Abstract
Chalcidoidea (Hymenoptera) are extremely diverse with more than 23,000 species described and over 500,000 species estimated to exist. This is the first comprehensive phylogenetic analysis of the superfamily based on a molecular analysis of 18S and 28S ribosomal gene regions for 19 families, 72 subfamilies, 343 genera and 649 species. The 56 outgroups are comprised of Ceraphronoidea and most proctotrupomorph families, including Mymarommatidae. Data alignment and the impact of ambiguous regions are explored using a secondary structure analysis and automated (MAFFT) alignments of the core and pairing regions and regions of ambiguous alignment. Both likelihood and parsimony approaches are used to analyze the data. Overall there is no impact of alignment method, and few but substantial differences between likelihood and parsimony approaches. Monophyly of Chalcidoidea and a sister group relationship between Mymaridae and the remaining Chalcidoidea is strongly supported in all analyses. Either Mymarommatoidea or Diaprioidea are the sister group of Chalcidoidea depending on the analysis. Likelihood analyses place Rotoitidae as the sister group of the remaining Chalcidoidea after Mymaridae, whereas parsimony nests them within Chalcidoidea. Some traditional family groups are supported as monophyletic (Agaonidae, Eucharitidae, Encyrtidae, Eulophidae, Leucospidae, Mymaridae, Ormyridae, Signiphoridae, Tanaostigmatidae and Trichogrammatidae). Several other families are paraphyletic (Perilampidae) or polyphyletic (Aphelinidae, Chalcididae, Eupelmidae, Eurytomidae, Pteromalidae, Tetracampidae and Torymidae). Evolutionary scenarios discussed for Chalcidoidea include the evolution of phytophagy, egg parasitism, sternorrhynchan parasitism, hypermetamorphic development and heteronomy.
Introduction
Chalcidoidea (Hymenoptera) are minute wasps that generally range in size from 1-4 mm, with the smallest only 0.11 mm and the largest up to 45 mm. With an estimated diversity of up to 500,000 morphologically distinct species and an even larger number of cryptic species possible [1], [2], [3], [4], this superfamily is likely the most diverse group of insects. While several families are phytophagous (e.g. all Agaonidae; some Eurytomidae, Eulophidae, Pteromalidae, Tanaostigmatidae and Torymidae), most chalcid wasps are parasitoids. They attack immature and adult stages of virtually all insect orders, but have their greatest diversification on the Hemiptera and Holometabola. Because the individual host is killed as a result of parasitoid development, many chalcid species are successfully used as biological control agents of agricultural and ornamental pests (e.g. Aphelinidae and Encyrtidae) [3]. Both economically and ecologically Chalcidoidea have tremendous importance in both natural and managed ecosystems.
Despite their importance, our understanding of their taxonomy and evolutionary relationships is clearly wanting. Partly because of their small size, they are difficult to collect and study, and only about 23,000 species have been described [4]. Nineteen families are currently recognized, with their diversity spread across as many as 80-89 subfamilies, in many cases without consensus on their higher-level placement.
Chalcidoidea and their proposed sister group Mymarommatoidea first appear in mid Cretaceous amber deposits (Mymaridae) [5], [6], [7]. Most extant lineages do not appear until the Eocene, suggesting an extremely rapid post-Cretaceous radiation [6]. However, the presence of Eulophidae and Trichogrammatidae in Late Cenomanian amber from Ethiopia pushes chalcidoid diversification back to the mid Cretaceous, about 93–95 Mya [8].
Synapomorphies uniting most of the members of Chalcidoidea include an exposed prepectus, positioning of the mesothoracic spiracle on the lateral margin of the mesoscutum, wing venation reduced to submarginal, marginal, stigmal, and postmarginal veins, and the presence of multiporous plate sensilla on one or more of the antennal flagellomeres [9], [10]. Molecular evidence places Chalcidoidea as a monophyletic group nested within a monophyletic Proctotrupomorpha and as the sister group to either Diaprioidea or Mymarommatoidea [11], [12], [13], but see Sharanowski et al. [14] for an alternate proposal for Ceraphronoidea as the sister group.
Both morphological and molecular evidence place Mymaridae as the sister group of the rest of Chalcidoidea [10], [11], [13]. A few intuitive hypotheses of relationships within the superfamily have been proposed based on limited morphological justification [5], [15], [16]. However, for relationships within Chalcidoidea, there has not been a morphology-based cladistic analysis across more than just a few inclusive families [9]. A few molecular analyses have addressed relationships broadly across the superfamily, but these have used relatively few taxa to represent such a diverse group [17], [18].
Herein we present the first comprehensive phylogenetic analysis of relationships within the Chalcidoidea using 18S rDNA and the 28S rDNA D2–D5 expansion regions sampled across 722 taxa. The diversity of the superfamily is addressed by the inclusion of 72 subfamilies and 343 genera. Data were aligned according to a secondary structural model, which allows for the unambiguous partitioning of data into conserved regions and regions of ambiguous alignment [19], [20], [21]. Different optimizations of the alignment using MAFFT [22] are analyzed to compensate for potential alignment artifacts and increase phylogenetic resolution. Our analysis provides a new framework for evaluating the composition and relationships of major groups and hopefully will lead to a better understanding of their evolution.
Materials and Methods
Taxonomic sampling and specimen vouchering
Sequences were obtained for 722 taxa, with 56 outgroups and 666 ingroups (Table S1). Chalcidoidea are represented by all 19 families, 72 subfamilies, 343 genera and 649 species. Most species are represented by a single specimen; however, to remove any doubt of sequencing error, additional individuals of some species that were difficult to place within any expected grouping (e.g., Idioporus, Cynipencyrtus and Diplesiostigma) were sequenced. Outgroup taxa included exemplars of Ceraphronoidea (Ceraphronidae and Megaspilidae), Cynipoidea (Cynipidae, Figitidae, Ibaliidae and Liopteridae), Diaprioidea (Diapriidae, Maamingidae and Monomachidae), Mymarommatoidea (Mymarommatidae), Platygastroidea (Platygastridae) and Proctotrupoidea (Heloridae, Pelecinidae, Proctotrupidae, Roproniidae and Vanhorniidae). In the present manuscript we follow the family and subfamily classification of Chalcidoidea of Noyes [4], with additional resolution from the following: Agaonidae follows Cruaud et al. [23], Aphelinidae follows Hayat [24], Chalcididae follows Bouček and Delvare [25] and Narendran [26]; Cleonyminae follows Gibson [27], Eucharitidae follows Heraty [28], Eulophidae follows Burks et al. [29]; Pteromalidae follows Bouček [30], Delucchi [31], Graham [32] and Hedqvist [33], Toryminae follows Grissell [34], and Trichogrammatidae follows Owen et al. [35].
The majority of taxa were sequenced and vouchered at the University of California Riverside (UCR). Additional sequences were provided by co-authors (AC and JYR: Agaonidae and some Pteromalidae; PJ: Torymidae), the HymAToL project (various outgroup taxa), Matt Yoder (NC State University; various outgroup taxa), and Andy Austin (University of Adelaide; various outgroup taxa). See Table S1 for a complete listing of contributed sequences and voucher locations. Taxa sequenced at UCR are represented by either a primary (remains of actual specimen sequenced) or secondary (compared specimen from same collection series) specimen voucher. UCR voucher specimens were each assigned a unique UCRC_ENT Museum identification number and barcode. Additional voucher information is housed in a FileMaker Pro database at UCR developed by JM, and is available on request. UCR vouchers were imaged using a GT-Vision automontage system, with images deposited on MorphBank 4.0 (http://www.morphbank.net/).
DNA Extraction, Amplification and Sequencing
Genomic DNA extraction at UCR followed a modified version of the Chelex® protocol [36]. Primer sequences for PCR amplification of 18S rDNA and the 28S rDNA D2, D3 and D4+D5 expansion regions are provided in Table 1. Herein, the amplified regions shall be referred to simply as 18Sa-c, D2, D3 and D4+D5. In some cases, a shorter version of 18Sb was amplified with internal primers (18Si, Table 1). Amplification and sequencing followed established protocols at UCR [37]. UCR sequencing was conducted at the San Diego State University Microchemical Core Facility or the UCR Genomics Core Facility. Protocols for the Rasplus lab sequences follow Cruaud et al. [23]. Sequence verification was conducted by comparing forward and reverse sequences. All sequences are deposited on Genbank (Table S1).
Table 1. Primer sequences.
| Primer Name | Primer Sequence | Reference |
| 28S D2-3551 F | 5′ - CGT GTT GCT TGA TAG TGC AGC - 3′ | [17] |
| 28S D3-4046 F | 5′ - GAC CCG TCT TGA AAC ACG GA - 3′ | [134] |
| 28S D2-4057 R | 5′ - TCA AGA CGG GTC CTG AAA GT - 3′ | [37] |
| 28S D3-4413 R | 5′ - TCG GAA GGA ACC AGC TAC TA - 3′ | [134] |
| 28S D5-4625 R | 5′ - CCC ACA GCG CCA GTT CTG CTT ACC - 3′ | [135] |
| 18Sa-1 F | 5′ - TAC CTG GTT GAT CCT GCC AGT AG - 3′ | [135] |
| 18Sb-441 F | 5′- AAA TTA CCC ACT CCC GGC A -3′ | [11] |
| 18Sa-591 R | 5′- G AAT TAC CGC GGC TGC TGG -3′ | [135] |
| 18Si-673 F | 5′- ATC GCT CGC GAT GTT TAA CT -3′ | [11] |
| 18Si-905 R | 5′- AGA ACC GAG GTC CTA TTC CA -3′ | [11] |
| 18Sc-1204 F | 5′ - ATG GTT GCA AAG CTG AAA C - 3′ | [135] |
| 18Sb-1299 R | 5′- TGG TGA GGT TTC CCG TGT T - 3′ | [11] |
| 18Sc-1991 R | 5′ - GAT CCT TCC GCA GGT TCA CCT AC - 3′ | [135] |
Secondary structure alignment
Sequences were manually aligned using secondary structure models following Deans et al. [38] and Gillespie et al. [20], [21], [39], [40]. The 18Sa fragment began three bases (TAC) prior to the core helix H9 and included the variable regions V1 and V2 and ended with helix H39'. Fragment 18Sb began four bases (AUAA) prior to the core helix H406a (CGAUACGGGACUC), and included the variable regions V3, V4 (expansion region E23-1 through E23-14) and V5, and ended with core helix H960', just prior to V6. 18Sc began with a conserved loop (AAACCTCA), which preceded H984 and ended with the conserved loop (TGA) between H1506 and H1506', and included regions V6–V9. Amplification of the 28S rDNA D2, D3 and D4+D5 expansion regions began a single base (C) prior to helix H375 (GGGUUGC) in the core region preceding D2 and terminated 2 bases following helix H976 (UGG), subsequent to D5. The final alignment contained 545 blocks of data, which accounted for base-pairing helices and their prime, ambiguously-pairing regions of expansion and contraction (REC), ambiguously-pairing regions of slipped-strand compensation (RSC), non-pairing yet highly conserved loops, and non-pairing and variable loop regions of ambiguous alignment (RAA). For the purposes of this paper, we treat all three of these regions together as RAA regions.
Comparison between secondary structure and algorithmically generated alignments
Two important aspects of the dataset led us to compare the results obtained with various alignment strategies. First, we are confident of the alignment in the conserved stem-based and core regions; however vagaries of the secondary structure model lead to some local alignments that might not be optimal based on exact pairing of compensatory base changes. Second, distribution and size of RAAs are variable across Chalcidoidea. For such a large matrix, by-eye alignment of these highly-variable ambiguous regions from distantly related taxa is hard to justify. However, these RAAs can be locally informative [11], [29] and we prefer not to exclude them from our analyses. To test different optimizations of our secondary structure alignment and the impact of RAAs, we created two submatrices: one including the conserved stem-based and core regions and another including the regions of ambiguous alignment.
The core secondary structure-derived (SS) submatrix was created by manually removing regions of ambiguous alignment (RAAs), leaving only the structurally aligned helices, core regions, and conserved blocks. As alluded to previously, not all loops are ‘highly variable’ and conserved non-pairing regions, including some loops found in the core, were retained in the SS submatrix.
The second submatrix (RAAs) included the regions of ambiguous alignment sensu lato (RAAs, REC, RSCs, and unnamed blocks). An initial 77 regions of ambiguous alignment were identified. Where RECs and their pairing primes bounded an RAA, the blocks were concatenated. Additionally, REC 4 H3q, RAA 24 loop 9, REC 4' H3q', and RAA 25 were concatenated into a single block. Concatenation reduced the number of isolated RAA regions from 77 to 55. Each of these regions was aligned independently and re-included in the corresponding gene region for each of the following datasets.
Sixteen datasets were constructed from these submatrices (Table 2) that can be grouped into four categories: 1) SS submatrix without RAAs; 2–7) SS combined with algorithm-aligned RAAs; 8–10) algorithm-aligned SS submatrix without RAAs; 11–13) algorithm-aligned SS submatrix and algorithm-aligned RAAs, and 14–16) algorithm-aligned dataset in which the SS and RAA submatrices were not treated separately, but with each of the 6 gene regions individually isolated and independently algorithm-aligned.
Table 2. Alignment strategies for use of secondary structure and MAFFT alignments of both core/stem (SS) and ambiguous (RAA) regions.
| dataset | core/stem | RAA | length | inform. | uninfo. | 18Sa | 18Sb | 18Sc | 28S | 28S | 28S | RAxML | No. of steps |
| alignment | alignment | D2 | D3 | D4-5 | best score | SSME data | |||||||
| SSNR | SS | no RAA | 2996 | 853 | 356 | 500 | 757 | 633 | 591 | 333 | 182 | -85277.62 | 32461 |
| SSGE | SS | guide tree+E-INS-i | 4369 | 1675 | 566 | 507 | 969 | 701 | 1302 | 519 | 371 | -144234.60 | 32236 |
| SSGL | SS | guide tree+L-INS-i | 4369 | 1676 | 565 | 507 | 969 | 701 | 1302 | 519 | 371 | -144255.37 | 32223 |
| SSGG | SS | guide tree+G-INS-i | 4536 | 1773 | 550 | 507 | 963 | 697 | 1451 | 531 | 387 | -144123.77 | 32220 |
| SSME | SS | no guide+E-INS-i | 3917 | 1408 | 483 | 506 | 906 | 693 | 993 | 450 | 369 | -150220.93 | 31951 |
| SSML | SS | no guide+L-INS-i | 3917 | 1408 | 487 | 506 | 906 | 693 | 993 | 450 | 369 | -150223.77 | 31957 |
| SSMG | SS | no guide+G-INS-i | 3906 | 1433 | 468 | 506 | 906 | 694 | 1023 | 450 | 327 | -147954.87 | 31951 |
| MENR | E-INS-i | no RAA | 3024 | 861 | 375 | 507 | 758 | 634 | 605 | 337 | 183 | -85889.86 | 32522 |
| MLNR | L-INS-i | no RAA | 3024 | 861 | 374 | 507 | 758 | 634 | 605 | 337 | 183 | -85852.51 | 32483 |
| MGNR | G-INS-i | no RAA | 3025 | 859 | 380 | 507 | 758 | 634 | 606 | 337 | 183 | -85953.75 | 32527 |
| MEME | E-INS-i | no guide+E-INS-i | 3944 | 1415 | 502 | 513 | 907 | 694 | 1007 | 453 | 370 | -150774.64 | 32247 |
| MLML | L-INS-i | no guide+L-INS-i | 3944 | 1415 | 501 | 513 | 907 | 694 | 1007 | 453 | 370 | -150775.39 | 32236 |
| MGMG | G-INS-i | no guide+G-INS-i | 3934 | 1438 | 492 | 513 | 907 | 695 | 1038 | 453 | 328 | -148553.26 | 32254 |
| MESR | E-INS-i (all data by partition) | 4133 | 1536 | 553 | 506 | 901 | 693 | 1196 | 531 | 306 | -145056.78 | 31983 | |
| MLSR | L-INS-i (all data by partition) | 4099 | 1507 | 545 | 506 | 901 | 693 | 1162 | 531 | 306 | -145084.06 | 32187 | |
| MGSR | G-INS-i (all data by partition) | 4139 | 1519 | 551 | 506 | 901 | 694 | 1201 | 531 | 306 | -145293.59 | 31997 | |
The guide tree was generated from a RAxML analysis of the SSNR dataset (no RAA). Except for the all data alignments (no submatrix partition), each of the 55 RAA blocks were aligned independently and reinserted into the appropriate gene partition for analysis. E-INS-i, G-INS-i and L-LINS-i are MAFFT alignment options. The RAxML best score was obtained from 10 independent runs using CIPRES v.2.0. The number of informative and uninformative sites and parsimony steps were calculated in PAUP 4.0* for each resulting tree using the SSME dataset.
Automated alignments were performed with MAFFT [22], [41], [42]. Both the online server (v.6) and the downloadable program (v.6.244b) were used to create initial alignments that utilized the following MAFFT algorithms: E-INS-i, G-INS-i and L-INS-i. Alignments for each partition (core region and each of the 55 regions of ambiguous alignment taken independently) were generated using the default settings (gap opening penalty = 1.53 and offset value = 0.00).
The RAAs were aligned both with and without a guide tree that was generated using the SSNR (core with no RAA) dataset. Our purpose for using a guide tree was to optimize local alignments for each of the RAAs within terminal clusters of independently recognized taxa grouped through analysis of the SSNR, thus aligning nearest neighbors, as opposed to aligning disparate taxa across the entire dataset without any prior grouping. Maximum likelihood (ML) analyses of this dataset were conducted with RAxML v.7.2.7 using a partitioned GTR+Γ model [43] on the Teragrid cluster, Abe [44] via the CIPRES portal V2.2 [45]. We used 1000 rapid bootstrap (BS) replicates for each run, with initial tests using the autoMRE criterion [46] showing 350 BS to be adequate. A GTRCAT approximation of models was used for ML bootstrapping [47]. Ten RAxML analyses utilizing different starting seeds were executed, followed by ML optimization to find the best-scoring tree. The 10 resulting trees were used to generate a strict consensus tree that was converted to a MAFFT-readable guide tree with the script newick2mafft.rb (http://mafft.cbrc.jp/alignment/software/treein.html). This guide tree was implemented in the MAFFT alignments of the isolated RAAs utilizing the E-INS-i, G-INS-i and L-INS-i algorithms (SSGE, SSGG and SSGL, Table 2).
The secondary structure-derived matrix with MAFFT-aligned RAA regions (SSME) is deposited on Texas A&M's Parasitic Hymenoptera Research Labs' jRNA Secondary Structure and its Phylogenetic Implications website (available through http://hymenoptera.tamu.edu/rna/) and as Supplemental Nexus File S1. The 15 remaining datasets, with and without RAA regions, are available from JMH upon request.
Dataset partitioning
Sequences were partitioned into six gene regions 18Sa, 18Sb, 18Sc, D2, D3, and D4+D5, with each partition including their respective aligned RAA regions. The 18Sa-c partitions were defined simply as the region sequenced, inclusive of the primers used. The 28S rDNA expansion regions are also contiguous, being bounded on either side by core sequence, which was amplified in the PCR reaction. The decision as to where to define the end of D2 and start of D3 and likewise, the end of D3 and start of D4+D5, was arbitrarily made to fall within the core regions between the expansion regions. The helix H1a' (UUUCAGG), was assigned to mark the end of D2; while the un-named, non-pairing block of sequence (AC), which follows helix H1a' and proceeds helix H563 (CCGU) marked the start of D3. Helix H812 (CCCUCC) was assigned to mark the end of D3, while the un-named, non-pairing block of sequence (GAAG), which follows helix H812 and precedes helix H822 (UUUCC), marks the start of D4+D5.
Phylogenetic analyses
Maximum Likelihood (ML) analyses and associated bootstrapping (BS) were conducted on the 16 datasets with RAxML v.7.2.7 using a partitioned GTR+Γ model [43] on the Teragrid cluster, Abe [44] via the CIPRES portal V2.2 [45]. A GTRCAT approximation of models was used for ML bootstrapping [47]. To accommodate parameter variation in separate runs [48], 10 analyses were conducted using different seed numbers and 1000 rapid bootstrap (BS) replicates, with the tree with the best known likelihood (BKL) score chosen from among these sets. For comparison of alignments strategies, we examined the number of parsimony informative and uninformative sites, overall length, and the number of step changes mapped with PAUP 4.0* [49] onto each tree using the SSME dataset. The SSME dataset was chosen for the Parsimony analysis, because it provided what we considered to be the optimal results in terms of clade retention and used both the SS and RAA submatrices.
The parsimony analysis of the SSME dataset was conducted with TNT v.1.1 [50], [51]. Heuristic searches were performed using a New Technology Search with default settings, except for using a sectorial search, ratchet weighting probability of 5% with 50 iterations, tree-drifting of 50 cycles, tree-fusing of 5 rounds, and best score hit of 10 times, followed by swapping to completion on all trees found. Nodal supports were calculated using 1000 standard bootstrap replicates.
To be consistent with our interpretation of bootstrap percentage (BP), we use the following scale: a bootstrap percentage of ≥90% is considered very strong, 80–89% means strong, 70–79% means moderate, and 50–70% means low bootstrap support.
To better track relationships, each taxon includes a prefix which is an abbreviation of it family-group (c.f. Table 3, S1), and the suffix includes the DNA voucher code and letters correponding to the gene regions sequenced, corresponding to the three regions of 18S (tuv), 28S-D2 (x), D3 (y) and D4-5 (z).
Table 3. Summary of traditional clades within Chalcidoidea, diversity sampled, and support from various datasets and analyses.
| core only | core and RAA | RAxML | TNT | ||||||||||
| Code | Taxonomy | gen | spp | SSNR | MENR | SSGE | SSME | MGMG | MGSR | MJR* | SSME | ||
| AG | Agaonidae (76/757) | 19 | 104 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 97 | ||
| AGA | ‘Agaoninae’a | 12 | 48 | – | – | – | – | – | – | – | – | ||
| AG4 | ‘Agaonidae group 4′ | 2 | 3 | – | par | 70 | 75 | 86 | 92 | 75 | – | ||
| AGB | ‘Blastophaginae’ | 3 | 24 | – | – | – | – | – | – | – | – | ||
| AGK | Kradibiinae | 2 | 25 | – | par | – | – | – | – | – | – | ||
| AGT | Tetrapusinae | 1 | 4 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| AP | Aphelinidae (33/1168) | 21 | 87 | – | – | – | – | – | – | – | – | ||
| API | Aphelinidae incertae sedis | 4 | 4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| APA | Aphelininae | 7 | 22 | 88b | 88b | 97b | 96b | 91b | 86b | 100b | 56b | ||
| APAY | Aphytini | 3 | 12 | par | par | par | 53 | par | par | par | + | ||
| APZ | Azotinae | 1 | 12 | 99 | 100 | 100 | 100 | 100 | 100 | 100 | 99 | ||
| APC | Coccophaginae | 6 | 43 | + | + | 81 | + | + | + | 94 | – | ||
| APCP | Pteroptricini | 5 | 31 | par | par | par | par | par | par | par | – | ||
| APE | Eretmocerinae | 1 | 5 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| APR | Euryischiinae | 2 | 2 | 100 | 100 | 100 | 89 | 100 | 100 | 100 | 100 | ||
| CAL | Calesinae (1/4) | 1 | 3 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| CH | Chalcididae (87/1464) | 20 | 37 | – | – | – | – | – | – | – | – | ||
| CHC | Chalcidinae | 8 | 19 | – | – | – | – | – | – | – | – | ||
| CHCB | Brachymeriini | 1 | 6 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| CHCC | Chalcidini | 2 | 8 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| CHCR | Cratocentrini | 3 | 3 | – | – | – | – | – | – | – | – | ||
| CHCP | Phasgonophorini | 2 | 2 | 98 | 100 | 100 | 100 | 100 | 99 | 100 | 100 | ||
| CHD | Dirhininae | 1 | 5 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| CHE | Epitranininae | 1 | 3 | + | 90 | 99 | 95 | 94 | 98 | 100 | 56 | ||
| CHH | Haltichellinae | 8 | 12 | 88 | 90 | 100 | 98 | 98 | 97 | 100 | + | ||
| CHHA | Haltichellini | 5 | 9 | + | + | + | par | – | 56 | + | – | ||
| CHHY | Hybothoracini | 3 | 3 | par | par | par | 93 | – | par | par | par | ||
| CHS | Smicromorphinae | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| EN | Encyrtidae (460/3735) | 12 | 14 | + | 50 | 81 | 72 | 73 | 78 | 100 | + | ||
| ENE | Encyrtinae | 8 | 9 | par | par | par | + | 72 | + | 89 | + | ||
| ENT | Tetracneminae | 4 | 5 | 72 | 69 | 87 | 77 | 97 | par | 65 | + | ||
| EU | Eucharitidae (55/423) | 22 | 46 | 100c | 100c | 100c | 100c | 100c | 100c | 100c | 100c | ||
| EUE | Eucharitinae | 16 | 27 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 96 | ||
| EUG | Gollumiellinae | 2 | 3 | 80 | 93 | 98 | 76 | 86 | 99 | 100 | par | ||
| EUO | Oraseminae | 4 | 16 | par | + | 71 | + | + | + | 75 | + | ||
| EL | Eulophidae (297/4472) | 27 | 28 | 89d | 92d | 99d | 98d | 97d | 98d | 100d | +d | ||
| ELI | Eulophidae i.s. | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| ELE | Entedoninae | 8 | 8 | – | + | 50 | + | 74 | 59 | 88 | + | ||
| ELN | Entiinae | 5 | 6 | – | – | 67 | par | + | 58 | 81 | + | ||
| ELU | Eulophinae | 9 | 10 | 66 | + | 96 | 95 | 91 | 85 | 100 | – | ||
| ELO | Opheliminae | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| ELT | Tetrastichinae | 3 | 3 | 98 | 98 | 100 | 100 | 100 | 100 | 100 | 99 | ||
| EP | Eupelmidae (45/907) | 19 | 25 | – | – | – | – | – | – | – | – | ||
| EPC | Calosotinae | 5 | 7 | – | – | – | – | – | – | – | – | ||
| EPE | Eupelminae | 12 | 14 | + | + | + | – | + | – | – | – | ||
| EPN | Neanastatinae | 2 | 4 | – | – | – | + | – | – | – | – | ||
| EY | Eurytomidae (88/1424) | 14 | 28 | – | – | – | – | – | – | – | – | ||
| EYE | Eurytominae | 9 | 14 | 100e | 99e | 100e | 100e | 100e | 100e | 100e | 100e | ||
| EYH | Heimbrinae | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| EYR | Rileyinae | 2 | 7 | + | + | 97 | 90 | 87 | 87 | 100 | + | ||
| LEU | Leucospidae (4/134) | 2 | 6 | 98 | 90 | 100 | 100 | 98 | 98 | 100 | 98 | ||
| MY | Mymaridae (103/1424) | 13 | 15 | 98 | 95 | 100 | 99 | 98 | 97 | 100 | 61 | ||
| MYI | Mymaridae i.s. | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| MYA | Alaptinae | 3 | 3 | – | – | – | – | – | – | – | – | ||
| MYE | Eubronchinae | 1 | 2 | 99 | 100 | 98 | 99 | 100 | 87 | 100 | 84 | ||
| MYM | Mymarinae | 8 | 9 | – | – | – | – | – | – | – | – | ||
| ORM | Ormyridae (3/125) | 2 | 3 | 66 | 56 | 67 | + | 61 | 52 | 100 | + | ||
| PE | Perilampidae (15/277) | 14 | 34 | +f | +f | – | – | – | – | – | – | ||
| PEI | Perilampidae i.s. | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PEA | Akapalinae | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PEM | Philomidinae | 3 | 3 | 99 | 98 | 100 | 100 | 100 | 100 | 100 | 97 | ||
| PEC | Chrysolampinae | 4 | 9 | 73 | 67 | 88 | 72 | 68 | 80 | 100 | – | ||
| PEP | Perilampinae | 5 | 20 | 96 | 98 | 100 | 100 | 100 | 99 | 100 | 76 | ||
| PT | Pteromalidae (588/3506) | 111 | 130 | – | – | – | – | – | – | – | – | ||
| PTI | Pteromalidae i.s. | 2 | 2 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PT01 | Asaphinae | 3 | 3 | – | – | – | – | – | – | + | – | ||
| PT02 | Ceinae | 1 | 2 | 93 | 93 | 100 | 98 | 98 | 99 | 100 | 98 | ||
| PT03 | Cerocephalinae | 3 | 3 | 99 | 99 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| PT04 | Chromeurytominae | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PT05 | Cleonyminae | 10 | 10 | – | – | – | – | – | – | – | – | ||
| PT05D | Chalcedectini | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PT05C | Cleonymini | 3 | 3 | 68 | 56 | 84 | 54 | + | 52 | 100 | + | ||
| PT05L | Lyciscini | 5 | 5 | + | + | 92 | 55 | + | + | 100 | + | ||
| PT05O | Ooderini | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PT06 | Coelocybinae | 4 | 4 | – | – | – | – | – | – | – | – | ||
| PT07 | Colotrechninae | 2 | 2 | – | – | – | – | – | – | – | – | ||
| PT08 | Cratominae | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PT09 | Diparinae | 6 | 8 | – | – | – | – | – | – | – | – | ||
| PT09D | Diparini | 4 | 4 | – | – | – | – | – | – | – | – | ||
| PT09N | Neapterolelapini | 1 | 2 | 57 | 55 | 96 | 73 | 63 | + | 81 | – | ||
| PT10 | Epichrysomallinae | 16 | 28 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 93 | ||
| PT11 | Eunotinae | 6 | 7 | – | – | – | – | – | – | – | – | ||
| PT11E | Eunotini | 4 | 5 | 52g | 75g | 90g | 86g | 93g | 98g | 100g | 61g | ||
| PT11M | Moranilini | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PT11T | Tomocerodini | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PT12 | Eutrichosomatinae | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PT13 | Herbertiinae | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PT14 | Leptofoeninae | 2 | 3 | – | – | – | – | – | – | – | – | ||
| PT15 | Macromesinae | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PT16 | Miscogasterinae | 9 | 10 | – | – | – | – | – | – | – | – | ||
| PT16M | Miscogasterini | 5 | 6 | – | – | – | – | – | – | – | – | ||
| PT16S | Sphegigasterini | 2 | 2 | – | – | – | – | – | – | – | – | ||
| PT16T | Trigonoderini | 2 | 2 | – | – | – | – | – | – | – | – | ||
| PT17 | Ormocerinae | 6 | 5 | – | – | – | – | – | – | – | – | ||
| PT17M | Melanosomellini | 3 | 3 | – | – | par | – | + | – | – | – | ||
| PT17S | Systasini | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PT18 | Otitesellinae | 3 | 4 | par | – | – | – | – | – | – | – | ||
| PT19 | Panstenoninae | 1 | 2 | 96 | 89 | 98 | 98 | 84 | 77 | 100 | 96 | ||
| PT20 | Pireninae | 4 | 4 | – | – | – | – | – | – | – | – | ||
| PT21 | Pteromalinae | 17 | 18 | – | – | – | – | – | – | – | – | ||
| PT21P | Pteromalini | 4 | 4 | – | – | – | – | – | – | par | – | ||
| PT22 | Spalangiinae | 1 | 3 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| PT23 | Sycoecinae | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| PT24 | Sycophaginae | 5 | 6 | 82 | 94 | 91 | 81 | 77 | 91 | 100 | + | ||
| PT25 | Sycoryctinae | 2 | 2 | – | – | – | – | – | – | – | – | ||
| ROT | Rotoitidae (2/2) | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| SI | Signiphoridae (4/76) | 8 | 26 | 81 | 80 | 95 | 98 | 97 | 97 | 100 | 52 | ||
| SIS | Signiphorinae | 1 | 9 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 99 | ||
| SIT | Thysaninae | 3 | 12 | par | par | par | par | par | par | par | par | ||
| TAN | Tanaostigmatidae (9/92) | 4 | 5 | 98h | 95h | 99h | 100h | 99h | 100h | 100h | 77h | ||
| TE | Tetracampidae (15/50) | 6 | 7 | – | – | – | – | – | – | – | |||
| TEM | Mongolocampinae | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| TEP | Platynocheilinae | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| TET | Tetracampinae | 4 | 5 | 100i | 100i | 100i | 100i | 100i | 100i | 100i | 97i | ||
| TO | Torymidae (68/986) | 29 | 41 | – | – | – | – | – | – | – | – | ||
| TOM | Megastigminae | 3 | 6 | 66 | 67 | 99 | 99 | 97 | 97 | 100 | 92 | ||
| TOT | Toryminae | 28 | 37 | – | + | 67 | + | + | 62 | 86 | + | ||
| TOTI | Toryminae i.s. | 3 | 4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| TOTM | Microdonteromerini | 6 | 8 | – | – | – | par | par | – | par | par | ||
| TOTN | Monodontomerini | 6 | 8 | 80 | par | 100 | 91 | 89 | 81 | 100 | 97 | ||
| TOTP | Palachiini | 2 | 2 | – | – | – | – | – | – | – | – | ||
| TOTO | Podagrionini | 4 | 4 | par | 57 | par | 90 | par | 55 | 62 | + | ||
| TOTT | Torymini | 3 | 6 | 75 | 74 | 66 | 87 | 68 | 66 | 100 | – | ||
| TOTY | Torymoidini | 4 | 5 | par | – | – | – | – | – | 88 | – | ||
| TR | Trichogrammatidae (83/839) | 12 | 21 | – | + | 61 | 65 | 64 | + | 94 | + | ||
| TRO | Oligositinae | 9 | 10 | 98 | 100 | 97 | 96 | 95 | 93 | 100 | + | ||
| TROI | Oligositinae i.s. | 3 | 4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| TROC | Chaeotostrichini | 2 | 3 | 99 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| TROO | Oligositini | 1 | 2 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| TROP | Paracentrobiini | 1 | 1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| TRT | Trichogrammatinae | 3 | 11 | + | par | par | par | par | par | par | par | ||
| TRTI | Trichogrammatinae i.s. | 3 | 5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | ||
| TRTT | Trichogrammatini | 2 | 6 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| Number of clades with positive support: | 56 | 59 | 60 | 58 | 58 | 59 | 62 | 52 | |||||
Dataset abbreviations explained in Table 4. RAxML majority rule (MJR) is a consensus across all 16 submatrices. Support values are bootstrap percentages. The number of clades with positive support is summed for all clades with either a + (presence) or numerical support; par = paraphyletic; – = not monophyletic. Estimated diversity (genera/species) after family group names from Noyes [4]. Taxa represented by a single OTU or incertae sedis (i.s.) were considered not applicable (n/a) for clade support.
= without Agaonidae Group 4 (Wiebesia and Blastophaga R1757);
= without Azotinae or Eretmocerus;
= excluding Akapalinae and Philomidinae;
= without Trisecodes;
= excluding Buresium;
= including Idioporus;
= excluding Idioporus;
= not including Cynipencyrtus;
= excluding Diplesiostigma.
Results
Alignment models, tree length and clade support
Summaries of the 16 datasets generated from the two submatrices are presented in Table 2. The core region (SS) was 2996 bp in length and only slightly shorter than the MAFFT alignment of the same data (3,024–3,025 bp), with the differences accumulated mostly in the 28S D2 region. The application of the guide tree to the RAAs produced the longest alignment (4,369–4,536 bp) with the greatest impact on the length of the 28S D2 and D3 regions. Application of the guide tree greatly increased the number of parsimony informative sites (1,675–1,773 bp), the number of uninformative (autapomorphic) sites (550–565 bp), and had the greatest impact on tree length using the SSME dataset as a metric (32,220–32,236 steps) (Table 2). The MAFFT aligned RAAs without a guide tree were added to both the core region (SSME, SSMG and SSML) and to the MAFFT alignment of the core region (MEME, MGMG and MLML). Using mapped state changes and the SSME metric, the core + no guide tree RAAs datasets produced the shortest tree topologies (31,951–31,957 steps). Both the alignment length, and the RAxML best score differed very little within the different MAFFT variants of each alignment model. The MAFFT alignment of all data without regard to partition (MESR, MGSR and MLSR) produced an alignment of intermediate length (4,099–4,139 bp).
Phylogenetic Analyses
A summary of supported clades across six of the 16 analyses is presented in Tables 3 and 4, along with a summary of the >50% majority rule consensus support (MJR) across all 16 best known likelihood (BKL) RAxML trees. We present the BKL tree from the SSME RAxML result (Figs 1– 7), with the caveat that this represents only one summary of relationships found within Chalcidoidea. The clade support tables are a better representation of the support for traditional subfamily and family groups (Table 3) and for some higher-level relationships (Table 4). When present, bootstrap support on Figures 1– 7 generally corresponds with support across all analyses. Surprisingly, there was little impact of alignment strategy (SS or MAFFT) on the results, except for a slight increase in support for various clades at all levels with the inclusion of RAAs (core and RAA, Tables 3, 4).
Table 4. Higher group relationships supported across various analyses.
| core only | core and RAA | RAxML | TNT | |||||
| Group Relationships | SSNR | MENR | SSME | SSGE | MGSR | MGMG | MJR | SSME |
| Pantolytomiya + Chalcidoidea | − | + | − | + | − | + | 62 | − |
| Diaprioidea (part) + Chalcidoidea | − | − | +a | − | − | − | 56 | − |
| ‘Diapriidae’ + Chalcidoidea | + | − | − | − | − | − | − | − |
| Mymarommatoidea + Chalcidoidea | − | − | − | − | + | − | − | − |
| (Proctotrupoidea + Diaprioidea) sister to Chalcidoidea | − | − | − | − | − | − | − | + |
| Chalcidoidea | 99 | 95 | 100 | 100 | 98 | 98 | 100 | 100 |
| remaining Chalcidoidea minus Mymaridae | 91 | 55 | 97 | 95 | 55 | 85 | 94 | + |
| remaining Chalcidoidea minus Rotoitidae and Mymaridae | + | + | + | 76 | + | + | 94 | − |
| Mymaridae: 4−segmented taxa | 74 | 78 | 75 | 87 | 57 | 80 | 88 | + |
| Mymaridae: 5-segmented taxa | + | + | 76 | 62 | 83 | + | 88 | + |
| Eulophidae: (Opheliminae + Perthiola) + Entiinae | − | − | + | + | − | − | 56 | + |
| Eucharitidae + Perilampidae | − | − | + | + | + | + | − | + |
| Perilampidae (with Akapalinae, Philomidinae and Idioporus) | + | + | par | + | + | + | − | − |
| Jambiya + Eucharitidae | − | − | + | + | + | + | − | + |
| Jambiya + Perilampidae | − | + | − | − | − | − | − | − |
| pteromaloid complexb | + | + | + | + | + | + | − | +c |
| Spalangiinae + Agaonidae | − | − | + | − | − | − | − | − |
| Sycophaginae + Agaonidae | + | − | − | − | − | − | − | − |
| remaining Agaonidae minus Tetrapusinae | + | 55 | + | − | − | + | − | + |
| Aphelininae + Coccophaginae | +d | − | − | − | − | − | − | − |
| Azotinae + Trichogrammatidae | + | + | + | − | + | + | 62 | − |
| Azotinae + Signiphoridae | − | − | − | − | − | − | − | + |
| Agaoninae + Blastophaginae (excluding group 4) | + | + | 65 | 61 | + | + | 62 | + |
a = Monomachidae + Diapriidae as sister groups;
b = includes Cratominae, Miscogastrinae, Otitesellinae, Panstenoninae, Pteromalinae and Sycoryctinae;
c = without Heterandrium (Otitesellinae);
d = including Platygerrhus (Microgasterinae: Trigonoderini).
Dataset abbreviations explained in Table 4. RAxML majority rule (MJR) is a consensus across all 16 submatrices. Support values are bootstrap percentages. Abbreviations: + refers to presence of clade but without numerical support; par = paraphyletic.
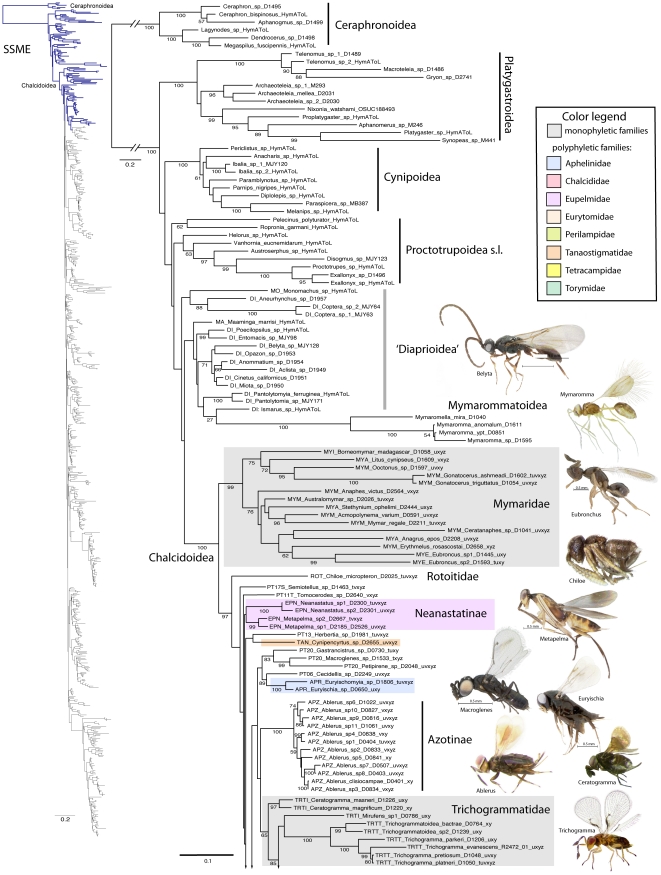
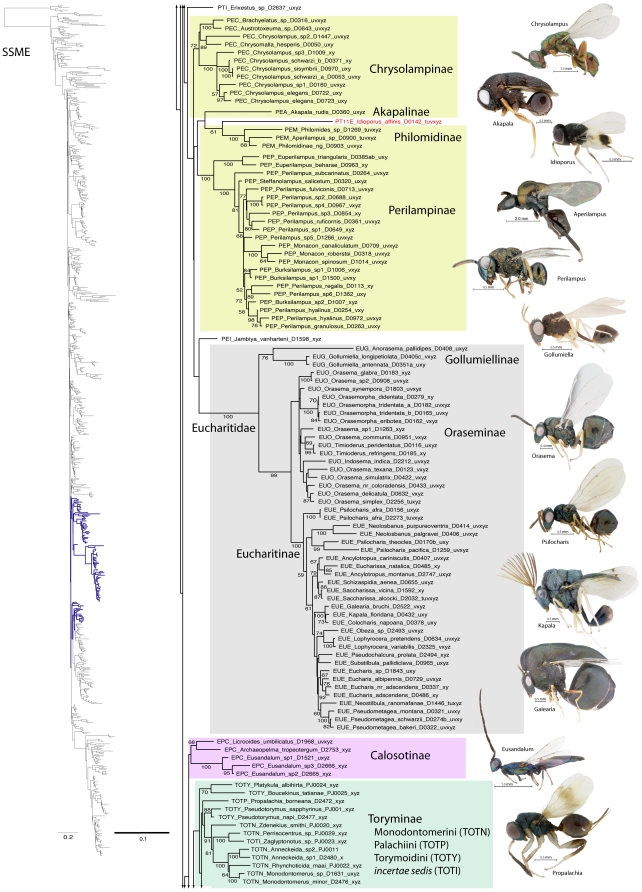
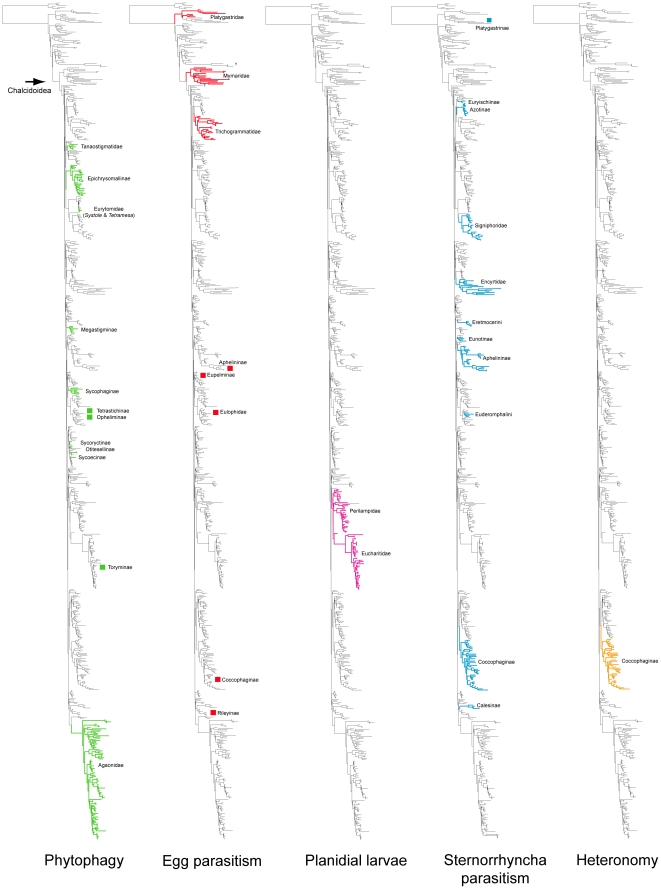
Figure 1. Phylogenetic tree from secondary structure alignment of stem data and E-INS-i alignment of RAAs (3917 aligned; SSME).
RAxML analysis with seed 38652 and 1000 rbs bootstrap replicates (support >50% above branches). Phylogram of entire tree on left colored to match inset. Taxon names with prefix indicating classification (see Table 3) and suffix indicating DNA voucher number and gene regions included for 18Sa-c (tuv) and D2 (x), D3 (y) and D4-5 (z). Monophyletic families indicated by gray shading; polyphyletic families other than Pteromalidae indicated according to inset color scheme.
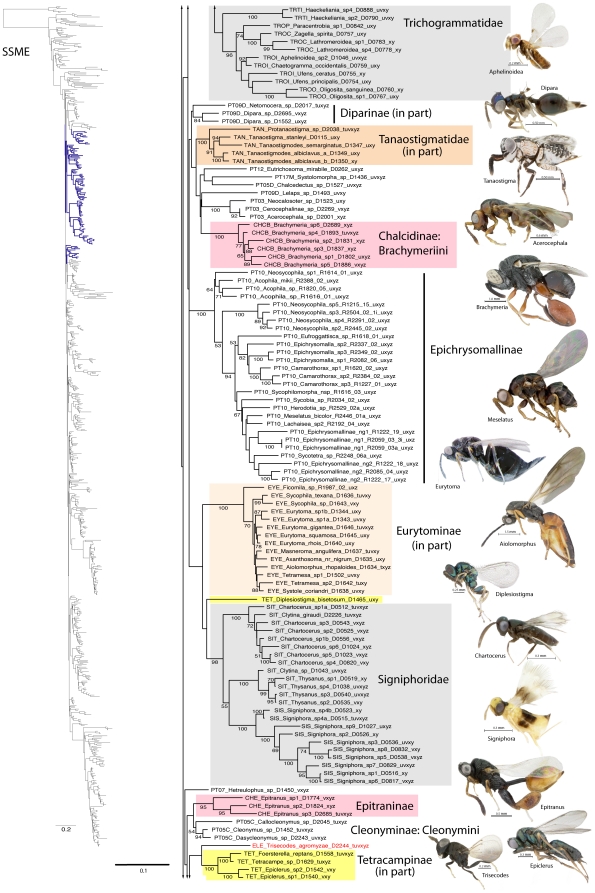
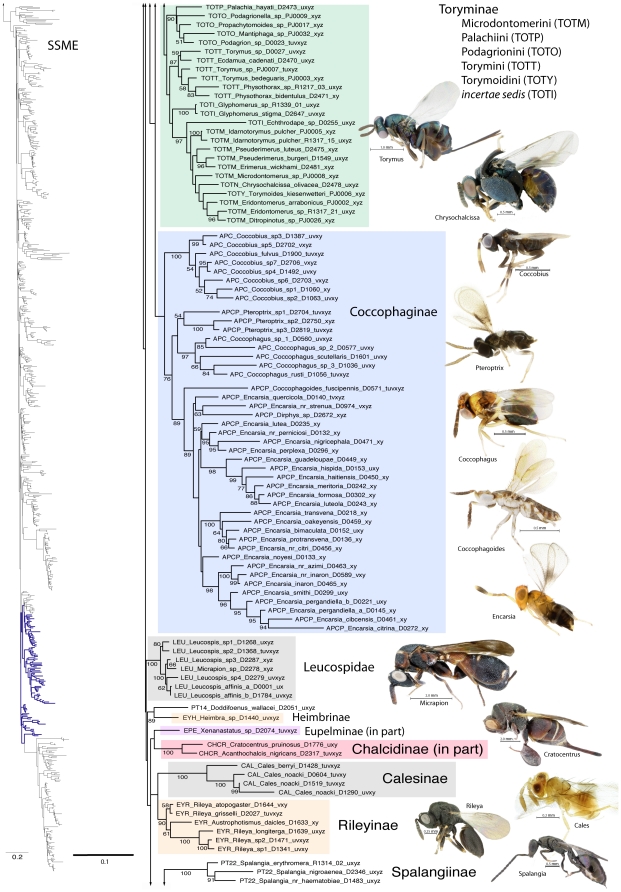
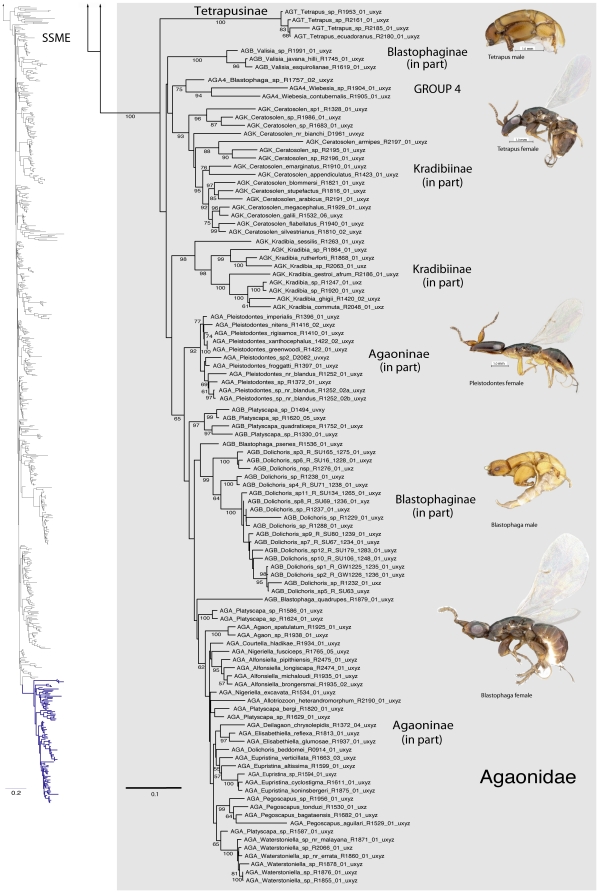
Figure 2. Phylogenetic tree of Chalcidoidea (continued).
Figure 3. Phylogenetic tree of Chalcidoidea (continued).
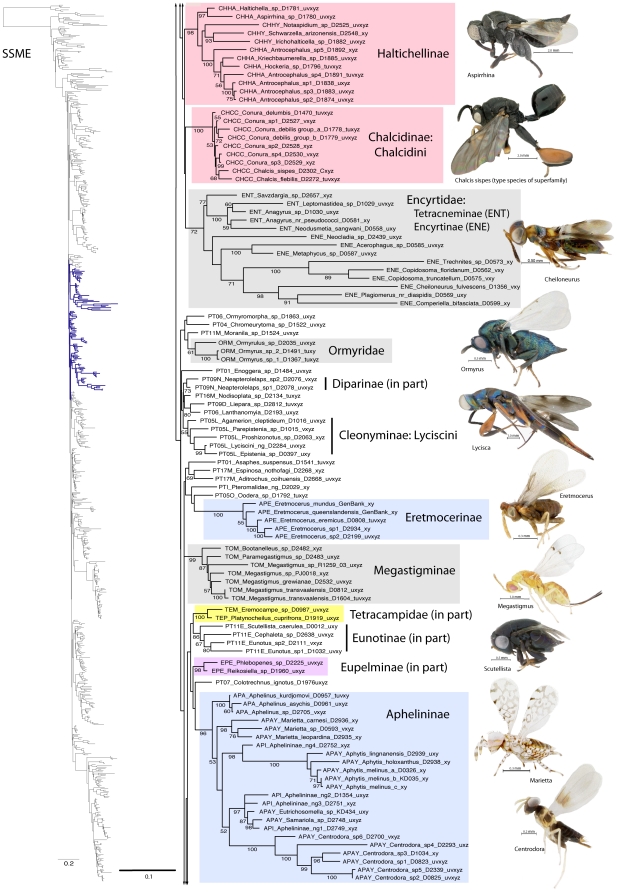
Figure 4. Phylogenetic tree of Chalcidoidea (continued).
Figure 5. Phylogenetic tree of Chalcidoidea (continued).
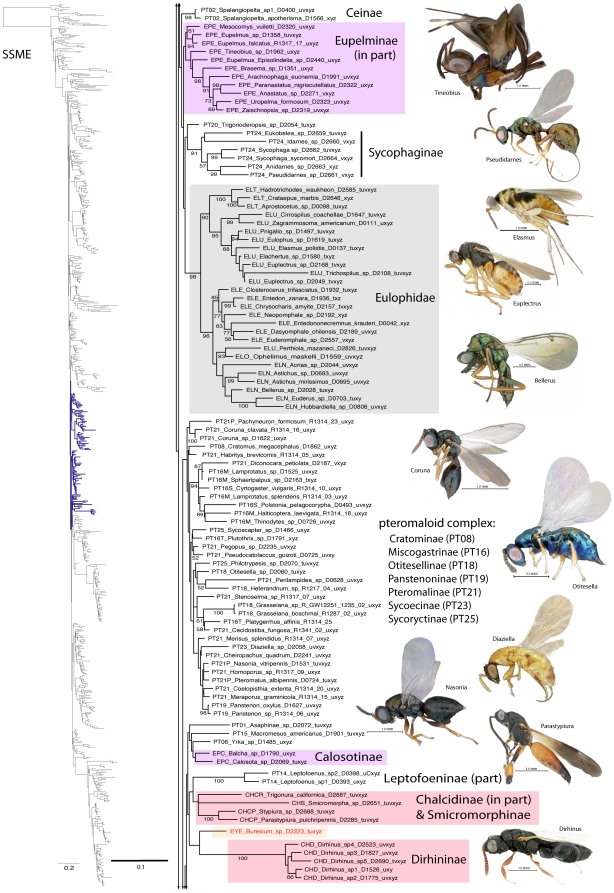
Figure 6. Phylogenetic tree of Chalcidoidea (continued).
Figure 7. Phylogenetic tree of Chalcidoidea (continued).
Interestingly, the automated (MAFFT) alignments of all data were comparable in clade support to any of the divided alignment strategies based on recognizing the core and stem data. There was slightly better clade support using G-INS-i when applied to data that included RAAs.
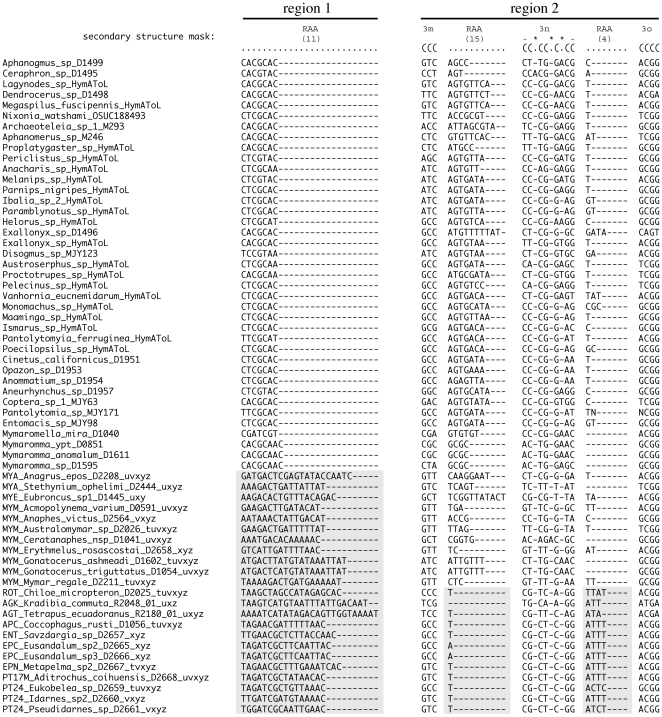
Informativeness of RAAs
Within 28S and 18S, distinct structural differences occur between RAA regions for the outgroups, Mymaridae, and the remaining Chalcidoidea taxa. For example, RAA(11) shows a pattern of increase in the number of bases and an associated decrease in degree of conservation for Chalcidoidea in comparison to the outgroup taxa (Fig. 8). Alternatively, RAA(15) reduces to a single nucleotide for Chalcidoidea, with the exclusion of Mymaridae. Within the same region, RAA(4) shows a slight but more subtle increase for Chalcidoidea excluding Mymaridae. RSC(4) and RSC(4′) both show support for Chalcidoidea excluding Mymaridae based on a respective increase to a 4 base motif (RSC 4), and an increase to a consistent AT or GT pattern (RSC 4′; not shown). These structural changes support both monophyly of Chalcidoidea and a sister group relationship between Mymaridae and the remaining Chalcidoidea. No RAA patterns were observed that would add support for relationships in the outgroup taxa. However within Chalcidoidea, additional structural changes within variable regions add support to some relationships (i.e., an increase in 18S loop(4) size in Perilampidae and Eucharitidae; and deletion of a contiguous variable region (RAAs 23-25) in Eulophinae + Tetrastichinae). Six variable regions in Agaonidae demonstrate substantial growth in size, both across and within the family, that distinguish them from all other Chalcidoidea. The different sizes of the variable regions might be expected to have the greatest impact on results from datasets contrasting the inclusion or exclusion of RAAs, or the MAFFT alignment without reference to the SS core structure; however, overall there appeared to be no impact, with all results consistently supporting monophyly of Chalcidoidea and a sister group relationship between Mymaridae and the remaining Chalcidoidea.
Figure 8. Examples of structural support from two sections of 28S-D2 (indicated by bar) for outgroups and a sampling of Chalcidoidea. RAA(11) shows an increase in the number of nucleotides and a decrease in the degree of conservation for Chalcidoidea including Mymaridae (highlighted).
In all Chalcidoidea excluding Mymaridae, RAA(15) undergoes a dramatic decrease to either 1 or no nucleotides and RAA(4) shows a slight increase in size. The bordering alignment around RAA(15) demonstrates compensatory changes in helices 3m, 3n and 3o.
Inclusion of the RAAs contributed to the monophyly of Encyrtinae, Entedoninae and Entiinae (Table 3). Their inclusion increased the BS support for a number of clades, including Agaoninae group 4, Encyrtidae, Eulophinae, Rileyinae, Lyciscini, Eunotini, Signiphoridae and Megastigminae (Tables 3, 4). At a higher group level, the inclusion of the RAA regions provided a greater amount of support for Eucharitidae + Perilampidae, and the genus Jambiya as the sister group of Eucharitidae. In no cases did the inclusion of RAAs result in a substantial decrease in support for a clade.
Phylogenetic Relationships
Relationships across the 16 ML analyses overall were the same regardless of alignment method or the inclusion or exclusion of RAAs (Figs. 1– 7, Tables 3, 4). The parsimony analysis of the SSME dataset produced more than 10,000 most parsimonious trees of 31,607 steps (RI = 0.62); however the strict consensus was well resolved (Supplementary Fig. S1) and in general accord with the likelihood results.
Outgroup relationships generally favored a paraphyletic Diaprioidea as sister group to Chalcidoidea (Fig. 1), but in a few cases Mymarommatoidea were the proposed sister group. A core Proctotrupomorpha clade of Proctotrupoidea sensu stricto, Diaprioidea, Mymarommatoidea and Chalcidoidea were supported in all results. Both Ceraphronoidea and Platygastroidea were distantly related in all analyses.
Chalcidoidea were always monophyletic with strong support, as was a sister group relationship between Mymaridae and the remaining Chalcidoidea (Table 4). Chiloe micropteron (Rotoitidae) was consistently supported in the likelihood results as the sister group of the remaining Chalcidoidea excluding Mymaridae (94% MJR), but with bootstrap support only in the SSGE results (BS 76). However, in the parsimony results Chiloe was deeply nested within Chalcidoidea (Supplementary Fig. S1).
Relationships within Chalcidoidea were highly variable along the backbone of the tree and should be regarded as a broad polytomy, but with consistent and sometimes strong support for many traditional taxon groupings at the family, subfamily, and tribe levels (Table 3). There is sometimes a lack of support for families that can be defined by several justifiable synapomorphies such as Chalcididae, and there is consistent support for some other families such as Eulophidae that are founded on what might be considered as weak loss or reductive features [9].
Discussion
Comparison of alignment strategies
Overall, there was little impact of the application of different MAFFT alignments to either the RAA regions, the core secondary structure data, or to the different gene regions without reference to secondary structure. This is optimistic for the future inclusion of new taxa to our data set where we can avoid the labor-intensive approach of having to align new taxa to our existing secondary structure model. Inclusion of the RAAs contributed to monophyly and clade support for a number of taxa, and also increased support at higher levels. Furthermore, structural differences found in various RAAs (Fig. 8) provide clear support for Chalcidoidea, a sister-group relationship between Mymaridae and other Chalcidoidea, and for some of the higher-level groups within Chalcidoidea. Clearly, RAAs do provide some phylogenetic signal and their inclusion in analyses is warranted despite some authors recommending complete [52] or partial [19] deleting of these regions.
Outgroup relationships
We found either Mymarommatoidea or Diaprioidea as the sister group of Chalcidoidea. These equivocal results were similar to results from a recent analysis of Hymenoptera that used more extensive molecular data from four gene regions and nearly complete 28S and 18S data [11]. Molecular data from both studies clearly support a monophyletic group of Diaprioidea, Mymarommatoidea and Chalcidoidea within the Proctotrupomorpha. With the inclusion of morphological data in a combined analysis, Mymarommatoidea is the sister group of Chalcidoidea [13], as hypothesized by Gibson [10]. Unfortunately, the biology of Mymarommatoidea remains unknown, making it difficult to compare with Chalcidoidea.
Phylogenetic relationships within Chalcidoidea
Chalcidoidea are well supported as monophyletic. Mymaridae are strongly supported as monophyletic and the sister group of the remaining Chalcidoidea. This hypothesis was first proposed by Gibson [10] based on morphology, and substantiated by Heraty et al. [11] and Sharkey et al. [13]. Chiloe micropteron (Rotoitidae) was the sister group of the remaining Chalcidoidea in all of the likelihood results, but not using parsimony. With more extensive gene sampling, Heraty et al. [11] recovered the same relationships in likelihood analyses of the eye-aligned data, and with parsimony only in the data aligned by eye. Mymaridae and Mymarommatidae are both common in early to mid Cretaceous amber deposits [5], [6], [8], which support their early origin and sister group relationships. Rotoitidae is unknown in any fossil deposits, but has a potentially archaic pattern of distribution, with genera known only in New Zealand and southern Chile [6], suggesting a late cretaceous origin [53].
After Rotoitidae, the relationships within Chalcidoidea become vague. The backbone of the chalcidoid tree has little support, with taxonomic groups shifting in different analyses from the base to somewhere more apical in the topology. As well, there are few consistent sister group relationships supported among the higher-level groups. One of the few relationships that can be substantiated based on larval morphology, Eucharitidae + Perilampidae [54], occurs in some but not all results, and never has bootstrap support. This is not simply an artifact of our ribosomal dataset; similar results with poor backbone support were also found by Desjardins et al. [18] using 4 nuclear protein coding genes and far fewer taxa. We do recover support for many of the traditional higher-level groups within Chalcidoidea, mostly at the subfamily and tribe level, but also for a few diverse family groups such as Agaonidae, Eulophidae, Eucharitidae and Trichogrammatidae. We also recovered consistent support for a novel pteromaloid complex that is a mix of morphologically very distinct subfamily groups. For some of the traditionally well-supported groups such as Chalcididae, the majority of the included taxa were monophyletic in only one analysis. A similar rare grouping was also found for a monophyletic Signiphoridae + Azotinae.
We found some taxa that could not be placed within any traditional higher-level group. There were also a few singleton taxa that defied placement, including Diplesiostigma, Cynipencyrtus and Idioporus. Interestingly, Idioporus is also difficult to place based on morphology, although neither Perilampidae (likelihood) or Rotoitidae (parsimony) were ever suggested as being related based on a morphological study by LaSalle et al. [55]. Calesinae are currently incertae sedis within Chalcidoidea [56], and our results to not offer any potential sister groups for this clade. Pteromalidae, as expected, is polyphyletic and affects greatly the composition and relationships of other taxa. Our results will be reevaluated in a combined morphological analysis, which is currently underway (Heraty et al. in prep), but it is clear that the family level relationships of Chalcidoidea are in need of major revision.
For the discussions below, some historical information on relationships is presented for each family group followed by the results of the current study. A more detailed review of classification history and biology can be found in Gibson et al. [9] and Hanson & Gauld [57]. We try not to discuss relationships of taxa within supported clades, but most often species within the same genera and species groups were monophyletic, and relationships within a clade were generally the same across different analyses (Figs 1– 7).
Agaonidae
Agaoninae and Sycophaginae (as Idarninae), once included in Torymidae, were moved to Agaonidae by Bouček [30]. Agaonidae sensu lato were comprised of Agaoninae, Epichrysomallinae, Otitesellinae, Sycoecinae, Sycophaginae and Sycoryctinae [58]. Bouček noted that there were no unique morphological characters to define Agaonidae sensu lato, yet argued against limiting the family to the pollinating group (Agaoninae) and suggested a sister-group relationship of at least Agaoninae + Sycophaginae. Grissell [34] suggested that Agaonidae (sensu lato) may form a derived clade within the Torymidae. Rasplus et al. [59] revised the Agaonidae, having determined that it was not monophyletic, limiting the family to include only Agaoninae (Agaonidae sensu stricto). Cruaud et al. [23] analyzed relationships within Agaonidae s.s. and proposed up to four subfamilies, Tetrapusinae, Agaoninae group 4 (potential subfamily), ‘Blastophaginae’ and ‘Agaoninae’, but with the latter two groups likely collapsing into a single subfamily Agaoninae.
Agaonidae (sensu stricto) was monophyletic in all analyses with likelihood BS values of 100% and parsimony support of 97%. Tetrapusinae were recovered with 100% BS in all analyses (Table 3), and were either sister group to the remaining Agaoninae, as reported in [23], or nested within Agaonidae (Table 4). Agaonidae Group 4 was monophyletic in all of the likelihood results, but not parsimony. Kradibiinae were never recovered as monophyletic, although both genera, Kradibia and Ceratosolen, were each monophyletic. Agaoninae were rendered paraphyletic in all analyses by Blastophaginae, but a monophyletic group of Agaonidae + Blastophaginae, excluding Agaonidae Group 4, was recovered in most results with low support (Table 4).
None of the other subfamilies previously placed in Agaonidae were placed near to Agaonidae, although in the SSNR dataset (core only), Sycophaginae were placed as the sister group of Agaonidae but without bootstrap support.
Aphelinidae
Woolley [60] suggested that monophyly of Aphelinidae was not certain, and noted the historical tendency to group all parasitoids of adult and nymphal Hemiptera into Aphelinidae without an understanding of relationships. Presently, most authors recognize that Aphelinidae may be paraphyletic if not polyphyletic [9], [17], [61]. Characters uniting the Aphelinidae may also not be apomorphic [24], [62]. Based on only a few taxa, Aphelinidae were paraphyletic in the molecular analysis of Campbell et al. [17]. Previous authors have placed aphelinids within various families, including Eulophidae [63], [64], Encyrtidae [65], [66], Pteromalidae [62] or as a distinct family [67]. Rosen and DeBach [68] noted that Aphelinidae share morphological affinities with both Encyrtidae (shape of the mesopleura and structure of the pro- and mesotibial spurs) and Eulophidae (thoracic sclerite morphology and antennal segmentation). Gibson [69] hypothesized an Aphelinidae + Signiphoridae relationship on the basis of the structure of the mesotrochantinal plate and metasternum, a relationship also proposed by Domenichini [70]. Woolley [71] found strong morphological evidence uniting Azotinae + Signiphoridae. Compere and Annecke [67] and Rosen and DeBach [68] considered Aphelinidae to be more closely related to Signiphoridae and Encyrtidae. Viggiani and Battaglia [72] proposed that Aphelinidae were morphologically allied with Eulophidae and Trichogrammatidae. Relationships within Aphelinidae are just as, if not more, complex [24], [63], [73], [74], [75], [76], [77], [78], [79], [80], [81], [82]. The most recent treatment of Aphelinidae [24] recognizes the following subfamilies and tribes; Aphelininae (tribes Aphelinini, Aphytini, Eretmocerini and Eutrichosomellini), Eriaphytinae, Azotinae, Coccophaginae (tribes Coccophagini, Physcini and Pteroptricini), Eriaporinae and Euryischiinae. Noyes [4] uses Eretmocerinae, which we follow herein. Calesinae were excluded from Aphelinidae by Hayat [24].
Our results lend support to the idea that Aphelinidae are not monophyletic (Figs 1– 6). At best, the two subfamilies Aphelininae (excluding Eretmocerus) + Coccophaginae were monophyletic in the SSNR analysis. Aphelininae, Azotinae (Ablerus), Eretmocerinae (Eretmocerus) and Euryischiinae were each recovered with very strong BS support in all analyses (Table 3). Coccophaginae were monophyletic in the majority (94%) of likelihood analyses, but Coccobius was excluded from the other taxa in the parsimony results (Table 3). In the majority of cases, the aphelinine tribes Aphelinini (Aphelinus), Aphytini, and Eutrichosomellini (all Aphelininae) are monophyletic, although Eutrichosomellini often renders Aphytini paraphyletic. Within Coccophaginae, Coccophagus consistently rendered Pteroptricini paraphyletic. Within Pteroptricini, Encarsia is consistently rendered paraphyletic by Dirphys.
There was no consistent or plausible sister group taxon for Aphelininae or Coccophaginae. In the majority of analyses, Euryischiinae is sister to Cecidellis sp. (Coelocybinae: Pteromalidae), which can be justified morphologically (RGB). The monogeneric Eretmocerinae is monophyletic with strong support in all results, but has no association with other aphelinid taxa. Azotinae were always monophyletic, with 100% bootstrap support, with former members of Azotus rendering Ablerus paraphyletic, which is an expected result. Azotinae were the sister group to Trichogrammatidae in the likelihood results, but without bootstrap support (Table 4). Monophyly of Azotinae + Signiphoridae is supported by several morphological synapomorphies [71], but this group was recovered only in the parsimony results (Table 4).
Calesinae (unplaced to family)
Cales (Calesinae) were excluded from Aphelinidae and left unplaced in Chalcidoidea by Hayat [83]. Mottern et al. [56] recently reviewed the Calesine, and discussed its unique morphology and potential relationships with various taxa, including Aphelinidae, Eretmocerinae, Eulophidae, Mymaridae and Trichogrammatidae.
Calesinae were monophyletic with 100% BS support in all analyses (Fig. 6). Included in our analysis are two morphological and geographically distinct species, Cales berryi from New Zealand, and Cales noacki from South America, including Chile. This same pattern of distribution was used as an argument for the archaic placement of Rotoitidae. Although Cales was intermediate between Mymaridae and other Chalcidoidea in Campbell et al. [17], it was always well nested within Chalcidoidea in all of our results. No consistent outgroups were identified in any of our results.
Chalcididae
Bouček and Halstead [84] noted that the classification of Chalcididae has changed little over the years. A sister-group relationship with Leucospidae or even the inclusion of Leucospidae within Chalcididae was suggested by Gibson [16], [85]. Monophyly of Chalcididae has not been previously doubted, largely based on four morphological synapomorphies [86], [87]. Traditional classifications have included Chalcidinae with the tribes, Chalcidini, Cratocentrini, Phasgonophorini and sometimes Brachymeriini, with other subfamilies including Dirhininae, Epitraninae, Haltichellinae and Smicromorphinae [30], [88]. In a phylogenetic analysis of the family, Wijesekara [86] proposed that Smicromorphinae were nested within Chalcidinae, with Chalcidinae including Smicromorphinae sister to the remaining chalcidids, followed by a sequence of Cratocentrinae, Brachymeriinae (Brachymeriini + Phasgonophorini), and finally Dirhininae (Dirhinini + Epitranini) + Haltichellinae (Haltichellini + Hybothoracini). Noyes [4] did not recognize Brachymeriinae, which is the convention followed herein.
Chalcididae were not monophyletic in any of our analyses. The MENR analysis produced the closest approximation to a monophyletic Chalcididae, with a grouping of Dirhinus (Dirhininae), Epitranus (Epitraninae), Chalcidinae, Brachymeria (Brachymeriinae), Phasgonophorini and Trigonura (Cratocentrini). However, this group surprisingly also included two pteromalid subfamilies (Macromesinae and Leptofoeninae) and excluded Cratocentrus and Acanthochalcis (Cratocentrini). Otherwise, the various groups were inconsistent in their grouping in the other analyses. At the subfamily level, Epitraninae, Dirhininae and Haltichellinae were all monophyletic with very strong BS support (Table 3). Smicromorphinae included only a single taxon, and was either independent from other chalcidids or it grouped with Cratocentrini or Phasgonophorini, but never with Chalcidini as proposed by Wijesekara. The subfamily Chalcidinae were never monophyletic, but the tribes Brachymeriini, Chalcidini and Phasgonophorini all had very high BS support across all analyses (Table 3). Interestingly, our Old World representatives of Chalcis (the type genus of the superfamily; occurring Worldwide) render the widespread New World genus Conura paraphyletic in all analyses. While monophyly of Haltichellinae was supported in all analyses, monophyly of the two tribes, Haltichellini and Hybothoracini, varied.
Our results do not offer much resolution for the relationships within Chalcididae, but do offer support for recognition of Brachymeriinae, Dirhininae, Epitraninae, Chalcidinae (as Chalcidini), Haltichellinae and Smicromorphinae. Both Phasgonophorini and Cratocentrini are less easily placed, and we could not recover the monophyly of the Cratocentrini (Trigonura and Acanthochalcis + Cratocentrus) in any of our analyses. Leucospidae never grouped with any of the chalcidid families, which contradicts hypotheses that they are the sister group of Chalcididae, or that they might render Chalcididae paraphyletic.
Encyrtidae
The monophyly of Encyrtidae is not questioned and there is strong morphological evidence to support this family [89]. An Encyrtidae + Tanaostigmatidae sister-group relationship has often been proposed, with this clade in turn being sister to Eupelmidae [69], [89], [90], [91]. Noyes et al. [89] followed the division of Encyrtidae into the subfamilies Tetracneminae and Encyrtinae [92], [93], [94] and noted that while Tetracneminae is undoubtedly monophyletic, Encyrtinae may represent a paraphyletic assemblage.
Encyrtidae were monophyletic across all analyses, with moderate to very strong BS support from the likelihood analyses with RAAs included (Table 3). Tetracneminae were monophyletic with moderate to very strong support across most analyses, with Encyrtinae forming either a paraphyletic or monophyletic sister group. The extraordinary branch lengths found within Encyrtidae (Fig. 3) occur in the results of both SS and SS + RAA analyses, and thus are not simply the result of having several taxa with long RAA inserts. Our results never supported a close relationship with Cynipencyrtus, Tanaostigmatidae or any of the eupelmid subfamilies.
Eucharitidae
Several morphological features support the monophyly of Eucharitidae [28]. Largely on the basis of the highly sclerotized first instar larva (planidium), Heraty and Darling [54] proposed a sister-group relationship for Eucharitidae and Perilampidae. Based on molecular and morphological evidence, Gollumiellinae form the sister group of Oraseminae + Eucharitinae [6], [37]. Akapalinae and Philomidinae were proposed as belonging to Eucharitidae by Bouček [30]. Philomidinae share planidial larvae with Eucharitidae [95], but immatures of Akapalinae are unknown.
Eucharitidae sensu stricto (Gollumiellinae, Oraseminae and Eucharitinae) were monophyletic with 100% BS support across all analyses. Akapalinae were grouped with Perilampinae in all of the likelihood results, but as the sister group of Eucharitidae s.s. in the parsimony analysis. Philomidinae were never grouped with Eucharitidae.
While Eucharitinae were always very strongly supported, Oraseminae was occasionally paraphyletic to Eucharitinae. Gollumiellinae was paraphyletic only in the parsimony analysis. Monophyly of Psilocharitini (Psilocharis and Neolosbanus) is not supported, which is similar to results from other molecular studies [37].
A Eucharitidae + Perilampidae sister group was retrieved in most of the likelihood analyses that included RAAs, and also in the parsimony analysis (Table 4); however, without bootstrap support. Morphological support for this group rests on the presence of a sclerotized planidial first-instar larvae [54], [95], and we place some degree of confidence in results that support their monophyly. With the inclusion of Philomidinae in this clade, it would support a single origin of planidia larvae within Chalcidoidea (Fig. 9). However, parsimony results supported a monophyletic Perilampidae + Eucharitidae, without Philomidinae, which was grouped instead with some Phasgonophorini (Chalcididae) and Rileyinae (Eurytomidae).
Figure 9. Five life history traits mapped onto SSME likelihood tree. Colored squares refer to presence of a trait in a clade, but not in a member sampled in this study.
Eulophidae
Monophyly of Eulophidae generally has not been challenged, although morphological support is based almost entirely on character reduction [29]. Based largely on molecular evidence, Elasmidae was synonymized with Eulophidae by Gauthier et al. [96]. At a higher level, Schauff et al. [97] suggested a grouping of Eulophidae, Elasmidae and Trichogrammatidae, but made note that there was no strong evidence for such a relationship. Eulophinae were suggested to be the most basal of the four subfamilies due to their “less-specialized features” [97]. In a combined analysis, Burks et al. [29] proposed that Eulophinae + Tetrastichinae were the sister group of (Opheliminae + Entiinae) + Entedoninae. The only eulophid with three-segmented tarsi, Trisecodes, was removed from Entedoninae and placed as incertae sedis within Eulophidae [29]. The whitefly parasitoid group Euderomphalini were sister group to Entedonini in Entedoninae, which was contrary to their placement in Entiinae by Gumovsky [98].
Eulophidae were monophyletic with strong to very strong support in all of our analyses (Fig. 4, Table 3), but with the exclusion of Trisecodes, which in all analyses was sister group to taxa outside Eulophidae. Support was consistently very high for Tetrastichinae, and increased with the inclusion of RAAs for Entedoninae, Entiinae and Eulophinae. As proposed by Gauthier et al. [96], Elasus (formerly Elasmidae) was always nested within Eulophinae. As well, Tetrastichinae and Eulophinae (including Elasmus) have a unique deletion of a contiguous variable region (RAAs 23-25). Perthiola (Anselmellini) was always the sister group Ophelimus with high bootstrap support. Anselmellini were placed outside of Eulophinae by Gauthier et al. [96]. With added resolution from the RAAs, Perthiola + Opheliminae grouped either with Entiinae (54% of likelihood trees and parsimony; Table 4) or with Entedoninae. Without the RAAs, these four groups were monophyletic but unresolved. Our results support the hypothesis of relationships suggested by Burks et al. [29], and substantiate the potential inclusion of Anselmellini within Opheliminae.
The exclusion of Trisecodes from Eulophidae as proposed by Burks et al. [29] is justified. This genus was usually placed (81% of likelihood analyses and parsimony), but without strong support, as the sister group of Tetracampinae (excluding Diplesiostigma), and was never grouped with other Eulophidae.
Importantly, there was no relationship supported for Eulophidae with any of the aphelinid subfamilies, including Calesinae, which have many similar reductive features [56]. The analyses without RAAs (SSNR, MENR) did support a Eulophidae + (Azotinae + Trichogrammatidae) clade, but otherwise there were no consistent outgroups, and never any groups that have been previously proposed in the literature.
Eupelmidae
While there is strong morphological support for the monophyly of each of the three subfamilies of Eupelmidae, it has been proposed that the family might represent a grade rather than a clade [9], [69], [99], [100]. The grade was implicated to include Encyrtidae and Tanaostigmatidae, and potentially Aphelinidae, which all share an expanded acropleuron and other associated features; however, there is also a possibility of closer relationships of one or more subfamilies to Cleonyminae (Pteromalidae) [69].
Eupelmidae were never monophyletic. Also, its subfamilies Calosotinae, Eupelminae, and Neanastatinae were almost never monophyletic. The SSME dataset was one of the rare instances in which Neanastatinae were monophyletic (Fig. 1), but in the same results both Calosotinae and Eupelminae occur twice in very different parts of the tree (Figs 3– 6). Eupelminae were monophyletic in some analyses, including both datasets that did not include the RAAs (Table 3). Calosotinae were never monophyletic, with Calosota and Balcha grouping distantly from Archaeopelma, Licrooides and Eusandalum. None of the Eupelmidae ever grouped with Tanaostigmatidae or Encyrtidae.
Eurytomidae
The monophyly of Eurytomidae was recently questioned as no synapomorphies defining the family are known [101]. Indeed, the molecular analyses of Campbell et al. [17] and Chen et al. [102] and the morphological analyses of Lotfalizadeh et al. [103] failed to recover a monophyletic Eurytomidae. Stage & Snelling [104] recognized Heimbrinae, Rileyinae and Eurytominae, with the latter including the previously recognized Buresiinae. Chen et al. [102] proposed elevating Rileyinae to family status, while Lotfalizadeh et al. [103] found Rileyinae to consist of two clades of unrelated taxa (Rileya and Macrorileya + Buresium). Both molecular and morphological investigations found Eurytoma to be polyphyletic [102], [103].
Eurytomidae was never recovered as monophyletic in any of our analyses. However, Eurytominae (excluding Buresium) were monophyletic in all of our analyses with very high support (Table 3). Rileya (Rileyinae) were monophyletic in all analyses, but with very high support only in the likelihood analyses when RAAs were included. Both Heimbra (Heimbrinae) and Buresium (Eurytominae) never grouped with the other eurytomid genera. No logical outgroups were identified.
Leucospidae
Leucospidae are generally recognized as a monophyletic group of four genera closely related to Chalcididae [86], [105]. However, characters proposed to support the monophyly of this combined lineage are all problematic and potentially convergent [9], [86].
Leucospidae were monophyletic and had greater than 90% support across all analyses. Our one species of Micrapion (South Africa) consistently rendered Leucospis (worldwide representation) paraphyletic. No close association with Chalcididae was found.
Mymaridae
Although there was some early doubt about the monophyly of Mymaridae [106], the family has been well substantiated based on morphology and molecular evidence [17], [107], [108]. Huber [108] noted that the higher classification of Mymaridae is unstable, and as per the advice of Huber and Triapitsyn (personal communication) Mymaridae subfamilies have been abandoned and genera grouped according to their number of tarsal segments. Gibson [10] was the first to propose morphological evidence that Mymaridae might be the sister group of the remaining Chalcidoidea, but without firm resolution.
Mymaridae were found to be monophyletic in all analyses with very strong support (Fig. 1, Table 3). The 4-segmented tarsi group, represented by the genera Borneomymar, Gonatocerus, Litus and Ooctonus, were consistently monophyletic across all analyses with moderate to strong support (Table 4). The remaining genera, Acmopolynema, Anagrus, Anaphes, Australomymar, Ceratanaphes, Erythmelus, Eubroncus, Mymar and Stethynium, formed the 5-segmented tarsi group. This group is supported in most analyses (88% of likelihood analyses), with moderate to strong BS support only when RAAs were included. There was no support for Mymarinae or Alaptinae. Eubronchinae were monophyletic, but these were represented by only a single genus. Mymaridae were strongly supported as the sister group of the remaining Chalcidoidea in all analyses.
Ormyridae
Hanson [109], noted that the status and relationships of Ormyridae are uncertain. Members of the family have been included as a subfamily in Pteromalidae [111], Torymidae [112], or as their own family [30].
The two genera, Ormyrus and Ormyrulus, were monophyletic in all of our analyses but with low to very strong BS support (Fig. 3). In 56% of the likelihood analyses, all based on use of the core SS alignment and with or without RAAs, supported a sister-group relationship with Moranila (Pteromalidae: Eunotinae: Moranilini), but otherwise there were no consistent outgroup associations, and never any close association with either of the torymid subfamilies.
Perilampidae
The limits of Perilampidae are not clear, with variable inclusion of the subfamilies Chrysolampinae, Philomidinae and Perilampinae, and treatment of each or all groups as a separate family or subfamily of Pteromalidae [9], [100], [113]. Akapala (Akapalinae) were initially placed in Perilampidae, but later transferred to Eucharitidae [30]. More recently, Jambiya was described and included within Perilampidae, but an association with either Chrysolampinae or Perilampinae could not be made [114]. Jambiya has an enlarged ovipositor, which is also a feature of basal lineages of Eucharitidae, and a relationship with that family cannot be rejected. A proposed relationship between Perilampidae, Philomidinae and Eucharitidae is based on presence of a planidial larva [54], [95].
In likelihood results, Perilampidae sensu stricto (Chrysolampinae + Perilampinae) was never recovered. With RAAs excluded, a monophyletic ‘Perilampidae’ was recovered with low support that included Chrysolampinae (67-73% BS), Perilampinae (96-98% BS), Akapalinae, Philomidinae and Jambiya. This group also included the pteromalid genus Idioporus (Pteromalidae: Eunotinae: Eunotini). In these analyses, Eucharitidae and Perilampidae were not monophyletic. With the inclusion of RAAs, the results are more variable, but often recover Perilampidae and Eucharitidae as monophyletic, Jambiya as sister group to Eucharitidae, but again with Philomidinae, Akapalinae and Idioporus nested within a paraphyletic or monophyletic Perilampidae, but still with Chrysolampinae and Perilampinae each monophyletic (Fig. 5). A monophyletic Perilampidae s.s. (Chrysolampinae + Perilampinae) was recovered only in the parsimony analysis. These results also supported Jambiya as the sister group of Akapalinae + Eucharitidae. Philomidinae were distantly placed with Phasgonophorini (Chalcididae) and Rileyinae (Eurytomidae). Thus, while Eucharitidae s.s. is well supported, there is conflicting support for the definition of Perilampidae and a definitive association with Eucharitidae.
Pteromalidae
Pteromalidae are essentially a dumping-ground for presumably monophyletic groups that cannot be assigned to established families and which lack family status in their own right [9]. Herein, we recognize the 30 subfamilies of Noyes [4], as well as the three non-pollinator fig-wasp associated subfamilies assigned to Pteromalidae (Otitesellinae, Sycoecinae and Sycoryctinae) or placed as incertae sedis (Epichrysomallinae and Sycophaginae) by Rasplus et al. [59]. Historically, many pteromalid subfamilies were elevated to family status, only to once again resume subfamily status within Pteromalidae [9]. There has been no comprehensive morphological analysis of the family. Molecular analyses have supported the concept of a polyphyletic assemblage, but even the most comprehensive studies have sampled relatively few taxa across the spectrum of the family [17], [18]. We were able to sample 25 of these 36 subfamilies, and where possible sample more extensively within groups (Table 3). We limit our discussion below to significant groupings or results. Notably, many of the taxa are ‘almost’ monophyletic, often with the exclusion of one or more taxa, and many of these cases will need to be evaluated elsewhere.
Pteromalidae were expected to be polyphyletic [9], [15], and were never retrieved as monophyletic. Several subfamilies were monophyletic and very strongly supported across all analyses including Ceinae (Spalangiopelta), Cerocephalinae, Epichrysomallinae, Panstenoninae (Panstenon), Pteromalinae, Spalangiinae (Spalangia) and Sycophaginae. In no case did support increase with the addition of RAAs. Of interest is the a novel grouping of the pteromalid subfamilies Cratominae (Cratomus), Miscogastrinae (except Nodisoplata), Otitesellinae, Panstenoninae, Pteromalinae, Sycoecinae (Diaziella) and Sycoryctinae. This grouping occurs in all analyses, including parsimony, but without bootstrap support. A clade of Miscogastrinae and Pteromalinae was strongly supported by Desjardins et al. [18], but none of these other subfamilies were included as part of that study. This ‘pteromalid complex’ is peculiar for its small amount of molecular divergence and high degree of morphological complexity, especially for the non-pollinating fig wasps Otitesellinae and Sycoryctinae. The low divergence and stability across various analyses suggest that the subfamilies in this group might eventually be synonymized under Pteromalinae. The taxononic placement of Nodisoplata, which was placed outside of this complex, needs to be reconsidered. The two other two fig-wasp associated subfamilies, Epichrysomallinae and Sycophaginae, were monophyletic but not associated with any consistent outgroup taxon. In one analysis without RAAs (SSNR), Sycophaginae were the sister group of Agaonidae, but without BS support. This result was proposed by Copland and King [115].
Coelocybinae, Ormocerinae, Pireninae and Pteromalinae were never monophyletic. Cleonyminae were polyphyletic. In all analyses, Cleonymini and Lyciscini were each monophyletic with low support in all analyses, with Lyciscini gaining increased support from the inclusion of RAAs. Chalcedectini (Chalcedectus) had variable relationships, but never with other Cleonyminae. Ooderini (Oodera) had sister-group relationships that varied from Leucospidae to Encyrtidae, and on two occasions, Lyciscini. Cratominae (Cratomus) had variable relationships throughout the analyses, but often occurred in the pteromalid complex as suggested by its morphology. Diparinae were never monophyletic, as also found by Desjardins et al. [18]. Eunotinae were never retrieved as monophyletic, and the tribes Moranilini and Tomocerodini, each represented by a single taxon, were inconsistently allied with other families. Eunotini were monophyletic and strongly supported in all of the analyses. Surprisingly, Leptofoeninae, which have strong morphological support, were never monophyletic. Ormocerinae were never monophyletic. Sycoryctinae and Otitesellinae were consistently polyphyletic which is a result supported by morphology [59]. Within Otitesellinae, the two Grasseiana species form a monophyletic group, while Heterandrium sp. and Otitesella sp. were inconsistently allied with other taxa. Panstenoninae were nested within Pteromalinae. Pireninae and Pteromalinae were never monophyletic. Spalangiinae were always monophyletic, but were never recovered with a consistent sister group.
For Pteromalidae, our results are similar to those of Desjardins et al. [18] based on an analysis of four protein coding genes. The family is polyphyletic with respect to most Chalcidoidea and few of the higher-level assemblages can be consistently grouped with other pteromalid or chalcidoid groups.
Rotoitidae
In their description of the family, Bouček and Noyes [116] noted that Rotoitidae may be the sister group of Tetracampidae and Eulophidae. Other potential associations have included Eulophidae, Mymaridae, Trichogrammatidae and Tetracampidae [15], [16]. Based on an analysis of both distribution and ovipositor morphology, Gibson & Huber [117] concluded that Rotoitidae might be the second most ancestral lineage of Chalcidoidea after Mymaridae, but noted that features of the antenna and mesosoma conflict with this conclusion.
Rotoitidae were represented by one species, Chiloe micropteron. In all but one of the likelihood analyses, it was basal and sister to the remaining Chalcidoidea after Mymaridae, with BS support for a monophyletic Chalcidoidea after Rotoitidae only in the SSGE results. The alternate likelihood result placed it as the sister group of Mymaridae, thus still basal within the superfamily. Parsimony results have Chiloe nested within Chalcidoidea as the sister group of Idioporus (Eunotinae: Eunotini) in a clade with Systolomorpha (Pteromalidae: Ormocerinae: Melanosomellini) and Trichogrammatidae. No morphological features would support this alternative hypothesis.
Signiphoridae
There is little doubt over the monophyly of Signiphoridae; however, Thysaninae may be paraphyletic with respect to Signiphorinae [71], [118]. Gibson [69] suggested a relationship between Signiphoridae and Aphelinidae, or members within Aphelinidae. Woolley [71] proposed a Signiphoridae + Azotinae sister group based on an unsegmented antennal club, presence of an epiproct [70] posterior to the syntergum in all female Azotinae and Signiphoridae, and apodemes projecting forward from the anterolateral angles of sterna 3 to 6 of the metasoma of females. Pedata and Viggiani [119] alluded to an azotine + signiphorid relationship with the discovery of tubercles above the spiracles of third instar Ablerus perspeciosus and Signiphora flavella larvae.
Signiphoridae and Signiphorinae (Signiphora) both monophyletic with very strong support across all analyses (Table 3). Thysaninae were paraphyletic in all of our results. The placement of Clytina was puzzling, with C. giraudi rendering Chartocerus paraphyletic in all analyses, while Clytina sp. D1023 was consistently the sister group of Thysanus.
Signiphoridae were not placed with Azotinae, or any logical outgroup, in any of the likelihood analyses. In these analyses, Azotinae was consistently the sister group of Trichogrammatidae. However, in the parsimony analysis, Azotinae and Signiphoridae were monophyletic and did not group with Trichogrammatidae.
Tanaostigmatidae
Tanaostigmatidae sensu LaSalle [90] is a distinct monophyletic group. LaSalle and Noyes [91] transferred Cynipencyrtus from Encyrtidae to Tanaostigmatidae, yet noted that this genus was morphologically and biologically distinct from other members of the family. It has been argued that Cynipencyrtus could be sister to Encyrtidae, sister to Tanaostigmatidae + Encyrtidae, or sister to Tanaostigmatidae alone [9], [69], [99]. There is strong morphological support for monophyly of the Tanaostigmatidae + Encyrtidae clade, but weaker support for the inclusion of Eupelmidae within this group [9].
Tanaostigmatidae sensu stricto (without Cynipencyrtus) was always monophyletic with strong support. Cynipencyrtus was variously allied with other taxa throughout the different analyses, and tanaostigmatids were never the sister group of Encyrtidae. This disparate grouping may be an artifact of the larger analysis, as we have been able to recover Tanaostigmatidae + (Cynipencyrtus + Encyrtidae) in a study with a smaller and more selective sampling of taxa (Mottern & Heraty, unpublished).
Tetracampidae
Tetracampidae probably represents a polyphyletic assemblage with three extant subfamilies [120]. There is considerable argumentation for placement of the different subfamilies as Aphelinidae, Eulophidae or Pteromalidae [9], [30], [55].
Tetracampidae were never monophyletic in our analyses. Excluding Diplesiostigma, Tetracampinae were monophyletic and very strongly supported. Diplesiostigma varied in placement in every analysis, but never occurred with other Tetracampidae. The two representatives of Mongolocampinae and Platynocheilinae were clustered in a monophyletic group in all analyses with very high support, and most likelihood results grouped them with Eunotini (Pteromalidae: Eunotinae; excluding Idioporus), however with low support.
Torymidae
Placement of Torymidae is uncertain, and it was proposed that the family arose from within the pteromalid lineage [121]. Historically, Torymidae have included Agaoninae and Sycophaginae ( = Idarninae), which were removed by Bouček [30]. Torymidae were revised by Grissell [34] and include only two subfamilies, the largely phytophagous Megastigminae and the mostly parasitic Toryminae, with the latter divided into seven tribes that encompassed the previously recognized Erimerinae, Monodontomerinae and Thaumatotoryminae and several taxa as incertae sedis. Campbell et al. [17] failed to find a monophyletic group, despite what they and Gibson et al. [9] noted to be strong morphological support for the family.
Torymidae were never monophyletic, but Megastigminae and Toryminae were each monophyletic with very strong support (Table 3). Support for tribes within Toryminae was variable. Torymini were monophyletic with low to very strong support in all analyses except parsimony, and Podagrionini were either monophyletic mostly with low support (62% of likelihood analyses) or paraphyletic. Monodontomerini were monophyletic with strong bootstrap support in all analyses, but with the inclusion of the unplaced Zaglyptonotus and exclusion of Chrysochalcissa which clusters deep within Microdontomerini. Echthrodape (Toryminae incertae sedis) was previously placed in Eucharitidae and Perilampidae and then Torymidae by Grissell [34]. This genus was recovered as the sister group of Microdontomerini. The unplaced Glyphomerus exemplars remained unplaced within Toryminae with no particular association with other tribes. The two representatives of Palachiini grouped either with Torymoidini or Podagrionini, but never together. None of the groups seemed to be impacted by the inclusion or exclusion or RAAs. No logical sister groups were identified for either subfamily.
Trichogrammatidae
Trichogrammatidae are well defined and according to Bouček and Noyes [116], are possibly the only monothetic family of Chalcidoidea. Owen et al. [35] assessed higher-level groups and generic relationships based on molecular and morphological evidence and recognized a paraphyletic Trichogrammatinae and monophyletic Oligositinae. Of the groups sampled herein, Ceratogramma (Trichogrammatinae; unplaced to tribe) were recognized as the sister group of the remaining Trichogrammatidae.
Trichogrammatidae were monophyletic in nearly all of our analyses (94% of the MJR consensus trees), but with low BS support in likelihood analyses only after the inclusion of RAAs. Ceratogramma was sister to the remaining Trichogrammatidae in all results, except for one analysis when it was excluded from the family (Table 3, SSNR). Our internal relationships mirror those of Owen et al. [35]. Trichogrammatidae were sister to Azotinae in all but the parsimony analysis, which placed them as a sister group of Idioporus, Rotoita and Systolomorpha.
Conclusions
Is the diverse and unsupported backbone of Chalcidoidea the product of a rapid radiation event [48], [122]? Mymaridae first appear in the early to mid Cretaceous [6]. Based on what appear to be valid fossils of Eulophidae and Trichogrammatidae, there are records of higher-level chalcidoids in only one mid-Cretaceous deposit [8], with records of the same age other than Mymaridae more questionable [6]. The diversification of chalcidoid families does not appear until the Eocene, with modern genera common in Oligocene and Miocene amber deposits [6]. Chalcidoids are mostly parasitoids, and their host groups in the Hemiptera and Holometabola were all undergoing an explosive radiation during the same period at the end of the Cretaceous [123], and a similar tracking of host diversification is not unexpected.
Using an array of nuclear protein coding genes but with fewer taxa, Desjardins et al. [18] found similar results that showed a weak backbone of relationships across their chalcidoid groups sampled. Given a scenario of explosive radiation of Chalcidoidea during a relatively short time period, it may be difficult to resolve higher group relationships with confidence [122]. However, the trees that we have recovered can help to evaluate some scenarios within a context of which groups are consistently supported and their relationships on the various tree topologies. These molecular results provide a unique perspective for examining relationships and hypotheses of chalcidoid evolution, especially in a group prone to morphological convergence.
What is the ancestral mode of host association for Chalcidoidea? Bouček [124] proposed Cleonyminae or some other wood-beetle parasitoids as having the most ancestral forms, but hypothesized that phytophagy could be plesiomorphic for the superfamily. This latter assumption was based on his observation that phytophagous species tend to be primitive within their respective groups. The placement of Chalcidoidea as sister group to either Diaprioidea or Proctotrupoidea sensu stricto and the basal sister group placement of Mymaridae argue against Bouček's hypothesis of a phytophagous ancestor. As well, the phytophagous groups are scattered across the tree and almost never basal within a particular lineage, as in with gall-forming Opheliminae derived from within Eulophidae, or seed-feeding Megastigminae, which are distantly placed from their proposed sister group, the Toryminae (Fig. 9).
Noyes [15] argued for a monophyletic Mymaridae + (Rotoitidae + Tetracampidae) as the sister group of the remaining Chalcidoidea. Our results somewhat support his hypothesis, placing Mymaridae and Rotoitidae at the base of the chalcidoid tree (Fig. 1), but with a different phylogenetic ordering, and with Tetracampidae both polyphyletic and placed more distally on the various topologies. Morphological evidence supports a sister group relationship between Mymaridae and the remaining Chalcidoidea [10], [16], [61]. Our results and more comprehensive analyses of Hymenoptera [11], [13] strongly support this hypothesis. Likelihood results place Rotoitidae as the sister group of the remaining Chalcidoidea after Mymaridae.
Mymaridae are virtually all egg parasitoids, primarily of Auchenorrhyncha, Heteroptera and Coleoptera [125]. The only known exception is for two species of Stethynium attacking larvae of Ophelimus (Eulophidae) [126]. We included S. ophelimi in our analysis, and its derived placement within the family suggests a secondary derivation of larval parasitism (Fig. 1). Egg parasitism is likely the ancestral trait for Mymaridae. Within the remaining Chalcidoidea, egg parasitism occurs in all Trichogrammatidae and a few other scattered taxa (Fig. 9). None of our results placed these chalcidoid egg parasitoids close to the root of Chalcidoidea. Is it possible for egg parasitism to be ancestral for the superfamily? Mymarommatoidea may be egg parasitoids of Psocoptera [127]. The small body size of Rotoitidae suggests that they also might be egg parasitoids, but there is not even a suspected host for this group [9]. Diaprioidea are primarily larval parasitoids of fly larvae or pupae with a few taxa hyperparasitic on Dryinidae or Formicidae [128]; none are egg parasitoids. Even if Mymarommatoidea are resolved as the sister group of Chalcidoidea (only in some of our results), the biology of these and Rotoitidae will need to be resolved before we can confidently consider egg parasitism as a basal trait for the superfamily.
Associated with an extreme diversity of host use, larval morphology is extremely diverse in Chalcidoidea [129]. Two types of hypermetamorphic development occur in Hymenoptera [130]. Type II involves deposition away from the host of a sclerotized planidiform first-instar larva that transforms in later instars to a typical weakly sclerotized sac-like hymenopteriform larva. Within Hymenoptera, this occurs only in one genus of Ichneumonidae (Euceros) and in Perilampidae (including Philomidinae) and Eucharitidae [95]. Although not recovered across all analyses, our results offer support for the single development of this trait within Chalcidoidea (Fig. 9).
Another important trait is the use of sessile Sternorrhyncha as hosts within Chalcidoidea, which ultimately leads to their importance in biological control programs. Mapping sternorrhynchan parasitism, either as primary parasitoids or hyperparasitoids, onto our current ‘best’ hypothesis shows a general scattering of host use that suggests multiple independent host shifts to this group. Probably most significant is the lack of grouping in any of our analyses of Encyrtidae and the aphelinid subfamilies Aphelininae, Azotinae, Coccophaginae, Eretmocerinae and Euryischiinae, which have in the past been treated as a single family [66]. Our results suggest that any traits associated with successful host use of Sternorrhyncha are independent events, and especially within Aphelinidae, should not be considered as phylogenetically linked. This is also important when we consider the single origin of heteronomy, or alternate host use by different sexes, which occurs only in the monophyletic Coccophaginae (Fig. 9).
Our results present the most comprehensive phylogenetic analysis of relationships Chalcidoidea based only on molecular data.. While not robust across the backbone of relationships within Chalcidoidea, they offer some firm insights into the origin and evolution of this important and highly diverse group of insects. Monophyly of many of the traditional groups is supported, and the secondary structure alignment and data set will be useful for future studies. Many changes in the higher classification of taxa within Chalcidoidea are suggested by these results. However, we reserve any judgment on these changes until our combined morphological and molecular analyses are complete.
Supporting Information
Parsimony analysis of SSME dataset using TNT (31,607 steps; r.i. 0.62, strict consensus of >10,000 trees). Bootstrap values plotted to nodes with values greater than 95% represented by dot.
(PDF)
Specimens sequenced and deposition information for specimen data and genebank accession numbers.
(XLS)
Chalcidoidea SSME dataset.
(NEX)
Acknowledgments
We thank Andrew Carmichael, Jan Kostecki, Andrew Ernst, Elizabeth Murray and Albert Owen for sequencing taxa. Help with imaging specimens was provided by Lisa Gonzalez, Jessica Ortiz, Christine Martinez, Maria Saleh, and Jasmine Soto. Sequence data for outgroups were provided by Andy Austin, Matt Buffington and Matt Yoder. Specimens were obtained from various sources, but in particular we would like to thank Chris Burwell, Terry Erwin, Lisa Foerster, Michael Gates, Gary Gibson, Tony van Harten, Yoshimitsu Higashiura, John Huber, Jung-Wook Kim, John LaSalle, Robert Luck, Lubomir Masner, John Pinto, Alain Roques, Mike Sharkey, Richard Stouthamer, Serguei Triapitsyn, Doug Yanega and Bob Zuparko. Help with identifications was provided by Chris Darling, Gérard Delvare, Gary Gibson, Michael Gates, John Huber and John Pinto.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding was provided by National Science Foundation grants TOL EF-0341149 and PEET DEB-0730616 to JMH, @-Speed-Id and ANR BioFigs to JYR, and MSM0021620828 to PJ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Heraty JM, Gates ME. Biodiversity of Chalcidoidea of the El Edén Ecological Reserve, Mexico. In: Gómez-Pompa A, Allen MF, Fedick SL, Jiménez-Osornio JJ, editors. Proceedings of the 21st Symposium in Plant Biology, “Lowland Maya Area: Three Millenia at the Human-Wildland Interface. New York: Haworth Press; 2003. pp. 277–292. [Google Scholar]
- 2.Noyes JS. Encyrtidae of Costa Rica (Hymenoptera: Chalcidoidea), 1. The subfamily Tetracneminae, parasitoids of mealybugs (Homoptera: Pseudococcidae). Memoirs of the American Entomological Institute. 2000;62:1–355. [Google Scholar]
- 3.Heraty JM. Parasitoid Biodiversity and Insect Pest Management. In: Foottit B, Adler P, editors. Insect Biodiversity: Science and Society. Hague, Netherlands: Springer-Verlag Press; 2009. pp. 445–462. [Google Scholar]
- 4.Noyes JS. Universal Chalcidoidea Database website. 2011 Available: www.nhm.ac.uk/entomology/chalcidoids/index.html. Accessed 2011 Sep 30. [Google Scholar]
- 5.Yoshimoto CM. Cretaceous chalcidoid fossils from Canadian amber. Canadian Entomologist. 1975;107:499–529. [Google Scholar]
- 6.Heraty JM, Darling DC. Fossil Eucharitidae and Perilampidae (Hymenoptera: Chalcidoidea) from Baltic Amber. Zootaxa. 2009;2306:1–16. [Google Scholar]
- 7.Poinar G, Jr, Huber JT. A new genus of fossil Mymaridae (Hymenoptera) from Cretaceous amber and key to Cretaceous mymarid genera. Zookeys. 2011;130:461–472. doi: 10.3897/zookeys.130.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt AR, Perrichot V, Svojtka M, Anderson KB, Belete KH, et al. Cretaceous African life captured in amber. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7329–7334. doi: 10.1073/pnas.1000948107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson GAP, Heraty JM, Woolley JB. Phylogenetics and classification of Chalcidoidea and Mymarommatoidea – a review of current concepts (Hymenoptera: Apocrita). Zoologica Scripta. 1999;28:87–124. [Google Scholar]
- 10.Gibson GAP. Evidence for monophyly and relationships of Chalcidoidea, Mymaridae and Mymarommatidae (Hymenoptera: Terebrantes). Canadian Entomologist. 1986;118:205–240. [Google Scholar]
- 11.Heraty JM, Ronquist F, Carpenter JC, Hawks D, Schulmeister S, et al. Hymenopteran relationships: structure of a megaradiation. Molecular Phylogenetics and Evolution. 2011;60:73–88. doi: 10.1016/j.ympev.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Castro LR, Dowton M. Molecular analyses of Apocrita (Insecta: Hymenoptera) suggest that the Chalcidoidea are sister to the diaprioid complex. Invertebrate Systematics. 2006;20:603–614. [Google Scholar]
- 13.Sharkey M, Carpenter JC, Vilhelmsen L, Heraty J, Dowling A, et al. Phylogenetic relationships among superfamilies of Hymenoptera. Cladistics. 2011;27:1–33. doi: 10.1111/j.1096-0031.2011.00366.x. [DOI] [PubMed] [Google Scholar]
- 14.Sharanowski BJ, Robbertse B, Walker J, Voss SR, Yoder R, et al. Expressed sequence tags reveal Proctotrupomorpha (minus Chalcidoidea) as sister to Aculeata (Hymenoptera: Insecta). Molecular Phylogenetics and Evolution. 2010;57:101–112. doi: 10.1016/j.ympev.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Noyes JS. A word on chalcidoid classification. Chalcid Forum. 1990;13:6–7. [Google Scholar]
- 16.Gibson GAP. A word on chalcidoid classification. Chalcid Forum. 1990;13:7–9. [Google Scholar]
- 17.Campbell B, Heraty JM, Rasplus J-Y, Chan K, Steffen-Campbell J, et al. Molecular systematics of the Chalcidoidea using 28S-D2 rDNA. In: Austin AD, Dowton M, editors. The Hymenoptera: Evolution, Biodiversity and Biological Control. Melbourne: CSIRO publishing; 2000. pp. 57–71. [Google Scholar]
- 18.Desjardins C, Regier JC, Mitter C. Phylogeny of pteromalid parasitic wasps (Hymenoptera: Pteromalidae): Initial evidence from four protein-coding nuclear genes. Molecular Phylogenetics and Evolution. 2007;45:454–469. doi: 10.1016/j.ympev.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Letsch HO, Kjer KM. Potential pitfalls of modelling ribosomal RNA data in phylogenetic tree reconstruction: Evidence from case studies in the Metazoa. BMC Evolutionary Biology. 2011;11:146. doi: 10.1186/1471-2148-11-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillespie JJ, Munro JB, Heraty JM, Yoder MJ, Owen AK, et al. A secondary structural model of the 28S rRNA expansion segments D2 and D3 for chalcidoid wasps (Hymenoptera: Chalcidoidea). Molecular Biology and Evolution. 2005;22:1593–1608. doi: 10.1093/molbev/msi152. [DOI] [PubMed] [Google Scholar]
- 21.Gillespie JJ, Johnson JS, Cannone JJ, Eickbush TH, Gutell RR. Characteristics of the nuclear (18S, 5.8S, 28S and 5S) and mitochondrial (12S and 16S) rRNA genes of Apis mellifera (Insecta: Hymenoptera): structure, organization, and retrotransposable elements. Insect Molecular Biology. 2006;15:657–686. doi: 10.1111/j.1365-2583.2006.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh MY, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cruaud A, Jabbour-Zahab R, Genson G, Cruaud C, Couloux A, et al. Laying the foundations for a new classification of Agaonidae (Hymenoptera: Chalcidoidea), a multilocus phylogenetic approach. Cladistics. 2010;26:359–387. doi: 10.1111/j.1096-0031.2009.00291.x. [DOI] [PubMed] [Google Scholar]
- 24.Hayat M. Aphelinidae of India (Hymenoptera: Chalcidoidea): A taxonomic revision. Memoirs on Entomology, International. 1998;13:1–416. [Google Scholar]
- 25.Bouček Z, Delvare G. On the New World Chalcididae. Memoirs of the American Entomological Institute. 1992;53:1–466. [Google Scholar]
- 26.Narendran TC. Kerala, India: University of Calicut; 1989. Oriental Chalcididae (Hymenoptera: Chalcididae).441 [Google Scholar]
- 27.Gibson GAP. Phylogenetics and classification of Cleonyminae (Hymenoptera: Chalcidoidea: Pteromalidae). Memoirs on Entomology, International. 2003;16:1–339. [Google Scholar]
- 28.Heraty JM. A revision of the genera of Eucharitidae (Hymenoptera: Chalcidoidea) of the World. Memoirs of the American Entomological Institute. 2002;68:1–359. [Google Scholar]
- 29.Burks RA, Heraty JM, Gebiola M, Hansson C. Combined molecular and morphological phylogeny of Eulophidae (Hymenoptera: Chalcidoidea), with focus on the subfamily Entedoninae. Cladistics. 2011;27:1–25. doi: 10.1111/j.1096-0031.2011.00358.x. [DOI] [PubMed] [Google Scholar]
- 30.Bouček Z. A Biosystematic Revision of Genera of Fourteen Families, with a Reclassification of Species: CAB International; 1988. Australasian Chalcidoidea (Hymenoptera). [Google Scholar]
- 31.Delucchi V. Résultats scientifiques des missions zoologiques de l'I.R.S.A.C. en Afrique orientale (P. Basilewsky et N. Leleup, 1957), 81. Hymenoptera Chalcidoidea. Annales du Musée Royal de l'Afrique Centrale (Série in 8°) Sciences Zoologique. 1962;110:363–392. [Google Scholar]
- 32.Graham MVRdV. The Pteromalidae of north–western Europe (Hymenoptera: Chalcidoidea). Bulletin of the British Museum (Natural History) (Entomology) Supplement. 1969;16:1–908. [Google Scholar]
- 33.Hedqvist KJ. Notes on Netomocera Bouc. with description of new species (Hym., Chalcidoidea, Pteromalidae). Entomologisk Tidskrift. 1971;92:237–241. [Google Scholar]
- 34.Grissell EE. Toryminae (Hymenoptera: Chalcidoidea: Torymidae): A redefinition, generic classification and annotated world catalogue of species. Memoirs on Entomology, International. 1995;2:1–474. [Google Scholar]
- 35.Owen A, George J, Pinto J, Heraty J. A molecular phylogeny of the Trichogrammatidae (Hymenoptera: Chalcidoidea), with an evaluation of the utility of their male genitalia for higher level classification. Systematic Entomology. 2007;32:227–251. [Google Scholar]
- 36.Walsh PS, Metzger DA, Higuchi R. Chelex® 100 as a medium for simple extraction of DNA for PCR–based typing from forensic material. Biotechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- 37.Heraty JM, Hawks D, Kostecki JS, Carmichael AE. Phylogeny and behaviour of the Gollumiellinae, a new subfamily of the ant-parasitic Eucharitidae (Hymenoptera: Chalcidoidea). Systematic Entomology. 2004;29:544–559. [Google Scholar]
- 38.Deans AR, Gillespie JJ, Yoder MJ. An evaluation of ensign wasp classification (Hymenoptera: Evaniidae) based on molecular data and insights from ribosomal RNA secondary structure. Systematic Entomology. 2006;31:517–528. [Google Scholar]
- 39.Gillespie JJ, McKenna CH, Yoder MJ, Gutell RR, Johnson JS, et al. Assessing the odd secondary structural properties of nuclear small subunit ribosomal RNA sequences (18S) of the twisted-wing parasites (Insecta: Strepsiptera). Insect Molecular Biology. 2005;14:625–643. doi: 10.1111/j.1365-2583.2005.00591.x. [DOI] [PubMed] [Google Scholar]
- 40.Gillespie JJ, Yoder MJ, Wharton RA. Predicted secondary structure for 28S and 18S rRNA from Ichneumonoidea (Insecta: Hymenoptera: Apocrita): impact on sequence alignment and phylogeny estimation. Journal of Molecular Evolution. 2005;61:114–137. doi: 10.1007/s00239-004-0246-x. [DOI] [PubMed] [Google Scholar]
- 41.Katoh MY, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh MY, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 43.Stamatakis A, Hoover P, Rougemount J. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 44.Pfeiffer W, Stamatakis A. Ninth IEEE International Workshop on High Performance Computational Biology (HiCOMB 2010) Atlanta, April 19, 2010; 2010. Hybrid MPI/Pthreads parallelization of the RAxML phylogenetics code. [Google Scholar]
- 45.Miller M, Holder MT, CVos R, Midford P, Liebowitz T, et al. CIPRES; 2010. The CIPRES Portals. pp. 2009–08-04. [Google Scholar]
- 46.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? . Journal of Computational Biology. 2010;17:337–354. doi: 10.1089/cmb.2009.0179. [DOI] [PubMed] [Google Scholar]
- 47.2006; Rhodes, Greece; Stamatakis A. Phylogenetic models of rate heterogeneity: a high performance computing perspective. [Google Scholar]
- 48.Regier JC, Zwick A, Cummings MP, Kawahara AY, Cho S, et al. Toward reconstructing the evolution of advanced moths and butterflies (Lepidoptera: Ditrysia): an initial molecular study. BMC Evolutionary Biology. 2009;9:280[221 pp.]. doi: 10.1186/1471-2148-9-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swofford DL. Version 4.0 ß10. Sunderland, Mass: Sinauer; 2002. PAUP*. [Google Scholar]
- 50.Goloboff PA, Farris JS, Nixon KC. T.N.T.: Tree analysis using New Technology. Københaven Universitets Zoologische Museum website. 2003 Available: http://www.zmuc.dk/public/phylogeny/tnt/. Accessed 2011 Oct 10. [Google Scholar]
- 51.Goloboff PA, Farris JS, Nixon KC. TNT, a free program for phylogenetic analysis. Cladistics. 2008;24:774–786. [Google Scholar]
- 52.Swofford DL, Olsen GJ. Phylogenetic Reconstruction. In: Hillis DM, Moritz C, editors. Molecular Systematics. Sunderland, Massachusetts: Sinauer Associates; 1990. 588 [Google Scholar]
- 53.McLoughlin S. The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Australian Journal of Botany. 2001;49:271–300. [Google Scholar]
- 54.Heraty JM, Darling DC. Comparative morphology of the planidial larvae of Eucharitidae and Perilampidae (Hymenoptera: Chalcidoidea). Systematic Entomology. 1984;9:309–328. [Google Scholar]
- 55.LaSalle J, Polaszek A, Noyes JS, Zolnerowich G. A new whitefly parasitoid (Hymenoptera: Pteromalidae: Eunotinae), with comments on its placement, and implications for classification of Chalcidoidea with partcular reference to the Eriaporinae (Hymenoptera: Aphelinidae). Systematic Entomology. 1997;22:131–150. [Google Scholar]
- 56.Mottern JL, Heraty JM, Hartop E. Cales (Hymenoptera: Chalcidoidea): morphology of an enigmatic taxon with a review of species. Systematic Entomology. 2011;36:267–284. [Google Scholar]
- 57.Hanson P, Gauld ID. Oxford Oxford University Press; 1995. The Hymenoptera of Costa Rica. [Google Scholar]
- 58.Bouček Z. Chapter 4. Agaonidae. In: Gibson GAP, Huber JT, Woolley JB, editors. Annotated keys to the genera of Nearctic Chalcidoidea (Hymenoptera) Ottawa: National Research Council of Canada Research Press; 1997. 794 [Google Scholar]
- 59.Rasplus J-Y, Kerdelhué C, Clainche Il, Mondor G. Molecular phylogeny of fig wasps. Agaonidae are not monophyletic. Comptes Rendus de l'Academie des Sciences, Paris (III) (Sciences de la Vie) 1998;321:517–527. doi: 10.1016/s0764-4469(98)80784-1. [DOI] [PubMed] [Google Scholar]
- 60.Woolley JB. Chapter 5. Aphelinidae. In: Gibson GAP, Huber JT, Woolley JB, editors. Annotated keys to the genera of Nearctic Chalcidoidea (Hymenoptera) Ottawa: National Research Council of Canada Research Press; 1997. 794 [Google Scholar]
- 61.Heraty JM, Woolley JB, Darling DC. Phylogenetic implications of the mesofurca in Chalcidoidea (Hymenoptera), with an emphasis on Aphelinidae. Systematic Entomology. 1997;22:45–65. [Google Scholar]
- 62.Hayat M. The genera of Aphelinidae (Hymenoptera) of the world. Systematic Entomology. 1985;8:63–102. [Google Scholar]
- 63.Ashmead WH. Classification of the chalcid flies, or the superfamily Chalcidoidea, with descriptions of new species in the Carnegie Museum, collected in South America by Herbert H. Smith. Memoirs of the Carnegie Museum. 1904;1:225–551. [Google Scholar]
- 64.Muesebeck CFW, Krombein KV, Townes HK, editors. Agriculture Monograph 2. Washington, D.C: United States Government Printing Office; 1951. Hymenoptera of America north of Mexico: Synoptic catalog.1420 [Google Scholar]
- 65.Girault AA. Australian Hymenoptera Chalcidoidea – IV. Memoirs of the Queensland Museum. 1915;4:1–184. [Google Scholar]
- 66.Gordh G. Catalog of Hymenoptera in America North of Mexico. Washington, DC: Smithsonian Press; 1979. Encyrtidae.1198 [Google Scholar]
- 67.Compere H, Annecke DP. Descriptions of parasitic Hymenoptera and comments (Hymenopt.: Aphelinidae, Encyrtidae, Eulophidae). Journal of the Entomological Society of Southern Africa. 1961;24:17–71. [Google Scholar]
- 68.Rosen D, DeBach P. Ectoparasites. In: Rosen D, editor. Armored scale insects: Their biology, natural enemies, and control. Amsterdam, New York: Elsevior; 1990. 688 [Google Scholar]
- 69.Gibson GAP. Phylogeny and classification of Eupelmidae, with a revision of the world genera of Calosotinae and Metapelmatinae (Hymenoptera: Chalcidoidea). Memoirs of the Entomological Society of Canada. 1989;149:1–121. [Google Scholar]
- 70.Domenichini G. Sulla morfologia e posizione sistematica dei Thysanidae ( = Signiphoridae) (Hym. Chalcidoidea). Bolletino di Zoologia Agraria e Bachicoltura. 1954;20:95–110. [Google Scholar]
- 71.Woolley JB. Phylogeny and classification of the Signiphoridae (Hymenoptera: Chalcidoidea). Systematic Entomology. 1988;13:465–501. doi: 10.1111/j.1365-3113.1988.tb00256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Viggiani G, Bataglia D. Male genitalia in the Aphelinidae (Hym. Chalcidoidea). Bolletino del Laboratorio de Entomologia Agraria ‘Filippo Silvestri’ di Portici. 1984;41:149–172. [Google Scholar]
- 73.Yasnosh VA. Classification of the parasitic Hymenoptera of the family Aphelinidae (Chalcidoidea). Entomological Review. 1976;55:114–120. [Google Scholar]
- 74.Khan MY, Shafee SA. Historical Review and classification of the family Aphelinidae (Hymenoptera: Chalcidoidea). Science and Environment (Aligarh) 1980;2:59–65. [Google Scholar]
- 75.Erdös J. Hymenoptera II, Chalcidoidea III. Fauna Hungariae. 1964;73:1–372. [Google Scholar]
- 76.DeSantis L. Taxonomia de la familia Aphelinidae (Hymenoptera, Chalcidoidea). Revista del Museo de la Plata (Nueve Serie) 1946;5:1–21. [Google Scholar]
- 77.Ferrière C. Paris: Masson; 1965. Hymenoptera Aphelinidae de l'Europe et du Bassin Méditerranéen.206 [Google Scholar]
- 78.Ghesquière J. Contribution à l'étude du genre Eriaporus Waterston et genres affins (Hym. Chalcidoidea Aphelinidae). Memoirs of the Royal Entomological Society of Belgium. 1955;27:217–238. [Google Scholar]
- 79.Hayat M. Family Aphelinidae. Subba Rao BR, Hayat M, editors. In Chalcidoidea (Insecta: Hymenoptera) of India and the adjacent countries. Part I. Reviews of families and keys to families and genera. Pages 226–232. Oriental Insects. 1985;19:163–310. [Google Scholar]
- 80.Mercet RG. Notas sobre afelinidos (Hym. Chalc.) 2a nota. EOS. 1929;5:111–117. [Google Scholar]
- 81.Nikol'skaya MN, Yasnosh VA. Aphelinids of the european part of the USSR and the Caucasas (Hemenoptera: Aphelinidae). Opredeliteli po Faune SSSR. 1966;91:1–296. [Google Scholar]
- 82.Shafee SA, Khan MY. Subfamilies and tribes of the family Aphelinidae (Hymenoptera: Chalcidoidea). Journal of Zoological Research, Aligarh. 1978;2:42–45. [Google Scholar]
- 83.Hayat M. Notes on some genera of the Aphelinidae (Hymenoptera: Chalcidoidea), with comments on the classification of the family. Oriental Insects. 1994;28:81–96. [Google Scholar]
- 84.Bouček Z, Halstead JA. Chapter 6. Chalcididae. In: Gibson GAP, Huber JT, Woolley JB, editors. Annotated keys to the genera of Nearctic Chalcidoidea (Hymenoptera) Ottawa: National Research Council of Canada Research Press; 1997. 794 [Google Scholar]
- 85.Gibson GAP. Leucospidae. In: Goulet H, Huber JT, editors. Hymenoptera of the World: An identification guide to families. Ottawa: Agriculture Canada Research Branch Publication 1894/E; 1993. 668 [Google Scholar]
- 86.Wijesekara GAW. Phylogeny of Chalcididae (Insecta: Hymenoptera) and its congruence with contemporary hierarchical classification. Contributions of the American Entomological Institute. 1997;29:1–61. [Google Scholar]
- 87.Gibson GAP. Sister-group relationships of the Platygastroidea and Chalcidoidea (Hymenoptera) – an alternative hypothesis to Rasnitsyn (1988). Zoologica Scripta. 1999;28:125–138. [Google Scholar]
- 88.Delvare G. Delvare G, Bouček Z, editors. A reclassification of the Chalcidini with a check list of the New World species. On the New World Chalcididae (Hymenoptera): Memoirs of the American Entomological Institute. 1992. pp. 119–459.
- 89.Noyes JS. Chapter 5. Encyrtidae. In: Gibson GAP, Huber JT, Woolley JB, editors. Annotated keys to the genera of Nearctic Chalcidoidea (Hymenoptera) Ottawa: National Research Council of Canada Research Press; 1997. 794 [Google Scholar]
- 90.LaSalle J. A revision of the New World Tanaostigmatidae (Hymenoptera: Chalcidoidea). Contributions to the American Entomological Institute. 1987;23:1–181. [Google Scholar]
- 91.LaSalle J, Noyes JS. New family placement for the genus Cynipencyrtus (Hymenoptera: Chalcidoidea: Tanaostigmatidae). Journal of the New York Entomological Society. 1985;93:1261–1264. [Google Scholar]
- 92.Triapitzin VA. The classification of the family Encyrtidae (Hymenoptera, Chalcidoidea). Part 1. Survey of the systems of classification. The subfamily Tetracneminae Howard, 1892. Entomological Review. 1973;57:118–125. [Google Scholar]
- 93.Triapitzin VA. The classification of the family Encyrtidae (Hymenoptera, Chalcidoidea). Part 1. Survey of the systems of classification. The subfamily Encyrtinae Walker, 1837. Entomological Review. 1973;57:287–295. [Google Scholar]
- 94.Triapitzin VA. Parasitic Hymenoptera of the fam. Encyrtidae of Palaearctics. Opredeliteli po Faune SSSR. 1989;158:1–489. [Google Scholar]
- 95.Darling DC. The life history and larval morphology of Aperilampus (Hymenoptera: Chalcidoidea: Phoilomidinae), with a discussion of the phylogenetic affinities of the Philominiidae. Systematic Entomology. 1992;17:331–339. [Google Scholar]
- 96.Gauthier N, LaSalle J, Quicke DLJ, Godfray HC. Phylogeny of Eulophidae (Hymenoptera: Chalcidoidea), with a reclassification of Eulophidae and the recognition that Elasmidae are derived eulophids. Systematic Entomology. 2000;25:521–539. [Google Scholar]
- 97.Schauff ME, LaSalle J, Coote L. Chapter 10. Eulophidae. In: Gibson GAP, Huber JT, Woolley JB, editors. Annotated keys to the genera of Nearctic Chalcidoidea (Hymenoptera) Ottawa: National Research Council of Canada Research Press; 1997. 794 [Google Scholar]
- 98.Gumovsky AV. Monophyly and preliminary phylogeny of Entedoninae (Hymenoptera, Chalcidoidea, Eulophidae): 28S D2 rDNA considerations and morphological support. In: Melika G, Thuroczy C, editors. Parasitic Wasps: Evolution, Systematics, Biodiversity and Biological Control. Budapest, Hungary: Agroinfrom; 2002. [Google Scholar]
- 99.Gibson GAP. Description of Leptoomus janzeni, n. gen. and n. sp. (Hymenoptera: Chalcidoidea) from Baltic amber, and discussion of its relationships and classification relative to Eupelmidae, Tanaostigmatidae and Encyrtidae. Zootaxa. 2008;1730:1–26. [Google Scholar]
- 100.Gibson GAP. Superfamilies Mymarommatoidea and Chalcidoidea. In: Goulet H, Huber JT, editors. Hymenoptera of the World: An identification guide to families. Ottawa: Agriculture Canada Research Branch Publication 1894/E; 1993. 668 [Google Scholar]
- 101.Gates ME. Species revision and generic systematics of world Rileyinae (Hymenoptera: Eurytomidae). University of California Publications in Entomology. 2008;127:1–332. [Google Scholar]
- 102.Chen Y, Xiao H, Fu J, Huang D-W. A molecular phylogeny of eurytomid wasps inferred from DNA sequence data of 28S, 18S, 16S, and COI genes. Molecular Phylogenetics and Evolution. 2004;31:300–307. doi: 10.1016/S1055-7903(03)00282-3. [DOI] [PubMed] [Google Scholar]
- 103.Lotfalizadeh H, Delvare G, Rasplus J-Y. Phylogenetic analysis of Eurytominae (Chalcidoidea : Eurytomidae) based on morphological characters. Zoological journal of the Linnean society. 2007;151:441–510. [Google Scholar]
- 104.Stage GI, Snelling RR. The subfamilies of Eurytomidae and systematics of the subfamily Heimbrinae (Hymenoptera: Chalcidoidea). Contributions in Science. 1986;375:1–17. [Google Scholar]
- 105.Bouček Z. A revision of the Leucospidae (Hymenoptera: Chalcidoidea). Bulletin of the British Museum (Natural History) (Entomology) Supplement. 1974;23:1–241. [Google Scholar]
- 106.Annecke DP, Doutt RL. The genera of Mymaridae (Hymenoptera: Chalcidoidea). Republic of South Africa, Department of Agriculture Technical Services, Entomology Memoirs. 1961;5:1–71. [Google Scholar]
- 107.Schauff ME. The Holarctic genera of Mymaridae (Hymenoptera: Chalcidoidea). Memoirs of the Entomological Society of Washington. 1984;12:1–67. [Google Scholar]
- 108.Huber JT. Chapter 14. Mymaridae. In: Gibson GAP, Huber JT, Woolley JB, editors. Annotated keys to the genera of Nearctic Chalcidoidea (Hymenoptera) Ottawa: National Research Council of Canada Research Press; 1997. 794 [Google Scholar]
- 109.Hanson P. The Nearctic species of Ormyrus Westwood (Hymenoptera: Chalcidoidea: Ormyridae). Journal of Natural History. 1992;26:1333–1365. [Google Scholar]
- 110.Hanson P. Chapter 15. Ormyridae. In: Gibson GAP, Huber JT, Woolley JB, editors. Annotated keys to the genera of Nearctic Chalcidoidea (Hymenoptera) Ottawa: National Research Council of Canada Research Press; 1997. 794 [Google Scholar]
- 111.Burks BD. Catalog of Hymenoptera in America North of Mexico. Washington, DC: Smithsonian Press; 1979. Ormyridae.1198 [Google Scholar]
- 112.Bouček Z, Watsham A, Wiebes JT. The fig wasp fauna of the receptacles of Ficus thonnongii (Hymenoptera Chalcidoidea). Tijdschrift voor Entomologie. 1981;124:149–233. [Google Scholar]
- 113.Darling DC. Chapter 16. Perilampidae. In: Gibson GAP, Huber JT, Woolley JB, editors. Annotated keys to the genera of Nearctic Chalcidoidea (Hymenoptera) Ottawa: National Research Council of Canada Research Press; 1997. 794 [Google Scholar]
- 114.Heraty JM, Darling DC. A new genus and species of Perilampidae (Hymenoptera: Chalcidoidea) with uncertain placement in the family. Journal of the Entomological Society of Ontario. 2007;138:33–47. [Google Scholar]
- 115.Copland MJW, King PE. The structure of the female reproductive system in the Agaonidae (Chacidoidea, Hymenoptera). Journal of Entomology (A) 1973;48:25–35. [Google Scholar]
- 116.Bouček Z, Noyes JS. Rotoitidae, a curious new family of Chalcidoidea (Hymenoptera) from New Zealand. Systematic Entomology. 1987;12:407–412. [Google Scholar]
- 117.Gibson GAP, Huber JT. Review of the family Rotoitidae (Hymenoptera: Chalcidoidea), with description of a new genus and species from Chile. Journal of Natural History. 2000;34:2293–2314. [Google Scholar]
- 118.Woolley JB. Chapter 18. Signiphoridae. In: Gibson GAP, Huber JT, Woolley JB, editors. Annotated keys to the genera of Nearctic Chalcidoidea (Hymenoptera) Ottawa: National Research Council of Canada Research Press; 1997. 794 [Google Scholar]
- 119.Pedata PA, Viggiani G. Preliminary morpho–biological observations on Azotus perspeciosus (Girault) (Hymenoptera: Aphelinidae), hyperparasitoid of Pseudaulacaspis pentagona (Targioni Tozzetti) (Homoptera: Diaspididae). Redia. 1991;74:343–350. [Google Scholar]
- 120.Gumovsky AV, Perkovsky EE. Taxonomic notes on Tetracampidae (Hymenoptera: Chalcidoidea) with description of a new fossil species of Dipricocampe from Rovno amber. Entomological Problems. 2005;35:123–130. [Google Scholar]
- 121.Grissell EE. Chapter 21. Torymidae. In: Gibson GAP, Huber JT, Woolley JB, editors. Annotated keys to the genera of Nearctic Chalcidoidea (Hymenoptera) Ottawa: National Research Council of Canada Research Press; 1997. 794 [Google Scholar]
- 122.Banks JC, Whitfield JB. Dissecting the ancient rapid radiation of microgastrine wasp genera using additional nuclear genes. Molecular Phylogenetics and Evolution. 2006;41:690–703. doi: 10.1016/j.ympev.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grimaldi DA, Engel MS. New York: Cambridge University Press; 2005. Evolution of the Insects.755 [Google Scholar]
- 124.Bouček Z. An overview of the higher classification of the Chalcidoidea (Parasitic Hymenoptera). In: Gupta VK, editor. Advances in Parasitic Hymenoptera Research. Leiden, The Netherlands: E. J. Brill; 1988. pp. 11–23. [Google Scholar]
- 125.Huber JT. Systematics, biology, and hosts of the Mymaridae and Mymarommatidae (Insecta: Hymenoptera). Entomography. 1986;4:185–243. [Google Scholar]
- 126.Huber JT, Mendel Z, Protasov A, LaSalle J. Two new Australian species of Stethynium (Hymenoptera: Mymaridae), larval parasitoids of Ophelimus maskelli (Ashmead) (Hymenoptera: Eulophidae) on Eucalyptus. Journal of Natural History. 2006;40:1909–1921. [Google Scholar]
- 127.Huber JT, Gibson GAP, Bauer LS, Liu HP, Gates M. The genus Mymaromella (Hymenoptera: Mymarommatidae) in North America, with a key to described extant species. Journal of Hymenoptera Research. 2008;17:175–194. [Google Scholar]
- 128.Masner L. The proctotrupoid families. In: Hanson P, Gauld ID, editors. Oxford Oxford University Press. pp; 1995. pp. 209–246. [Google Scholar]
- 129.Parker HL. Recherches sur les formes postembryonaires de chalcidiens. Annales de la Société Entomologique de France. 1924;93:1–210. [Google Scholar]
- 130.Pinto J. Hypermetamorphosis. In: Resh V, Cardé R, editors. Encylopedia of Insects. New York: Academic Press; 2003. pp. 546–549. [Google Scholar]
- 131.Linares AR, Hancock JM, Dover GA. Secondary structure constraints on the evolution of Drosophila 28S ribosomal RNA expansion segments. Journal of Molecular Biology. 1991;219:381–390. doi: 10.1016/0022-2836(91)90178-9. [DOI] [PubMed] [Google Scholar]
- 132.Hancock JM, Tautz D, Dover GA. Evolution of the secondary structures and compensatory mutations of the ribosomal RNAs of Drosophila melanogaster. Molecular Biology and Evolution. 1988;5:393–414. doi: 10.1093/oxfordjournals.molbev.a040501. [DOI] [PubMed] [Google Scholar]
- 133.Tautz D, Hancock JM, Webb DA, Tautz C, Dover GA. Complete sequences of the ribosomal-RNA genes of Drosophila melanogaster. Molecular Biology and Evolution. 1988;5:366–376. doi: 10.1093/oxfordjournals.molbev.a040500. [DOI] [PubMed] [Google Scholar]
- 134.Nunn GB, Theisen F, Christensen B, Arctander P. Simplicity-correlated size growth of the nuclear 28S ribosomal RNA D3 expansion segment in the crustacean order Isopoda. Journal of Molecular Evolution. 1996;42:211–223. doi: 10.1007/BF02198847. [DOI] [PubMed] [Google Scholar]
- 135.Schulmeister S. Simultaneous analysis of basal Hymenoptera (Insecta): introducing robust-choice sensitivity analysis. Biological Journal of the Linnean Society. 2003;79:245–275. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Parsimony analysis of SSME dataset using TNT (31,607 steps; r.i. 0.62, strict consensus of >10,000 trees). Bootstrap values plotted to nodes with values greater than 95% represented by dot.
(PDF)
Specimens sequenced and deposition information for specimen data and genebank accession numbers.
(XLS)
Chalcidoidea SSME dataset.
(NEX)