Abstract
In 2008, an unusual strain of methicillin-sensitive Staphylococcus aureus (MSSA68111), producing both Panton-Valentine leukocidin (PVL) and toxic shock syndrome toxin-1 (TSST-1), was isolated from a fatal case of necrotizing pneumonia. Because PVL/TSST-1 co-production in S. aureus is rare, we characterized the molecular organization of these toxin genes in strain 68111. MSSA68111 carries the PVL genes within a novel temperate prophage we call ФPVLv68111 that is most similar, though not identical, to phage ФPVL – a phage type that is relatively rare worldwide. The TSST-1 gene (tst) in MSSA68111 is carried on a unique staphylococcal pathogenicity island (SaPI) we call SaPI68111. Features of SaPI68111 suggest it likely arose through multiple major recombination events with other known SaPIs. Both ФPVLv68111 and SaPI68111 are fully mobilizable and therefore transmissible to other strains. Taken together, these findings suggest that hypervirulent S. aureus have the potential to emerge worldwide.
Introduction
Staphylococcus aureus infections range in severity from superficial abscesses to complicated deep soft tissue infections and toxic shock syndrome (TSS) [1], [2]. The pathogenesis of these diverse infections is attributed to multiple extracellular toxins including the Panton-Valentine leukocidin (PVL) and Toxic Shock Syndrome Toxin 1 (TSST-1). PVL has been epidemiologically linked to severe community-associated MRSA (CA-MRSA) infections [3], [4] while TSST-1 clearly mediates shock and organ failure in staphylococcal TSS [5], [6].
Historically, a single strain of S. aureus rarely produced both PVL and TSST-1. However, in 2005, one British report documented the TSST-1 gene in 4 of 30 PVL-positive isolates [7], one of which was associated with severe pneumonia [7]. Subsequently, twenty S. aureus isolates (15 MSSA; 5 MRSA) harboring both PVL and TSST-1 toxin genes were reported in the United Kingdom [8]. Of these, seventeen strains were from one of three clonal complex (CC) lineages: twelve (60%) belonged to lineage CC30 and five were either CC5 or CC22. The other three strains could not be assigned to any known clonal complex. Four of the 5 MRSA strains (80%) were multi-drug resistant. Eight of these isolates (40%) were associated with serious diseases including pneumonia, empyema, deep-seated abscesses and toxic shock. Nine patients presented with abscess or other skin infections [9]. One fatal case of necrotizing pneumonia was also reported in a 14-year-old child who presented initially with sore throat and pyrexia, and then deteriorated rapidly, developing hypotension and multiple organ failure [9].
In this report, we describe the genetic makeup of the hypervirulent TSST-1/PVL co-producing MSSA isolated from this fatal case (MSSA68111; CC30) [9]. Our results demonstrate that MSSA68111 produces both PVL and TSST-1 toxins. Further, its PVL-carrying phage and TSST-1-carrying pathogenicity island (SaPI) are both unique and not heretofore reported in S. aureus. Both novel genetic elements are fully mobilizable, suggesting that the emergence and dissemination of hypervirulent S. aureus may be forthcoming worldwide.
Materials and Methods
S. aureus
MSSA68111 was from a fatal case of necrotizing pneumonia in a 14-year-old child who presented initially with sore throat and pyrexia, and then deteriorated rapidly, developing hypotension and multiple organ failure [9]. It is penicillin-resistant but sensitive to erythromycin, tetracycline, vancomycin and linezolid [8]. It is of the MLST-CC30 lineage (sequence type ST776, a triple locus variant of ST30;spa type t399), harbors agr type 3 and is gene-positive for sec but negative for sea, seb, seg, seh and sei [8] (and unpublished data). TSST-1-positive MSSA strains were reported as being highly associated with the CC30 lineage, which is a very common genotype worldwide and is the most predominant lineage reported in the United Kingdom, Ireland, and Switzerland [10]–[13]. Laboratory strain ATCC 49775 (American Type Culture Collection) harbors the PVL genes (herein referred to as lukF-PV and lukS-PV) and produces LukF/S-PV [14], [15]. A clinical CA-MRSA isolate, strain 04-014, produces TSST-1 but not PVL [16]. Two clinical USA300 MRSA strains, 934814 and 09-301-02119, were from patients with septic arthritis and fatal post-influenza pneumonia, respectively. MSSA laboratory strain RN4220 is a restriction-deficient derivative of the 8325-4 strain [17]. MRSA strain MW2 (USA400) was originally isolated from a 16-month-old girl with fatal septicaemia [18].
S. aureus were routinely cultured in Mueller-Hinton II broth; no differences in growth rates were observed. For analysis of toxin production, washed S. aureus from overnight cultures were diluted to 1-3×106 CFU/mL in fresh media and grown at 37°C in 5% CO2 with shaking (200 rpm) for 20 h (yields 1-4×109 CFU/mL). Culture supernatant fluids were filter sterilized and frozen at −70°C until assayed for toxins.
Toxin assays
TSST-1 and PVL were measured by ELISA [16], [19]. Alpha-hemolysin activity was measured by lysis of rabbit erythrocytes [19]. All assays were run in duplicate or triplicate and included 2–3 biological replicates.
PVL induction and Phage DNA Purification
Exponential phase S. aureus (OD600 = 0.8) in CYPG (Casamino acids 10 g; yeast extract, 10 g; NaCl, 5 g;0.5% glucose and 0.06 M phosphoglycerate in 1 L) were treated with 1 µg/ml mitomycin C and cultured for 3.5 hrs at 32°C with slow shaking (80 rpm). The culture was centrifuged (8,000Xg, 20 min, 4°C) and the filtered (0.22 um) supernatant was treated with DNase I (0.5 U/ml) and RNase A (0.25 mg/ml) at 37°C for 1 hr. After centrifugation (20,000Xg, 1 hr, 4°C), the pellet was resuspended in TES Buffer (100 mM Tris pH 8, 100 mM EDTA, 1% SDS) in the presence of proteinase K (1 mg/ml). After 30 min at 55°C, the DNA was extracted twice with phenol-chloroform and precipitated with two volumes of cold 100% ethanol. The DNA pellet was washed twice with 70% ethanol and resuspended in 100 ul H2O. The junction sequence of induced PVL phage in MSSA68111 was PCR-amplified using an inverse primer set targeting the integrase (int) (PVL-int: AGCAATTATGAAAAGGGT-AGGACA) and PVL (lukF-PV: AGTTGATTGGGAAAATCATACAGTT) genes.
Southern Blot Analysis of PVL Phage DNA
Restriction enzyme digestion of PVL phage DNA and Southern blot analysis was performed using standard methods. Specific DNA fragments were detected using a chemiluminescence system (North2South, Thermo Fisher, Rockford, IL). lukS-PV probe was PCR-amplified using the primers lukS-left: TGTATCTCCTGAGCCTTTTTCA and lukS-right: CAGACAATGAATTACCCCCATT.
Chromosomal Localization of PVL Phage
Bacteria were grown in CYPG to OD625 = 0.6. Genomic DNA was extracted with a commercial kit (Qiagen). The chromosomal insertion site of PVL phage was PCR-verified with the primer pair phiPVL-reverse (to the lukF-PV gene: AGTTGATTGGGAAAATCATACAGTT) and phiPVL-forward (to a conserved hypothetical protein: TCCATCGTTTGAATTGCTTG).
Tst-SaPI induction and Genomic DNA Purification
Bacteria were grown in CYPG to OD625 = 0.15, centrifuged, resuspended in CYPG-Phage Buffer (1∶1) at a density of OD625 = 0.05 and infected with phage 11 or phage 80α at a multiplicity of 6∶1 or 3∶1. Cultures were grown at 32°C with slow shaking (80 rpm) for 1 hr. Samples (8 ml) were centrifuged and the bacterial pellet resuspended in 100 ul GET buffer (0.1 M glucose, 0.01 M EDTA and 0.05 M Tris-HCl, pH 8) and lysed with lysostaphin (0.5 mg/ml) in the presence of RNaseA (0.5 mg/ml) at 37°C for 1 hr. The lysate was further digested with proteinase K (0.5 mg/ml) at 55°C for 3 hrs. Genomic DNA was then extracted with a commercial kit (Qiagen). The junction sequence of induced SaPI68111 was PCR-amplified using an inverse primer set targeting the sel (CTGGACCTACATCGCCGTTA) and SaPI-integrase (GTGTTGGATGAGCAATTACCAA) genes. The chromosomal insertion site of SaPI68111 was further PCR-verified with the primer pair ssrP-up:TAGCGGAAAATCGTAAAGCAAG and integrase-down:GTGTTGGATGAGCAAT-TACCAA.
Quantitative Real-Time PCR
DNase-treated total RNA (1 µg) was reverse transcribed using M-MuLV enzyme (New England Biolab, Ipswich, MA) and random hexamer primers and dNTPs (Invitrogen, Carlsbad, CA). cDNA was diluted fifty-fold in nuclease-free water and used for real-time PCR performed with a 7500 Fast Real-time PCR System (Applied Biosystems, Carlsbad, CA) using the RT2 real-time SYBR green/Rox PCR master mix (SuperArray, Frederick, MD) and primer sets: (1) LukS-PV-up:TGTATCTCCTGAGCCTTTTTCA and LukS-PV-down: CAGACAATGAATTACCCCCATT (Accession: YP_494079); (2) RecA-up:TGCAGAACA-GCTTAAACAAGGA and RecA-down:ACTCTAAATGGTGGTGCCACTT (Accession: L25893); (3) gyrase-up:ATGTAGCAAGCCTTCCAGGTAA and gyrase-down:CAGAGTCCCCTT-CGACTAAGAA (Accession: YP_492727); (4) TSST-1-up:TCGCTACAGATTTTACCCCTGT and TSST-1-down:CGTTTGTAGATGCTTTTGCAGT (Accession: NC_002745). Relative gene expression was determined using the 2-ÄÄCt method [20]. The fold change in expression of the gene of interest was normalized to the internal control gene (Gyrase B) and made relative to the calibrator sample (Time = 0).
Results
1. Toxin Production
In our initial studies, we compared by ELISA the levels of PVL and TSST-1 in 20-hr culture supernatant from S. aureus strain 68111 with known PVL- (ATCC49775 and a clinical MRSA isolate 934814) or TSST-1-producing (04-014) isolates (see details in Materials and Methods section) [14] –[16]. Our analysis showed that MSSA68111 produces 160.24 ng/mL PVL in 20-hr culture, which is significantly lower than that from the well-characterized PVL-producing MSSA strain ATCC49775 (Table 1), but comparable to that obtained from a clinical strain 934814 (USA300) (Table 1). Similarly, the production of TSST-1 in MSSA68111 is also relatively low, yielding 716.5 ng/mL, whereas the TSST-1 positive clinical isolate, strain 04-014, had a strongly positive ELISA result, producing 6739 ng/mL (Table 2).
Table 1. PVL in 20-hr bacterial culture supernatants.
| Strain | Concentration of LukF-PV (ng/mL)±Std Dev |
| ATCC 49775 | 852.7±207.2 |
| 934814 (USA300) | 113.74±15.4 |
| 68111 | 160.24±28.8 |
Table 2. TSST-1 in 20-hr bacterial culture supernatants.
| Strain | Concentration of TSST-1 (ng/mL) ±Std Dev |
| 04–014 | 6739±533.2 |
| 68111 | 716.5±14.8 |
Further, various hemolytic activities of MSSA68111 were assessed. When 20-hr culture supernatant of MSSA68111 was assayed for alpha-toxin with rabbit erythrocytes, a titer of 246 HU/ml was obtained (Table 3). With the same supernatant, no “hot-cold” hemolysis could be detected on sheep erythrocytes, indicating that staphylococcal beta-toxin is either absent or present only in a small amount in this strain (data not shown). Last, for the delta-hemolysin, we adapted a quick-screening methodology established by Herbert [21] and Traber [22] which takes advantage of the fact that delta-hemolysin activity is enhanced by beta-hemolysin. Thus, by streaking test strains perpendicularly to strain RN4220, which produces only beta-hemolysin, one can visually identify delta-hemolysin production in test strains as an enhanced area of hemolysis where the two strains come in near to one another. As shown in Figure 1, MSSA68111 produces a significant amount of delta-hemolysin, indicated by the increased area of hemolytic activity when it nears strain RN4220. This same assay also can be used to detect alpha-hemolysin production in test strains since alpha-hemolysin activity is inhibited by beta-hemolysin. Using this technique, alpha-hemolysin production by MSSA68111 was also demonstrated (Figure 1). The moderate alpha- and delta-hemolysin activities, as well as production of TSST-1 (albeit in low amounts) suggested MSSA68111 carries an intact Agr/RNAIII operon.
Figure 1. Alpha- and delta-hemolysin activities of S. aureus strain 68111.
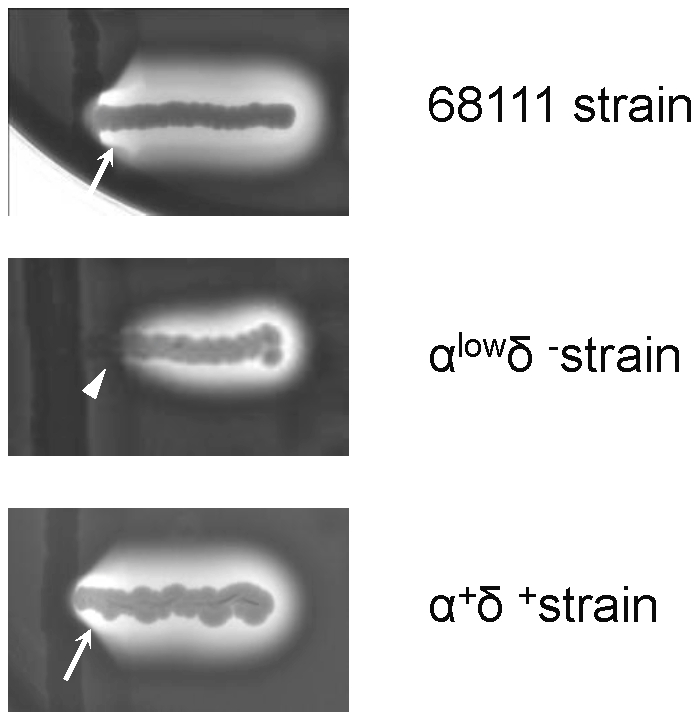
A plate-based screening method that capitalizes on beta-hemolysin-mediated effects on alpha- and delta-hemolysin activities was used to examine extracellular production of these toxins in MSSA68111. For this, beta-hemolysin-producing S. aureus strain RN4220 was streaked vertically on sheep blood agar. MSSA68111 (top) and two laboratory control strains (α-hemolysinlow/δ-hemolysin- [middle] and α-hemolysin+/δ-hemolysin+ [bottom]) were then streaked perpendicularly to RN4220. Hemolysis was assessed after 24 hrs incubation at 37°C (no cold shock applied). At the junction between the test strains and RN4220, alpha-hemolysin activity is inhibited (arrowhead); the presence of delta-hemolysin is observed as an enhanced area of hemolysis (arrows).
Having confirmed that MSSA68111 does indeed produce both TSST-1 and PVL – a phenomenon that is rare among S. aureus strains - we investigated the genetic organization of these genes in this unique clinical isolate.
2. Characterization of PVL-Carrying Phage in MSSA68111
2.1. PVL phage lineage and mobility
The genes for PVL are commonly carried by one of six lysogenic bacteriophages [18], [23] –[27] having two distinct phage-head morphologies: icosahedral (ФPVL and Ф108PVL) or elongated (ФSLT, ФSa2 mw, ФSa2958 and ФSa2usa) [18], [23] –[27]. Ma et al. recently reported a 2-step PCR-based scheme for characterizing S. aureus PVL-encoding phages [28]. The first round of PCR identifies the morphological group using primers specific to the gene lineage between lukS-PV and the tail gene. The second round of PCR establishes the phage type using primers recognizing phage-specific structures. With this approach, we determined that MSSA68111 carries an icosahedral-head type PVL-encoding phage, because PCR-1 (amplifying phage portal and tail genes) and PCR-3 (amplifying lukS-PV to group-specific tail gene) were positive, whereas PCRs-2 and -4 (amplifying genes specific for elongated-head type phage) were negative. Further, our PCR-5 result (targeting to phage specific structures) showed a PCR product of 1411 bps suggesting that the PVL-carrying phage in MSSA68111 belongs to the ФPVL, but not Ф108 PVL, lineage.
To assess the mobility of this PVL phage, we treated MSSA68111 with mitomycin C since it induces the lytic cycle of many temperate phages. After 2–4 hrs, the culture visibly cleared, suggestive of phage-meditated bacterial lysis. To confirm this, phage DNA was purified and amplified with an inverse primer set which targets the integrase and lukF-PV genes. The resultant PCR product (Figure 2A) suggested that the PVL-carrying phage in MSSA68111 did indeed become lytic following mitomycin C treatment. Commercial sequencing of the PCR product with subsequent GenBank BLAST searching demonstrated that the attP junction sequence of MSSA68111's PVL-carrying phage was similar to that in other known PVL-phage lineages (Figures 2B, 2C), demonstrating that the nucleotide sequences of the integrase as well as the chromosomal integration site are highly conserved among different PVL-carrying phages.
Figure 2. DNA sequence of the attP junction of MSSA68111 PVL phage.
(A.) PCR was performed with an inverse primer set using mitomycin C-elicited phage genomic DNA from MSSA68111 as a template. PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide. (B.) Nucleotide sequence of the 68111 PCR product was determined. The sequence originating from PVL phage integrase is printed in upper-case letters, and that derived from lukF-PV gene is shown in italic letters, and twenty-nine base pairs of the core attP sequence are bolded and underlined. (C.) This attP sequence of MSSA68111 PVL phage was compared to that from other published PVL-carrying phages. Nucleotide variations were underlined.
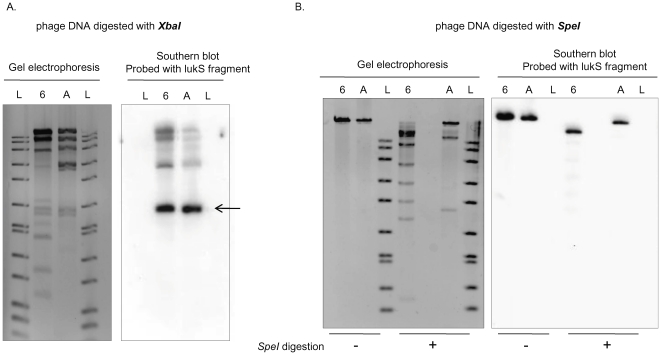
We next compared the restriction profiles of the mitomycin C-elicited PVL-phage genomic DNA from MSSA68111 and ATCC49775 (carries ФPVL [25]) by Southern blot analysis using a lukS-PV-specific probe. Identical hybridization patterns were seen following XbaI, but not SpeI, digestion (Figures 3A, 3B). Thus, the PVL-carrying bacteriophage in MSSA68111 has a similar, but not identical, restriction profile to ФPVL. We propose the name ФPVLv68111 for this ФPVL variant. Last, sequence analysis of LukSF-PV genes in MSSA68111 indicated they belonged to the H1 variant with an H176R amino acid substitution. This information is consistent with the work of O'Hara et al [29], showing that H variants of LukSF-PV genes were mostly carried by mecA - S. aureus isolates found mainly outside the United States.
Figure 3. Southern blot analysis of PVL phage genomic DNA from strain 68111 and strain ATCC49775.
Phage DNA from MSSA68111 (Lanes designated “6”) and from strain ATCC49775 (Lanes “A”) was digested with either XbaI (A.) or SpeI (B.). In both A and B, the panels on the left are photographs of ethidium bromide-stained gels; the panels on the right are Southern blots hybridized with the lukS-PV probe. 1 kb DNA ladders (Lanes “L”) are shown. The lukS-PV probe hybridized to identical 2 kb fragments in XbaI -digested phage DNA from both strains (A, arrow) whereas SpeI-digestion produced different sized lukS-PV fragments (B.).
2.2. Characterization of ФPVLv68111 integration site
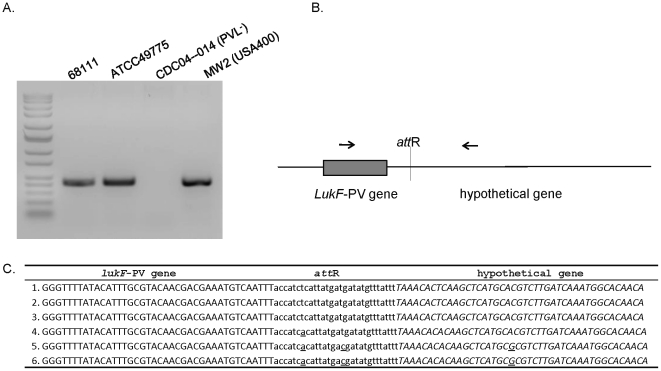
To investigate the chromosomal insertion site of ФPVLv68111, we performed PCR based on the known phage PVL insertion sites in the other strains (e.g., ATCC49775, MW2) using primers that hybridize to the lukF-PV gene and to a hypothetical gene (Figures 4A, 4B). Commercial sequencing of the resultant PCR product revealed a 29 nucleotide region of the ФPVLv68111 attR site (lysogenic form) that was identical to attP sequence of ФPVLv68111 (lytic form; Figure 2C), and the position at which the phage inserts into the hypothetical gene of the host chromosome was well conserved among currently known PVL-producing strains (Figure 4C).
Figure 4. DNA sequences of integration site of PVL phage in bacterial genome.
(A.) PCR was performed with primers, as shown in (B.) using bacterial genomic DNA as templates. PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide. (C.) The sequence of PCR product from MSSA68111 was determined and is shown relative to other strains carrying PVL phage. The sequence originating from the phage PVL genomic DNA including lukF-PV gene is printed in upper-case letters, and that derived from the bacterial chromosome, a conserved hypothetical protein, is shown in italic letters. Twenty-nine base pairs of core attR sequence are bolded and underlined. Nucleotide variations are underlined. 1.ФPVLv68111; 2. ΦPVL (strain ATCC49775 [25]); 3. ΦPVL108 (Accession: AB243556); 4. Φ2958 PVL (Accession:AP009363); 5. ΦSLT (Accession: CP000255); 6. ΦSa2 MW (Accession: BA000033).
3. Characterization of Tst-Carrying SaPI in MSSA68111
3.1. Gene organization of sel-sec-tst
Several S. aureus exotoxin genes cluster within pathogenicity islands (SaPIs) [30], [31]. MSSA68111 was reported to be gene-positive for the exotoxins SEL, SEC and TSST-1 [9] (coded for by sel, sec and tst, respectively) which are commonly clustered in SaPI1 [32], SaPI2 [30], [33], SaPIn1/m1 [34] or SaPIbov [35], [36]. To investigate the organization of these toxin genes in MSSA68111, PCR was performed using primers based on the known SaPIn1 and SaPIbov sequences. The size of the PCR products and corresponding sequencing data demonstrated that sel-sec-tst were clustered sequentially in MSSA68111. Like SaPIn1 or SaPIm1, the sec and tst genes are approximately 2 kb apart and in opposite orientations (data not shown).
3.2. Mobility of tst-carrying SaPI by helper phage
In the excision-replication-packaging (ERP) cycle, temperate bacteriophages can induce excision and replication of SaPIs and can package the ensuing DNA into special small phage-like particles using proteins supplied by the helper phage. Using genomic DNA electrophoresis [32], we assessed two common staphylococcal helper phages, phage 80α or phage 11, for their ability to mobilize the tst-carrying SaPI in MSSA68111. We did not observe the typical SaPI-specific band on a stained agarose gel containing bacterial genomic DNA following helper phage induction (data not shown), suggesting that this SaPI did not undergo massive helper phage-mediated ERP.
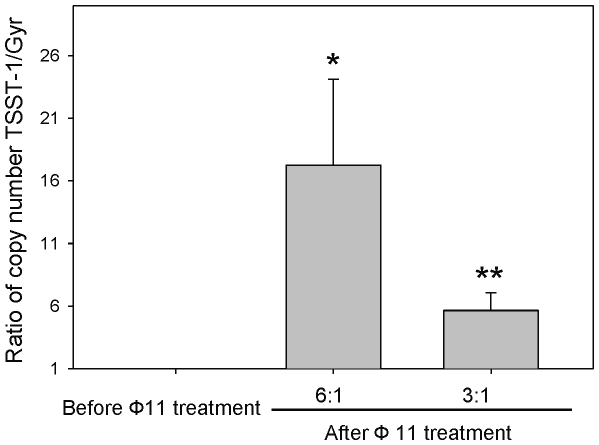
To increase the sensitivity of detection of ERP, we used real-time qPCR-based methods to quantify the copy number of tst-SaPI within MSSA68111 host cells (Figure 5). Tst and gyr were the target and the reference genes, respectively. Preliminary studies (data not shown) demonstrated that S. aureus genome contained only one copy each of tst and gyr and that the amplification efficiencies of these genes (calculated from the slope of the standard curve generated with serial dilutions of cDNA template for each gene) were equivalent (1.92 vs 1.90, respectively). Thus, the copy ratio of tst to gyr before SaPI induction was 1. In contrast, following superinfection with phage 11, the tst amplification curve was left-shifted indicating increased copy number of the tst gene compared to the gyr gene. The tst-carrying SaPI copy number relative to the reference gene gyr could be given by the ratio: [(Etst) ΔCttst(control-treated)]/[(Egyr) ΔCgyr(control-treated)] [37]. As shown in Figure 5, after 1.5 hrs of exposure to helper phage 11 at MOI of 3∶1 or 6∶1, the relative tst-carrying SaPI copy number in MSSA68111 was dose-dependently increased 5.6-fold and 17.2-fold, respectively.
Figure 5. Induction (excision/replication) of tst-SaPI elements by phage11.
Following superinfection with phage 11 as described in Materials and Methods, total genomic DNA was purified and analyzed for tst and gyr genes by real-time PCR. Relative copy numbers of tst versus gyr are presented. Data are means±SD (n = 4); p*<0.05; p**<0.01 vs control (before Ф11 treatment) (paired t-test).
In contrast, no excised form of tst-carrying SaPI was found following UV- or mitomycin C treatment or phage 80α induction either by electrophoretic analysis of sheared whole cell lysates or by the above qPCR method (data not shown). We conclude that the excision of tst-SaPI in MSSA68111 is highly phage 11-specific.
3.3. Characterization of the integrase of tst-carrying SaPI
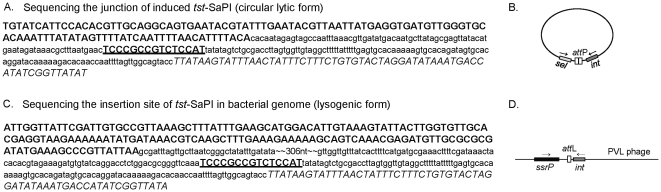
To determine the integrase sequence and chromosomal integration site of the tst-carrying SaPI in MSSA68111, an outward-directed PCR on genomic DNA was conducted following helper phage 11 induction (Figure 6A). Sequencing of the PCR product showed a 15-bp direct repeat occurs at the junction between sel and integrase as indicated in Figure 6. The att core (5′-TCCCGCCGTCTCCAT-3′) matches the core att site for several other SaPIs (e.g., SaPIm4, SaPImw2). Further comparison of the integrase sequence as well as the chromosomal integration site of the tst-carrying SaPI in MSSA 68111 with those from the previously characterized SaPIm4 and SaPImw2 showed 96% and 87% identity of intergrase genes, respectively, and the same chromosomal insertion site of all three, at 3′ end of ssrP (Figure 6B). In contrast, the organization of accessory genes in SaPI68111 including sel, sec, and tst genes, is most closely related to that of SaPIn1.
Figure 6. Sequence analyses of tst-carrying SaPI in MSSA68111.
(A.) Junctional sequence of circulated SaPI (lytic form) following helper phage 11 induction. Sel coding region is bolded; integrase of tst-SaPI is shown in italic letter and 15 base pairs of core attP are bolded and underlined. Other intergenic regions are presented in lower-case font. A schematic illustration of the PCR strategy used to amplify this junctional region is shown in (B.). (C.) Sequence of integration site of tst-SaPI (lysogenic form) in bacterial genome. SsrP coding region is bolded, integrase of tst-SaPI is shown in italic letter and 15 base pairs of core attL are bolded and underlined. Other intergenic regions are presented in lower-case font. A schematic illustration of the PCR strategy used to amplify the integration site of tst-SaPI is shown in (D.).
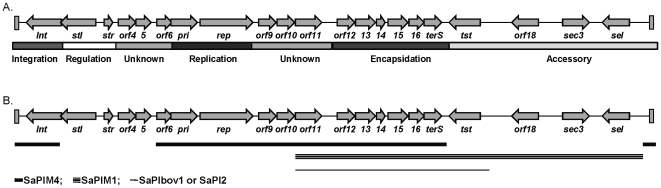
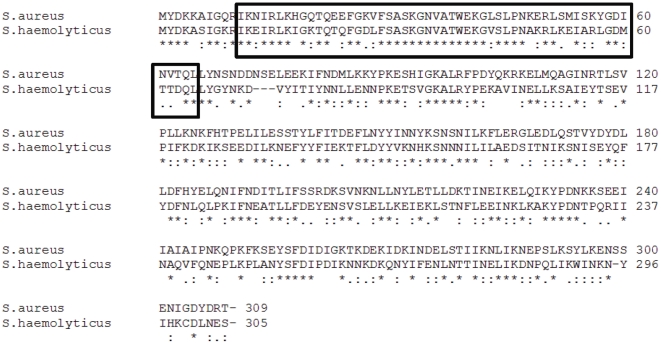
Because of these findings, a complete genome sequencing of the tst-carrying SaPI in MSSA68111 was conducted using a PCR and bi-directional primer walking-based approach. In all, the 16,422-bp SaPI68111 (Accession: JN689383) was determined to have 21 ORFs potentially encoding proteins over 50 amino acids in length, 3 of which encoded staphylococcal exotoxins (sel, sec, and tst), and many of which have homologs in SaPIM4, SaPIM1, SaPIbov1 and SaPI2 as indicated in Figure 7A, 7B and Table 4. These data suggest that the evolutional history of SaPI68111 probably includes at least one major recombination event with these SaPIs or other similar elements. Also interestingly, we found that the predicated product of stl in SaPl68111, a SaPI master repressor, shares 64% similarity (over 298 amino acids) to a hypothetical phage repressor protein of Staphylococcus haemolyticus bacteria (Accession: YP_254016; Figure 8). Particularly, the putative N-terminal HTH (helix-turn-helix) motif of SaPI68111-stl repressor belongs to the HTH -XRE-family of proteins and shows a high degree of 80% similarity (as much as 65% identity) to the HTH motifs of repressor protein of Staphylococcus haemolyticus but not to HTH motifs of most S. aureus (Figure 8). These data have led to the supposition that this gene was acquired by S. aureus through horizontal transfer from Staphylococcus haemolyticus during co-colonization of human skin [38].
Figure 7. Illustrative representation of pathogencity island 68111 (SaPI68111).
Arrows represent the location and orientation of open reading frames (OFRs), which were determined using the NCBI-GLIMMER program and by the relative position of ORFs within the island and/or similarity of the predicted gene products. (A.) An illustrative diagram of the SaPI68111 genome with different functional annotations presented at the bottom of the figure. (B.) A summary diagram of the homology regions between SaPI68111 and other close-related SaPIs. SaPIM4 (thick line), SaPIM1 (triple lines) and SaPIbov1 or SaPI2 (thin line).
Table 3. alpha-hemolysin in 20-hr bacterial culture supernatants.
| Strain | Alpha Hemolytic Activity (HU/mL) |
| 09–301–02119 (USA300) | 310 |
| ATCC 49775 | 98 |
| 68111 | 246 |
Figure 8. Amino acid alignment of predicted stl from MSSA68111 and hypothetical phage repressor protein SH2101 from Staphylococcus haemolyticus (Accession: YP_254016.1).
The protein sequences were aligned by using the CLUSTAL alignment program. The Helix-turn-helix XRE-domain is highlighted in the black box. The asterisk denotes a position at which the two sequences have the same amino acid. Dots indicate the degree of homology when there is not complete sequence conservation.
Table 4. Relationships between SaPI68111 genes and those of other SaPIs.
| SaPI68111 | Mu50 | RF122 | RN3984 | MW2 | Annotation or Function | ||
| SaPIM4 | SaPIm1 | SaPIbov1 | SaPI2 | SaPIMW2 | |||
| orf1 | int | 96(98) | 87(100) | Integrase | |||
| orf2 | stl | SaPI master repressor | |||||
| orf3 | str | 88(91) | 88(91) | Regulatory protein | |||
| orf4 | HP | 82(92) | 83(91) | Hypothetical protein | |||
| orf5 | HP | 97(88) | 95(88) | 96(88) | Hypothetical protein | ||
| orf6 | HP | 96(100) | 93(100) | 96(100) | 90(100) | Hypothetical protein | |
| orf7 | pri | 96(100) | 93(69) | 93(69) | 93(69) | Similar to DNA primase | |
| orf8 | rep | 99(100) | Similar to replication initiation protein | ||||
| orf9 | HP | 99(100) | Hypothetical protein | ||||
| orf10 | HP | 99(100) | Hypothetical protein | ||||
| orf11 | HP | 96(100) | 96(100) | 96(100) | 95(100) | Hypothetical protein | |
| orf12 | HP | 97(100) | 97(100) | 97(100) | 96(100) | Hypothetical protein | |
| orf13 | cp | 98(100) | 98(100) | 94(100) | 98(100) | Capisid size determinant | |
| orf14 | cp | 99(100) | 99(100) | 96(100) | 100(100) | Capisid size determinant | |
| orf15 | cp | 96(100) | 96(100) | 97(100) | 95(100) | Capisid size determinant | |
| orf16 | HP | 96(100) | 96(100) | 98(100) | 96(100) | Hypothetical protein | |
| orf17 | terS | 98(100) | 98(100) | 94(100) | 97(100) | 90(27) | Terminase small subunit |
| orf18 | tst | 100(100) | 100(100) | 100(100) | Toxic shock syndrome toxin-1 | ||
| orf19 | HP | 100(100) | 77(82) | Hypothetical protein | |||
| orf20 | sec3 | 99(100) | 95(100) | 97(100) | Staphylococcal enterotoxin type C3 | ||
| orf21 | sel | 100(100) | 100(100) | 99(100) | Staphylococcal enterotoxin L | ||
Note: The similarities of SaPI68111 with other close-related SaPIs were determined with BLAST. Percentage of identity to corresponding gene in SaPI68111 and percentage of query coverage (in the bracket) were given in the table. Blank area indicated no corresponding genes.
4. Transcription of PVL and TSST-1
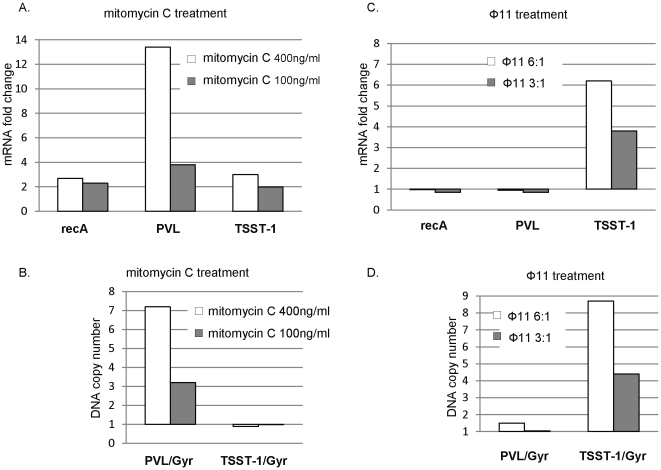
Increased PVL transcription after mitomycin C treatment has been attributed to an increase of phage copy number following phage excision and replication [39]. Consistent with this, our quantification of lukS-PV mRNA and DNA in MSSA68111 by real-time PCR confirmed that mitomycin C treatment resulted in a marked increases of both PVL transcript (Figure 9A) and PVL-DNA copy number (Figure 9B). Mitomycin C also activates recA-mediated autocleavage of phage repressors and resumption of the lytic cycle [40], [41]. Thus increased recA expression following DNA damage temporally coincides with increase phage-encoded gene transcription. Consistent with this notion, we observed a modest increase of recA expression in MSSA68111 following mitomycin C treatment (Figure 9A).
Figure 9. Comparative study of phage induction versus gene transcription in response to mitomycin C or phage 11 treatment.
(A. and B.) Bacteria were treated with mitomycin C (400 ng/ml or 100 ng/ml) for 1 hr. (C. and D.) Freshly prepared phage 11 was added for 1 hr. One half of samples were used for RNA preparation followed by a quantitative RT-PCR analysis. Fold changes in gene expression of recA, PVL and TSST-1 were normalized to the internal control gene gyrase (Gyr) and relative to no treatment control sample. The other half of samples were used for total genomic DNA preparation. Relative copy number of TSST-1 or PVL gene versus Gyr were calculated as described in Materials and Methods. Shown is one representative out of three independent experiments, all of which gave comparable results.
In contrast to PVL, mitomycin C treatment induced only a slight increase in TSST-1 mRNA and no excised form of tst-SaPI68111 was detected (Figures 9A, 9B). However, the abundance of both tst transcripts as well as the tst gene copy number were markedly and dose-dependently increased by helper phage 11 (Figures 9C, 9D). These results were observed in the absence of a recA/SOS response (Figure 9C). Similarly, PVL genes at both the DNA and mRNA levels also remained unchanged by phage 11 treatment (Figures 9C, 9D). Thus, tst-SaPI68111 induction (excision and replication) is positively correlated with TSST-1 transcription and does not involve the recA/SOS response.
Discussion
Co-production of SEC, TSST-1 and PVL in MSSA68111 was proposed to contribute to the rapid demise of the young British patient with hemorrhagic pneumonia [9]. Further, the recent report of severe infections due to three separate clonal lineages of S. aureus co-producing TSST-1 and PVL [8] portends a possible emergence of hypervirulent S. aureus worldwide.
In addition to its epidemiologic association with fatal pneumonia, MSSA68111 is also of considerable interest from a purely genetic point of view. For unknown reasons, certain combinations of staphylococcal toxins are rarely present in the same clinical isolate. This is true for TSST-1 with staphylococcal enterotoxin B (SEB) and for TSST-1 with PVL. Thus MSSA68111 is a significant deviation from the norm. As such, it provides a unique opportunity to investigate the mechanisms responsible for this phenomenon and to gain insight into likely emergence and dissemination of the next S. aureus “superbug”.
We confirmed that MSSA68111 produced both TSST-1 and PVL. PVL production by MSSA68111 was comparable to other S. aureus clinical isolates we have tested [16], however the level of TSST-1 was low. The relatively low level of TSST-1 in MSSA68111 could not be explained by a non-functional Agr/RNAIII system since both alpha- and delta-hemolysins were produced. In addition, real-time RT-PCR showed that RNAIII transcripts in MSSA68111 were comparable to those from MRSA strains FPR3757 (USA300) and MW2 (USA400) (data not shown). Expression of RNAIII was inversely related to expression of spa gene in wild-type MSSA68111 but not in its isogenic RNAIII mutant (data not shown). The low levels of TSST-1 and PVL are also not an artifact of the ELISA-based detection system since their respective nucleotide sequences in MSSA68111 are highly similar to those of other strains (data not shown).
Although extensive studies have been conducted by others, the exact mechanism responsible for the wide variation in the amount of TSST-1 produced by different strains of S.aureus,is still unknown. A study of 152 TSST-1-positive MRSA isolates by Nagao et al showed that differences in TSST-1 production (up to 170-fold) is not directly correlated with the allelic variations of the agr [42]. In addition, their sequencing of the promoter region of the tst gene, the entire sigma factor B and sar loci revealed no relevant nucleotide changes among these strains [42]. Thus it is likely that the low level of TSST-1 in MSSA68111 may not simply be explained by one mechanism, but rather multiple known or unknown complex interconnected regulatory systems are operative. In addition, the low level of TSST-1 production in MSSA68111 could be a consequence of co-production of both TSST-1 and PVL toxins, as other investigators have shown that some toxins function as autorepressors or negative regulators of other exoproteins [43].
Bacteriophages mediate dissemination of genes and hence bacterial diversity. We show that the PVL-carrying phage from MSSA68111 (ФPVLv68111) is a variant of icosahedral-head type phage ФPVL. The basis for this conclusion is that ФPVL and ФPVLv68111 share similar phage structural genes, identical XbaI restriction profiles and phage integration sites, but different SpeI restriction fragment profiles. Among clinical isolates that have been studied, ФPVL is relatively rare in the United States but is more prevalent in Europe [28]. In contrast, the elongated-head type PVL-phage, represented by ФSa2usa, is currently dominant worldwide [44]. These findings suggest that ФPVL and ФPVLv68111 might have evolved from a common ancestor and that genetic drift may have occurred in one or both. Features unique to ФPVLv68111 may have permitted MSSA68111 to acquire the genes for TSST-1 production.
Phage-encoded toxin genes are not always transmissible often because integrated prophages are defective. However, ФPVLv68111 is fully mobilized as part of a recA-mediated SOS response and its induction results in enhanced lukS-PV transcription. These findings indicate functional ФPVLv 68111 can be readily transmitted to other S. aureus strains.
Staphylococcal pathogenicity islands (SaPIs) are other mobile genetic elements responsible for inter- as well as intra-genic spread of exotoxin genes. MSSA68111 carries a unique SaPI which we have designated SaPI68111. Like SaPIn1/m1, SaPI68111 carries a central region of sequence identity encompassing sel-sec-tst in the classical arrangement. However, unlike all known tst-carrying SaPIs, SaPI68111 is inserted in an att site close to the ssrP gene with a 15-bp repeat. Further, the integrase gene in SaPI68111 appears related to SaPImw2 and SaPIm4, but not to SaPIn1/m1 or any other known tst-carrying SaPIs. Most interestingly, the predicated stl gene in SaPI68111, a master regulator for SaPI EPR cycle, does not share any homology with any published sequence in S. aureus, but has 64% similarity with a hypothetical phage repressor of Staphylococcus haemolyticus. This unique combination of genetic characteristics suggests that SaPI68111 probably arose following at least one major recombination event with several SaPIs or other similar elements, whereas the regulatory gene (e.g., stl gene) might be acquired by S. aureus through horizontal transfer from other species (e.g., Staphylococcus haemolyticus) during co-colonization or infections.
Induction of SaPIs excision/replication requires specific helper phages, and in MSSA68111, chromosomal excision of tst-SaPI68111 was driven solely by helper phage 11. This finding is unique in that most SaPIs are specifically induced by phage 80α alone or in combination with phage 11. To our knowledge, none is induced solely by phage 11. Like PVL, excision/replication of tst-SaPI68111 was positively correlated with increased TSST-1 gene transcription. Whether features of SaPI68111 contributed to the emergence of PVL/TSST-1 co-producing S. aureus remains to be determined.
Several hypotheses exist to explain the observed specific paired toxin exclusion in S. aureus. First, exclusion results from competition for a single chromosome insertion site [45]. However, this cannot fully explain the mutual exclusion of TSST-1 and PVL in nearly all S. aureus since, as described above, the chromosomal location of PVL-carrying phages does not overlap with any currently reported S. aureus SaPIs including SaPI68111 and induction of tst-carrying SaPIs requires specific helper phages that are genetically distinct from PVL-encoding phages. Second, unknown “exclusion factors” encoded by the first genetic element block acquisition of the second genetic element [46]. If such factors exist and mediate toxin exclusion, it would imply that MSSA68111 is exclusion factor-deficient. Third, one mobile genetic element commandeers a key bacterial resource essential for transduction. Fourth, membrane depolarization after infection by the first element prevents secondary infection [47], [48]. Lastly, defects in the Sau1 restriction modification system may permit multiple transduction events as has been suggested in S. aureus RN4220 [49]. Our genetic characterization of MSSA68111 offers new tools to investigate these possibilities.
In summary, our genetic analysis of a hypervirulent TSST-1/PVL co-producing CA-MSSA demonstrated that these potent virulence factors are each carried on unique and fully transmissible genetic elements. Together these findings portend a possible worldwide emergence of new hypervirulent S. aureus. Whether these elements are common among other recently described TSST-1/PVL co-producing S. aureus, and what their roles may be in overcoming mutual toxin exclusion, are under investigation.
Acknowledgments
The authors thank Dr. Frank DeLeo, Rocky Mountain Laboratory, Hamilton, MT for the generous gift of S. aureus strain MW2.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported, in part, by grants from the United States Department of Veterans Affairs, Research and Development Service, Biomedical Laboratory Research Program, VA Medical Center, Boise, ID (DLS and AEB) and from St. Luke's Mountain States Tumor and Medical Research Institute, Boise, ID (ZL). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Archer GL. Staphylococcus aureus: a well-armed pathogen. Clin Infect Dis. 1998;26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 2.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. 10.1056/NEJM199808203390806 [doi] [DOI] [PubMed] [Google Scholar]
- 3.Kaneko J, Kamio Y. Bacterial two-component and hetero-heptameric pore-forming cytolytic toxins: structures, pore-forming mechanism, and organization of the genes. Biosci Biotechnol Biochem. 2004;68:981–1003. doi: 10.1271/bbb.68.981. [DOI] [PubMed] [Google Scholar]
- 4.Gillet Y, Issartel B, Vanhems P, Fournet JC, Lina G, et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359:753–759. doi: 10.1016/S0140-6736(02)07877-7. S0140-6736(02)07877-7 [pii];10.1016/S0140-6736(02)07877-7 [doi] [DOI] [PubMed] [Google Scholar]
- 5.See RH, Kum WW, Chang AH, Goh SH, Chow AW. Induction of tumor necrosis factor and interleukin-1 by purified staphylococcal toxic shock syndrome toxin 1 requires the presence of both monocytes and T lymphocytes. Infect Immun. 1992;60:2612–2618. doi: 10.1128/iai.60.7.2612-2618.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraser J, Arcus V, Kong P, Baker E, Proft T. Superantigens - powerful modifiers of the immune system. Mol Med Today. 2000;6:125–132. doi: 10.1016/s1357-4310(99)01657-3. S1357-4310(99)01657-3 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Holmes A, Ganner M, McGuane S, Pitt TL, Cookson BD, et al. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. J Clin Microbiol 43/5/2384. 43:2384–2390. doi: 10.1128/JCM.43.5.2384-2390.2005. [pii];10.1128/JCM.43.5.2384-2390.2005 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kearns AM, Smith IM, Ganner M, Perry C, Warner M, et al. Staphylococcus aureus encoding both TSST-1 and PVL in the UK.Abstract of International Symposium on Staphylococci and Staphylococcal Infections 2008 [Google Scholar]
- 9.Mushtaq F, Hildrew S, Okugbeni G, Ellis RW, Deshpande S. Necrotizing haemorrhagic pneumonia proves fatal in an immunocompetent child due to Panton-Valentine Leucocidin, toxic shock syndrome toxins 1 and 2 and enterotoxin C-producing Staphylococcus aureus. Acta Paediatr. 2008;97:985–987. doi: 10.1111/j.1651-2227.2008.00797.x. APA797 [pii];10.1111/j.1651-2227.2008.00797.x [doi] [DOI] [PubMed] [Google Scholar]
- 10.Hallin M, Denis O, Deplano A, De MR, De RR, et al. Genetic relatedness between methicillin-susceptible and methicillin-resistant Staphylococcus aureus: results of a national survey. J Antimicrob Chemother. 2007;59:465–472. doi: 10.1093/jac/dkl535. dkl535 [pii];10.1093/jac/dkl535 [doi] [DOI] [PubMed] [Google Scholar]
- 11.Peacock SJ, Moore CE, Justice A, Kantzanou M, Story L, et al. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect Immun. 2002;70:4987–4996. doi: 10.1128/IAI.70.9.4987-4996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruimy R, Armand-Lefevre L, Barbier F, Ruppe E, Cocojaru R, et al. Comparisons between geographically diverse samples of carried Staphylococcus aureus. J Bacteriol. 2009;191:5577–5583. doi: 10.1128/JB.00493-09. JB.00493-09 [pii];10.1128/JB.00493-09 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holtfreter S, Grumann D, Schmudde M, Nguyen HT, Eichler P, et al. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J Clin Microbiol. 2007;45:2669–2680. doi: 10.1128/JCM.00204-07. JCM.00204-07 [pii];10.1128/JCM.00204-07 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finck-Barbancon V, Prevost G, Piemont Y. Improved purification of leukocidin from Staphylococcus aureus and toxin distribution among hospital strains. Res Microbiol. 1991;142:75–85. doi: 10.1016/0923-2508(91)90099-v. 0923-2508(91)90099-V [pii] [DOI] [PubMed] [Google Scholar]
- 15.Prevost G, Cribier B, Couppie P, Petiau P, Supersac G, et al. Panton-Valentine leucocidin and gamma-hemolysin from Staphylococcus aureus ATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infect Immun. 1995;63:4121–4129. doi: 10.1128/iai.63.10.4121-4129.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton SM, Bryant AE, Carroll KC, Lockary V, Ma Y, et al. In vitro production of panton-valentine leukocidin among strains of methicillin-resistant Staphylococcus aureus causing diverse infections. Clin Infect Dis. 2007;45:1550–1558. doi: 10.1086/523581. 10.1086/523581 [doi] [DOI] [PubMed] [Google Scholar]
- 17.Fairweather N, Kennedy S, Foster TJ, Kehoe M, Dougan G. Expression of a cloned Staphylococcus aureus alpha-hemolysin determinant in Bacillus subtilis and Staphylococcus aureus. Infect Immun. 1983;41:1112–1117. doi: 10.1128/iai.41.3.1112-1117.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. S0140673602087135 [pii] [DOI] [PubMed] [Google Scholar]
- 19.Stevens DL, Ma Y, Salmi DB, McIndoo E, Wallace RJ, et al. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J Infect Dis. 2007;195:202–211. doi: 10.1086/510396. JID36388 [pii];10.1086/510396 [doi] [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. 10.1006/meth.2001.1262 [doi];S1046-2023(01)91262-9 [pii] [DOI] [PubMed] [Google Scholar]
- 21.Herbert S, Ziebandt AK, Ohlsen K, Schafer T, Hecker M, et al. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun. 2010;78:2877–2889. doi: 10.1128/IAI.00088-10. IAI.00088-10 [pii];10.1128/IAI.00088-10 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Traber KE, Lee E, Benson S, Corrigan R, Cantera M, et al. agr function in clinical Staphylococcus aureus isolates. Microbiology. 2008;154:2265–2274. doi: 10.1099/mic.0.2007/011874-0. 154/8/2265 [pii];10.1099/mic.0.2007/011874-0 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. S0140-6736(06)68231-7 [pii];10.1016/S0140-6736(06)68231-7 [doi] [DOI] [PubMed] [Google Scholar]
- 24.Kaneko J, Kimura T, Kawakami Y, Tomita T, Kamio Y. Panton-valentine leukocidin genes in a phage-like particle isolated from mitomycin C-treated Staphylococcus aureus V8 (ATCC 49775). Biosci Biotechnol Biochem. 1997;61:1960–1962. doi: 10.1271/bbb.61.1960. [DOI] [PubMed] [Google Scholar]
- 25.Kaneko J, Kimura T, Narita S, Tomita T, Kamio Y. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage phiPVL carrying Panton-Valentine leukocidin genes. Gene. 1998;215:57–67. doi: 10.1016/s0378-1119(98)00278-9. S0378111998002789 [pii] [DOI] [PubMed] [Google Scholar]
- 26.Ma XX, Ito T, Chongtrakool P, Hiramatsu K. Predominance of clones carrying Panton-Valentine leukocidin genes among methicillin-resistant Staphylococcus aureus strains isolated in Japanese hospitals from 1979 to 1985. J Clin Microbiol. 2006;44:4515–4527. doi: 10.1128/JCM.00985-06. JCM.00985-06 [pii];10.1128/JCM.00985-06 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narita S, Kaneko J, Chiba J, Piemont Y, Jarraud S, et al. Gene. Vol. 268. 195-206. S0378111901003900 [pii]; 2001. Phage conversion of Panton-Valentine leukocidin in Staphylococcus aureus: molecular analysis of a PVL-converting phage, phiSLT. [DOI] [PubMed] [Google Scholar]
- 28.Ma XX, Ito T, Kondo Y, Cho M, Yoshizawa Y, et al. Two different Panton-Valentine leukocidin phage lineages predominate in Japan. J Clin Microbiol. 2008;46:3246–3258. doi: 10.1128/JCM.00136-08. JCM.00136-08 [pii];10.1128/JCM.00136-08 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Hara FP, Guex N, Word JM, Miller LA, Becker JA, et al. A geographic variant of the Staphylococcus aureus Panton-Valentine leukocidin toxin and the origin of community-associated methicillin-resistant S. aureus USA300. J Infect Dis. 2008;197:187–194. doi: 10.1086/524684. 10.1086/524684 [doi] [DOI] [PubMed] [Google Scholar]
- 30.Subedi A, Ubeda C, Adhikari RP, Penades JR, Novick RP. Sequence analysis reveals genetic exchanges and intraspecific spread of SaPI2, a pathogenicity island involved in menstrual toxic shock. Microbiology. 2007;153:3235–3245. doi: 10.1099/mic.0.2007/006932-0. 153/10/3235 [pii];10.1099/mic.0.2007/006932-0 [doi] [DOI] [PubMed] [Google Scholar]
- 31.Novick RP, Subedi A. The SaPIs: mobile pathogenicity islands of Staphylococcus. Chem Immunol Allergy. 2007;93:42–57. doi: 10.1159/000100857. 10.1159/0000100857 [pii];10.1159/0000100857 [doi] [DOI] [PubMed] [Google Scholar]
- 32.Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol Microbiol. 1998;29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- 33.Ruzin A, Lindsay J, Novick RP. Molecular genetics of SaPI1 – a mobile pathogenicity island in Staphylococcus aureus. Mol Microbiol. 2001;41:365–377. doi: 10.1046/j.1365-2958.2001.02488.x. mmi2488 [pii] [DOI] [PubMed] [Google Scholar]
- 34.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. S0140673600044032 [pii] [DOI] [PubMed] [Google Scholar]
- 35.Fitzgerald JR, Monday SR, Foster TJ, Bohach GA, Hartigan PJ, et al. Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J Bacteriol. 2001;183:63–70. doi: 10.1128/JB.183.1.63-70.2001. 10.1128/JB.183.1.63-70.2001 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ubeda C, Tormo MA, Cucarella C, Trotonda P, Foster TJ, et al. Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol Microbiol. 2003;49:193–210. doi: 10.1046/j.1365-2958.2003.03577.x. 3577 [pii] [DOI] [PubMed] [Google Scholar]
- 37.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, et al. Whole-genome sequencing of staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J Bacteriol. 2005;187:7292–7308. doi: 10.1128/JB.187.21.7292-7308.2005. 187/21/7292 [pii];10.1128/JB.187.21.7292-7308.2005 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wirtz C, Witte W, Wolz C, Goerke C. Transcription of the phage-encoded Panton-Valentine leukocidin of Staphylococcus aureus is dependent on the phage life-cycle and on the host background. Microbiology. 2009;155:3491–3499. doi: 10.1099/mic.0.032466-0. mic.0.032466-0 [pii];10.1099/mic.0.032466-0 [doi] [DOI] [PubMed] [Google Scholar]
- 40.Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–74. doi: 10.1038/nature02241. 10.1038/nature02241 [doi];nature02241 [pii] [DOI] [PubMed] [Google Scholar]
- 41.Galkin VE, Yu X, Bielnicki J, Ndjonka D, Bell CE, et al. Cleavage of bacteriophage lambda cI repressor involves the RecA C-terminal domain. J Mol Biol. 2009;385:779–787. doi: 10.1016/j.jmb.2008.10.081. S0022-2836(08)01362-4 [pii];10.1016/j.jmb.2008.10.081 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagao M, Okamoto A, Yamada K, Hasegawa T, Hasegawa Y, et al. Variations in amount of TSST-1 produced by clinical methicillin resistant Staphylococcus aureus (MRSA) isolates and allelic variation in accessory gene regulator (agr) locus. BMC Microbiol. 2009;9:52. doi: 10.1186/1471-2180-9-52. 1471-2180-9-52 [pii];10.1186/1471-2180-9-52 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vojtov N, Ross HF, Novick RP. Global repression of exotoxin synthesis by staphylococcal superantigens. Proc Natl Acad Sci U S A. 2002;99:10102–10107. doi: 10.1073/pnas.152152499. 10.1073/pnas.152152499 [doi];152152499 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boakes E, Kearns AM, Ganner M, Perry C, Hill RL, et al. Distinct bacteriophages encoding PVL among international clones of PVL-MRSA. J Clin Microbiol.JCM. 2010 01917-10 [pii];10.1128/JCM.01917-10 [doi] [Google Scholar]
- 45.De Boer ML, Chow AW. Toxic shock syndrome toxin 1-producing Staphylococcus aureus isolates contain the staphylococcal enterotoxin B genetic element but do not express staphylococcal enterotoxin B. J Infect Dis. 1994;170:818–827. doi: 10.1093/infdis/170.4.818. [DOI] [PubMed] [Google Scholar]
- 46.Delbruck MA. Interference between bacterial viruses; the mutual exclusion effect and the depressor effect. J Bacteriol. 1945;50:151–170. doi: 10.1128/JB.50.2.151-170.1945. [DOI] [PubMed] [Google Scholar]
- 47.Parma DH, Snyder M, Sobolevski S, Nawroz M, Brody E, et al. The Rex system of bacteriophage lambda: tolerance and altruistic cell death. Genes Dev. 1992;6:497–510. doi: 10.1101/gad.6.3.497. [DOI] [PubMed] [Google Scholar]
- 48.Li BH, Bockrath R. Photolyase-dimer-DNA complexes and exclusion stimulation in Escherichia coli: depolarization of the plasma membrane. Mol Gen Genet. 1993;240:450–454. doi: 10.1007/BF00280400. [DOI] [PubMed] [Google Scholar]
- 49.Waldron DE, Lindsay JA. Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J Bacteriol. 2006;188:5578–5585. doi: 10.1128/JB.00418-06. 188/15/5578 [pii];10.1128/JB.00418-06 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]