Abstract
The chloroplast-localized SIB1 protein was previously identified by its interaction with SIGMA FACTOR 1 (SIG1), a component of the RNA polymerase machinery responsible for transcription of plastid genes. The physiological function of SIB1 is little known. We found that expression of SIB1 is induced by infection with Pseudomonas syringae, suggesting its possible involvement in the defence response. The sib1 loss-of-function mutation compromises induction of some defence-related genes triggered by pathogen infection and the treatments with salicylic acid (SA) and jasmonic acid (JA), two key signalling molecules in the defence response. Conversely, constitutive over-expression of SIB1 causes the plants to hyper-activate defence-related genes following pathogen infection or the SA and JA treatments, leading to enhanced resistance to infection by P. syringae. SIB1 is a member of the large plant-specific VQ motif-containing protein family, and might act as a link to connect defence signalling with chloroplast function.
Keywords: disease resistance, jasmonic acid, salicylic acid, SIB1, sigma factors
INTRODUCTION
Plants use multi-layered defence systems to protect themselves against pathogen attack. In addition to pre-formed physical and biochemical barriers, the induced defence response is triggered when a host receptor recognizes an invading pathogen through microbe-associated molecular patterns (MAMPs) (Ausubel 2005; Jones & Dangl 2006). Successful pathogens have evolved various mechanisms to counteract the first layer of host defence. One of the virulence mechanisms employed by pathogens is the delivery of a variety of effector proteins into host cells that suppress the defence response, and modify host physiology to favour pathogen propagation (Abramovitch & Martin 2004; Jones & Dangl 2006; da Cunha, Sreerekha & Mackey 2007). In turn, plants have evolved resistance (R) proteins, each of which directly or indirectly recognizes a cognate effector. This recognition triggers the hypersensitive response (HR) [also known as effector-triggered immunity (ETI)], which often leads to programmed cell death of plant tissue at the attempted invasion site to limit pathogen spread (Jones & Dangl 2006). A pathogen strain expressing a recognized effector, thus triggering the plant’s HR, is often called an avirulent strain, whereas the strain that evades recognition, thus causing disease, is called a virulent strain.
Detection of an invading pathogen through its MAMPs and effectors triggers a series of signalling pathways leading to the expression of appropriate plant defence mechanisms. Salicylic acid (SA) and jasmonic acid (JA) are two of the best known defence signalling molecules that are accumulated in response to pathogen infection to activate different sets of defence-related genes (Glazebrook 2005). The SA response pathway is mediated by the protein NON-EXPRESSOR OF PATHOGENESIS-RELATED GENE 1 (NPR1), a key regulator of both local and systemic acquired resistance (SAR) (Cao et al. 1994). SA accumulation alters the redox state of NPR1, which then shuttles from the cytosol to the nucleus where it activates transcription factors (Mou, Fan & Dong 2003; Tada et al. 2008). JA mediates a signalling pathway through COI1, an SKP/Cullin/F-box (SCF) E3 ubiquitin ligase (Xie et al. 1998), to regulate transcriptional repressors (JAZ proteins) and the transcriptional activator (MYC2), leading to transcriptional reprogramming (Boter et al. 2004; Chini et al. 2007; Thines et al. 2007).
Growing evidence indicates that the chloroplast plays an important role in host-pathogen interaction. Functional chloroplasts are essential for some effector-triggered defence pathways and for the SA-mediated induction of PATHOGENESIS-RELATED (PR) genes (Genoud et al. 2002; Karpinski et al. 2003; Zeier et al. 2004; Roberts & Paul 2006). Furthermore, it was found that the MAPKcascade mediates the defence response through alteration of chloroplast metabolic activities, leading to generation of reactive oxygen species (ROS) from the chloroplast source (Liu et al. 2007). Not surprisingly, pathogens have evolved mechanisms to manipulate chloroplast function to suppress host defence and promote virulence. Many effectors contain chloroplast localization signals (Greenberg & Vinatzer 2003). Several effectors have been reported to target chloroplast proteins (Abbink et al. 2002; Fu et al. 2007; Jelenska et al. 2007; Caplan et al. 2008).These findings underline the importance of the chloroplast as a battlefield in host-pathogen interaction.
The plastid genome of a higher plant generally contains 60-200 open reading frames (ORFs) (Leister 2003). Plastid genes are transcribed by either the plastid-encoded RNA polymerase (PEP) or the nucleus-encoded RNA polymerase (NEP) (Kanamaru & Tanaka 2004; Lysenko 2007). NEP is believed to be responsible for overall transcription of the plastid genome, whereas PEP is a prokaryotictype enzyme principally involved in transcription of photosynthesis-related genes. The function of PEP is regulated by the nuclear-encoded sigma subunits (SIGs), which confer promoter recognition specificity and are required for transcription initiation (Kanamaru & Tanaka 2004; Lysenko 2007). Six SIG genes (SIG1-6) have been identified from the Arabidopsis nuclear genome. The sig2 mutation leads to chlorophyll deficiency (Shirano et al. 2000; Privat et al. 2003), possibly through its effects on transcription of psbD, psbJ and D1, as well as several tRNA-encoding plastid genes (Kanamaru et al. 2001; Nagashima et al. 2004a). SIG3 and SIG4 regulate transcription of psbN (Zghidi et al. 2007) and ndhF (Favory et al. 2005), respectively. SIG5 is induced by various abiotic stresses, including blue light irradiation, and controls transcription of psbD (Tsunoyama et al. 2002, 2004; Nagashima et al. 2004b). SIG6 functions in light-dependent chloroplast development, and its loss-of-function mutation affects expression of several plastid genes (Ishizaki et al. 2005; Loschelder et al. 2006). SIG1 is one of the most abundant sigma factors in Arabidopsis (Tanaka et al. 1997), but its function has not been defined. The rice sig1 mutant was found to have reduced chlorophyll content, an increase in transcript levels of at least 10 plastid genes and the reduction in transcript levels of at least 12 other plastid genes (Tozawa et al. 2007).
Arabidopsis SIGMA FACTOR-BINDING PROTEIN 1 (SIB1) was previously identified through yeast two-hybrid screening as an interacting protein of AtSIG1, and the SIG1-SIB1 interaction was further verified through a pulldown assay (Morikawa et al. 2002). SIB1 contains a putative plastid targeting signal, and the SIB1-GFP fusion was found to be localized in chloroplasts (Morikawa et al. 2002). The finding suggests that SIB1 may regulate the function of SIG1. Recently, the Arabidopsis SIB1 gene was found to be induced by SA (Narusaka et al. 2008); however, its biological function remains unclear. Here, we report data suggesting a role of SIB1 in the disease resistance pathway in Arabidopsis.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis thaliana (ecotype Columbia) plants were grown in a growth room with the following conditions: 22 °C, 50% humidity, a 9/15 h day/night cycle at a light intensity of 125 mol m−2 s−1 provided by cool white fluorescent bulbs. The plants were fertilized every 2 weeks. Plants used for pathogen inoculation and chemical treatments were approximately 5 weeks old. For seed production, 5-week-old plants were transferred from the growth room to an air-conditioned greenhouse with the same growth conditions, except the light was provided by natural sunlight and supplemented by the fluorescent light bulbs at a 15/9 h day/night cycle.
Pathogens and plant inoculation
Culturing and preparation of Pseudomonas syringae, plant inoculation and the in planta bacterial growth assay were carried out as previously described (Ge et al. 2007). For in planta bacterial growth assay, leaves were infiltrated with a bacterial suspension of 5 × 104 cfu mL−1. For preparing pathogen-challenged leaf tissues for RNA extraction, leaves were infiltrated with bacterial suspension of 1 × 107 cfu mL−1.
Botrytis cinerea was maintained in the maltose agar medium (1.5% maltose, 0.3% peptone, 3% glucose and 12 g L−1 agar). For spore production, the potato dextrose medium was used (24 g potato dextrose broth and 12 g agar for 1 L medium) for culturing. Leaves were pricked with a needle, and the punched site of each leaf was inoculated by placing 20 μL of spore suspension (with a concentration of 5 × 105 spores mL−1). After the inoculums were dry, the plants were covered for 3 d to maintain high humidity. Symptoms were scored 5-6 d post-inoculation. Alternaria brassicicola culturing and inoculation were performed in a similar way, except that the fungus was grown on the V8 medium (163 mL V8 juice, 1.63 g CaCO3 and 12 g agar for 1 L medium) for spore production.
PCR analysis of the sib1 T-DNA insertion alleles
The T-DNA flanking sequence of SALK_063337 was amplified using the primer pair LBb1 (5′-AGTTGCAGCAAGCGGTCCACGC-3′) and SIB1p1 (CTCTTAC AGGAACCGAACATGGAG). The wild-type (wt) fragment of this locus was amplified using SIB1p1 and SIB1p2r (TTACGATGAGAACTCGATAACCTGA).
RNA isolation and RNA blot analysis
RNA was isolated using the TRIzol reagent according to the manufacturer’s instruction (Invitrogen, Carlsbad, CA, USA). RNA electrophoresis, transfer onto nylon membrane and hybridization were performed according tostandard protocols (Sambrook, Fritsch & Maniatis 1989). The probes were labelled using the Ready-to-Go DNA labelling kit (Amersham Biosciences, Piscataway, NJ, USA). The following primers were used to amplify the DNA fragments from genomic DNA for using as the probes in the RNA blotting analysis: 5′-GGTACCGAACATGGAGTCATCATC-3′ and 5′-TCTAGACATAGAATCG ATGCTTCCA-3′ for SIB1; 5′-GATCAGTCATCATCAA CGTTGCTC-3′ and 5′-CAGAGAGAACCAATGCTTC CTAAAG-3′ for At2g41180 (T3K9.5). Hybridization and washes were carried out under the high stringent conditions at 65 °C.
Treatments with salicylate (SA) and jasmonate (JA)
SA and JA were purchased from Sigma-Aldrich (St Louis, MO, USA). Plants were sprayed with the SA solution at the concentration of 1 mm, and leaf samples were harvested at different time-points after the treatment. For the JA treatment, plants were sprayed with 100 μM methyl jasmonate.
Construction of 35S∷SIB1 transgenic lines
To construct 35S∷SIB1, the SIB1 genomic fragment without the putative promoter region was amplified through PCR with the primer pair SIB1Kpn (5′-GGTACCGAACATGGAGTCATCATCG-3′) and SIB1Pst (5′-CTGCAGC AGTAACGGGTACATTGGG-3′). The PCR product was cloned into pCR-BluntII-TOPO. The KpnI/PstI fragment was cut from the TOPO clone, and inserted into downstream of the 35S promoter in the binary vector pCHF3 to generate 35S∷SIB1.
Chlorophyll determination
The relative amount of chlorophyll in leaves was determined using Chlorophyll Meter SPAD-502 (Spectrum Technologies, Plainfield, IL, USA) according to the manufacturer’s instruction. For each genotype, eight fully expanded leaves from four 6-week-old plants were measured.
Quantitative real-time reverse transcription PCR (qPCR) analysis
The primers used for the qPCR analysis were designed through Universal ProbeLibrary Assay Design Center, Roche Applied Science (http://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp?id=uplct_030000). Transcript levels were normalized against constitutively expressed AtACT2. The primer sequences are listed in Supporting Information Table S1. Total RNA samples for the analysis were isolated using the TRIzol reagent as described earlier, and purified by RNeasy MinElute Cleanup Kit (Qiagen, Valencia, CA, USA, cat. no. 74204), and treated with DNase. cDNA was synthesized with iScript cDNA Synthesis Kit from Bio-Rad (Hercules, CA, USA, cat. no.170-8891). PCR was performed in a 20 μL reaction mixture with SYBR GreenER (Invitrogen, cat. no. 11762) using the following program: 95 °C for 3 min, 40 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, followed by one cycle of 95 °C for 1 min, 55 °C for 30 s and 95 °C for 30 s.The instrument used for qPCR was the Mx3005P system from Stratagene (La Jolla, CA, USA).
Affymetrix GeneChip analysis
Leaves of 5-week-old wt plants were inoculated with Pst avrRpm1 (1 × 107 cfu mL−1 suspension), and collected for RNA isolation 6 h post-inoculation. RNA was isolated using the TRIzol extraction method followed by RNA purification using RNeasy MiniElute Cleanup kit (Qiagen). Six Affymetrix ATH1 arrays were used to hybridize the RNA samples including three biological replicates for the pathogen-treated wt and sib1 samples. The original microarray data (.cel files) were background corrected, quantile normalized and summarized for each probe set using the affy package (Irizarry et al. 2003) in BioConductor with default settings. The summary scores were then analysed probe set by probe set using the MAANOVA package (Wu et al. 2003) in BioConductor. A shrinkage-based t-test was conducted to compare the two groups (Cui et al. 2005). An empirical P value was obtained for each probe set based on permutation of the observed data (Yang & Churchill 2007). The fold changes are shown to the genes only if the difference is statistically significant (with a P value of <0.05).
RESULTS
Pathogen-triggered induction of SIB1 is partially dependent on NPR1 and SA accumulation
We have previously carried out a defence transcriptome analysis using the Affymetrix GeneChip to identify Arabidopsis genes that are quickly induced by infection with the bacterial pathogen P. syringae (Ge et al. 2007). One of the pathogen-responsive genes identified in the analysis is At3g56710, which encodes the nuclear-encoded and chloroplast-localized SIB1 (Morikawa et al. 2002). SIB1’s strong induction following pathogen infection suggests it could be a component in a defence response pathway. The result prompted us to examine the role of SIB1 in disease resistance.
We further examined expression patterns of SIB1 during the host-pathogen interaction through RNA blot analysis. RNA samples were isolated from Arabidopsis wt plants (Col-1 ecotype) and the sib1 mutant (see below) at different time-points following infection with either the virulent P. syringae strain DC3000 (Pst) or the avirulent Pst strain that expresses the type III effector gene avrRpm1 (Pst avrRpm1). A single 0.85 kb band was detected from the wt plants infected with Pst avrRpm1, but not from the sib1 mutant, indicating that the probe only detects the SIB1transcripts (Fig. 1a). As shown in Fig. 1b, SIB1 displayed a similar induction pattern following infection by both Pst and Pst avrRpm1. The SIB1 transcripts were barely detectable in uninfected (M) tissues, but started to increase within 1.5 h (h) after infection. The SIB1 transcripts reached a much higher level at 5 h post-infection (hpi), and its level remained elevated at 24 hpi.
Figure 1.
Pathogen-triggered expression of the SIB1 gene. (a) An RNA blot probed with the SIB1 gene. RNA was isolated from the uninfected and Pst avrRpm1-infected plants. The pathogen-infected wild-type (wt) plants accumulated a 0.85 kb SIB1 transcript, whereas the sib1 mutant did not produce a detectable level of the normal SIB1 transcripts. Col: the Columbia ecotype used as wt plants. A portion of the ethidium bromide-stained agarose gel is shown as an RNA loading control. (b) An RNA blot revealed that SIB1 expression was induced by both Pst and Pst avrRpm1, and its expression was compromised in npr1, enhanced disease susceptibility 5 (eds5) and NahG plants. (c) The transcript levels of SIB1 in uninfected and Pst avrRpm1-infected wt, npr1, eds5 and NahG plants determined by qPCR analysis. (d) An RNA blot showing SIB1 transcript levels following infection with the Pst hrcC mutant strain and Pst avrRpm1. (e and f) Transcript levels of At2g41180 in wt, sib1 and SIB1 over-expression line (see below) determined by RNA blotting (e) and qPCR analysis (f). Leaves were inoculated with Pst avrRpm1. Mock (M) represents RNA samples from leaves after mock (H2O) inoculation.
To investigate whether the pathogen-triggered SIB1 expression is dependent on SA and NPR1, we examined SIB1’s expression patterns in npr1, enhanced disease susceptibility 5 (eds5) and NahG plants.The eds5 is defective in SA accumulation (Nawrath et al. 2002). The NahG plants express the bacterial enzyme salicylate hydroxylase, which not only degrades SA, making the plants defective in disease resistance (Delaney et al. 1994), but also produces the degradation product catechol, causing a defect in basal resistance (van Wees & Glazebrook 2003). The RNA blot result showed that pathogen-triggered accumulation of the SIB1 transcripts was compromised but not abolished in npr1, eds5 and NahG plants (Fig. 1b), indicating that its induction is partially dependent on SA accumulation and NPR1. The result from qPCR analysis further confirmed that pathogen-triggered induction of SIB1 was compromised by the npr1 and eds5 mutation, and by the NahG over-expression (Fig. 1c).
In a separate experiment, we compared induction of SIB1 by Pst avrRpm1, and by the P. syringae mutant hrcC. The strain Pst hrcC is defective in delivering type III effectors into host cells, and therefore does not cause disease (Boch et al. 2002). The Pst hrcC mutant strain was also able to induce SIB1 expression (Fig. 1d). SIB1 induction by Pst hrcC was slightly weaker than that by Pst avrRpm1 at 5 hpi; however, SIB1 transcripts were accumulated at a higher level in the Pst hrcC-treated leaves than in the Pst avrRpm1-treated leaves at 24 hpi, raising a possibility that some type III effector(s) might attenuate SIB1 expression. The above result also suggests that the pathogen-mediated induction of SIB1 does not require the effector-triggereddefence response, but is largely triggered by the MAMP-triggered defence response.
T3K9.5 (At2g41180) was previously identified as a homolog of SIB1 that shares over 50% sequence identity at the amino acid level with SIB1, and was also found to interact with SIG1 (Morikawa et al. 2002). Transcript levels of At2g41180 were slightly higher following infection with Pst avrRpm1 in the wt plants as revealed by the RNA blotting and qPCR analysis (Fig. 1e,f). However, in the sib1 mutant, the At2g41180 transcript level was found to be more significantly increased than in the wt plants following the pathogen infection (Fig. 1e,f).
SIB1 is a member of the plant-specific VQ motif-containing protein family
The SIB1 gene contains a single exon, and encodes a polypeptide with 151 residues. BLAST searches revealed that SIB1 does not share a significant sequence similarly to other proteins with known functions or to any non-plant proteins in the GenBank sequence databases, suggesting that it is a plant-specific protein. SIB1 contains the plant-specific VQ motif that is found in at least 34 predicted Arabidopsis proteins (Fig. 2 and Supporting Information Table S2). A group of VQ motif-containing proteins from a variety of plant species were described previously (Andreasson et al. 2005). The supplementary data section provides additional information on sequence analysis of VQ motif-containing proteins, and lists the 34 such proteins identified from the Arabidopsis sequence database. A large majority of these proteins are small (100-250 amino acids) and share little sequence similarity to other proteins in the GenBank databases. It remains to be determined whether the VQ motif-containing proteins have descended from a common ancestor. Interestingly, genes encoding VQ domain proteins are found in the genomes of higher plants and mosses, but not in the algal genomes, indicating that the VQ domain-containing proteins are unique to land plants.
Figure 2.
A sequence alignment of the VQ domain-containing regions from 25 plant proteins. Sequences were aligned using the ClustalW algorithm of the VectorNTI software. Identical residues are in black background, and similar/conserved residues are in grey background. Included in the alignment are 17 Arabidopsis proteins, two rice proteins, two grape proteins (CAO63044 and CAN64541), two moss proteins (XP_001760459 and XP_001771471) and two spike moss sequences (SmSeq1 and SmSeq2).
Among the members of this VQ motif-containing family in Arabidopsis, At2g41010 was previously identified as a calmodulin-binding protein (AtCAMBP25) and functions as a negative regulator of osmotic stress tolerance (Perruc et al. 2004).Another known member of this family is MKS1 (At3g18690), which was originally identified as a substrate of Arabidopsis MAP kinase 4 (MAPK4) (Andreasson et al. 2005). MKS1 interacts with both MAPK4 and WRKY tran-scription factors WRKY25 and WRKY 33, and is implicated in the SA-dependent defence pathway by coupling MAPK4 to the WRKY transcription factors (Andreasson et al. 2005). In our defence transcriptome results, at least 14 of the Arabidopsis genes encoding members of this family were found significantly induced by pathogen infection, and onewas suppressed (Supporting Information Table S2), suggesting that many of them are likely involved in stress responses.
Figure 2 shows a sequence alignment of the VQ motif-containing regions from 25 plant proteins including 17 Arabidopsis proteins and two proteins each from rice, grape, moss (Physcomitrella patens) and spike moss (Selaginella moellendorffii). In the Conserved Domain Database (CDD) (Marchler-Bauer et al. 2007), the consensus sequence of the VQ motif is listed as FXhVQChTG (http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?uid=pfam05678) where X is any amino acid and h is a hydrophobic amino acid. However, based on our sequence alignment results (Fig. 2 and data not shown), there are generally three residues between the F and VQ residues, and VQ is not often followed by a C residue. In addition, the two hydrophobic residues are not always conserved. Therefore, we suggest the consensus sequence for the VQ motif to be changed to FXXXVQXXTG.
Over-expression of SIB1 enhances resistance to P. syringae
We carried out mutational analyses to determine the role of SIB1 in plant defence.
A putative T-DNA insertion line (SALK_063337) for SIB1 was obtained from the Arabidopsis Biological Resource Center (ABRC, Columbus, OH, USA). A homozygous line for the T-DNA insertion allele was identified from the pooled T4 seedlings through PCR analysis. This line has a single T-DNA insertion just upstream of the stop codon of the SIB1 ORF. In the RNA blot analysis using RNA isolated from leaves with or without pathogen infection, the normal SIB1 transcripts were not detected in the homozygous insertion line (Fig. 1a), although a weak band with an abnormal size (approximately 1.2 kb) was detected, indicating that the insertion resulted in knock-out or severe knock-down of the SIB1 gene. We named this insertion line sib1-1 (abbreviated as sib1). We later obtained another homozygous T-DNA insertion line (SALK_127478C) from the ARBC. SALK_127478C has a T-DNA insertion in the promoter region approximately −200 from the ATG start codon of the SIB1 ORF. Similarly, SIB1 transcripts were undetectable from this line (Fig. 3a). SALK_127478C was named sib1-2. The sib1 mutant plants are morphologically indistinguishable from wt plants, although the mutant plants were slightly larger in size (Fig. 3b).
Figure 3.
Phenotype analyses of loss- and gain-of-function mutations of SIB1. (a) The RNA blot shows the SIB1 transcript was undetectable from the sib1-2 mutant. The RNA samples were extracted from uninfected leaves (0 h) and leaves inoculated with Pst avrRpm1 at 6 hpi. (b) Morphological phenotypes of 4-week-old plants. (c) In planta growth of Pst in wild-type (wt), sib1 and 35S∷SIB1-1 leaves. Each data point represents the average of three replicates ± SD. (d) The RNA blot result showing the SIB1 transcript levels in uninfected leaves of the two SIB1 over-expression lines and wt. (e) The relative amount of chlorophyll in leaves of wt, sib1 and the SIB1 over-expression lines. Values are the average of eight replicates ± SD.
An in planta bacterial growth assay was performed to determine whether the sib1 mutation results in any alteration in resistance to Pst. Leaves of 5-week-old plants were inoculated with Pst, and leaf discs were collected at 2 and 4 d post-infection (dpi) for counting bacterial numbers. In multiple independent assays, bacterial growth rates in the sib1 mutant were 1.3- to 3.4-fold higher than in wt plants; however, the differences were found statistically insignificant in three out of four independent experiments (Fig. 3c and data not shown).
Transgenic Arabidopsis lines were generated that express SIB1 under the control of the cauliflower mosaic virus 35S promoter (35S∷SIB1). Among 20 independent SIB1over-expression lines generated for this study, at least 12 lines exhibited various degrees of growth retardation compared with wt plants (Fig. 3b). We choose two transgenic lines (35S∷SIB1-1 and 35S∷SIB1-2) for further analyses. 35S∷SIB1-1 plants showed a more pronounced growth retardation, whereas 35S∷SIB1-2 plants displayed a moderate growth retardation (Fig. 3b). The RNA blotting result revealed that these two lines constitutively accumulate high levels of SIB1 transcripts with a higher transcript level in 35S∷SIB1-1 than in 35S∷SIB1-2 (Fig. 3d). The leaves of the over-expression lines appeared darker than wt leaves, and contained higher chlorophyll content (Fig. 3e).
The results from the in planta bacterial growth assay revealed that SIB1 over-expression leads to enhanced resistance to Pst (Fig. 3c). In multiple independent assays, bacterial growth in the 35S∷SIB1-1 plants were found to be 2.0- to 6.7-fold lower than in wt plants. The 35S∷SIB1-2 line was also found to be more resistant than wt plants with the bacterial growth rate reduced by 1.8- to 4.2-fold; however, in three out of five independent assays, the difference between 35S∷SIB1 and two wt plants was not found to be statistically significant. We did not find any obvious difference between wt, sib1 and the SIB1 over-expression lines in appearance of hypersensitive cell death following inoculation with Pst avrRpm1.
Mutations in SIB1 alter induction of defence-related genes triggered by pathogen infection and the treatments with SA and JA
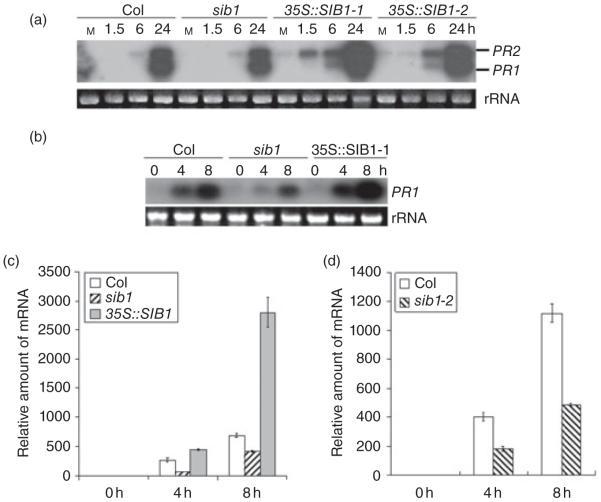
RNA blot analysis was carried out to examine whether the SIB1 loss- and gain-of-function mutations cause alteration in pathogen-triggered induction of the defence-related genes, PR1, PR2 and avrRpt2-INDUCED GENE 1 (AIG1). RNA samples were isolated from leaves inoculated with Pst avrRpm1, and collected at different time-points following the pathogen infection. We did not find any obvious difference in transcript profiles of AIG1 between wt and the loss- and gain-of-function mutants of SIB1 (data not shown). However, the two PR genes were induced more rapidly and more strongly in the SIB1 over-expression lines than in wt plants, whereas in the sib1 loss-of-function mutant, the transcript levels of PR1 and PR2 were slightly lower than in the wt plants at 24 hpi (Fig. 4a). Because PR1 and PR2 are SA-responsive genes, we further determined whether the mutations of SIB1 alter the plant response to the SA treatment. As shown in Fig. 4b, the PR1 gene was strongly induced within 4 h of SA application; however, the sib1 mutant showed weaker PR1 induction than wt. This result was confirmed from an independent qPCR analysis (Fig. 4c). Similarly, sib1-2 was also compromised in SA-triggered PR1 induction (Fig. 4d). Conversely, induction of PR1 by SA was significantly enhanced in the SIB1 over-expression line (Fig. 4b,c).Although PR2 induction by SA was enhanced in the SIB1 over-expression line, we did not detect a significant difference in SA-mediated induction patterns of PR2 between the sib1, sib1-2 and wt plants (data not shown).
Figure 4.
Loss- and gain-of-function mutations of SIB1 affect expression of defence-related genes triggered by pathogen infection and the SA treatment. (a) An RNA blot probed with PR1 and PR2 genes. Leaves were inoculated with Pst avrRpm1. SIB1 over-expression led to quicker and stronger induction of these genes. The sib1 mutant accumulated a slightly lower level of these transcripts at 24 hpi. (b, c) PR1 transcript levels in wild-type (wt), sib1 and the SIB1 over-expression lines following the SA treatment determined by RNA blot analysis (b) and qPCR analysis (c). (d) The qPCR result revealed that sib1-2 was also defective in SA-mediated induction of PR1.
We then carried out a transcriptome analysis to reveal genes whose pathogen-mediated induction might be affected by the sib1 mutation. Leaves of wt and sib1 plants were inoculated with Pst avrRpm1, and the inoculated leaves were collected at 6 hpi. Total RNA extracted from these leaf samples was used for labelling and for hybridization of the Affymetrix’s Arabidopsis ATH1 GeneChip. Three biological replicates were included in the experiment. The transcript profiles of the pathogen-infected leaves of wt and sib1 plants were compared to identify differentially expressed genes between the wt and the sib1 plants. PR1 and PR2 transcript levels were not found to be significantly different between the wt and sib1 plant at 6 hpi (less than 1.3-fold with a P value of >0.05). However, 64 other genes were expressed over twofold lower in the sib1 mutant than in the wt plants. Supporting Information Table S3 lists the genes whose transcript levels differ more than twofold between the wt and sib1 mutant (with a P value of <0.05).
Although infection with biotrophic Pst strains generally triggers the SA signalling pathway, some well-known JA-responsive genes, such as pdf1.2a and pdf1.2b, are also induced by the Pst avrRpm1 infection. However, the result from the GeneChip analysis showed that the transcript levels of these two genes were 5.4- and 2.3-fold lower, respectively, in the sib1 mutant than in the wt plants (Supporting Information Table S3). qPCR analysis further confirmed that the sib1, as well as sib1-2, mutations compromise the pathogen-mediated induction of pdf1.2a and pdf1.2b (Fig. 5a). In contrast, the SIB1 over-expression line accumulated higher transcript levels of these two genes in both uninfected and pathogen-infected plants (Fig. 5a). To further reveal whether the loss- and gain-of-function mutants of SIB1 affect the JA signalling pathway, we used qPCR analysis to compare induction patterns of pdf1.2a and pdf1.2b following the JA treatment. As shown in Fig. 5b, JA-mediated induction of these two genes was significantly reduced in the sib1 and sib1-2 mutants, but was stronger in the SIB1 over-expression line. Because the JA signalling pathway plays a crucial role in a plant’s resistance to necrotrophic pathogens, we challenged the sib1 mutant, the SIB1 over-expression line and wt plants with the necrotrophic fungal pathogens B. cinerea and A. brassicicola; however, we did not find any obvious difference between the loss- and gain-of-function mutants of SIB1 and wt plants in resistance to these fungal pathogens.
Figure 5.
Loss- and gain-of-function mutations of SIB1 alter expression of jasmonic acid (JA)-responsive genes following pathogen infection and the JA treatment. RNA was extracted from leaves taken at different time-points after inoculation with Pst avrRpm1 (a) and the JA treatment (b), and the transcript levels of pdf1.2a and pdf1.2b were determined by qPCR analysis.
Does SIB1 regulate the function of SIG1?
Disruption of the SIG1 gene in rice (OsSIG1) is not fatal, and the study on the rice sig1 mutants has revealed that OsSIG1 regulates transcription of the plastid genes psaA and psaB (Tozawa et al. 2007). The specific function of SIG1 (At1G64860) in Arabidopsis remains unknown. We obtained two T-DNA insertion lines (Salk_000235 and Salk_147985) (AT1G64860) for the AtSIG1 gene from the ABRC. Salk_147985 has a T-DNA insertion at 80 bp upstream of the stop codon. However, we could not obtain any homozygous insertion line from its progenies, suggesting that the insertion at this gene is lethal. Salk_000235 has a T-DNA insertion at 47 bp after the stop codon.A homozygous insertion line for Salk_000235 was obtained; however, this insertion apparently does not affect AtSIG1 expression as RNA blot analysis showed that there was no detectable difference in AtSIG1 transcript accumulation between wt and the homozygous insertion line.
Expression of AtSIG1 was found to be suppressed following infection with Pst avrRpm1: its transcript level was 3.3-fold lower at 6 hpi in the wt plants than in theuninfected plants (Fig. 6a). If AtSIG1 is also a transcriptional regulator of psaA and psaB in Arabidopsis, we would expect that psaA and psaB transcript levels might also be reduced following the pathogen infection. However, the transcript levels of these two plastid genes were slightly higher in the infected tissues (Fig. 6b). A possible explanation for the inconsistence in the level of AtSIG1, and psaA and psaB is that the AtSIG1 protein level might not be reduced in the infected tissues, although we could not rule out the possibility that AtSIG1 might not control expression of psaA and psaB in Arabidopsis. The transcript levels of AtSIG1, psaA and psaB in the sib1 mutant were not significantly different from those in the wt plants; however, in the SIB1 over-expression line, the transcript levels of all these three genes were slightly reduced (Fig. 6a-c). It remains to be determined whether this reduction was directly or indirectly regulated by SIB1 over-expression.
Figure 6.
Determination of transcript levels of SIG1, psaA and psaB. RNA samples were extracted from uninfected and Pst avrRpm1-infected leaves (1.5 and 6 hpi) of wild-type (wt), sib1 and the SIB1 over-expression line. The transcript levels of SIG1 (a), psaA (b) and psaB (c) were determined by qPCR.
DISCUSSION
SIB1 was initially identified by its interaction with SIG1, and was speculated to function as an inhibitor of SIG 1 (Morikawa et al. 2002). We found that the SIB1 gene is up-regulated, but the SIG1 gene is down-regulated by pathogen infection, suggesting that both SIG1 and SIB1 could be involved in host-pathogen interactions. The sib1 mutation impairs pathogen-triggered induction of a subset of defence-related genes. Further analyses revealed that the sib1 mutations cause a defect in the SA- and JA-mediated defence response. Conversely, SIB1 constitutive over-expression makes the plants hyper-responsive to pathogen infection, and the SA and JA treatments, leading to enhanced disease resistance. However, the sib1 mutant did not show a significant alteration in resistance to P. syringae or to the necrotrophic fungal pathogens B. cinerea and A. brassicicola, which could be caused by functional redundancy with other defence pathways. The homolog of SIB1, At2g41180, could also have an overlapping function with SIB1. However, the transcript level of At2g41180 was not significantly changed following pathogen infection. The attempt to obtain a knock-out line for At2g41180 was not successful as the T-DNA insertion line from ABRC (Salk_152005C) which carries the insertion at approximately 90 bp upstream of the start codon does not appear to be a knock-out or knock-down mutant (data not shown).
SIB1 is a member of the plant-specific VQ domain-containing protein family. At least 34 predicted proteins in Arabidopsis contain the VQ motif. These proteins are generally small and share little significant sequence similarity to other proteins in the GenBank sequence databases. Other than the regions containing the conserved VQ motif, their primary structures are highly diverse. The land plant lineage-specific evolution and expansion of this family suggest that they might function in land plant-specific biological processes. Two other VQ motif-containing proteins have been characterized to date, and were found to be involved in signalling pathways in abiotic stress tolerance (Perruc et al. 2004) and in disease resistance (Andreasson et al. 2005). Interestingly, these two proteins and SIB1 were all identified initially by their interaction with other proteins. It is intriguing to speculate that many of the VQ domain-containing proteins might act as protein scaffolds that couple signalling components (such as calmodulin and MAPK4) to their downstream effectors (such as transcriptional regulators).
Recently, SIB1 was reported as an SA-induced gene (Narusaka et al. 2008). Narusaka et al. (2008) also generated SIB1 over-expression lines which were found to accumulate elevated transcript levels of ROS-related genes. However, the over-expression lines did not show any significant difference from wt plants in resistance to P. syringae (Narusaka et al. 2008). We found that, among the 35S∷SIB1 lines, only those with higher levels of the SIB1 transcripts exhibited moderately enhanced resistance to infection by Pst, whereas the other over-expression lines did not show any obvious difference from wt plants in resistance to Pst.
Although the sib1 loss-of-function mutation does not lead to significant change in resistance to Pst or to B. cinerea and A. brassicicola, the transcriptome analysis revealed that expression of a subset of defence-related genes was affected by the sib1 mutation. Many of the differentially expressed genes are known to be SA and/or JA responsive.Analysis of expression patterns of several SA- or JA-responsive genes following the treatments of SA and JA further indicated that both SA- and JA-responsive pathways are affected by the sib1 mutations.
Chloroplasts play an important role in the defence response (see reviews by Karpinski et al. 2003; Roberts & Paul 2006). Changes in abundance of genes and proteins involved in photosynthesis and other chloroplast functions following pathogen infection have been well documented (Bunker et al. 1995; Lehto et al. 2003; Zou et al. 2005; Jones et al. 2006; Thilmony, Underwood & He 2006). Phytopathogens have evolved various mechanisms to manipulate chloroplast function. Ptr ToxA, a host-selective proteinaceous toxin from the wheat pathogen Pyrenophora tritici-repentis, targets a chloroplast protein with an undefined function (Manning, Hardison & Ciuffetti 2007). Several virulence effector proteins have been determined to target chloroplast proteins, including HopI1 (Jelenska et al. 2007), HopU1 (Fu et al. 2007), the TMV p50 helicase (Abbink et al. 2002) and NRIP1 (Caplan et al. 2008). In tobacco, NRIP1 and another chloroplast protein, an FtsH-like metalloprotease, are also required for the N-mediated HR (Seo et al. 2000; Caplan et al. 2008). The fact that pathogens target chloroplast proteins with different activities indicates that the virulence strategies used by pathogens to manipulate chloroplast are diverse.
We have not found obvious alteration in the transcript levels of psaA or psaB by the sib1mutation. psaA And psaB are two of the chloroplast genes regulated by SIG1 in rice; however, it is not clear whether these chloroplast genes are also regulated by SIG1 in Arabidopsis. We could not rule out the possibility that SIB1 might function in the defence response through a mechanism other than its binding to and regulating SIG1. Determination of the SIB1’s protein levels, subcellular localization and its interaction with SIG1 during the defence response will help to define the precise mechanism by which SIB1 regulates the defence pathway. Nevertheless, our study indicates that SIB1 is involved in both the SA- and JA-mediated defence responses, and might act as a link between chloroplast function and defence signalling.
Supplementary Material
ACKNOWLEDGMENTS
We thank Barbara Kunkel for all Pst strains including Pst hrcC, and Xinnian Dong for the npr1 mutant seeds. We thank Anita Snyder for editing the manuscript. This work was supported by the National Institutes of Health (grant no. GM076420 to Y.X.), the International Collaboration Program of China (grant no. 2007FA314600 to M.P.), the National Natural Science Foundation of China (grant nos. 30870220 and 30721062 to D.R.), the Institute of Tropical Bioscience and Biotechnology, CATAS (institutional special research funds grant no. ITBBZX0811 to W.L.) and the Hong Kong UGC AoE Plant Agricultural Biotechnology Project (grant no. AoE B-07/09 to D.G.).
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors.Any queries (other than missing material) should be directed to the corresponding author for the article.
REFERENCES
- Abbink TEM, Peart JR, Mos TNM, Baulcombe DC, Bol JF, Linthorst HJM. Silencing of a gene encoding a protein component of the oxygen-evolving complex of photosystem II enhances virus replication in plants. Virology. 2002;295:307–319. doi: 10.1006/viro.2002.1332. [DOI] [PubMed] [Google Scholar]
- Abramovitch RB, Martin GB. Strategies used by bacterial pathogens to suppress plant defenses. Current Opinion in Plant Biology. 2004;7:356–364. doi: 10.1016/j.pbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Andreasson E, Jenkins T, Brodersen P, et al. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO Journal. 2005;24:2579–2589. doi: 10.1038/sj.emboj.7600737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nature Immunology. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- Boch J, Joardar V, Gao L, Robertson TL, Lim M, Kunkel BN. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Molecular Microbiology. 2002;44:73–88. doi: 10.1046/j.1365-2958.2002.02877.x. [DOI] [PubMed] [Google Scholar]
- Boter M, Ruiz-Rivero O, Abdeen A, Prat S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes & Development. 2004;18:1577–1591. doi: 10.1101/gad.297704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker TW, Koetje DS, Stephenson LC, Creelman RA, Mullet JE, Grimes HD. Sink limitation induces the expression of multiple soybean vegetative lipoxygenase mRNAs while the endogenous jasmonic acid level remains low. The Plant Cell. 1995;7:1319–1331. doi: 10.1105/tpc.7.8.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. The Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell. 2008;132:449–462. doi: 10.1016/j.cell.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Cui X, Hwang JT, Qiu J, Blades NJ, Churchill GA. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6:59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- da Cunha L, Sreerekha M-V, Mackey D. Defense suppression by virulence effectors of bacterial phytopathogens. Current Opinion in Plant Biology. 2007;10:349–357. doi: 10.1016/j.pbi.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, et al. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Favory JJ, Kobayshi M, Tanaka K, Peltier G, Kreis M, Valay JG, Lerbs-Mache S. Specific function of a plastid sigma factor for ndhF gene transcription. Nucleic Acids Research. 2005;33:5991–5999. doi: 10.1093/nar/gki908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Guo M, Jeong B-R, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–288. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- Ge X, Li G-J, Wang S-B, Zhu H, Zhu T, Wang X, Xia Y. AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiology. 2007;145:204–215. doi: 10.1104/pp.107.103374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud T, Buchala AJ, Chua N-H, Métraux J-P. Phytochrome signalling modulates the SA-perceptive pathway in Arabidopsis. The Plant Journal. 2002;31:87–95. doi: 10.1046/j.1365-313x.2002.01338.x. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Vinatzer BA. Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Current Opinion in Microbiology. 2003;6:20–28. doi: 10.1016/s1369-5274(02)00004-8. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T. A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. The Plant Journal. 2005;42:133–144. doi: 10.1111/j.1365-313X.2005.02362.x. [DOI] [PubMed] [Google Scholar]
- Jelenska J, Yao N, Vinatzer BA, Wright CM, Brodsky JL, Greenberg JT. A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Current Biology. 2007;17:499–508. doi: 10.1016/j.cub.2007.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jones AME, Thomas V, Bennett MH, Mansfield J, Grant M. Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiology. 2006;142:1603–1620. doi: 10.1104/pp.106.086231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru K, Tanaka K. Roles of chloroplast RNA polymerase sigma factors in chloroplast development and stress response in higher plants. Bioscience, Biotechnology, and Biochemistry. 2004;68:2215–2223. doi: 10.1271/bbb.68.2215. [DOI] [PubMed] [Google Scholar]
- Kanamaru K, Nagashima A, Fujiwara M, Shimada H, Shirano Y, Nakabayashi K, Shibata D, Tanaka K, Takahashi H. An Arabidopsis sigma factor (SIG2)-dependent expression of plastid-encoded tRNAs in chloroplasts. Plant & Cell Physiology. 2001;42:1034–1043. doi: 10.1093/pcp/pce155. [DOI] [PubMed] [Google Scholar]
- Karpinski S, Gabrys H, Mateo A, Karpinska B, Mullineaux PM. Light perception in plant disease defence signalling. Current Opinion in Plant Biology. 2003;6:390–396. doi: 10.1016/s1369-5266(03)00061-x. [DOI] [PubMed] [Google Scholar]
- Lehto K, Tikkanen M, Hiriart J-B, Paakkarinen V, Aro E-M. Depletion of the photosystem II core complex in mature tobacco leaves infected by the flavum strain of tobacco mosaic virus. Molecular Plant-Microbe Interactions. 2003;16:1135–1144. doi: 10.1094/MPMI.2003.16.12.1135. [DOI] [PubMed] [Google Scholar]
- Leister D. Chloroplast research in the genomic age. Trends in Genetics. 2003;19:47–56. doi: 10.1016/s0168-9525(02)00003-3. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ren D, Pike S, Pallardy S, Gassmann W, Zhang S. Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. The Plant Journal. 2007;51:941–954. doi: 10.1111/j.1365-313X.2007.03191.x. [DOI] [PubMed] [Google Scholar]
- Loschelder H, Schweer J, Link B, Link G. Dual temporal role of plastid sigma factor 6 in Arabidopsis development. Plant Physiology. 2006;142:642–650. doi: 10.1104/pp.106.085878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko EA. Plant sigma factors and their role in plastid transcription. Plant Cell Reports. 2007;26:845–859. doi: 10.1007/s00299-007-0318-7. [DOI] [PubMed] [Google Scholar]
- Manning VA, Hardison LK, Ciuffetti LM. Ptr ToxA interacts with a chloroplast-localized protein. Molecular Plant-Microbe Interactions. 2007;20:168–177. doi: 10.1094/MPMI-20-2-0168. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Research. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa K, Shiina T, Murakami S, Toyoshima Y. Novel nuclear-encoded proteins interacting with a plastid sigma factor, Sig1, in Arabidopsis thaliana. FEBS Letters. 2002;514:300–304. doi: 10.1016/s0014-5793(02)02388-8. [DOI] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- Nagashima A, Hanaoka M, Motohashi R, Seki M, Shinozaki K, Kanamaru K, Takahashi H, Tanaka K. DNA microarray analysis of plastid gene expression in an Arabidopsis mutant deficient in a plastid transcription factor sigma, SIG2. Bioscience, Biotechnology, and Biochemistry. 2004a;68:694–704. doi: 10.1271/bbb.68.694. [DOI] [PubMed] [Google Scholar]
- Nagashima A, Hanaoka M, Shikanai T, Fujiwara M, Kanamaru K, Takahashi H, Tanaka K. The multiple-stress responsive plastid sigma factor, SIG5, directs activation of the psbD blue light-responsive promoter (BLRP) in Arabidopsis thaliana. Plant & Cell Physiology. 2004b;45:357–368. doi: 10.1093/pcp/pch050. [DOI] [PubMed] [Google Scholar]
- Narusaka M, Kawai K, Izawa N, Seki M, Shinozaki K, Seo S, Kobayashi M, Shiraishi T, Narusaka Y. Gene coding for SigA-binding protein from Arabidopsis appears to be transcriptionally up-regulated by salicylic acid and NPR1-dependent mechanisms. Journal of General Plant Pathology. 2008;74:345–354. [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Metraux J-P. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. The Plant Cell. 2002;14:275–286. doi: 10.1105/tpc.010376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perruc E, Charpenteau M, Ramirez BC, Jauneau A, Galaud J-P, Ranjeva R, Ranty B. A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. The Plant Journal. 2004;38:410–420. doi: 10.1111/j.1365-313X.2004.02062.x. [DOI] [PubMed] [Google Scholar]
- Privat I, Hakimi MA, Buhot L, Favory JJ, Mache-Lerbs S. Characterization of Arabidopsis plastid sigma-like transcription factors SIG1, SIG2 and SIG3. Plant Molecular Biology. 2003;51:385–399. doi: 10.1023/a:1022095017355. [DOI] [PubMed] [Google Scholar]
- Roberts MR, Paul ND. Seduced by the dark side: integrating molecular and ecological perspectives on the influence of light on plant defence against pests and pathogens. New Phytologist. 2006;170:677–699. doi: 10.1111/j.1469-8137.2006.01707.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. 2nd edn Cold Spring Harbor Laboratory Press; New York, NY, USA: 1989. [Google Scholar]
- Seo S, Okamoto M, Iwai T, Iwano M, Fukui K, Isogai A, Nakajima N, Ohashi Y. Reduced levels of chloroplast FtsH protein in tobacco mosaic virus-infected tobacco leaves accelerate the hypersensitive reaction. The Plant Cell. 2000;12:917–932. doi: 10.1105/tpc.12.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirano Y, Shimada H, Kanamaru K, et al. Chloroplast development in Arabidopsis thaliana requires the nuclearencoded transcription factor sigma B. FEBS Letters. 2000;485:178–182. doi: 10.1016/s0014-5793(00)02216-x. [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Dong X. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321:952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Tozawa Y, Mochizuki N, Shinozaki K, Nagatani A, Wakasa K, Takahashi H. Characterization of three cDNA species encoding plastid RNA polymerase sigma factors in Arabidopsis thaliana: evidence for the sigma factor heterogeneity in higher plant plastids. FEBS Letters. 1997;413:309–313. doi: 10.1016/s0014-5793(97)00906-x. [DOI] [PubMed] [Google Scholar]
- Thilmony R, Underwood W, He SY. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. The Plant Journal. 2006;46:34–53. doi: 10.1111/j.1365-313X.2006.02725.x. [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. see comment. [DOI] [PubMed] [Google Scholar]
- Tozawa Y, Teraishi M, Sasaki T, Sonoike K, Nishiyama Y, Itaya M, Miyao A, Hirochika H. The plastid sigma factor SIG1 maintains photosystem I activity via regulated expression of the psaA operon in rice chloroplasts. The Plant Journal. 2007;52:124–132. doi: 10.1111/j.1365-313X.2007.03216.x. [DOI] [PubMed] [Google Scholar]
- Tsunoyama Y, Morikawa K, Shiina T, Toyoshima Y. Blue light specific and differential expression of a plastid sigma factor, Sig5 in Arabidopsis thaliana. FEBS Letters. 2002;516:225–228. doi: 10.1016/s0014-5793(02)02538-3. [DOI] [PubMed] [Google Scholar]
- Tsunoyama Y, Ishizaki Y, Morikawa K, Kobori M, Nakahira Y, Takeba G, Toyoshima Y, Shiina T. Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3304–3309. doi: 10.1073/pnas.0308362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees SCM, Glazebrook J. Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. The Plant Journal. 2003;33:733–742. doi: 10.1046/j.1365-313x.2003.01665.x. [DOI] [PubMed] [Google Scholar]
- Wu H, Kerr MK, Cui X, Churchill GA. MAANOVA, a software package for the analysis of spotted cDNA microarray experiments. In: Parmigiani G, Garret ES, Irizarry RA, Zeger SL, editors. The Analysis of Gene Expression Data:An Overview of Methods and Software. Springer; NewYork,NY, USA: 2003. pp. 313–341. [Google Scholar]
- Xie D-X, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Yang H, Churchill G. Estimating P-values in small microarray experiments. Bioinformatics. 2007;23:38–43. doi: 10.1093/bioinformatics/btl548. [DOI] [PubMed] [Google Scholar]
- Zeier J, Pink B, Mueller MJ, Berger S. Light conditions influence specific defence responses in incompatible plantpathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta. 2004;219:673–683. doi: 10.1007/s00425-004-1272-z. [DOI] [PubMed] [Google Scholar]
- Zghidi W, Merendino L, Cottet A, Mache R, Lerbs-Mache S. Nucleus-encoded plastid sigma factor SIG3 transcribes specifically the psbN gene in plastids. Nucleic Acids Research. 2007;35:455–464. doi: 10.1093/nar/gkl1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Rodriguez-Zas S, Aldea M, Li M, Zhu J, Gonzalez DO, Vodkin LO, DeLucia E, Clough SJ. Expression profiling soybean response to Pseudomonas syringae reveals new defense-related genes and rapid HR-specific downregulation of photosynthesis. Molecular Plant-Microbe Interactions. 2005;18:1161–1174. doi: 10.1094/MPMI-18-1161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.