Abstract
Holocarboxylase synthetase (HCS) plays an essential role in catalyzing the biotinylation of carboxylases and histones. Biotinylated carboxylases are important for the metabolism of glucose, lipids, and leucine; biotinylation of histones plays important roles in gene regulation and genome stability. Recently, we reported that HCS activity is partly regulated by subcellular translocation events and by miR-539. Here we tested the hypothesis that the HCS 3’-UTR contains binding sites for miR other than miR-539. A binding site for miR-153 was predicted to reside in the HCS 3’-UTR by using in silico analyses. When miR-153 site was overexpressed in transgenic HEK-293 human embryonic kidney cells, the abundance of HCS mRNA decreased by 77% compared with controls. In silico analyses also predicted three putative cytosine methylation sites in two miR-153 genes; the existence of these sites was confirmed by methylation-sensitive PCR. When cytosines were demethylated by treatment with 5-aza-2’-deoxycytidine, the abundance of miR-153 increased by more than 25 times compared with untreated controls, and this increase coincided with low levels of HCS and histone biotinylation. Together, this study provides novel insights into the mechanisms of novel epigenetic synergies among folate-dependent methylation events, miR, and histone biotinylation.
Keywords: cytosine methylation, holocarboxylase synthetase, human kidney cells, miR-153
1 INTRODUCTION
Holocarboxylase synthetase (HCS) plays a pivotal role in biotin-dependent pathways. In humans, HCS catalyzes the covalent binding of biotin to two families of proteins, i.e., five distinct carboxylases [1-3] and histones H1, H3, H4 and, to a lesser extent, H2A [4-9]. Biotinylated carboxylases are key enzymes in essential metabolic pathways in the metabolism of glucose, fatty acids, and leucine [10]. Biotinylated histones play roles in the transcriptional repression of genes and repeats [11, 12]. Importantly, evidence suggests that K12-biotinylated histone H4 (H4K12bio) contributes towards transcriptional repression of retrotransposons, and that low abundance of H4K12bio in HCS- or biotin-deficient cells is linked with activation of retrotransposons and chromosomal abnormalities [13].
Consistent with the important roles of HCS in intermediary metabolism and gene regulation, no living HCS null individual has ever been reported, suggesting embryonic lethality. HCS knockdown studies (~30% residual activity) produced phenotypes such as decreased life span and heat resistance in Drosophila melanogaster [14]. Numerous mutations in the human HCS gene have been identified and characterized at both the enzymatic and clinical level; these mutations cause a substantial decrease in HCS activity [15, 16]. Unless diagnosed and treated early, HCS deficiency appears to be uniformly fatal [17].
HCS can be detected in both nuclear and extranuclear compartments [7, 18]; nuclear HCS is a chromatin protein [14] and its binding to chromatin might be mediated by physical interactions with histones H3 and H4 [9]. Knowledge about regulation of HCS is limited to the following observations: (i) Both the expression and nuclear translocation of HCS depend on biotin [19]. (ii) The human HCS promoter has been tentatively identified [20], but has not been characterized in great detail. (iii) The expression of HCS is repressed by miR-539 [21]. (iv) Preliminary evidence has been provided for epigenetic synergies between cytosine methylation and HCS-dependent histone biotinylation. Specifically, we reported that histone biotinylation is substantially impaired when cytosine methylation marks are erased by treating cells with 5-aza-2’-deoxycytidine (AZA) [13]. In these previous studies we proposed that biotinylation of histones depends on prior methylation of cytosines (“cross-talk theory”).
Here we investigated an alternative pathway, by which cytosine methylation might alter histone biotinylation. We tested the hypothesis that low levels of cytosine methylation in the promoters of two miR-153 genes enhance mir-153 transcript abundance compared with normal methylation levels, and that mir-153 binds to a target sequence in the 3’-UTR of HCS mRNA, thereby decreasing HCS expression (“promoter methylation theory”). If proven true, mir-153 would be the second miR known to participate in the regulation of HCS expression. Note that the cross-talk and promoter methylation theories to explain epigenetic synergies between methylation and biotinylation events are not mutually exclusive.
2 MATERIALS AND METHODS
2.1 Cell culture
HEK293 human embryonic kidney cells were obtained from ATCC (Mansassas, VA) and were cultured in Dulbecco's Modified Eagle's Medium following ATCC's recommendations.
2.2 Identification of miR targets in the HCS 3’-UTR
The 3’-UTR of human HCS was screened in silico for miR target sequences by using TargetScan [22] and microRNA.org [23]. Both softwares predicted a miR-153 target site 2918-2924 basepairs (bp) downstream from the HCS stop codon.
2.3 Plasmids Ecd-miR153-CMV-Luc-HCS3UTR and CMV-miR153-CMV-GFP
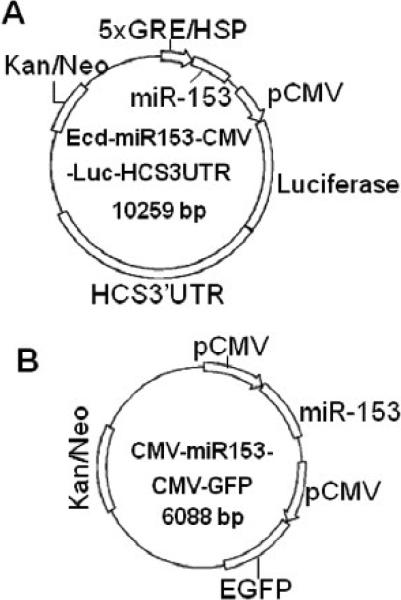
Plasmids were generated by using protocols analogous to those described in our previous publication [21]. Briefly, miR-153-2 and its flanking sequence (587bp) was ligated into the multiple cloning site (MCS) 1 in plasmid Ecd-MCS1-CMV-Luc-HCS3UTR [21] to produce plasmid Ecd-miR153-CMV-Luc-HCS3UTR (Fig. 1). In this plasmid, ecdysone (Ecd)-inducible response elements in the heat shock protein (HSP) promoter regulate the expression of human miR-153 (see below). This plasmid also contains a sequence coding for the luciferase reporter gene fused to the 3’UTR from human HCS; the luciferase/3’-UTR element is located downstream of a constitutively active CMV promoter. Expression of miR-153 in cells transfected with Ecd-mir153-CMV-Luc-HCS3UTR depends on the dimerization of retinoid X receptor (RXR) with Ecd receptor, and binding of the synthetic Ecd receptor ligand PonA [24]. HEK-293 cells were stably transformed with plasmid pVgRXR to produce RXR and Ecd receptor [21]. Activation of Ecd elements with 5 μmol/L ponasterone A (PonA) for 48 h causes transcription of miR-153, which then alters luciferase expression, depending on binding of miR-153 to the HCS 3’UTR [21].
FIGURE 1.
Vector maps. Panel A = Ecd-miR153-CMV-Luc-HCS3UTR; panel B = CMV-miR153-CMV-GFP.
Briefly, the miR-153 overexpression plasmid CMV-miR153-CMV-GFP was generated as follows. The miR-153-2 hairpin precursor and its native flanking sequence (587 bp) were PCR amplified using Jurkat cell genomic DNA as template, and primers 5’- ATGTCGACCTCTGGCCTGGCGCGTTC -3’ (forward) and 5’- AAGGGCCCGTGACACAGCCGTCTCATTTGAC-3’ (reverse). The PCR product was inserted into MCS1 of plasmid CMV-MCS1-CMV-GFP [21] by using SalI and ApaI to produce plasmid CMV-miR153-CMV-GFP (Fig. 1). Expression of green fluorescent protein (GFP) was used to confirm successful transfection (not shown).
The above plasmids were sequenced to confirm identity; all plasmids mediate resistance to G418, permitting selection for stably transformed clones.
2.4 Luciferase assay
Luciferase activity was assayed in triplicate, using independent samples; empty vectors and β-galactosidase vectors were used as baseline and transfection controls, respectively [25].
2.5 Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated by using RNA Spin Mini isolation kits (GE Health). The abundance of miR-153 was quantified by stem-loop qRT-PCR [26]. Briefly, primer 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGATCAC -3’ was used for reverse transcription of miR-153 by using the ImProm-II Reverse Transcription System (Promega). miR-153 was quantified by qRT-PCR, using forward primer 5’-CGGCGGTTGCATAGTCACAA-3’ and reverse primer 5’- GTGCAGGGTCCGAGGT-3’. The relative expression of miR-153 was normalized using U6 snRNA as control; U6 snRNA was reverse transcribed using 5’-GTCAGGCAGCGTGCAGGGTCCGAGGTATTCGCACGCTGCCTGACAAAAAT-3’, and PCR amplified using forward primer 5’-CGCAAGGATGACACGCAAATT-3’and reverse primer 5’-GTGCAGGGTCCGAGGT-3’. All reverse transcriptions were conducted using 2 μg of total RNA. The qRT-PCR thermocycler was set at 95°C for 15 s and 55°C for 60 s per cycle for a total 40 cycles. Relative changes in mRNA abundances were quantified by using the Ct method; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA and U6 RNA were used as reference amplicons for data normalization [27, 28]. Luciferase and HCS transcripts were quantified by qRT-PCR as described [21].
2.6 Western blots
The abundance of luciferase, HCS, and GAPDH (loading control) was quantified as described before [21]. Biotinylation of histone H4 was probed using anti-biotin as described [21]; equal loading of histone extracts was confirmed using anti-human histone H3. Proteins were visualized using fluorophore-labeled secondary antibodies and an Odyssey Infrared Imaging system (LICOR).
2.7 Methylation analyses of miR-153 promoters
The upstream regions of the miR153-1 and miR153-2 genes (-5,000 to +1 bp from mature miR-153) were analyzed with CpG Island Searcher to identify putative CpG islands [29, 30]. The in silico predictions were experimentally confirmed by methylation-sensitive PCR [31]. PCR primers were designed to minimize GC content while maintaining >2 HpaII sites and ≥1 MrcBC site by using the software Primo [32]. Five-hundred nanograms of genomic DNA from HEK-293 cells were digested consecutively with 30 units of HpaII and McrBC overnight in 50 μL of buffers recommended by the manufacturer (New England Biolabs). Thirty nanograms of DNA were PCR-amplified in a 10-μL reaction volume, using the following primers: 5’-AGGTGAACCTGGTGTCGGAG-3’ (forward) and 5’- AGAGGGAAGAAGGGAGAGGG-3’ (reverse) for a 931-bp sequence spanning the predicted CpG island in the miR-153-1 gene; 5’-TGCTAGCCACCAGCAAGTGAG-3’ (forward) and 5’-ACCACGCTGTGAGGCGAATG-3’ (reverse) for a 852-bp sequence spanning the predicted CpG island 1 in the miR-153-2 gene; and 5’- GTTCACATCAGCGCGACTCC-3’ (forward) and 5’- ACCACCTAGCCAATGCGGAC-3’ (reverse) for a 1331-bp sequence spanning the predicted CpG island 2 in the miR-153-2 gene. PCR products were visualized by staining a 1% agarose gel with ethidium bromide.
Methylation-sensitive PCR was conducted using DNA from both HEK-293 cells treated with AZA to remove methylation marks and control cells not treated with AZA. Briefly, ~106 HEK-293 cells were treated with 0.25 μmol/L of AZA in 12-well plates as described [33, 34]. Briefly, after 24 h of treatment with AZA, the medium was replaced with AZA-free medium and culturing was continued for 24 h. Cells were treated with AZA again for another 24 h, before being allowed to recover in AZA-free medium for another 24 h. Finally, cells were cultured in AZA medium for another 72 h before being collected for transcript analysis and for analysis of cytosine methylation. Previous studies suggest that this protocol is effective in erasing methylation marks while not causing cell death to a meaningful extent [13]. Viability was monitored and confirmed at timed intervals by using the trypan blue exclusion test.
2.8 Statistics
Variances among groups were homogenous, as judged by Bartlett's test [35]. The statistical significance of differences between groups was analyzed by unpaired t-test. StatView 5.0.1 (SAS Institute, Cary, NC) was used for statistical analyses. Differences were considered significant if P < 0.05. Data are reported as mean ± S.D.
3 RESULTS
3.1 Regulation of HCS expression by miR-539
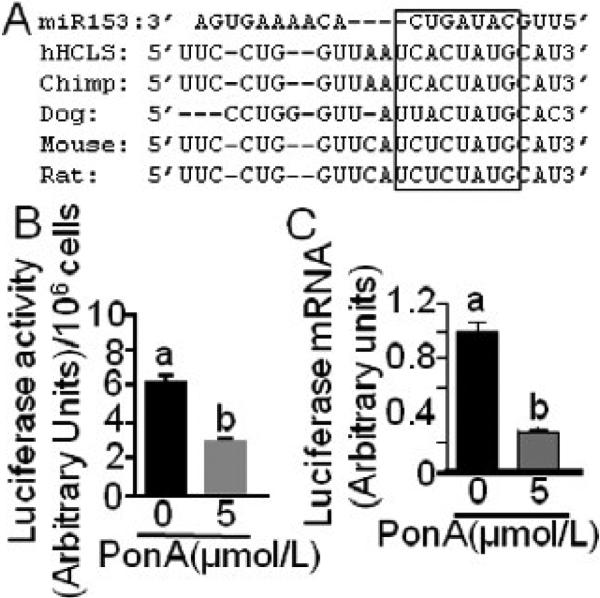
In silico analysis by miR search engines predicted a miR-153 binding site (5’-TCACTATG-3’) 2918-2924 bp downstream from the stop codon in human HCS mRNA. The miR-153 target site in human HCS is evolutionary conserved in mammals, particularly the CUAUGC sequence (Fig. 2A). The in silico prediction was confirmed experimentally in pVgRXR-transformed HEK-293 cells that were also transfected with Ecd-mir153-CMV-Luc-HCS3UTR. When cells were treated with 5 μmol/L PonA for 48 h, the activity of luciferase was 55 ± 1.2% lower than in untreated controls (Fig. 2B); times and concentrations of PonA treatment were optimized in previous studies [21]. Likewise, when cells were treated with PonA the abundance of luciferase mRNA decreased by 73 ± 1.1% compared with untreated controls (Fig. 2C).
FIGURE 2.
The 3’UTR of HCS mRNA contains a miR-153 binding site. (A) Sequence alignment of miR-153 (top row) and its putative binding site in the 3’UTR of HCS mRNA in humans and other species (rows 2-6). The box denotes the most highly conserved region. Luciferase activity (B) and mRNA abundance (C) was quantified in HEK-293 cells that were stably transformed with Ecd-mir153-CMV-Luc-HCS3UTR and treated with PonA for 48 h; controls were cultured in the absence of PonA. Values are means ± SD, n= 3. Means without a common letter differ, P < 0.05.
3.2 Effects of miR-153 on HCS expression and activity
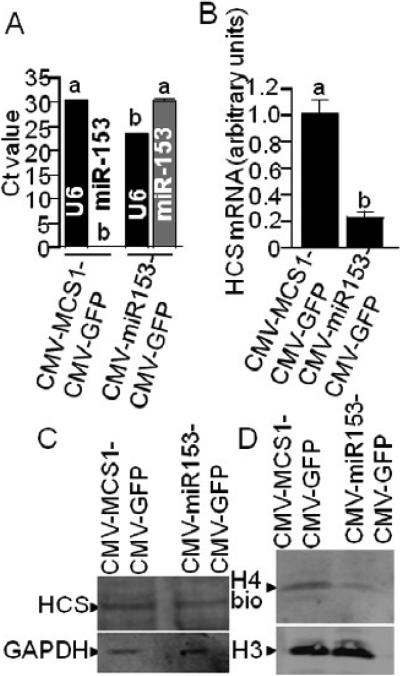
miR-153 impairs the expression of HCS to an extent that affects histone biotinylation. When HEK-293 cells were stably transformed with CMV-miR153-CMV-GFP, the abundance of mature miR-153 was significantly greater compared with cells transformed by using the CMV-MCS1-CMV-GFP empty control vector (Fig. 3A). Note that no endogenous, mature miR-153 was detectable in CMV-MCS1-CMV-GFP, unless the promoters of endogenous miR-153 genes were activated by treatment with AZA (see below). The abundance of U6 snRNA (control) was moderately lower in cells transfected with CMV-miR153-CMV-GFP compared with CMV-MCS1-CMV-GFP cells (Fig. 3A).
FIGURE 3.
Overexpression of miR-153 causes low HCS expression and low abundance of biotinylated histones in HEK-293 cells. HEK-293 cells were stably transformed with CMV-miR539-CMVGFP or empty vector (CMV-MCS1-CMV-GFP), and the abundance of U6 and miR-153 (A), HCS mRNA (panel B), HCS protein (C, upper gel), and biotinylated histone H4 (H4bio) (D, upper gel) was quantified. Equal loading was confirmed by using anti-GAPDH (C, lower gel) and anti-H3 (D, lower gel). Note that statistical analysis of panel A data was conducted by comparing U6 vs. U6 and miR-153 vs. miR-153 in cells transfected with the two different plasmids. Values are means ± SD, n=3. Means without a common letter differ, P < 0.05.
High levels of miR-153 coincided with low expression of HCS in HEK-293 cells. First, the abundance of endogenous HCS mRNA was 77 ± 4.7% lower in CMV-miR153-CMVGFP-transformed cells than in CMV-MCS1-CMV-GFP controls when normalized for qRTPCR efficiency by using GAPDH (Fig. 3B). Second, the abundance of HCS protein was lower in CMV-miR153-CMV-GFP-transformed cells than in controls (Fig. 3C), particularly considering that the lane with extracts from CMV-miR153-CMV-GFP-transformed cells was moderately overloaded compared with CMV-MCS1-CMV-GFP cells (compare GAPDH control). Assays were repeated five times by using biological replicates, and a representative example is shown. Third, the levels of biotinylated histone H4 were lower in CMV-miR153-CMV-GFP-transformed cells than in controls (Fig. 3D); equal loading was confirmed using histone H3 as a marker.
3.3 Activation of endogenous miR-153 genes by cytosine demethylation
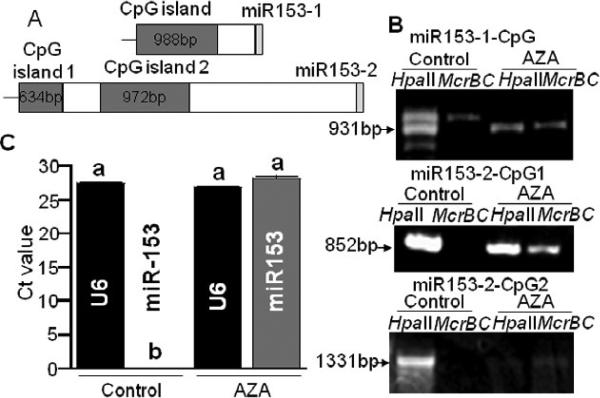
Mature miR-153 was not detectable in non-transformed HEK-293 cells (Fig. 3A), Jurkat human lymphoblast cells, Caco-2 human colon carcinoma cells, and IMR-90 primary human lung fibroblasts (data not shown). We hypothesized that the silencing of the miR-153-1 and miR153-2 genes might be mediated by cytosine methylation in the promoters of these genes. In silico analysis of the upstream regions of the genes predicted the following three CpG islands (Fig. 4A): one CpG island with a length of 988 bp is located 434 bp upstream of mature mir-153 in the miR-153-1 gene; a second CpG island with a length of 634 bp is located 4028 bp upstream of mature mir-153 in the miR-153-2 gene; and a third CpG island with a length of 972 bp is located 2721 bp upstream of mature mir-153 in the miR-153-2 gene.
FIGURE 4.
Cytosine methylation in CpG islands in the upstream regions of miR-153-1 and miR-153-2 genes. (A) CpG islands and mature miR-153 in miR-153-1 and miR-153-2 genes. (B) Analysis of cytosine methylation in the CpG islands in miR-153-1 and miR-153-2 in AZA-treated and AZA-free (control) HEK-293 cells. (C) Abundance of miR-153 abundance; U6 snRNA = internal standard in stem-loop qRT-PCR of miR-153. Note that statistical analysis of panel C data was conducted by comparing U6 vs. U6 and miR-153 vs. miR-153 in AZA-treated cells and untreated controls. Values are means ± SD, n=3. Means without a common letter differ, P < 0.05.
We tested whether miR-153 genes could be de-repressed by erasing cytosine methylation marks in CpG islands by treating wild-type HEK-293 cells with AZA. Note that the endonuclease HpaII cuts only unmethylated CCGG sites, whereas McrBC digests methylated sequences with the RmCN40-80RmC motif, where R = purine, mC = methylated cytosine, and N = nucleotide [36, 37]. When genomic DNA from untreated cells was digested with HpaII or McrBC and amplified with locus-specific PCR primers, the HpaII-digested samples produced a strong signal if probed with ethidium bromide (Fig. 4B); McrBC-digested samples produced a weak signal for the CpG island in the miR-153-1 gene and no signal for the CpG islands in the miR-153-2 gene. In contrast, when cytosine methylation marks were erased by AZA treatment before analysis by methylation-sensitive PCR, strong signals were obtained with both HpaII and McrBC digests for the CpG island in miR-153-1 and for island 1 in miR153-2; the signal for island 2 in miR153-2 was faint but detectable. Thus, AZA treatment caused a partial or complete demethylation of CpG islands upstreams of miR-153 genes in HEK-293 cells. Consistent with our theory, endogenous miR-153 became easily detectable upon treatment of HEK-293 cells with AZA (Fig. 4C).
4 DISCUSSION
To the best of our knowledge, this is the first report to demonstrate that miR-153 genes are repressed by cytosine methylation in HEK-293 cells. This is also the first report of a miR-153 binding site in the HCS 3’-UTR in humans and other species. These findings are important for the following reasons. First, repression of HCS by miR-153 is a novel mechanism of HCS regulation with many possible downstream effects in macronutrient metabolism, gene regulation, and genome stability [10-13, 19]. Second, methylation events depend on folate and other methyl donors. Therefore, this report generates possible mechanistic insights in epigenetic synergies between folate and biotin in mediating gene repression. Both cytosine methylation and histone biotinylation are known to play roles in gene repression [12, 13, 19, 38]; cytosine methylation marks are known to co-localize with H4K12bio in retrotransposons, mediating their repression and maintaining genome stability [13]. Third, the extent of HCS repression by miR-153 is sufficiently strong to have downstream effects on protein biotinylation such as biotinylation of histone H4.
There is precedence for the regulation of HCS by miR. In a recent report, we demonstrated that HCS is also repressed by miR-539 and that miR-539 can be detected in HEK-293 cells, Jurkat cells, Caco-2 cells and IMR-90 fibroblasts without AZA treatment [21]; the expression of miR-539 in some of these cells depends on biotin. Interestingly, the recently identified HCS promoter also appears to depend on epigenetic mechanisms for activation, i.e., trichostatin A-mediated inhibition of histone deacetylases [20]. We currently do not know the contributions of the various regulatory pathways to the overall regulation of HCS, but speculate that many of these pathways are tissue specific. For example, miR-153 is a brain-specific miR [39].
The existence of biotinylated histones was recently questioned [40], thereby indirectly questioning the possibility of epigenetic cross-talk between biotin and folate. Please note that three independent laboratories [41-43], in addition to ours [4-6, 8, 9, 12], confirmed that biotinylation is a natural histone modification. Histone biotinylation is a comparably rare event (<0.1% of histones are biotinylated) [4, 41], but the abundance of an epigenetic mark is no indicator for its importance. For example, only ~3% of cytosines are methylated, but the role of DNA methylation in gene regulation is undisputed [38]. Likewise, serine-14 phosphorylation in histone H2B and histone poly(ADP-ribosylation) are detectable only after induction of apoptosis and major DNA damage, respectively, but the role of these epigenetic marks in cell death is unambiguous [44-46]. Moreover, low abundance of an epigenetic mark in bulk histone extracts compared with its high enrichment at specific loci is consistent with regulatory function. Evidence suggests that about one out of three histone H4 molecules are biotinylated at K12 in telomeric repeats [47].
Taken together, we report the elucidation of a novel pathway of HCS regulation that ties cytosine methylation to histone biotinylation. It remains to be determined whether this pathway plays a particularly important role in the human brain where miR-153 is expressed in the absence of AZA.
Acknowledgments
Supported in part by funds provided through the Hatch Act. Additional support was provided by NIH grants DK063945, DK077816, DK082476 and ES015206, USDA CSREES grant 2006-35200-17138, NSF grant EPS 0701892, and by Shanghai Municipal Education Commission grants S30701 and 11ZZ146.
Abbreviations
- AZA
5-aza-2’-deoxycytidine
- bp
basepair
- Ecd
ecdysone
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- H4K12bio
K12-biotinylated histone H4
- HCS
holocarboxylase synthetase
- HSP
heat shock protein
- meC
methylated cytosine
- MCS
multiple cloning site
- miR
microRNA
- N
nucleotide
- P
purine
- PCR
polymerase chain reaction
- PonA
ponasterone A
- qRT-PCR
quantitative real-time PCR
- RXR
retinoid X receptor
- 3’-UTR
3’-untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Suzuki Y, Aoki Y, Ishida Y, Chiba Y, Iwamatsu A, Kishino T, et al. Isolation and characterization of mutations in the human holocarboxylase synthetase cDNA. Nat Genet. 1994;8:122–28. doi: 10.1038/ng1094-122. [DOI] [PubMed] [Google Scholar]
- 2.Campeau E, Gravel RA. Expression in Escherichia coli of N- and C-terminally deleted human holocarboxylase synthetase. Influence of the N-terminus on biotinylation and identification of a minimum functional protein. J Biol Chem. 2001;276:12310–16. doi: 10.1074/jbc.M009717200. [DOI] [PubMed] [Google Scholar]
- 3.Wolf B. Disorders of Biotin Metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York, NY: 2001. pp. 3935–62. [Google Scholar]
- 4.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur J Biochem. 2001;268:5424–29. doi: 10.1046/j.0014-2956.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 5.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur J Biochem. 2004;271:2257–63. doi: 10.1111/j.1432-1033.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 6.Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, et al. K4, K9, and K18 in human histone H3 are targets for biotinylation by biotinidase. FEBS J. 2005;272:4249–59. doi: 10.1111/j.1742-4658.2005.04839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chew YC, Camporeale G, Kothapalli N, Sarath G, Zempleni J. Lysine residues in N- and C-terminal regions of human histone H2A are targets for biotinylation by biotinidase. J Nutr Biochem. 2006;17:225–33. doi: 10.1016/j.jnutbio.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobza K, Sarath G, Zempleni J. Prokaryotic BirA ligase biotinylates K4, K9, K18 and K23 in histone H3. BMB Reports. 2008;41:310–15. doi: 10.5483/bmbrep.2008.41.4.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bao B, Pestinger V, I. HY, Borgstahl GEO, Kolar C, Zempleni J. Holocarboxylase synthetase is a chromatin protein and interacts directly with histone H3 to mediate biotinylation of K9 and K18. J Nutr Biochem. 2011 doi: 10.1016/j.jnutbio.2010.04.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zempleni J, Wijeratne SS, Hassan YI. Biotin. BioFactors (Oxford, England) 2009;35:36–46. doi: 10.1002/biof.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camporeale G, Oommen AM, Griffin JB, Sarath G, Zempleni J. K12-biotinylated histone H4 marks heterochromatin in human lymphoblastoma cells. J Nutr Biochem. 2007;18:760–68. doi: 10.1016/j.jnutbio.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pestinger V, Wijeratne SSK, Rodriguez-Melendez R, Zempleni J. Novel histone biotinylation marks are enriched in repeat regions and participate in repression of transcriptionally competent genes. J Nutr Biochem. 2011 doi: 10.1016/j.jnutbio.2010.02.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chew YC, West JT, Kratzer SJ, Ilvarsonn AM, Eissenberg JC, Dave BJ, et al. Biotinylation of histones represses transposable elements in human and mouse cells and cell lines, and in Drosophila melanogaster. J Nutr. 2008;138:2316–22. doi: 10.3945/jn.108.098673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camporeale G, Giordano E, Rendina R, Zempleni J, Eissenberg JC. Drosophila holocarboxylase synthetase is a chromosomal protein required for normal histone biotinylation, gene transcription patterns, lifespan and heat tolerance. J Nutr. 2006;136:2735–42. doi: 10.1093/jn/136.11.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki Y, Yang X, Aoki Y, Kure S, Matsubara Y. Mutations in the holocarboxylase synthetase gene HLCS. Human Mutation. 2005;26:285–90. doi: 10.1002/humu.20204. [DOI] [PubMed] [Google Scholar]
- 16.National Center for Biotechnology Information [7/18/2008];Online Mendelian Inheritance in Man, National, Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov/sites/entrez?db=omim.
- 17.Thuy LP, Belmont J, Nyhan WL. Prenatal diagnosis and treatment of holocarboxylase synthetase deficiency. Prenat Diagn. 1999;19:108–12. doi: 10.1002/(sici)1097-0223(199902)19:2<108::aid-pd476>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 18.Narang MA, Dumas R, Ayer LM, Gravel RA. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum Mol Genet. 2004;13:15–23. doi: 10.1093/hmg/ddh006. [DOI] [PubMed] [Google Scholar]
- 19.Gralla M, Camporeale G, Zempleni J. Holocarboxylase synthetase regulates expression of biotin transporters by chromatin remodeling events at the SMVT locus. J Nutr Biochem. 2008;19:400–08. doi: 10.1016/j.jnutbio.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warnatz HJ, Querfurth R, Guerasimova A, Cheng X, Haas SA, Hufton AL, et al. Functional analysis and identification of cis-regulatory elements of human chromosome 21 gene promoters. Nucleic Acids Res. 2010;38:6112–23. doi: 10.1093/nar/gkq402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bao B, Rodriguez-Melendez R, Wijeratne SS, Zempleni J. Biotin regulates the expression of holocarboxylase synthetase in the miR-539 pathway in HEK-293 cells. J Nutr. 2010;140:1546–51. doi: 10.3945/jn.110.126359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.TargetScan [5/8/2009]; http://www.targetscan.org/index.html.
- 23.microRNA.org [5/8/2009]; www.microrna.org/microrna/getGeneForm.do.
- 24.Rangasamy D, Tremethick DJ, Greaves IK. Gene knockdown by ecdysone-based inducible RNAi in stable mammalian cell lines. Nat Protoc. 2008;3:79–88. doi: 10.1038/nprot.2007.456. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Melendez R, Camporeale G, Griffin JB, Zempleni J. Interleukin-2 receptor g-dependent endocytosis depends on biotin in Jurkat cells. Am J Physiol Cell Physiol. 2003;284:C415–C21. doi: 10.1152/ajpcell.00365.2002. [DOI] [PubMed] [Google Scholar]
- 26.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–08. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–08. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 29.Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–40. [PubMed] [Google Scholar]
- 30.Takai D, Catherall S. [10/22/2010];CpG Island Searcher. 2004 http://cpgislands.usc.edu/
- 31.Yamada Y, Watanabe H, Miura F, Soejima H, Uchiyama M, Iwasaka T, et al. A comprehensive analysis of allelic methylation status of CpG islands on human chromosome 21q. Genome Res. 2004;14:247–66. doi: 10.1101/gr.1351604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang B. [10/22/2010];Primo Pro 3.4. 2010 http://www.changbioscience.com/primo/primo.html.
- 33.Wozniak RJ, Klimecki WT, Lau SS, Feinstein Y, Futscher BW. 5-Aza-2'-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 dimethylation levels are linked to tumor suppressor gene reactivation. Oncogene. 2007;26:77–90. doi: 10.1038/sj.onc.1209763. [DOI] [PubMed] [Google Scholar]
- 34.Liu ZJ, Zhang XB, Zhang Y, Yang X. Progesterone receptor gene inactivation and CpG island hypermethylation in human leukemia cancer cells. FEBS Lett. 2004;567:327–32. doi: 10.1016/j.febslet.2004.04.044. [DOI] [PubMed] [Google Scholar]
- 35.Abacus Concepts . StatView. Abacus Concepts Inc.; Berkeley, CA: 1996. [Google Scholar]
- 36.Sutherland E, Coe L, Raleigh EA. McrBC: a multisubunit GTP-dependent restriction endonuclease. J Mol Biol. 1992;225:327–48. doi: 10.1016/0022-2836(92)90925-a. [DOI] [PubMed] [Google Scholar]
- 37.Stewart FJ, Raleigh EA. Dependence of McrBC cleavage on distance between recognition elements. Biol Chem. 1998;379:611–16. [PubMed] [Google Scholar]
- 38.Li E, Bird A. DNA methylation in mammals. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 341–56. [Google Scholar]
- 39.Xu J, Liao X, Wong C. Downregulations of B-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int J Cancer. 2010;126:1029–35. doi: 10.1002/ijc.24823. [DOI] [PubMed] [Google Scholar]
- 40.Healy S, Perez-Cadahia B, Jia D, McDonald MK, Davie JR, Gravel RA. Biotin is not a natural histone modification. Biochim Biophys Acta. 2009;1789:719–33. doi: 10.1016/j.bbagrm.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 41.Bailey LM, Ivanov RA, Wallace JC, Polyak SW. Artifactual detection of biotin on histones by streptavidin. Anal Biochem. 2008;373:71–77. doi: 10.1016/j.ab.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Takechi R, Taniguchi A, Ebara S, Fukui T, Watanabe T. Biotin deficiency affects the proliferation of human embryonic palatal mesenchymal cells in culture. J Nutr. 2008;138:680–84. doi: 10.1093/jn/138.4.680. [DOI] [PubMed] [Google Scholar]
- 43.Ghosh S. School of Biological Sciences. Ph.D. University of Nebraska-Lincoln; Lincoln, NE: 2009. Physiology, regulation, and pathogenesis of nitrogen metabolism in opportunistic fungal pathogen Candida albicans. [Google Scholar]
- 44.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–17. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 45.Boulikas T. At least 60 ADP-ribosylated variant histones are present in nuclei from dimethylsulfate-treated and untreated cells. EMBO J. 1988;7:57–67. doi: 10.1002/j.1460-2075.1988.tb02783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boulikas T. DNA strand breaks alter histone ADP-ribosylation. Proc Natl Acad Sci USA. 1989;86:3499–503. doi: 10.1073/pnas.86.10.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wijeratne SS, Camporeale G, Zempleni J. K12-biotinylated histone H4 is enriched in telomeric repeats from human lung IMR-90 fibroblasts. J Nutr Biochem. 2010;21:310–16. doi: 10.1016/j.jnutbio.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]