Abstract
Background
Porphyra yezoensis Ueda is one of the most important edible seaweed, with a dimorphic life cycle which consists of gametophyte as macroscopical blade and sporophyte as microscopic filamentous. Conspicuous differences exist in the two generations, such as morphology, cell structure, biochemistry, physiology, and so on. The developmental process of Porphyra yezoensis has been studied thoroughly, but the mechanism is still ambiguous and few studies on genetic expression have been carried out.
In this study, the suppression subtractive hybridization (SSH) method conducted to generate large-scale expressed sequence tags (EST) is designed to identify gene candidates related to the morphological and physiological differences between the gametophytic and sporophytic generations of Porphyra yezoensis Ueda.
Findings
Each 300 clones of sporophyte and gametophyte cells were dipped onto the membrane for hybridization. The result of dot-blot suggested there were 222 positive clones in gametophyte library and 236 positive clones in sporophyte library. 383 positive clones of strongest signals had been sequenced, and 191 EST sequences of gametophyte and 192 of sporophyte were obtained.
A total of 196 genes were obtained, within which 104 genes were identified from the gametophyte and 92 from the sporophyte. Thirty-nine genes of the gametophyte and 62 genes of the sporophyte showed sequence similarity to those genes with known or putative functions which were classified according to their putative biological roles and molecular functions. The GO annotation showed about 58% of the cellular component of sporophyte and gametophyte cells were mainly located in cytoplasm and nucleus. The special genes were located in Golgi apparatus, and high expression in plastid, ribosome and endoplasmic reticulum. The main biological functions of gametophyte cells contributed to DNA repair/replication, carbohydrate metabolism, transport and transcription, especially in response to heat and oxidative stress. The sporophyte cell expresses more genes in transcription, transport, carbohydrate metabolism, particularly in signal transduction, DNA and protein modification, protein and nucleotide metabolism. Four genes are expressed on both gametophyte and sporophyte cells and eighteen genes have not been annotated.
Conclusion
According to the information of GO annotation, the gametophyte tends to growth and self- protection while the sporophyte tends to be more active in development. Interpretation of the differentially expressed genes revealed new insights into the molecular processes of the generation alternation of Porphyra yezoensis. Further investigation are needed due to insufficiency of functional genes research and indeterminancy of the functions of many sequences.
Background
Porphyra yezoensis is one of the most important edible seaweeds commercially cultivated mainly in China, Japan and Korea. It has a dimorphic life cycle which consists of macroscopical haploid foliose blade phase and microscopic diploid filamentous conchocelis phase [1]. Conspicuous differences exist in the two generations, such as morphology, cell structure, biochemistry, physiology, and so on. Seed-stock material for Porphyra cultivation consists of carpospores (also called zygotespores) [2] spread into oyster shells in the spring, conchocelis germinated and grow in the shells at the onset of conchospores release in the autumn. The conchospores settle on net twine, cultivate in the sea water and develop into blade thalli mainly in winter and early spring. In a mature thallus of Porphyra yezoensis, four kinds of cells are developed. From the basal to the top, there are rhizoid cells, vegetative cells, male gametes and female gamete [3].
The developmental process of Porphyra yezoensis has been studied thoroughly, but the mechanism is still ambiguous. Very little of the genetic expression has been noticed. What is the process and essential happened in the period from vegetative cells to format sexual cells? What is the difference between the male gametes and female gametes? What is the difference between the foliose blade phase and conchocelis phase?
Nikaido et al (2000) first constructed the normalized and size-selected cDNA libraries of leafy gametophyte Porphyra yezoensis and generated a total of 10,154 expressed sequence tags (EST) [4]. Only 934 EST groups were classified based on the annotations of their homologous protein entries in the public databases. The reason for this feature could be that fewer sequences from red and green algae have been registered in the public DNA databases compared to those from higher plants. Asamizu et al (2003) compared the expressed sequence tags (EST) differences between the gametophytic and sporophytic generations of Porphyra yezoensis Ueda [5]. Among the 10,625 EST groups, only 1013 (22.5%) groups were classified as ESTs that commonly occurred in both generations, whereas a large proportion of EST groups were identified as being unique to either the gametophyte (43.1%) or the sporophyte (34.3%). A statistical significance test revealed 89 and 112 highly expressed gene candidates in the gametophyte and the sporophyte respectively.
Kakinuma et al (2006) have constructed subtracted cDNA libraries enriched for differentially expressed transcripts in vegetative and reproductive thalli of Porphyra, and randomly selected 1,152 cDNAs from each subtracted library [6]. BLAST analysis showed that the cDNAs represented 63 and 59 unique clones for the vegetative and reproductive subtracted libraries respectively. Interestingly, some of the cDNAs isolated from the reproductive subtracted library were homologous to genes encoding protein kinases, GTP-binding protein, and heat shock proteins involved in signal transduction and the molecular chaperon system.
The SSH method is designed to selectively amplify differentially expressed transcripts while suppressing the amplification of abundant transcripts, thus eliminating the need to separate single- and double-stranded molecules. In addition, SSH normalizes target transcripts to approximately equal abundance [7,8].
To identify gene candidates related to the morphological and physiological differences between the gametophytic and sporophytic phases of Porphyra yezoensis, suppression subtractive hybridization (SSH) method is conducted to generate large-scale expressed sequence tag (EST). In this study, the carpospores instead of the filamental conchocelis were chosen as sporophyte material. The vegetative (gametophyte) and carpospores (sporophyte) cells live in the same blade of thallus so that the difference between the samples could be neglected.
Findings
Suppression subtractive hybridization (SSH)
Extraction of RNA
The plant RNA Extraction Reagent was adopted to extract the total RNA from two kinds of Porphyra yezoensis cells. Due to the thickness of Porphyra yezoensis frond, it contained such large amount of polysaccharose that ordinary RNA Extraction Reagent can not extract the RNA in high quality and the separation of carpospores and blade vegetative cells of P. yezoensis were difficult, so it was harder to get much experiment material. For all the reasons, we got the cDNA under the condition of trace total RNA by choosing the SMARTTM PCR cDNA Synthesis kit, it can enrich the Poly A+RNA selectively and reduce the product of cDNA according to the ribosome RNA. Beside these, it can also avoid the cumbersome process of Poly A+RNA enrichment and the samples losses and contamination problems. The value of OD260/OD280 which estimated by UV spectrophotometer is 1.82. It suggested that there is nearly no protein, phenol, and polysaccharose contamination and no degradation exist through 1% agarose gel electrophoresis. The results suggested that the purity and integrity of total RNA meet the requirement of the SSH experiment.
Detection of ligation reaction efficiency
The ligation reaction efficiency of front adaptor would affect not only the result of SSH but also the success ratio of cDNA library. The cDNA mixed with adaptor 1and 2R were respectively reacted by Nest PCR primer1 and PCR primer 2R. According to the result of electrophoresis, the bands are larger than 500 bp and these sequences ligated with the adaptor1 and adaptor 2R also had a good efficiency.
Detection of subtractive hybridization efficiency
The subtractive hybridization efficiency of related gene's cDNA was tested by comparing the abundance differences between their subtractive hybridization and without subtractive hybridization.
Construction of Subtracted cDNA Library
The reciprocal carpospores and blade vegetative cells were to be tester and driver to construct the forward and reverse Subtracted cDNA library which contained 1000 clones by SSH. Each subtracted cDNA was ligated to Target vector (TOYOBO) and transformed into the component cells (E. coli DH5α strain). Picking one clone randomly and using Nest Primer as primer to run the colony PCR to detect the situation of gene inserted into library showed that the size of the products was mainly concentrated on 300-1000 bp which suggested the insert efficiency was nearly 98%.
Dot-blot
From the result of dot-blot, the signal of forward subtractive cDNA clone hybridization probe was apparently higher than that of the reverse subtractive one. These clones were the specific expression gene or enhance expression gene in organization. The signal intensity which was similar to the two hybridization probes was thought to be background while no hybridization with two probes was thought to be the lowest abundance cDNA or false positive recombinants. The positive clones were selected by analyzing the image results by optiquant software.
In the experiment, each 300 clones were dipped onto the membrane for hybridization of carpospores and blade vegetative cells. The result of dot-blot suggested that there were 458 positive clones, the positive rate was 76.3% within which 222 positive clones in blade vegetative cells library and 236 positive clones in carpospore library.
Sequence Analyses
There 383 successes after 408 positive clones of strongest signals were sequenced so that 191 EST sequences of vegetative cells and 192 ESTs of carpospores were obtained. The EST sequences of corresponding clones were trimmed by removing the vector sequences and fuzzy sequences from the former sequences of bases and got 360 in all. All sequences were submitted to the GenBank databases. The assigned GenBank Accession Numbers of leafy carpospores are HS572581- HS572762, and those of leafy vegetative cells are HS572763- HS572942.
Bioinformatics analysis
The cleaned unigenes/non-redundant sequences were assemblied with more than 20,000 EST sequences deposited in GenBank. Total 196 genes with 104 from vegetative cells and 92 from carpospores were obtained,. Some genes were represented by multiple ESTs as shown in the Additional file 1and Additional file 2 that included the contigs with more than 170 ESTs. Database searches and similarity analyses of cDNA nucleotide sequences were carried out with the BLASTN and BLASTX programs against public nucleotide, EST, and protein databases [9,10]. Thirty-nine of the vegetative cells and 62 of the carpospores that showed sequence similarities to known or putative function proteins were classified according to their putative biological roles and molecular functions (Table 1).
Table 1.
The list of gene name annotated by Gene Ontology based on the homologous protein entries in the public databases
| Gene symbol | Full Name | vegetative cells | carpospores |
|---|---|---|---|
| NeoTet | pNeo4 NeoTet | √ | |
| fglh1 | lysyl hydroxylase 1 | √ | |
| response regulator | response regulator | √ | |
| sensor histidine kinase | sensor histidine kinase | √ | |
| Peb1 | response regulator Rr1 genes | √ | |
| rr1 | response regulator Rr1 | √ | |
| sigma-54 DNA-binding response regulator | √ | ||
| sigma 54 modulation protein | √ | ||
| RPL41 | ribosomal protein L41 | √ | √ |
| RPL27A | ribosomal protein L27A | √ | |
| RPL30 | ribosomal protein L30 | √ | |
| RPL23a | ribosomal protein L23a | √ | |
| RPL9 | ribosomal protein L9 | √ | √ |
| RPS3 | ribosomal protein S3 | √ | |
| RPS11 | ribosomal protein S11 | √ | √ |
| EEF1A1 | eukaryotic translation elongation factor 1 alpha 1 | √ | |
| RSIP | ribosomal subunit interface protein | √ | |
| GTP1/OBG family | translation-associated GTPase | √ | |
| RNA helicase | DEAD-box ATP dependent DNA helicase | √ | |
| Snf2 family protein | Snf2 family protein-related Helicase | √ | |
| tRNA synthase | tRNA pseudouridine synthase | √ | |
| RecQ | ATP-dependent DNA helicase RecQ | √ | |
| MutT | putative mutator protein | √ | |
| NUDIX hydrolase | MutT/nudix family protein | √ | |
| uvrA | excinuclease ABC, subunit A | √ | |
| integrase | Transposase from transposon Tn916 | √ | |
| DNA-directed RNA polymerase subunit | √ | ||
| recF | recombination protein F | √ | |
| RadA | DNA repair protein | √ | |
| hsdM | type I restriction enzyme M protein | √ | |
| RE | restriction endonuclease | √ | |
| GidA | tRNA uridine 5-carboxymethylaminomethyl modification enzyme |
√ | |
| GidA | glucose-inhibited division protein A | √ | |
| fruA/fruB | frutan hydrolase (fruA), FruB (fruB) | √ | |
| ek435 | cyplasin S | √ | |
| hsp68 | heat shock protein 68 | √ | |
| hrcA | repressor of heat shock gene expression HrcA | √ | |
| dnaK | molecular chaperone DnaK | √ | |
| grpE | molecular chaperone | √ | |
| PUM1 | pumilio 1 | √ | |
| CYTH2 | cytohesin 2 | √ | |
| CBX7 | protein binding | √ | |
| PSCD2 | pleckstrin homology, Sec7 and coiled domains 2 | √ | |
| APOBEC3A | √ | ||
| Acac | nucleolin-related protein, transcript variant | √ | |
| INHBA | inhibin beta A | √ | |
| RBM9 | RNA binding motif protein 9 | √ | |
| SLC25A5L | solute carrier family 25 | √ | |
| PAX1 | paired box gene 1 | √ | |
| E1BP1 | √ | ||
| g6pd | glucose-6-phosphate-1-dehydrogenase | √ | |
| thiol peroxidase | thiol peroxidase | √ | |
| SOD1 | superoxide dismutase 1 | √ | |
| BTG1 | B-cell translocation gene 1, anti-proliferative | √ | |
| ORFA | √ | ||
| ctc | √ | ||
| aab | aromatic amino acid biosynthetic gene cluster | √ | |
| Ndfip1 | Nedd4 family interacting protein 1 | √ | |
| RGS7 | regulator of G-protein signalling 7 | √ | |
| LAPTM4A | lysosomal-associated protein transmembrane 4α | √ | |
| ABC transporter | putative osmoprotectant ABC transporter | √ | √ |
| nitrite transporter | nitrite transporter | √ | |
| p76 | Transmembrane 9 superfamily member 2 precursor |
√ | |
| IBP | iron-binding protein genes | √ | |
| eep | √ | ||
| EF2015 | minor head protein | √ | |
| HAD | HAD superfamily hydrolase | √ | |
| methyltransferase | putative methyltransferase | √ | |
| carboxylesterase | carboxylesterase | √ | |
| Gfo/Idh/MocA | family oxidoreductase | √ | |
| SEPHCHC synthase | carboxylic acid synthase | √ | |
| PTS system | phosphotransferase system | √ | |
| SAG0894 family | CRISPR associated protein | √ | |
| SecY | preprotein translocase subunit SecY | √ | |
| signal peptidase I | signal peptidase I | √ | |
| clpC | stress response-related Clp protease | √ | |
| NDUFA9 | NADH dehydrogenase (ubiquinone) 1 alpha | √ | |
| ATP citrate synthase | ATP citrate synthase | √ | |
| ATP citrate lyase | ATP citrate lyase | √ | |
| atpA/B/C/D/E/F/G/H | H+ ATPase subunits | √ | |
| COX4I1 | cytochrome c oxidase subunit IV isoform 1 | √ | |
| pykF | pyruvate kinase | √ | |
| Rmet_0948 | ubiquinol oxidase, subunit II | √ | |
| FH | fumarate hydratase | √ | |
| ADPGlc Ppase | Glucose-1-phosphate adenylyltransferase | √ | |
| glycerol dehydrogenase | glycerol dehydrogenase | √ | |
| dapB | dihydrodipicolinate reductase | √ | |
| cmk | Cytidine monophosphate kinase | √ | |
| arcABCRD | arginine deiminase operon | √ | |
| argR1/argR2 | arginine repressors | √ | |
| pI | √ | ||
| h1 | √ | ||
| NGLY1 | N-glycanase 1 | √ | |
| prs | phosphoribosyl pyrophosphate synthetase | √ | |
| ldh | L-lactate dehydrogenase | √ | |
| gcaD | UDP-N-acetylglucosamine pyrophosphorylase | √ | |
| ghfp | glycosy hydrolase family protein | √ | |
| EF2591 | glyoxalase family protein | √ | |
| adk | adenylate kinase(AK) | √ | |
| YgfK | pyridine nucleotide-disulfide family | √ | |
| ApbE | thiamin biosynthesis ApbE | √ | |
| IPP isomerase | phosphomevalonate kinase | √ | |
| Gelsolin precursor | Gelsolin precursor | √ | |
| IVNS1ABP | influenza virus NS1A binding protein | √ | |
| holin | holin | √ | |
| psbO1 | oxygen-evolving enhancer 1 precursor | √ | |
| PBP 1B | membrane carboxypeptidase | √ | |
| MDRP | multidrug resistance protein | √ | |
| MFSP | major facilitator superfamily protein | √ |
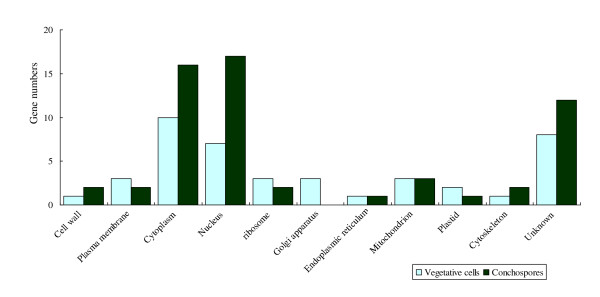
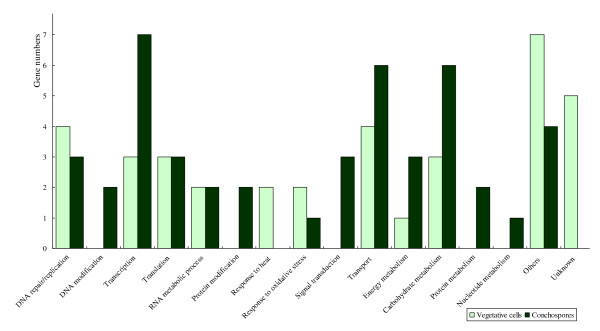
The GO annotation showed the cellular component of carpospores were mainly located in cytoplasm and nucleus, reaching to 58%; while vegetative cells also mainly located in cytoplasm and nucleus, especially located in Golgi apparatus, and abundance in plastid, ribosome and endoplasmic reticulum (Figure 1). The main biological functions of vegetative cells were related to DNA repair/replication, carbohydrate metabolism, transport and transcription, particularly in response to heat and oxidative stress. The carpospores expressed more transcripts in transcription, transport, carbohydrate metabolism, especially in signal transduction, DNA and protein modification, protein and nucleotide metabolism (Figure 2). According to the information of GO annotation, the vegetative cells tend to growth and self- protection, while the carpospores tend to more active in development. Four genes namely ribosomal protein L41, 50S ribosomal protein L9, ribosomal protein S11 and ATP-binding protein are expressed on both vegetative cells and carpospores. Eighteen genes have not been annotated. Because in this study functional genes are still not enough and the functions of many sequences are not clear, so further investigations are needed.
Figure 1.
The GO annotations of Cellular Component.
Figure 2.
The GO annotations of Biological Process and Molecular Function.
Discussion
The morphological and physiological differences between the leafy gametophyte and the filamentous sporophyte in the life cycle of Porphyra species are so great that many studies have been performed. Gametophyte- and sporophyte-specific cDNAs of Porphyra purpurea which encode proteins such as elongation factors, serine protease-like proteins, polysaccharide-binding proteins, and lipoxygenases had been isolated by differential screening and subtraction of phase-specific cDNA libraries (Liu et al., 1994, 1996) [11-15].
For single direct partial sequencing of anonymous cDNA clones of Porphyra purpurea performed by Liu et al was not available in obtaining enough genetic information. Lee et al. constructed the EST library of P. yezoensis thullus [16]. Among 190 ESTs generated, 81 sequence tags carried partial cDNA with significant amino acid sequence similarity to known genes, but the other 109 clones were not homologous to previously identified genes. At the same time, EST analysis has been performed to identify candidate genes related to the morphological and physiological differences between the gametophytic and sporophytic generations, and 20779 cDNAs have been identified [4,5]. However, because these are mainly new genes, without homologous to known ones, the genes which regulate the life history process of Porphyra thalli remain poorly understood.
Gametogenesis of Porphyra thalli was induced by changing the photoperiod and water temperature. Kakinuma et al. (2006) have constructed subtracted cDNA libraries enriched for differentially expressed transcripts in vegetative and reproductive thalli, and randomly selected 1,152 cDNAs from each subtracted library [17]. Results of the dot blot analyses used for identification of differentially expressed cDNAs and BLAST analysis of nucleotide and deduced amino acid sequences showed that the cDNAs represented 63 and 59 unique clones for the vegetative and reproductive. Some of the cDNAs isolated from the reproductive subtracted library were homologous to genes encoding protein kinases, GTP-binding protein, and heat shock proteins involved in signal transduction and the molecular chaperon system. However, it is possible that changes in expression of these genes may only be a response to the change in temperature and photoperiod and are unrelated to gametogenesis.
To identify genes involved in the Porphyra yezoensis asexual sporulation, Kitade, et al. (2008) compared the gene expression profiles derived from gametophytes and sporophytes using cDNA macroarray, which includes 4,896 nonredundant expressed sequence tag (EST) groups. Candidate genes were screened by two different macroarray data analyses combined with reverse transcription-PCR (RT-PCR) analysis and Northern analysis. RT-PCR analysis revealed that nine genes were expressed in gametophyte specific manner, and two genes were expressed only in gametophytes that formed archeospores [18].
The cDNA microarray provides a high capacity and credibility system for the investigation of Porphyra yezoensis genes expression profile. For example, the different generations of P. yezoensis 467 functional gene clones were obtained and dotted onto the slides by cDNA microarray which coated by poly-lysine. The analysis of the normalized data showed that 55 genes were excessively expressed in gametophyte generation, 21 of which were similar to the genes of known or putative function; 86 genes were excessively expressed in sporophyte generation, and among them, 24 were similar to the genes of known or putative functions [19].
Compared with previous works on Porphyra yezoensis generation related gene expression, we constructed the difference expression libraries by the suppression subtractive hybridization (SSH) method The GO annotation showed that 58% cellular components of carpospores were located in cytoplasm and nucleus; while vegetative cells were mainly located in cytoplasm and nucleus, especially located in Golgi apparatus, and high expression on plastid, ribosome, endoplamic reticulum. In the thallus of vegetative growth, photosynthesis and carbohydrate assimilation, RNA transcription and protein translation, and material transport are the main biological processes. According to the cultivation manner of P. yezoensis in China, the thalli are aerial exposure on every six hours during low tide. The extreme tolerance to desiccation of high intertidal P. yezoensis is demonstrated and blades survive a loss of up to 85-95% of their water during the daytime low tide to become crisp sheets. Temperature (winter freezing stress at high latitudes and heat stress in summer) and highlight stress often accompanies the cultivation period. The genes in response to heat and oxidative stress are expressed at large amount.
The carpospores express more genes in transcription, transport, carbohydrate metabolism, particular in signal transduction, DNA and protein modification, protein and nucleotide metabolism. The carpospores tends to be more active in development, rather in growth so that the signal transduction and a series of metabolisms happened in order to get enough energy for releasing and drilling into calcium carbonate shell. The expressed sequence tags of filamentous sporophyte of Porphyra haitanensis analysis studied by Pang et al. also found the transport and transcription, responsing to oxidation genes expression, and so on, similar to this research [24].
Subtractive hybridization is the first technique to be widely used for the purpose of identifying differentially expressed genes on a global scale. Advantages of the technique include the ability to isolate genes with no prior knowledge of their sequence or identity and the use of common molecular biology techniques that do not require specialized equipment or analyses. Several limitations of the original protocols, such as requirements of large quantities of RNA and bias toward abundant genes, have been overcome by incorporation of PCR into the suppression subtractive hybridization (SSH) technique. However, SSH remains applicable only on pair-wise treatment comparisons and must be replicated with the tester and driver reversed to identify gene expression changes in both directions. Additionally, subtractive hybridization does not provide a quantitative measure of expression differences and is most efficient at identifying genes that are completely absent, rather than expressed less abundantly, in the driver sample.
Though the suppression subtractive hybridization (SSH) technique can overcome the bias toward abundant genes more effective than subtractive hybridization, there are still four genes obtained in this research, such as ribosomal protein L41(HS), 50S ribosomal protein L9(HS), ribosomal protein S11 (HS572889 and HS572733) and ATP-binding protein (HS572892 and HS572609) which are identified on both vegetative cells and carpospores. These genes are too abundant to remove their mRNAs so that they can be amplified by the subsequent PCR process. This is another limitation of SSH.
Eighteen genes have not been annotated in this study. Due to the insufficient research on functional genes and unclear function of many sequences, further investigations are needed.
Conclusion
According to the information of GO annotation, the vegetative cells tend to growth and self- protection, while the carpospores tends to be more active in development. Interpretation of the differentially expressed genes revealed new insight into the molecular processes of the generation alternation of Porphyra yezoensis. Due to the insufficient research on functional genes and unclear function of many sequences, further investigations are needed.
Methods
Suppression subtractive hybridization (SSH)
The separation of Porphyra yezoensis cells
The mature blades of Porphyra yezoensis were collected from the culture nets in the sea located in the Dongtai City, Jiangsu Province. Carpospore and blade vegetative cells of Porphyra yezoensis were separated under the dissecting microscope to ensure its correctness. After that, the separated cells were collected in the freezing tubes which had pretreated with DEPC solution separately and were preserved in liquid nitrogen for future use.
The extraction of total RNA and synthesis of full-length cDNA
About 0.1 g separated cells that preserved in liquid nitrogen were put into the pre-cold mortar with liquid nitrogen. Then the cells were grounded into powder and were added 1 ml plant RNA Extraction Reagent to extract the total RNA. There was no degradation observed through 1% agarose gel. The RNA purity and quantities were measured by UV spectrophotometer method. The extracted RNAs were stored at -80°C for future use. The synthesis of cDNA was carried out according to the Super SMART PCR cDNA Synthesis Kit.
SSH process
According to the protocol of Clontech PCR-Select cDNA Subtraction Kit, the adaptor was connected and the ligation efficiency was estimated after the products were digested by Rna I (or RsaI) enzyme and purified. Carpospores were used as tester and blade vegetative cells were as driver, they were hybridized twice at 68°C for 8 h and 16 h respectively. Then the specific amplification of forward subtractive cDNA was gotten by twice inhibition PCR. At the same time, the reverse subtractive cDNA was performed with the tester and driver exchanged
The construction of subtractive cDNA library of differential expression between generations
The differentially expressed cDNAs between generations were ligated into the Target Vector (TOYOBO), then transformed into the component E. coli DH5α and the white-blue plaque selection were used to get the positive clones.
The detection of dot- blot
Fifty nanogram carpospore and 50 ng blade vegetative cells of Porphyra yezoensis were treated with Rsa I enzyme. The random primed labeling method was adapted on the double strands DNA and the radionuclide α-32P dATP was used to label the probes.
The 480 clones were picked randomly from library, and the amplification of colony PCR was constructed by using nested primer. The products larger than 800 bp were dipped on the nitrocellulose membrane (double membrane prepared), and then the dot-blot was carried on. Fifty nanogram forward and reverse subtractive cDNA groups were marked as probe to hybrid with the two repeated copies membranes of the same cDNA library.
EST processing
The two groups of EST sequences were cleaned with the CAP3 software.
Low quality sequences and vector sequences were removed from each EST clone by using this application. The cleaned EST sequences were used to find overlap assembly of contiguous sequences. Cap3 was used to identify overlaps between sequences and assemble sequence fragments into a larger sequence [20]. Samples that could be joined together were assembled into contiguous sequences "contigs". Those sequences from a cluster allowing the establishment of a consensus sequence were included in a contig. In this process, we defined singlets as clustered sequences that could not be included in a consensus sequence and singletons as sequences that were not grouped in a cluster. The unigenes were then the sum of singletons, singlets, and contigs.
EST comparative analysis and functional assignment
We performed Homologous search by Blastn tool with the unique sequences (including assembled contigs and singletons). A local implementation of BLAST server was used to search against the NCBI's non-redundant nuclear sequence database. We set up a cut off value and discarded hits with an E-value < 10-5. Parameters were as follow: Max target sequences: 10 (Descriptions:10, Alignments:10); Expect threshold: 0.01; Wordsize: 11; Match Score: 2; Mismatch Score: -3; Gap Open Penalty Score: 5; Gap Extension Penalty Score: 2.
The homology searches were made again with the program Blastx, with a threshold e-value of 1e-20. Because the conserved of amino acids sequence is higher than that of nuclear sequence, the ESTs sequences can be translated into amino acid sequences and searched the homology with Blastx for those ESTs without obvious results in Blastn. Parameters were as follow: Max target sequences: 10 (Descriptions:10, Alignments:10); Expect threshold: 0.01; Wordsize: 3; Matrix: BLOSUM62; Gap Open Penalty Score: 11; Gap Extension Penalty Score: 1.
To assign putative functions to the unique sequences, we employed blast2go to extract the GO hierarchical terms of their homologous genes from the protein databases [21]. Meanwhile, we also mapped the unique sequences to metabolic pathways in accordance with the KEGG [22]. Enzyme commission (EC) numbers [23] were obtained and used to putatively map unique sequences to specific biochemical pathways.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SS and PX initiated the study. YL and LW performed the RNA isolation, suppression subtractive hybridization (SSH) and sequencing. SS, GZ and LY designed the framework for data analysis and interpretation of data.
SS drafted the original manuscript. All authors have participated in the final editing and have read and approved the final manuscript.
All authors read and approved the final manuscript.
Supplementary Material
The identification of non-redundent singlets and contigs of vegetative cells from SSH library. The file contains SSH clone ID, cDNA insert size, putative identify, E-value, cellular component, biological process and molecular function annotation from the vegetative cells.
The identification of non-redundent singlets and contigs of carpospores from SSH library. The file contains SSH clone ID, cDNA insert size, putative identify, E-value, cellular component, biological process and molecular function annotation from the carpospores.
Contributor Information
Songdong Shen, Email: shensongdong@suda.edu.cn.
Gaochuan Zhang, Email: zhanggaochuan@suda.edu.cn.
Yanyan Li, Email: 84955549@qq.com.
Li Wang, Email: 24090010@qq.com.
Pu Xu, Email: xupu66@sina.com.
Lefei Yi, Email: yilefei@yahoo.com.cn.
Acknowledgements
This work was supported by the National Natural Sciences Foundation of China (NSFC 40576074).
References
- Cole K, Conway E. Phenetic implications of structural features of the perennating phase in the life history of Porphyra and Bangia (Bangiophyceae, Rhodophyta) Phycologia. 1975;14:239–245. doi: 10.2216/i0031-8884-14-4-239.1. [DOI] [Google Scholar]
- Polne-Fuller M, Gibor A. Development studies in Porphyra. I. Blade differentiation in Porphyra perforata as expressed by morphology, enzymatic digestion and protoplast regeneration. Journal of Phycology. 1984;20:609–616. doi: 10.1111/j.0022-3646.1984.00609.x. [DOI] [Google Scholar]
- Nelson WA, Brodie J, Guiry MD. Terminology used to describe reproduction and life history stages in the genus Porphyra (Bangiales, Rhodophyta) Journal of Applied Phycology. 1999;11:407–410. doi: 10.1023/A:1008174307352. [DOI] [Google Scholar]
- Nikaido I, Asamizu E, Nakajima M, Nakamura Y, Saga N, Tabata S. Generation of 10,154 Expressed Sequence Tags from a Leafy Gametophyte of a Marine Red Alga, Porphyra yezoensis. DNA Research. 2000;7:223–227. doi: 10.1093/dnares/7.3.223. [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakajima M, Kitade Y, Saga N, Nakamura Y, Tabata S. Comparison of RNA expression profiles between the two generations of Porphyra yezoensis (Rhodophyta), based on expressed sequence tag frequency analysis. Journal of Phycology. 2003;39(5):923–930. doi: 10.1046/j.1529-8817.2003.03003.x. [DOI] [Google Scholar]
- Kakinuma M, Kaneko I, Coury DA, Suzuki T, Amano H. Isolation and identification of gametogenesis-related genes in Porphyra yezoensis (Rhodophyta) using subtracted cDNA libraries. Journal of Applied Phycology. 2006;18:489–496. doi: 10.1007/s10811-006-9052-8. [DOI] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Pukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellog DE, Lukyanov KA, Lukyanov SA. An improved method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller V, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QY, van der Meer JP, Reith ME. Isolation and characterization of phase-specific complenentary DNAs from sporophytes and gametophytes of Porphyra purpurea (Rhodophyta) using subtracted complrmentary DNA libraries. Journal of Phycology. 1994;30(3):513–520. doi: 10.1111/j.0022-3646.1994.00513.x. [DOI] [Google Scholar]
- Liu QY, Reith ME. Isolation of a gametophyte-specific cDNA encoding a lipoxygenase from the red alga Porphyra purpurea. Mol Mar Biol Biotechnol. 1994;3:206–209. [PubMed] [Google Scholar]
- Liu QY, Baldauf SL, Reith ME. Elongation factor 1α genes of the red alga Porphyra purpurea include a novel, developmentally specialized variant. Plant Mol Biol. 1996;31:77–85. doi: 10.1007/BF00020608. [DOI] [PubMed] [Google Scholar]
- Liu QY, Ross N, Lanthier P, Reith ME. A gametophyte cell wall protein of the red alga Porphyra purpurea (Rhodophyta) contains four apparent polysaccharide-binding domains. Journal of Phycology. 1996;32:995–1003. doi: 10.1111/j.0022-3646.1996.00995.x. [DOI] [Google Scholar]
- Liu QY, Reith ME. A sporophyte cell wall protein of the red alga Porphyra purpurea (Rhodophyta) is a novel member of the chymotrypsin family of serine proteases. Journal of Phycology. 1996;32:1003–1009. doi: 10.1111/j.0022-3646.1996.01003.x. [DOI] [Google Scholar]
- Lee EK, Seo SB, Kim TH, Sung SK, An G, Lee CH, Kim YJ. Analysis of expressed sequence tags of Porphyra yezoensis. Mol Cell. 2000;3(10):338–342. [PubMed] [Google Scholar]
- Kakinuma M, Kaneko I, Coury DA, Suzuki T Amano H. Isolation and identification of gametogenesis-related genes in Porphyra yezoensis (Rhodophyta) using subtracted cDNA libraries. Journal of Applied Phycology. 2006;18:489–496. doi: 10.1007/s10811-006-9052-8. [DOI] [Google Scholar]
- Kitade Y, Asamizu E, Fukuda S, Nakajima M, Ootsuka S, Endo H, Tabata S, Saga N. Identification of genes preferentially expressed during asexual sporulation in Porphyra yezoensis gametophytes (Bangiales, Rhodophyta) Journal of Phycology. 2008;44:113–123. doi: 10.1111/j.1529-8817.2007.00456.x. [DOI] [PubMed] [Google Scholar]
- Zhou X, Mao Y, Wang M, Sui Z, Zhang X. Preparation of Porphyra yezoensis cDNA microarray for analyzing the expression profile differences between sporophyte and gametophyte. High Technology Communication. 2006;16(12):1300–1305. [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30(1):42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera M, Okuno Y, Hattori M, Goto S, Kanehisa M. Computational assignment of the EC numbers for genomic-scale analysis of enzymatic reactions. J Am Chem Soc. 2004;126(50):16487–16498. doi: 10.1021/ja0466457. [DOI] [PubMed] [Google Scholar]
- Pang G, Wang G, Hu S, Tseng CK. Acquirement and analysis of expressed sequence tags of filamentous sporophyte of Porphyra haitanensis. Oceanologia et Limnologia Sinica. 2005;36(5):452–458. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The identification of non-redundent singlets and contigs of vegetative cells from SSH library. The file contains SSH clone ID, cDNA insert size, putative identify, E-value, cellular component, biological process and molecular function annotation from the vegetative cells.
The identification of non-redundent singlets and contigs of carpospores from SSH library. The file contains SSH clone ID, cDNA insert size, putative identify, E-value, cellular component, biological process and molecular function annotation from the carpospores.