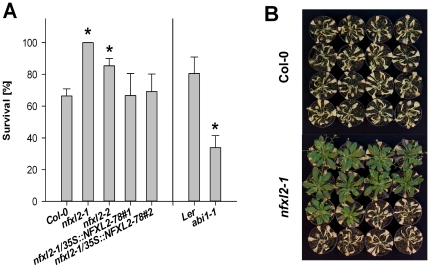
Survival experiments were performed in order to test for drought stress resistance. Plants were subjected to water withdrawal for seven days. The percentage of surviving plants was determined seven days later after water resupply. Five independent experiments were performed. The nfxl2-1 and nfxl2-2 mutants grew significantly better than wild-type plants. All nfxl2-1 plants survived the drought period; the survival rates of the nfxl2-2 and wild-type plants accounted for 85 and 66%, respectively (Figure 3A). Water withdrawal for nine days with subsequent rehydration resulted in wilting of all wild-type plants, but approximately 60% of the nfxl2-1 plants survived and were still turgescent (Figure 3B).
Figure 3. Drought stress tolerance.
A. 4-week-old soil-grown plants were withheld from water for 7 d. Survival rates were determined 7 d after water resupply. The number of surviving plants in five independent experiments is represented as the percentage (mean±SE) of the number of total plants (approximately 35 plants per genotype and experiment). Mutant values denoted with an asterisk are significantly different from those of their wild type (t test, P<0.01). B. For drought treatment, 4-week-old soil-grown plants were withheld from water for 9 d. Photos were taken 7 d after water resupply. Plants were arranged according to the visual phenotype. Wild-type plants were not able to survive the prolonged drought period, but approximately 60% of nfxl2-1 plants were still turgescent and viable.
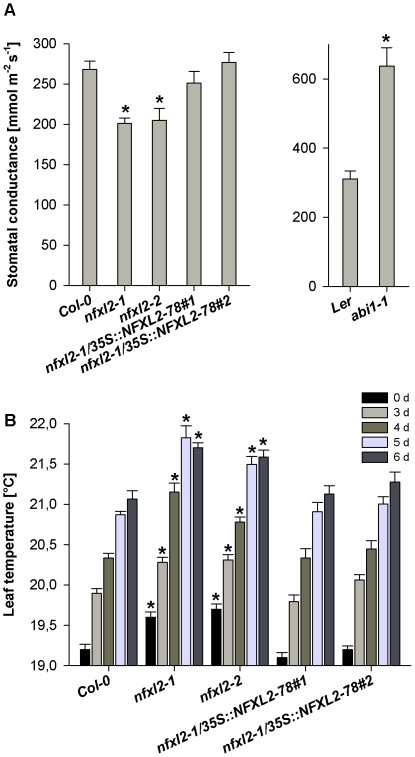
The ABA-insensitive abi1-1 mutant was used as stress-sensitive control [48]. It exhibited strong wilting and low survival rates (Figure 3A). ABA-induced stomatal closing is impaired in abi1-1 [49]–[51]. Water vapour flux at day 3 of the drought experiment was clearly elevated (215% in comparison to the wild type [Ler]) (Figure 4A). In contrast, the nfxl2-1 and nfxl2-2 mutants showed significantly lower stomatal conductance (75 and 76% of wild type [Col-0], respectively) (Figure 4A). Reduced stomatal conductance was associated with significantly higher leaf temperatures (Figure 4B). These initial findings posed the question whether the NFXL2 proteins control ABA responses.
Figure 4. Stomatal conductance and leaf temperature under drought stress.
A. Stomatal conductance was determined with a Decagon SC-1 porometer at day 3 of a drought stress experiment. Data are given as mean±SE (n = 10 to 15 leaves per genotype). Mutant values denoted with an asterisk are significantly different from those of their wild type (t test, P<0.001).B. Leaf temperature calculated from the quantification of infrared images (means±SE; n = 10 to 15 leaves per genotype). Values denoted with an asterisk are significantly different from the wild type (t test, P<0.001).
Stomatal aperture in nfxl2-1 leaves is reduced under various environmental conditions
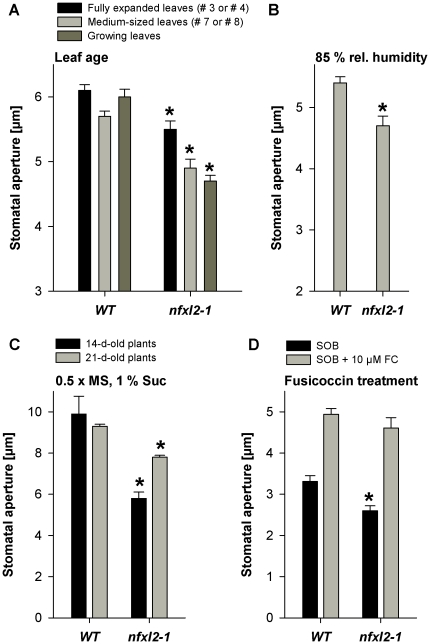
Stomatal aperture of well-watered plants was determined microscopically under different environmental conditions. Plants were grown in soil under long day conditions in a greenhouse or in a controlled growth chamber under ambient humidity, or in a controlled growth chamber under high humidity (85%), or in synthetic medium in jars. The nfxl2-1 mutant displayed significantly smaller stomatal aperture under all conditions (Figure 5A, Figure 6A–C). Reduced stomatal aperture was most pronounced in growing leaves (Figure 6A). In line with the reduced stomatal aperture, stomatal conductance was reduced in plants grown in soil under long day conditions (Figure 5B). Fusicoccin treatments of epidermal peels revealed that nfxl2-1 stomata were able to open as wide as wild-type stomata (Figure 6D), suggesting that the cellular mechanisms for stomatal opening were intact.
Figure 5. Stomatal aperture and stomatal conductance of well-watered plants grown under long day growth conditions.
A. Stomatal aperture was determined from images of silicone rubber imprints of the abaxial surfaces of rosette leaves of five-week-old plants. Plants were grown in a greenhouse under standard conditions (see Methods for details) or in a controlled growth chamber (16 h light, 21°C, 120 µmol m−2 s−1, 60% humidity; 8 h dark, 19°C, 75% humidity). Data are given as mean±SE of at least 60 stomata per genotype and condition. Values denoted with an asterisk are significantly different from those of their wild type (t test, P<0.001). B. Stomatal conductance of rosette leaves of five-week-old plants was determined with a Decagon SC-1 porometer. Plants were grown in a greenhouse under standard greenhouse conditions. Data are given as mean±SE (n = 10 to 15 leaves per genotype). Mutant values denoted with a positive sign are significantly different from their wild type (t test, P<0.05).
Figure 6. Stomatal aperture of wild-type and nfxl2-1 plants under different environmental conditions.
Stomatal aperture was determined from images of silicone rubber imprints of the abaxial surfaces of rosette leaves (A to C) or determined directly from epidermal peels using a microscope (D). Data are given as mean±SE of at least 60 stomata per genotype and condition. Mutant values denoted with an asterisk are significantly different from their wild type (t test, P<0.001). A. Stomatal aperture of 37-d-old plants grown in soil in a greenhouse under standard conditions. B. Stomatal aperture of 26-d-old wild-type and nfxl2-1 plants grown in a controlled growth chamber at 85% relative humidity (further climate parameters as given in Figure 5A). C. Stomatal aperture of 14-and 21-d-old plants grown in jars (0.5xMS medium supplemented with 1% sucrose). D. Stomatal response to fusicoccin. Epidermal peels were dissected from rosette leaves of soil-grown plants and incubated for 4 h in darkness either in stomata opening buffer (SOB) or SOB plus 10 µM fusicoccin (SOB+10 µM FC).
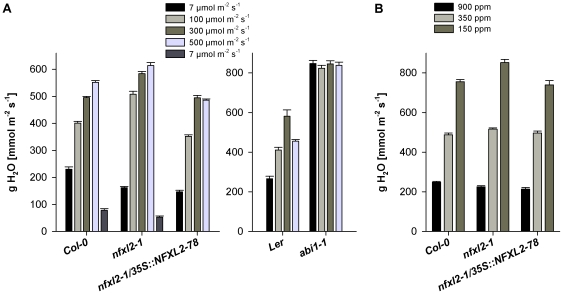
Light and CO2 are key factors in the control of stomatal movement [16], [52]. Low atmospheric CO2 concentrations and light induce stomatal opening, high CO2 concentrations and darkness cause stomatal closing. In the experiments depicted in Figure 7A, photon flux density (PFD) was increased sequentially from 7 µmol m−2 s−1 (quasi darkness) to 500 µmol m−2 s−1. Stomatal conductance of fully expanded rosette leaves of 5-week-old soil-grown plants was determined by infrared gas exchange measurements. The nfxl2-1 mutant exhibited less transpiration (71% of wild type) at 7 µmol m−2 s−1, but increasing PFDs caused an elevated stomatal conductance. For example, the increase in PFD from 7 to 500 µmol m−2 s−1 caused 140% increase of stomatal conductance in the wild type and 287% increase in nfxl2-1 plants (Figure 7A). Similarly to increasing PFD, a decrease of CO2 levels from 900 to 150 ppm resulted in an elevated stomatal conductance in the mutant (Figure 7B). The decrease from 900 to 150 ppm CO2 caused 204 and 279% increase of stomatal conductance in wild-type and nfxl2-1 plants, respectively. These findings indicate that the nfxl2-1 mutant is fully able to perceive and respond to light and CO2. The nfxl2-1 mutation did not cause a general defect in stomatal functioning, but modified short-term stomatal movements. Although short-term responses to light and CO2 were altered, the stomatal aperture under long-term conditions was invariably reduced under all tested conditions (Figure 5A, Figure 6).
Figure 7. Stomatal conductance in response to light and CO2.
4-week-old soil-grown plants were investigated by gas exchange measurements. Graphs represent H2O conductance rates obtained during a stepwise change of PFD (A) or CO2 concentration (B). Each data point represents the average of 5-10 individual plants±SE.
The nfxl2-1 mutant shows enhanced responses to ABA
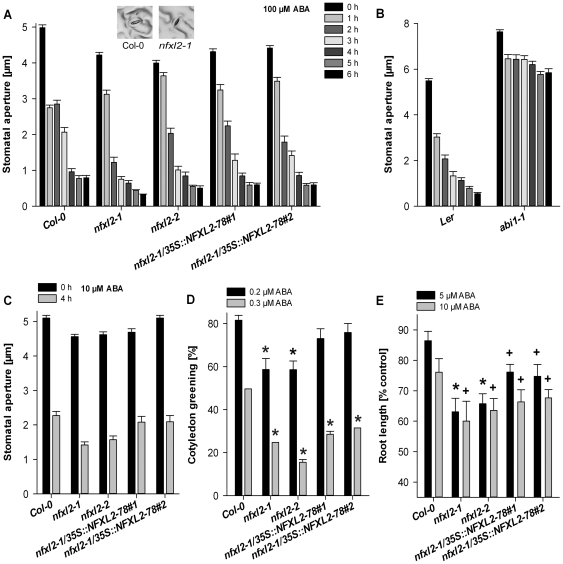
The effect of exogenous ABA on stomatal aperture was tested by application of synthetic ABA to leaf petioles. Stomata of nfxl2-1 and nfxl2-2 mutants exhibited a pronounced response to 100 µM ABA and were nearly fully closed after 6 h treatment (Figure 8A). Percent change (calculated as [[y2 – y1]/y1]*100) of stomatal aperture at the beginning (y1: 0 h) and end of treatment (y2: 6 h) was higher in both mutants in comparison to the wild type (Col-0: −84%; nfxl2-1: −92%; nfxl2-2: −87%). The exaggerated ABA-induced stomatal closing may represent the physiological basis for reduced stomatal aperture under various conditions (Figure 5A, Figure 6) and contribute to the enhanced drought tolerance (Figure 3). The abi1-1 mutant was grown in parallel and barely responded to ABA (Figure 8B). The nfxl2-1 and nfxl2-2 mutants also responded stronger to application of 10 µM ABA (percent change after 4 h: Col-0 −56%; nfxl2-1: −69%; nfxl2-2: −66%) (Figure 8C).
Figure 8. Stomatal aperture in rosette leaves, cotelydon greening, and root growth in the presence of exogenous ABA.
A. Stomatal aperture after ABA application. Stomatal aperture was determined from images of silicone rubber imprints of the abaxial surfaces of rosette leaves. Three droplets of an ABA solution (100 µM in SOB, solidified with 0.1% agarose) were supplied to leave petioles. Inset: Imprint of representative wild-type and nfxl2-1 stomata after 6 h ABA treatment (each photo represents a width of 50 µm).B. Stomatal aperture after ABA application. Three droplets of an ABA solution (100 µM in SOB, solidified with 0.1% agarose) were supplied to leave petioles. Details as in A.C. Stomatal aperture after ABA application. Three droplets of an ABA solution (10 µM in SOB, solidified with 0.1% agarose) were supplied to leave petioles. Further details as in A.D. Cotyledon greening in the presence of exogenous ABA. Seeds were allowed to germinate on half-concentrated MS medium supplemented with 1% sucrose at 4°C. 4-d-old seedlings were transferred to the same medium supplemented with 0.2 and 0.3 µM ABA and cultured at 22°C, with 14 h light (140 µmol m−2 s−1)/10 h dark photoperiod. After 5 d, the percentage of seedlings with green cotyledons was scored (mean±SE). Values denoted with an asterisk are significantly different from that of their wild type (t test, P<0.001).E. 4-d-old seedlings were transferred to half-concentrated MS medium supplemented with 1% sucrose and 0, 5, and 10 µM ABA. Root length of 14-d-old plants was determined and is given as percentage (mean±SE) of control plants. Values denoted with an * or+are significantly different from that of their wild type (t test, P<0.001 or P<0.03).
The enhanced response of nfxl2-1 stomata towards exogenous ABA prompted the analyses of additional ABA responses. First, inhibition of seed germination was tested by supplementation of 0.2 to 1 µM ABA to synthetic medium. ABA dose-response curves of wild-type and nfxl2-1 plants were similar (data not shown). Thus, the loss of NFXL2 did not affect ABA-sensitivity of seed germination under the tested conditions. Second, postgerminative growth was tested by scoring cotyledon greening in the presence of ABA. 82 and 50% of wild-type seedlings showed cotyledon greening in the presence of 0.2 and 0.3 µM ABA, respectively, whereas 59 and 25% of the nfxl2-1 seedlings developed green cotyledons under these conditions (Figure 8D). Third, root elongation was tested in the presence of 5 and 10 µM ABA. The nfxl2-1 mutant displayed approximately 25% less root growth in the presence of ABA compared with the wild type (Figure 8E). Thus, NFXL2 suppresses several ABA responses.
ABA-levels are increased in the nfxl2-1 mutant
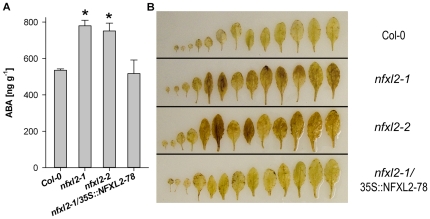
ABA levels were determined in plant material grown in a greenhouse under well-watered or drought conditions. Under standard greenhouse conditions, four-week-old wild-type, nfxl2-1, and nfxl2-2 plants accumulated 29±1, 33±2, and 32±1 ng ABA per gram fresh weight (mean±SE), respectively. Drought stress caused a 19-, 24-, and 24-fold increase of ABA levels in wild-type, nfxl2-1, and nfxl2-2 plants, respectively (ABA levels: 535, 780, and 751 ng ABA per gram fresh weight, Figure 9A). Thus, the nfxl2-1 and nfxl2-2 plants accumulated 46% and 40% more ABA under drought stress in comparison to the wild type.
Figure 9. Endogenous ABA levels and imaging of hydrogen peroxide production.
A. For drought treatment, soil-grown plants were withheld from water for 3 d. Two independent experiments were analyzed. Results are mean ng ABA per gram fresh weight±SE (n = 3 pools of 10 to 15 plants per genotype and experiment). Values denoted with an * are significantly different from that of their wild type (t test, P<0.001).B. Rosette leaves were infiltrated with 3,3-diamino-benzidine (DAB). Formation of brown polymerisation product indicates H2O2 formation [75]. Plants were grown in parallel to the plants used in A. Supplemental data are given in Figure S1.
Water stress also induces accumulation of reactive oxygen species (ROS) such as H2O2 [53], [54]. ROS can exacerbate cellular damage during stress, but also function as signalling compounds in plants. ROS modify the activity of protein kinases, transcription factors, and Ca2+ channels [55], [56]. Production of H2O2 was imaged in rosette leaves of drought-stressed plants. The nfxl2-1 and nfxl2-2 mutants displayed stronger staining in comparison to the wild type under mild drought stress conditions (Figure 9B, Figure S1). Thus, the loss of NFXL2 enhanced H2O2 production. Water withdrawal for 5 d resulted in massive H2O2 production in all genotypes (Figure S1).
The NFXL2-78 isoform largely complements the nfxl2-1 mutant
Introduction of the NFXL2-78 coding sequence under control of the CaMV35S promoter into the nfxl2-1 mutant resulted in the specific expression of the NFXL2-78 mRNA (Figure 2B). The NFXL2-97 and NFXL2-100 mRNAs were absent in the nfxl2-1/35S::NFXL2-78 lines. Stomatal aperture was increased in comparison to the mutant under favourable growth conditions (Figure 5A), stomatal conductance was increased to the wild-type level under drought stress (Figure 4), ABA-sensitivity of stomata, cotyledons, and roots was lower in comparison to the mutant (Figure 8), dry weight content (Table S1), survival rate after drought stress (Figure 3A), ABA-content (Figure 9A) and H2O2 levels (Figure 9B) were reduced to the wild-type level. Thus, the NFXL2-78 protein largely complemented the mutant phenotype.
Stable introduction of the NFXL2-97 and NFXL2-100 coding sequences into the nfxl2-1 mutant (transgenic lines termed nfxl2-1/35S::NFXL2-97 and nfxl2-1/35S::NFXL2-100, respectively) resulted in the expression of the NFXL2-97/NFXL2-78 and NFXL2-100/NFXL2-97/NFXL2-78 mRNAs, respectively (Figure 2B). Thus, the NFXL2-97 and NFXL2-100 sequences allowed formation of the transcripts of two and three isoforms, respectively. This is a consequence of the presence of the corresponding donor and acceptor splice sites. The NFXL2-97 protein presumably can act both synergistically and antagonistically to NFXL2-78 in ABA responses. For example, simultaneous expression of the NFXL2-78 and NFXL2-97 mRNAs caused ABA-hyposensitivity of stomata, but ABA-hypersensitivity in the cotyledon greening assay (Figure S2, Figure S3). The nfxl2-1/35S::NFXL2-100 plants were hyposensitive to exogenous ABA (Figure S2, Figure S3). However, future analyses of the specific functions of the NFXL2-97 and NFXL2-100 isoforms will require mutagenesis of the donor and acceptor splice sites in the NFXL2-97 and NFXL2-100 coding sequences in order to allow specific expression of the single mRNAs.
Discussion
Structural features of NFXL2 protein isoforms
The NFXL2-100, NFXL2-97, and NFXL2-78 proteins are characterized by the major structural features that also characterize the human NFX1 protein, namely the Cys-rich region and the C4HC3 RING finger motif. This suggests common mechanisms of action. The human NFX1 protein is the only member of the NFX1/NFXL protein family that has been characterized at the molecular level. One isoform, NFXL1-91, was identified as a transcriptional repressor that binds to X-box elements at the hTERT promoter [34]. NFXL1-91 interacts with the corepressor mSin3A/HDAC to maintain the deacetylated status at the hTERT promoter [57]. Recently it was shown that NFX1-91 also binds to the p105 promoter and activates p105 expression [35]. Thus, NFX1-91 functions as a dual regulator, a transcriptional repressor, and also a transcriptional activator, when bound to DNA. The Arabidopsis NFXL2 proteins presumably also act as transcriptional regulators, because they form the Cys-rich region (Figure 1A) and are targeted to the nucleus (Figure 1B).
The RING finger motif of the NFX1/NFXL proteins suggests E3 ubiquitin ligase activity [36]. Nuclear E3 activity could control transcription or the destruction of proteins [41], [44]. However, in vivo targets of the NFX1/NFXL proteins are unknown, and the functional relevance of the RING finger has not been shown for any NFX1/NFXL protein. E3 activity of the NFXL2 proteins could not be tested so far due to insufficient amounts of recombinant protein (data not shown).
The human NFX1 and Arabidopsis NFXL2 proteins also display structural differences, which presumably result in functional divergence. The second analysed NFX1 isoform, NFX1-123, interacts with cytoplasmic poly(A) binding proteins (PABPs). Cytoplasmic PABPs stabilize RNA and promote translation. A site-directed mutagenesis approach demonstrated that the putative PAM2 motif [58] of the NFX1-123 isoform was critical to bind cytoplasmic PABPs [59]. However, the NFXL2 proteins lack a PAM2 motif, and also do not form other known PABP recognition motifs. Furthermore, the NFXL2 proteins do not contain the R3H domain which is present in NFX1 and many non-plant NFXL proteins. The R3H motif is supposed to mediate interactions with single stranded nucleic acids [60]. Thus, the NFXL2 proteins presumably do not play a role in the control of mRNA stability.
NFXL2 controls levels of ABA and hydrogen peroxide
The loss of NFXL2 function caused an increase of approximately 10%ABA under favourable growth conditions (see above), and an increase of about 45% ABA after 3 d water withdrawal (Figure 9A). The increase of ABA is in line with phenotypic changes such as reduced transpiration (Figure 4A) and increased survival rates under water stress (Figure 3) and salt stress [31].
In addition to ABA, the nfxl2-1 mutant also produced more hydrogen peroxide than the wild type under favourable growth conditions [31] and under mild drought stress conditions (Figure 9B, Figure S1). Water withdrawal for 5 d was accompanied by massive ROS production in all genotypes (Figure S1). ROS function as important signal transduction molecules. ABA-dependent stomata closure is partially dependent on hydrogen peroxide production [61]–[63]. ABA induces the activity of the plasma membrane NADPH oxidase [61], and H2O2 activates specific ion channels [64] and is essential for stomatal closing [62], [63].
Ascorbic acid is the major antioxidant that scavenges H2O2 [65]. Addition of ascorbate to epidermal peels or overexpression of dehydroascorbate reductase causes an increase in stomatal aperture and stomatal conductance [62], [63]. The elevated H2O2 levels in the nfxl2-1 and nfxl2-2 mutants may be a consequence of elevated ABA levels and presumably contribute to the reduced stomatal aperture. In line with this interpretation, application of 10 mM ascorbate significantly reduced ABA-induced closing of nfxl2-1 stomata (151% aperture in comparison to ABA alone), whereas a minor effect was observed in the wild type (110% aperture) (Figure S4).
NFXL2 suppresses ABA responses
Responses to exogenous ABA were enhanced in the nfxl2-1 mutant (stomatal closure: Figure 8A,C; inhibition of cotyledon greening: Figure 8D; and inhibition of root growth: Figure 8E). Introduction of the NFXL2-78 isoform largely complemented the nfxl2-1 mutant phenotypes. The NFXL2-100 isoform may play a similar role as the NFXL2-78 protein, but action of the NFXL2-97 protein could be antagonistic in a subset of ABA responses (Figure S2, Figure S3).
NFXL2 gene products suppress both ABA accumulation (Figure 9A) and ABA responses (Figure 8). The molecular basis of this dual role is unknown, and different models are conceivable. Experimental evidence indicates that NFXL2 could control ABA levels and ABA responses via transcriptional control of the plant clock. NFXL2 (also termed EARLY BIRD [EBI]) was shown to associate with ZEITLUPE (ZTL) and to regulate the expression of components of the circadian clock [66]. A large number of ABA-responsive genes oscillate diurnally [67], and ABA metabolic genes are also clock-regulated [68]. Clock-dependent circadian modulation (‘gating’) of ABA function is important for cellular homeostasis under dehydration stress [69]–[71]. Alternatively, NFXL2 could control the level or activity of other nuclear factors involved in the control of ABA responses, or NFXL2 could control regulatory mechanisms upstream of ABA signalling (e.g. desensitize osmosensors or repress signalling events downstream of the osmosensors).
NFXL1 and NFXL2 supposedly play antagonistic roles in the control of abiotic stress responses. NFXL1 promotes the adaptation to salt stress [31] and drought stress (data not shown), NFXL2 suppresses the adaptation to salt stress [31] and drought stress (Fig. 3). Future work will address the specific roles of the NFXL2 proteins, the interplay between the NFXL2 isoforms, and the interplay between the NFXL1 and NFXL2 genes.
Materials and Methods
Screen for mutants and establishment of transgenic lines
The GABI-Kat line 668B09 [72] carries a T-DNA insertion in an NFXL2 intron (Figure 1A) and was named nfxl2-2. The DNA insertion site was confirmed by sequencing. Homozygosity of T-DNA insertions was confirmed by PCR on genomic DNA using T-DNA border-specific and gene-specific primers. The NFXL2-78, NFXL2-97, and NFXL2-100 coding sequences were amplified using the primers NFXL2-78/97/100-fw 5’ CAC CAT GAC TAA TAT GGC CGG AAC CG 3’, NFXL2-78-rev 5’ TTA ACA CCG ATT CAG CCA CCT GTA G 3’, and NFXL2-97/100-rev 5’ TTA GAT TCG AGG GTA TCT TCT AGA C 3’. The PCR fragments were cloned into the pENTR/D-TOPO (Invitrogen, Karlsruhe, Germany) vector, and used to establish GFP fusion constructs using the pK7FWG2 vector [73]. The NFXL2-78, NFXL2-97, and NFXL2-100 sequences were also inserted into the pH7WG2 vector for expression under control of the 35S promoter [73]. Sequence analysis of all cloned PCR products revealed 100% identity to the respective cDNA sequence. All constructs were transformed into Arabidopsis plants using the floral-dip method.
Growth conditions
Plants were established in soil. Seeds were allowed to germinate and grew for two weeks in controlled growth chambers (7 days: 16 h light, 140 µmol m−2 s−1, 20°C, 75% relative humidity; 8 h night, 6°C, 75% relative humidity; thereafter 7 days: 8 h light, 140 µmol m−2 s−1, 20°C, 60% relative humidity; 16 h night, 16°C, 75% relative humidity). Subsequently, plants were transferred to long day conditions in a greenhouse with artificial light (16 h light [high pressure sodium and metal halide lamps], 21°C, 50% relative humidity; 8 h night, 19°C, 50 % relative humidity) or to specific conditions as indicated in the text. All genotypes were grown side by side in a randomized manner. Alternatively, plants were grown under aseptic conditions as described before [31]. For ABA induction, a few droplets of 10 or 100 µM ABA (Duchefa, Haarlem, The Netherlands) dissolved in stomata opening buffer (SOB: 5 mM MES, 10 mM KCl, 50 µM CaCl2, pH 6.15) were added to the petioles of fully expanded rosette leaves.
Gene expression analysis
The NFXL2 transcripts could not be detected by means of Northern-blot analysis and were analyzed by RT-PCR as described [31]. Sequences of primers used for RT-PCR analysis were as follows: NFXL2-78-fw 5’ AAG GCG CGC TCC TCC CTT GT 3’, NFXL2-78-rev 5’ CAC CGA TTC AGC CAC CTG TA 3’, NFXL2-97-fw 5’ CCG TGG ACC TTG TCA CAG AAA 3’, NFXL2-97-rev 5’ CGA ACA ACC ACC TTT TTA CCA CA 3’, NFXL2-100 fw 5’ ATA TAT CCA TTT GGG ATG CTG TAT CT 3’, and NFXL2-100-rev 5’ GCA GCT AGC ATC GCC ACT AA 3’. The eIF1α, PDF1, and LRS1 genes were used to normalize the expression levels. Sequences of primers were as follows: eIF1α-fw 5’ TTG ACA GGC GTT CTG GTA AGG 3’, eIF1α-rev 5’ CAG CGT CAC CAT TCT TCA AAA A 3’, PDF1-fw 5’ ACG TCG CTA AAG TAC TTC AAT CCC 3’, PDF1-rev 5’ CGA ATC GTC TTC TCC ACA ACC G 3’, LRS1-fw 5’ ATG GGC ATT TGA CGA GGA TGC G3’, and LRS1-rev 5’ CGT CGT TCA CCC AGT CAA CAT GAG 3’. The amplification efficiency of each PCR was computed with the LinRegPCR software [74] and was used to correct the readout for each primer pair and run.
Measurement of ABA levels and DAB staining
ABA extraction was done at 4°C with dimmed light. 50 mg powdered tissue were subjected to lyophilisation for 24 h. Tissue was suspended in 0.5 ml extraction buffer (MeOH containing 2.5 mM citric acid monohydrate and 0.5 mM butylated hydroxytoluene). The extract was incubated for at least 20 h in the dark at 4°C under shaking conditions and centrifuged at 1500×g for 15 min at 4°C. The supernatants were recovered and 2 ml 62.5% extraction buffer/28.5% MeOH was added. C18 Sep-Pak cartridges (Waters, Eschborn, Germany) were equilibrated with 2 ml extraction buffer and subsequently with 1 ml 70% extraction buffer/30% MeOH. Supernatants were passed through a C18 Sep-Pak cartridge. 1 ml 70% MeOH was loaded onto the cartridge and flow-throughs were united. Additional elutions were analyzed exemplarily to check for residual ABA on the cartridge. The eluates were dried in a lyophilizer. Dried samples were resuspended in 1 ml 50 mM Tris buffered saline pH 7.5 (TBS)/MeOH (10 :1). Dilutions in TBS of each sample extract or ABA standard (Duchefa) were subjected to analysis by an enzyme immunoassay using the PGR1 kit (Sigma-Aldrich) according to the manufacturer' instruction. The ABA standard was assumed to be a mixture of equal amounts of the (S)-2-cis and (S)-2-trans form of ABA.
The production of H2O2 was imaged in plant material infiltrated with 3,3-diaminobenzidine (DAB) [75]. A method described in [76] was used for quantitative H2O2 measurements.
Microscopy
GFP-fluorescence was visualized in roots of 14-day-old transgenic plants grown in sterile media using a Leica TCS SP5 confocal microscope.
Stomatal aperture, stomatal conductance, leaf temperature, and gas exchange measurements
Stomatal aperture width was determined by light microscopy of nail polish images from silicone rubber imprints of abaxial surfaces of rosette leaves as described in [77]. Alternatively, epidermal peels were prepared from 5-week-old plants grown in a greenhouse, mounted on a microscope slide, and analyzed by light microscopy. Stomatal conductance of rosette leaves was determined abaxially using the Decagon SC-1 porometer after adjusting the clamp pressure of the sensor head to measurements with Arabidopsis according to the manufacturer’s instructions. Infrared gas exchange measurements were performed as described before [78]. 10 µM fusicoccin (Sigma-Aldrich) was added to SOB and applied to epidermal peels. Leaf temperature was measured by an infrared thermal imaging camera (PIR uc 180 InfraTec, Dresden, Germany). Images were recorded online by application of the IRBIS-3 thermography software (InfraTec, Dresden, Germany).
Supporting Information
Hydrogen peroxide levels. A. For drought treatment, soil-grown plants were withheld from water for 5 d. Rosette leaves were infiltrated with 3,3-diamino-benzidine (DAB). Formation of brown polymerisation product indicates H2O2 formation. B. For drought treatment, soil-grown plants were withheld from water for 1, 3, and 5 d. H2O2 levels were determined as described in [76].
(PDF)
Cotyledon greening in the presence of exogenous ABA. For experimental details see Fig. 8D.
(PDF)
Stomatal aperture in the presence of ABA and ascorbate. Epidermal peels were dissected from rosette leaves of soil-grown plants and incubated for 5 h in light (120 µmol m−2 s−1) either in stomata opening buffer supplemented with 10 µM ABA or in stomata opening buffer supplemented with 10 µM ABA+10 mM ascorbate. Aperture of untreated stomata is shown as reference. Aperture of ABA+ascorbate treated nfxl2-1 stomata is significantly different from ABA treated stomata (t test, P<0.001). Percent difference (ABA+ascorbate vs. ABA) was significantly larger in nfxl2-1 in comparison to the wild type (t test, P<0.001).
(PDF)
Dry weight content.
(PDF)
Acknowledgments
We thank Heidemarie Held for technical assistance in carrying out the gas exchange and leaf temperature measurements.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a grant from the Deutsche Forschungsgemeinschaft (MU 1738/6-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zhu J-K. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signalling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 3.Hubbard KE, Nishimura N, Hitomi.
- 4.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 5.Park S-Y, Fung P, Nishimura N, Jensen DR, Fujii H, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, et al. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009;60:575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 7.Nishimura N, Sarkeshik A, Nito K, Park S-Y, Wang A, et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii H, Zhu J-K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signalling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- 11.Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, et al. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21:3170–3184. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sirichandra C, Davanture M, Turk BE, Zivy M, Valot B, et al. The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14-3-3 binding site involved in its turnover. PLoS One. 2010;5(11):e13935. doi: 10.1371/journal.pone.0013935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pandey S, Zhang W, Assmann SM. Roles of ion channels and transporters in guard cell signal transduction. FEBS Let. 2007;581:2325–2336. doi: 10.1016/j.febslet.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Geiger D, Scherzer S, Mumm P, Stange A, Marten I, et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim T-H, Böhmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vahisalu T, Puzörjova I, Brosche M, Valk E, Lepiku M, et al. Ozone-triggered rapid stomatal response involves the production of reactive oxygen species, and is controlled by SLAC1 and OST1. Plant J. 2010;62:442–453. doi: 10.1111/j.1365-313X.2010.04159.x. [DOI] [PubMed] [Google Scholar]
- 18.Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124:509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- 19.Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, et al. Arabidopsis basic leucine zipper transcription factors involved in abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang J-Y, Choi H-I, Im M-Y, Kim SY. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signalling. Plant Cell. 2002;14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Kang J-Y, Cho D-I, Park JH, Kim SY. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004;40:75–87. doi: 10.1111/j.1365-313X.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- 22.Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, et al. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17:3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signalling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee S-J, Kang J-Y, Park H-J, Kim MD, Bae MS, et al. DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 2010;153:716–727. doi: 10.1104/pp.110.154617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez DH, Siahpoosh MR, Roessner U, Udvardi M, Kopka J. Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiol Plant. 2008;132:209–219. doi: 10.1111/j.1399-3054.2007.00993.x. [DOI] [PubMed] [Google Scholar]
- 27.Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, et al. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 2009;57:1065–1078. doi: 10.1111/j.1365-313X.2008.03748.x. [DOI] [PubMed] [Google Scholar]
- 28.Verslues PE, Agarwal M, Katiyar-Agarwal K, Zhu J, Zhu J-K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006;45:523–539. doi: 10.1111/j.1365-313X.2005.02593.x. [DOI] [PubMed] [Google Scholar]
- 29.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 30.Boudsocq M, Lauriere C. Osmotic signalling in plants. Multiple pathways mediated by emerging kinase families. Plant Physiol. 2005;138:1185–1194. doi: 10.1104/pp.105.061275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lisso J, Altmann T, Müssig C. The AtNFXL1 gene encodes a NFX1 type zinc finger protein required for growth under salt stress. FEBS Let. 2006;580:4851–4856. doi: 10.1016/j.febslet.2006.07.079. [DOI] [PubMed] [Google Scholar]
- 32.Müssig C, Schröder F, Usadel B, Lisso J. Structure and putative function of NFX1-like proteins in plants. Plant Biol. 2010;12:381–394. doi: 10.1111/j.1438-8677.2009.00303.x. [DOI] [PubMed] [Google Scholar]
- 33.Song Z, Krishna S, Thanos D, Strominger JL, Ono SJ. A novel Cys-rich sequence-specific DNA-binding protein interacts with the conserved X-box motif of the human major histocompatibility complex class II genes via a repeated Cys-His domain and functions as a transcriptional repressor. J Exp Med. 1994;180:1763–1774. doi: 10.1084/jem.180.5.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gewin L, Myers H, Kiyono T, Galloway DA. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 2004;18:2269–2282. doi: 10.1101/gad.1214704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu M, Katzenellenbogen RA, Grandori C, Galloway DA. NFX1 plays a role in human papillomavirus type 16 E6 activation of NFκB activity. J Virol. 2010;84:11461–11469. doi: 10.1128/JVI.00538-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Stone SL. E3 ubiquitin ligases and abscisic acid signaling. Plant Signal Behav. 2011;6:344–348. doi: 10.4161/psb.6.3.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Yang C, Li Y, Zheng N, Chen H, et al. SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell. 2007;19:1912–1929. doi: 10.1105/tpc.106.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ko J-H, Yang SH, Han K-H. Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 2006;47:343–355. doi: 10.1111/j.1365-313X.2006.02782.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Xu W, Li Z, Deng XW, Wu W, et al. F-box protein DOR functions as a novel inhibitory factor for abscisic acid-induced stomatal closure under drought stress in Arabidopsis. Plant Physiol. 2008;148:2121–2133. doi: 10.1104/pp.108.126912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 42.Lee J-H, Kim WT. Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol Cell. 2011;31:201–208. doi: 10.1007/s10059-011-0031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van der Horst A, de Vries-Smits AMM, Brenkman AB, van Triest MH, van den Broek N, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 44.Conaway RC, Brower CS, Conaway JW. Emerging roles of ubiquitin in transcription regulation. Science. 2002;296:1254–1258. doi: 10.1126/science.1067466. [DOI] [PubMed] [Google Scholar]
- 45.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, et al. Adv Bioinformatics . Vol. 2008. 420747; 2008. Genevestigator V3: A reference expression database for the meta-analysis of transcriptomics.420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, et al. An ‘electronic fluorescent pictograph’ browser for exploring and analyzing large-scale biological data sets. PloS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods. 2008;4:6. doi: 10.1186/1746-4811-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant. 1984;61:377–383. [Google Scholar]
- 49.Leung J, Bouvier-Durand M, Morris P-C, Guerrier D, Chefdor F, et al. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- 50.Meyer K, Leube MP, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- 51.Pei Z-M, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI. Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell. 1997;9:409–423. doi: 10.1105/tpc.9.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimazaki K, Doi M, Assmann SM, Kinoshita T. Light regulation of stomatal movement. Annu Rev Plant Biol. 2007;58:219–247. doi: 10.1146/annurev.arplant.57.032905.105434. [DOI] [PubMed] [Google Scholar]
- 53.Jiang M, Zhang J. Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J Exp Bot. 2002;53:2401–2410. doi: 10.1093/jxb/erf090. [DOI] [PubMed] [Google Scholar]
- 54.Luna CM, Pastori GM, Driscoll S, Groten K, Bernard S, et al. Drought controls on H2O2 accumulation, catalase (CAT) activity and CAT gene expression in wheat. J Exp Bot. 2004;56:417–423. doi: 10.1093/jxb/eri039. [DOI] [PubMed] [Google Scholar]
- 55.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 56.Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stress. Plant Cell Env. 2010;33:453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- 57.Xu M, Luo W, Elzi DJ, Grandori C, Galloway DA. NFX1 interacts with mSin3A/histone deacetylase to repress hTERT transcription in keratinocytes. Mol Cell Biol. 2008;28:4819–4828. doi: 10.1128/MCB.01969-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Albrecht M, Lengauer T. Survey on the PABC recognition motif PAM2. Biochem Biophys Res Commun. 2004;316:129–138. doi: 10.1016/j.bbrc.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 59.Katzenellenbogen RA, Vliet-Gregg P, Xu M, Galloway DA. NFX1-123 increases hTERT expression and telomerase activity posttranscriptionally in human papillomavirus type 16 E6 keratinocytes. J Virol. 2009;83:6446–6456. doi: 10.1128/JVI.02556-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grishin NV. The R3H motif: a domain that binds single-stranded nucleic acids. Trends Biochem Sci. 1998;23:329–330. doi: 10.1016/s0968-0004(98)01258-4. [DOI] [PubMed] [Google Scholar]
- 61.Kwak JM, Mori IC, Pei Z-M, Leonhardt N, Torres MA, et al. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003;22:2623–2633. doi: 10.1093/emboj/cdg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Z, Gallie DR. The ascorbic acid redox state controls guard cell signaling and stomatal movement. Plant Cell. 2004;16:1143–1162. doi: 10.1105/tpc.021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X, Zhang L, Dong F, Gao J, Galbraith DW, et al. Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 2001;126:1438–1448. doi: 10.1104/pp.126.4.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pei Z-M, Murata Y, Benning G, Thomine S, Klüsener B, et al. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature. 2000;406:731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- 65.Jubany-Mari T, Munné-Bosch S, Alegre L. Redox regulation of water stress responses in field-grown plants. Role of hydrogen peroxide and ascorbate. Plant Physiol Biochem. 2010;48:351–358. doi: 10.1016/j.plaphy.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 66.Johansson M, McWatters HG, Bako L, Takata N, Gyula P, et al. Partners in time: EARLY BIRD associates with ZEITLUPE and regulates the speed of the Arabidopsis clock. Plant Physiol. 2011;155:21080–2122. doi: 10.1104/pp.110.167155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Minuzo T, Yamashino T. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 2008;49:481–487. doi: 10.1093/pcp/pcn008. [DOI] [PubMed] [Google Scholar]
- 68.Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9:R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Legnaioli T, Cuevas J, Mas P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009;28:3745–3757. doi: 10.1038/emboj.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mikkelsen MD, Thomashow MF. A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 2009;60:328–339. doi: 10.1111/j.1365-313X.2009.03957.x. [DOI] [PubMed] [Google Scholar]
- 71.Robertson FC, Skeffington AW, Gardner MJ, Webb AAR. Interactions between circadian and hormonal signalling in plants. Plant Mol Biol. 2009;69:419–427. doi: 10.1007/s11103-008-9407-4. [DOI] [PubMed] [Google Scholar]
- 72.Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, et al. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- 73.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 74.Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 75.Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 1997;11:1187–1194. [Google Scholar]
- 76.Kotchoni SO, Kuhns C, Ditzer A, Kirch H-H, Bartels D. Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ. 2006;29:1033–1048. doi: 10.1111/j.1365-3040.2005.01458.x. [DOI] [PubMed] [Google Scholar]
- 77.Berger D, Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 2000;14:1119–1131. [PMC free article] [PubMed] [Google Scholar]
- 78.Muschak M, Willmitzer L, Fisahn J. Gas-exchange analysis of chloroplastic fructose-1,6-bisphosphatase antisense potatoes at different air humidities and at elevated CO2. Planta. 1999;209:104–111. doi: 10.1007/s004250050611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hydrogen peroxide levels. A. For drought treatment, soil-grown plants were withheld from water for 5 d. Rosette leaves were infiltrated with 3,3-diamino-benzidine (DAB). Formation of brown polymerisation product indicates H2O2 formation. B. For drought treatment, soil-grown plants were withheld from water for 1, 3, and 5 d. H2O2 levels were determined as described in [76].
(PDF)
Cotyledon greening in the presence of exogenous ABA. For experimental details see Fig. 8D.
(PDF)
Stomatal aperture in the presence of ABA and ascorbate. Epidermal peels were dissected from rosette leaves of soil-grown plants and incubated for 5 h in light (120 µmol m−2 s−1) either in stomata opening buffer supplemented with 10 µM ABA or in stomata opening buffer supplemented with 10 µM ABA+10 mM ascorbate. Aperture of untreated stomata is shown as reference. Aperture of ABA+ascorbate treated nfxl2-1 stomata is significantly different from ABA treated stomata (t test, P<0.001). Percent difference (ABA+ascorbate vs. ABA) was significantly larger in nfxl2-1 in comparison to the wild type (t test, P<0.001).
(PDF)
Dry weight content.
(PDF)