Abstract
Autophagy has been established as a player in host defense against viruses. The mechanisms by which the host induces autophagy during infection are diverse. In the case of herpes simplex virus type 1 (HSV-1), dsRNA dependent protein kinase (PKR) is essential for induction of autophagy in fibroblasts through phosphorylation of eukaryotic initiation factor 2α (eIF2α). HSV-1 counteracts autophagy via ICP34.5, which dephosphorylates eIF2α and inhibits Beclin 1. Investigation of autophagy during HSV-1 infection has largely been conducted in permissive cells, but recent work suggests the existence of an eIF2α-independent autophagy-inducing pathway in non-permissive cells. To clarify and further characterize the existence of a novel autophagy-inducing pathway in non-permissive cells, we examined different HSV and cellular components in murine myeloid cells for their role in autophagy. We demonstrate that HSV-1-induced autophagy does not correlate with phosphorylation of eIF2α, is independent of functional PKR, and is not antagonized by ICP34.5. Autophagy was activated independent of viral gene expression but required viral entry. Importantly, we found that the presence of genomic DNA in the virion was essential for induction of autophagy and, conversely, that transfection of HSV-derived DNA induced LC3 II formation, a marker of autophagy. This occurred through a mechanism dependent on STING, an essential component for the IFN response to intracellular DNA. Finally we observed that HSV-1 DNA was present in the cytosol devoid of capsid material following HSV-1 infection of DCs. Thus, our data suggest that HSV-1 genomic DNA induces autophagy in non-permissive cells in a STING dependent manner.
Introduction
Macroautophagy (hereafter termed “autophagy”) is a highly conserved vacuolar, degradation and recycling pathway. Autophagy involves formation of so-called autophagosomes, in which a double membrane (the isolation membrane) encircles intracellular components, followed by degradation of the sequestered material by fusion with lysosomes. Autophagy has long been known to play important roles in e.g. cell death, starvation, and cellular development, but is now also appreciated to be induced during infections and to be important for host-defense against pathogens (1–3). Autophagy has traditionally been viewed as a non-specific degradation mechanism, but in recent years it has become clear that the process, at least in some cases, is selective, e.g. in the targeting of invading viruses and bacteria (4–6).
In the last few years, autophagy has been linked to both the innate and the adaptive immune response. Three of the main antiviral pathways are; (i) simple engulfment of the virion, resulting in degradation, thus limiting viral accumulation (4,7). (ii) Connection to the adaptive immune response by translocation of endogenous antigens from the cytosol to the major histocompatibility complex (MHC) class I and class II, thereby leading to activation of T cells (8–11). (iii) Promotion of the proinflammatory response, by engulfment and delivery of viral components to endosomal toll-like receptors (TLRs), resulting in e.g. type I IFN induction (12). In addition to classical autophagy, individual autophagy related genes (ATGs) have also been demonstrated to regulate the innate immune response. The ATG5–ATG12 conjugate has been found to directly interact with the cytosolic RNA sensor retinoic-acid inducible gene I and its adaptor molecule mitochondrial anti-viral signaling protein (MAVS), resulting in decreased type I IFN induction (13). Also ATG9a, but not ATG7, is involved in negative regulation of stimulator of IFN gene (STING), a transmembrane protein essential for type I IFN and pro-inflammatory cytokine induction mediated by cytosolic DNA receptors of which several have been linked to HSV recognition (14,15).
Conversely, many viruses, primarily of the Herpesviridae family, have evolved evasion strategies to suppress autophagic defense, such as targeting the autophagy protein Beclin-1 (11,16,17). The importance of autophagy is also illustrated by the observation that autophagy-deficient mice infected with HSV-2 and Sindbis virus-, and Drosophila infected with vesicular stomatitis virus (VSV) exhibit increased lethality (4,10,18).
The alpha-herpesvirus HSV-1 is a ubiquitous human dsDNA virus replicating in epithelial cells and establishing lifelong latency in sensory neurons. HSV-1 is known to first induce and subsequently block autophagy in murine fibroblast and neurons (7,19). It has been demonstrated that dsRNA dependent protein kinase (PKR), via phosphorylation of the alpha subunit of eukaryotic initiation factor 2 (eIF2α), is essential for HSV-1-activated autophagy (19,20). The HSV-1 neurovirulence protein ICP34.5 blocks autophagy by recruiting the host phosphatase PP1α to dephosphorylate eIF2α, and by inhibiting Beclin-1 (16,21). An HSV-1 mutant lacking ICP34.5 is unable to block autophagy stimulation and autophagic degradation of virions in permissive cells, and is less neurovirulent in mice (7,16,19). The late HSV-1 protein Us11 directly binds PKR or PACT, an activator of PKR, thus antagonizing the PKR/eIF2α pathway (22,23). Unlike what has been observed in fibroblasts and neurons, English et al. have recently shown that wild type HSV-1 is able to induce autophagy in macrophages in an apparently eIF2α-independent manner (9). Hence, there are data to suggest the existence of an alternative HSV-1 autophagy-stimulating pathway in non-permissive myeloid cells.
Here we have used bone-marrow derived dendritic cells (BM-DCs) and the macrophage-like cell line RAW267.4 to demonstrate that HSV-1 induces autophagy, as measured by microtubule-associated protein 1 light chain 3 (LC3) foci and LC3 II accumulation, in myeloid cells in a PKR/eIF2α-independent manner. Instead we identify a novel mechanism in which cytosolic viral DNA triggers autophagy in a mechanism dependent on STING. Furthermore, we demonstrate that blockage of autophagy by both a chemical and genetic approach reduces IFN-β and TNF-α induction during HSV-1 infection. Thus, autophagy can now be included in the list of cellular innate immunological processes activated by cytosolic DNA in a STING dependent mechanism.
Materials and Methods
Reagents
DMEM and RPMI 1640 (both from BioWhittaker) were supplemented with antibiotics (penicillin and streptomycin) and LPS-free FCS (BioWhittaker). GM-CSF was purchased from R&D Systems. Pam3Csk4, and ODN1826 were obtained from InvivoGen. LNAC and DPI were from CalBiochem. 3-methyladenine (3-MA), DAPI and Poly L-Lysine were from Sigma-Aldrich. Lipofectamine 2000, ProLong® Gold, G418, and rabbit Alexa Fluor 568- and anti-mouse Alexa Flour 647-labeled secondary antibodies were from Invitrogen. Antibodies used were mouse anti-VP5 (3B6, Virusys) and rabbit anti-LC3 (PM036, MBL), anti-eIF2α (phosphor-Ser52) (KAP-CP131-E), anti-Beclin 1 (3738, Cell Signaling), and anti-GAPDH (FL-335, Santa Cruz Biotechnology).
Mice
Inbred, specific pathogen-free mice used in this study were 8–12-wk-old female C57BL/6J, TLR2/9−/−, MAVS−/−, conditionally hematological Atg7 knockout Vav-iCre; Atg7Flox/Flox and Vav-iCre−; Atg7Flox/Flox control mice (24), and a mutant mouse strain, Goldenticket (Gt), that harbors a point mutation in STING (STINGGt/Gt) which fails to produce detectable protein (25). All knockout mice strains were on a C57BL/6J background. C57BL/6J, and TLR2/9−/− mice were bred at Taconic M&B. Bones from MAVS−/− mice were kindly provided by Z. J. Chen, University of Southwestern Medical Centre, Dallas, Texas.
Cells
BM-DCs were obtained as described elsewhere (26). Bone marrow-derived macrophages (BMMs) were obtained as BM-DCs, except 10 ng/ml GM-CSF was used for differentiation, and adherent cells were harvest. For isolation of whole cell extracts and total RNA BM-DCs and BMMs were seeded at a density of 2 × 106 cells/well in a 12-well tissue culture plate, and for confocal microscopy BM-DCs were seeded on coverslips in a 24-well tissue culture plate at a cell concentration of 8 × 105 cells/well. To improve cell adherence, coverslips were left in Poly L-Lysine in PBS (0,1 mg/ml) for 1 hour at 37°C before use. After seeding the cells were left overnight at 37°C and 5% CO2 to allow cells to settle prior to treatment. RAW 267.4 cells stably transfected with either the empty vector pBK-CMV or PKR-M7, expressing PKR mutant M7, which lacks the first dsRNA-binding domain, in the pBK-CMV vector (both generously provided by J. A. Corbett, St. Louis University School of Medicine, Missouri, USA) (27) were grown in DMEM with 10% FCS, supplemented with penicillin (200 IU/ml) and streptomycin (200 µg/ml), and for selection G418 (400 µg/ml). For isolation of whole cell extracts RAW267.4-pBK-CMV and RAW267.4-PKR-M7 were cultured at a cell concentration of 2.5 × 106 cells/well in a 6-well tissue culture plate and left overnight at 37°C and 5% CO2.
Viruses
The wild type (wt) viruses used in this study were the F and KOS strains of HSV-1 (where nothing else is mentioned, HSV-1 strain KOS has been used). The mutants ΔICP34.5 and ΔUL15ExII (ΔUL15) are on a F genetic background, whereas mutants lacking gB (ΔgB) and gH (ΔgH) are on a KOS genetic background. ΔICP34.5 was constructed by restoration of the tk gene in KY0234 (28). Other viruses used, were Pseudorabies virus (PrV) strain Kaplan (29), and VSV Indiana strain. Inactivated HSV-1 was obtained by UV treatment for 3 min as previously described (30). In all experiments the control was either mock or untreated. A marginal induction of LC3 II formation was observed in mock-treated cells as compared to untreated cells. Light (L)-particles were isolated from HSV-1 ARΔUL36 infected cells essentially as described elsewhere (31).
DNA oligonucleotide transfection
Transfection of BM-DCs with HSV-1 60mer oligonucleotide (sequence: 5’-TAAGACACGATGCGATAAAATCTGTTTGTAAAATTTATTAAGGGTACAAATTGCCCTAGC-3’, from DNA Technology) was achieved using Lipofectamine 2000 at 2 µg/ml, as recommended by the manufacturer.
Fluorescent in Situ hybridization
Fluorescent in Situ hybridization (FISH) with FITC-labelled oligonucleotide probe (1 ng/µl) and stained with anti-VP5 (1:200 dilution) was carried out as described elsewhere (32).
Confocal microscopy
Cells grown on glass coverslips were fixed for 15 min in 4% paraformaldehyde in PBS and were made permeable in 0.1% Triton X-100 for 90 seconds. Coverslips were preincubated in PBS with 1% BSA for 15–30 min, stained for 1 h at room temperature with primary antibodies (1:200 dilution) and for 1h with Alexa Fluor 568- and Alexa Flour 647-labeled secondary antibodies (1:500 dilution) in PBS with 1% BSA. Next, coverslips were incubated with DAPI (5 mg/ml) for 2 min and mounted in ProLong Gold. Images were obtained with a Zeiss LSM 710 scanning confocal microscope.
Autophagy Assays
Evaluation of autophagy by fluorescence microscopy was performed by immunofluorescence of LC3, a marker of autophagy, using LC3 antibody to detect LC3 foci. Quantitation of autophagy was done based on the percentage of cells with more than two LC3 dots, thereby excluding the low background level of LC3 foci most cells exhibit. A minimum of 100 cells per sample were counted. In evaluating autophagy by western blotting, levels of lipidated LC3 II, the only reliable protein marker for completed autophagosomes, were determined (33). LC3 I (not always visible) and LC3 II, were segregated by 4 to 20% gradient SDS-PAGE. For identification of LC3 II, whole cell extracts from wt mouse embryo fibroblasts (MEFs) and Atg5−/− MEFs, the latter incapable of forming LC3 II, were loaded in wells next to samples in all experiments in this work. For quantification of LC3 II, computer-assisted densitometry (ImageJ) was used to quantify the bands that were captured on a Luminescent Image Analyzer (FUJI-FILM), and after normalization to GAPDH, the results were depicted as change compared to control.
Western blotting
Nuclear extracts were denatured in XT Sample Buffer and XT Reducing Agent (Bio-Rad), heated to 90°C for 10 min, and subjected to SDS PAGE (Bio-Rad). The proteins were blotted onto a PVDF membrane and blocked for 30 min with TBS (10 mM Tris, 140 mM NaCl) supplemented with 0.05% Tween 20 and 5% skim milk powder. Primary antibodies were added for overnight incubation at 4°C. The membrane was washed four times for 10 min each in washing buffer (TBS with 0.05% Tween 20), followed by incubation for 50 min at room temperature with secondary polyclonal horseradish peroxidase-conjugated antibody. The membrane was washed as described above, and the horseradish peroxidase-conjugated antibody (Dako) was visualized by using enhanced chemiluminescence.
Quantitative RT-PCR
Total RNA was extracted with the High Pure RNA isolation kit (Roche Applied Science) according to the recommendations of the manufacturer. Murine IFN-β mRNA was quantified by real-time PCR with Taqman one-step RT-PCR kit (Applied Biosystems). β-actin + TNF-α mRNA was quantified by Brilliant II SYBR Green QRT-PCR Mix Kit (STRATAGENE, primers; 5′-TAGCACCATGAAGATCAAGAT-3′ (forward) and 5′-CCGATCCACACAGAGTACTT-3′ (reverse). All protocols were performed according to manufacturers’recommendations.
Reproducibility of data
The results shown in this work are derived from data that are representative for the results obtained. For each series of experiments, two to four independent repetitions were performed.
Results
Autophagy is induced in BM-DCs during HSV-1 infection
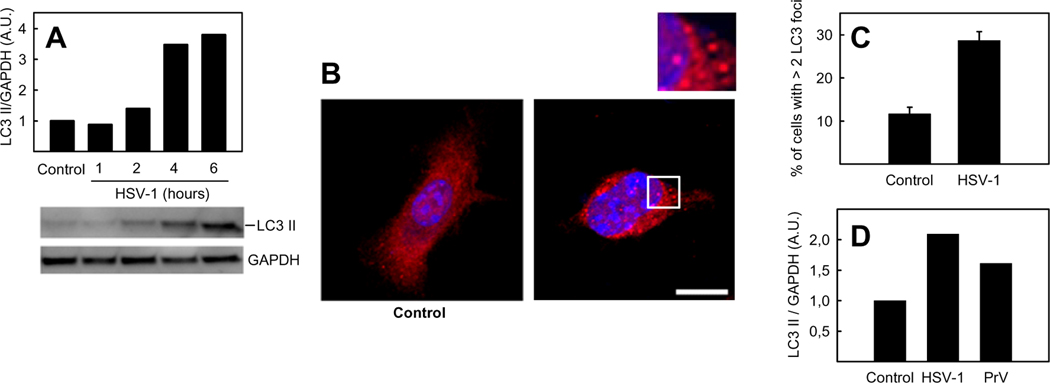
Recently, English and colleagues have shown that HSV-1 can induce autophagy in macrophages (9). To investigate whether autophagy can be observed in other myeloid cells, BM-DCs were examined for formation of the autophagy marker LC3 II following HSV-1 infection. BM-DCs were infected with HSV-1 for 1, 2, 4, and 6 h and LC3 II accumulation was analyzed by western blotting. For identification of LC3 II, whole cell extracts from wt MEFs and Atg5−/− MEFs, the latter incapable of forming LC3 II, were loaded in wells next to samples in all experiments in this work (not shown). We observed an increase in LC3 II levels starting from 2–4 hours post infection (Fig. 1A). To establish further the ability of HSV-1 to induce autophagy in BM-DCs, infected cells were stained with anti-LC3 antibody and foci indicating autophagosomes were visualized by confocal micoscopy (Fig 1B+C). Fig 1A–C show that LC3 II formation and the number of LC3 foci increased in response to HSV-1 in BM-DCs. To investigate whether the stimulation of autophagy in BM-DCs is specific for HSV-1, we also infected BM-DCs with another alpha-herpesvirus, pseudorabies virus (PrV). As seen in Fig 1D, PrV also activated autophagy in BM-DCs as measured by formation of LC3 II. These data show that HSV-1 and PrV infection stimulate autophagy in BM-DCs.
FIGURE 1.
Induction of autophagy in BM-DCs during HSV-1 infection. A, D, BM-DCs were infected with HSV-1 or PrV (both with a multiplicity of infection (MOI) of 3). Whole-cell extracts were harvested at 6 hours or at indicated time points and LC3 II and GAPDH protein levels were measured by western blotting. LC3 II was normalized to GAPDH and shown as bar charts. Western blots representing panel A are shown. B, Confocal microscopy of BM-DCs grown on coverslips and infected with HSV-1 (3 MOI) for 6 hours, then fixed and stained with antibody to LC3 (red) and with DAPI (blue) for visualization of nuclei. Inset, magnification of area outlined in main image. Images are a maximum projection of a z-stack. Scale bar, 10 µm. C, Percentage of BM-DCs stained with anti-LC3 with more than 2 LC3 dots, counted by confocal microscopy. The data shown represent mean ± S.D. from three independent experiments. A.U., arbitrary unit.
HSV-1-induced autophagy does not correlate with increased phosphorylation of eIF2α and is independent of ICP34.5 and PKR
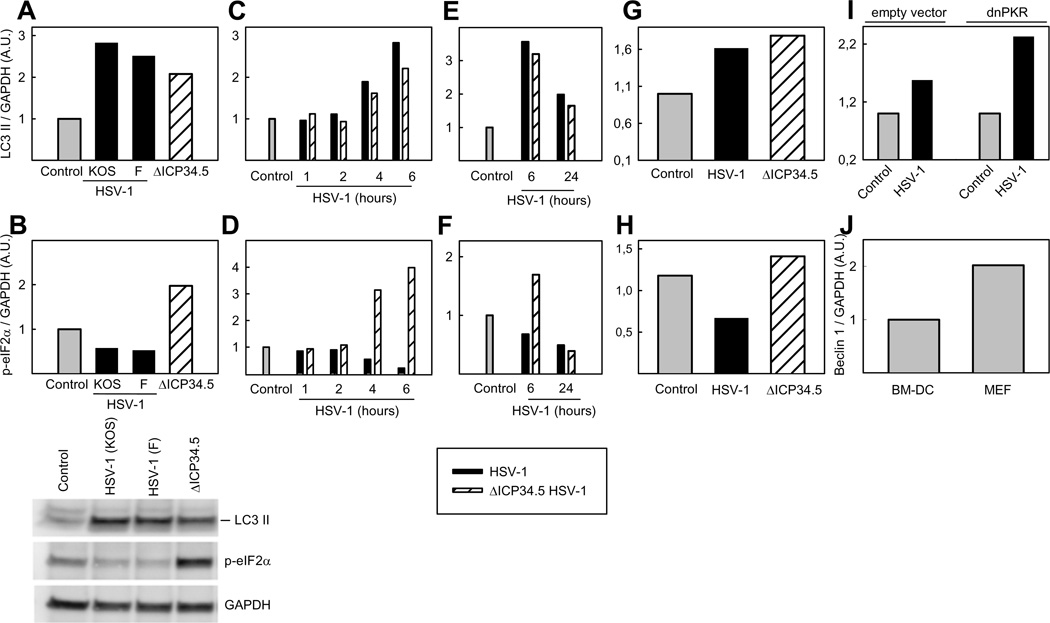
To establish whether HSV-1-activated autophagy in BM-DCs is a result of increased phosphorylation of eIF2α, and thus affected by ICP34.5, p-eIF2α levels in whole cell extracts, from cells infected with HSV-1 ICP34.5 mutant (ΔICP34.5) or two wt strains of HSV-1, KOS and F, were determined by western blotting. We observed an increase in LC3 II formation following infection with all viruses, but an increase in p-eIF2α levels were determined only following HSV-1 ΔICP34.5 infection, whereas p-eIF2α levels reduced following infection with the two wt strains of HSV-1 (Fig 2A and 2B). This reduction and increase of p-eIF2α in BM-DCs infected with wt HSV-1 and HSV-1 ΔICP34.5, respectively, started 2–4 h post infection (Fig 2C–D). Interestingly, HSV-1 ΔICP34.5-induced LC3 II formation did not reach a level higher than that observed in cells infected with wt viruses expressing ICP34.5. As shown in Fig 2E–F, the levels of p-eIF2α in cells infected for 24 h with the ΔICP34.5 mutant was decreased to a level comparable to that in wt infected cells. In addition, at 24 h the amount of LC3 II in both wt- and mutant virus-infected BM-DCs had peaked and were lower than at 6 h. The macrophage-like cell line, RAW264.7, also exhibited LC3 II formation independently of p-eIF2α (Fig 2G and 2H). These data demonstrate there is no correlation between p-eIF2α and LC3 II level during HSV1 infection, thus suggesting that induction of autophagy takes place in a p-eIF2α- and possibly PKR-independent manner. To examine the role of PKR in HSV-1 mediated induction of autophagy in myeloid cells, a RAW264.7 cell line expressing dominant negative (dn)PKR was used (27). LC3 II formation was unaltered in dnPKR-expressing RAW264.7 cells as compared to cells stably transfected with an empty vector (Fig 2I), further indicating that HSV-1 stimulates autophagy in myeloid cells by a PKR/eIF2α independent pathway. Our observations may indicate cell-type specificity in activation of autophagy by HSV-1. For instance, in neurons ICP34.5 was found to inhibit Beclin 1 and thereby preventing autophagy (16). To test whether the levels of Beclin 1 are particularly high in myeloid cells, we compared the level of this protein in BM-DCs and MEFs. As shown in Fig 2J, the levels of Beclin 1 were lower in BM-DCs than in MEFs. Collectively, HSV-1 induces autophagy in myeloid cells independent of the phosphorylation status of eIFaα and PKR.
FIGURE 2.
The influence of viral ICP34.5, functional PKR, level of Beclin 1, and phosphorylation of eIF2α on induction of autophagy. BM-DCs (A–F), RAW264.7 cells (G–I) stably transfected with an empty plasmid (pBK-CMV) or dnPKR (PKR-M7) were infected with HSV-1 strain KOS (A–I), HSV-1 strain F (A and B), or ΔICP34.5 HSV-1 strain F (A, B, G and H). For all viruses an MOI of 3 was used. Whole-cell extracts were harvest at 6 hours or at indicated time points and LC3 II (A, C, E, G, and I), p-eIF2α (B, D, F, and H) and GAPDH (A–I) protein levels were measured by western blotting. LC3 II and p-eIF2α were normalized to GAPDH and shown as bar charts. Western blots representing panels A and B are shown. J, Whole-cell extracts were harvest from BM-DCs and MEFs, and Beclin 1 protein levels were measured by western blots and normalized to GAPDH and shown as bar charts. A.U., arbitrary unit.
HSV-induced autophagy is independent of TLRs and ROS
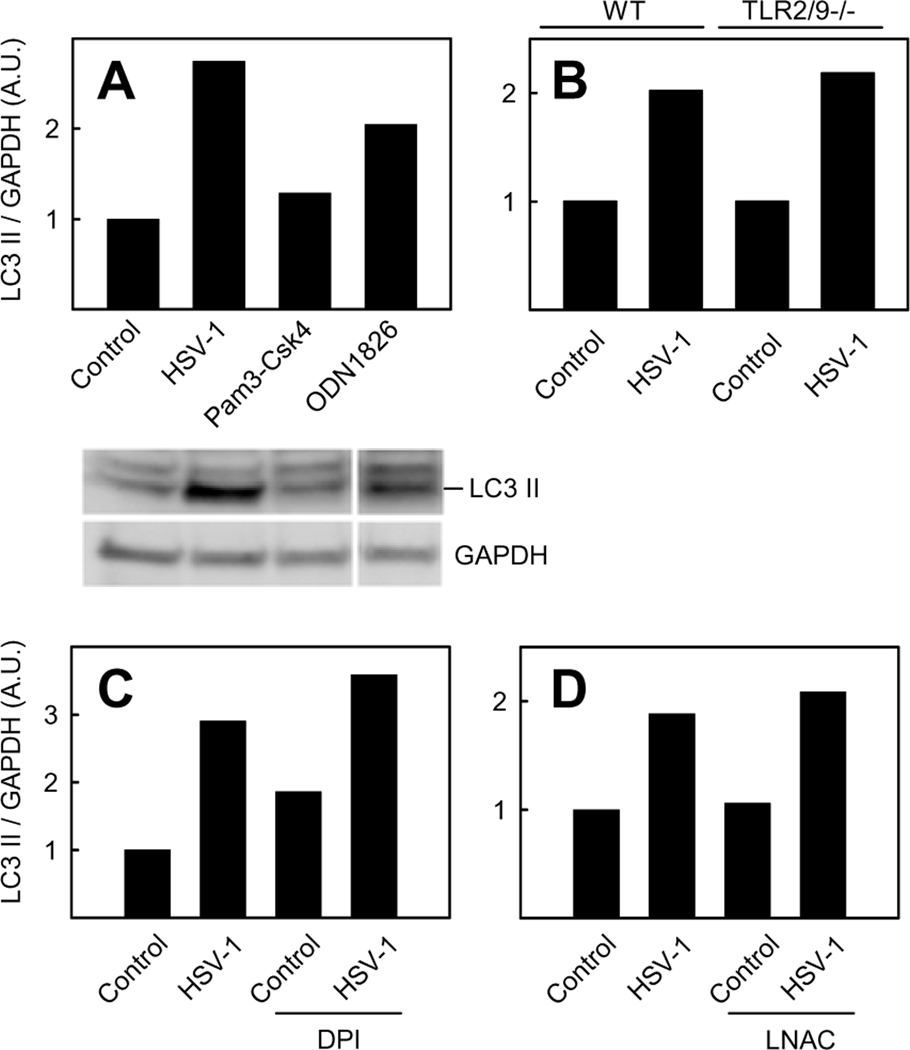
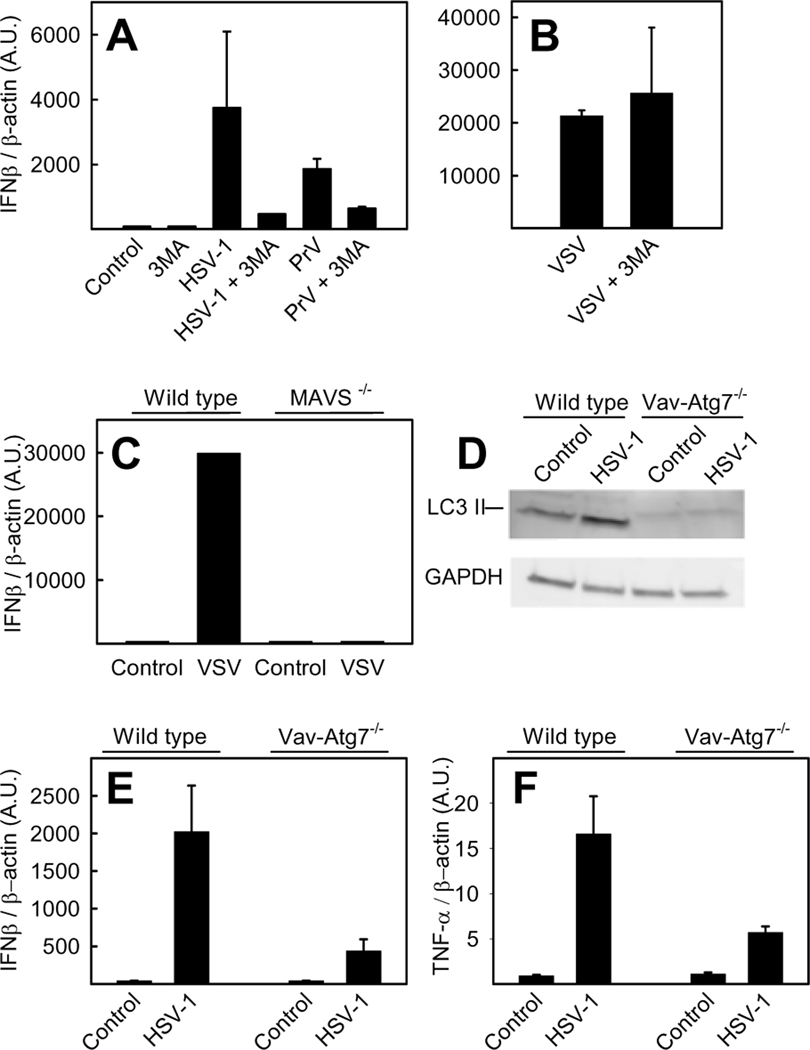
As observed, HSV-1-activated autophagy in BM-DCs appears to take place in a PKR/eIF2α independent manner, suggesting a role for one or more alternative receptors. One such receptor could be a member of the TLR family which previously has been shown to induce autophagy upon binding to PAMPs (34,35). TLR2 and TLR9 are two PRRs involved in the innate immune defense against HSV (36,37). To determine whether TLR2 or TLR9 can induce autophagy, BM-DCs were treated with Pam3Csk4 (TLR1/2 ligand) or ODN1826 (TLR9 ligand) and levels of LC3 II were determined. As seen in Fig 3A, a robust induction of LC3 II was observed only following treatment with ODN1826. However, although activation of TLR9 is capable of inducing autophagy in BM-DCs, this receptor seems not to be involved in HSV-1-stimulated autophagy, since LC3 II formation in TLR2/9−/− BM-DCs was comparable to what was observed in wt cells (Fig 3B).
FIGURE 3.
Role of TLR2/9 and ROS on HSV-1-induced autophagy. Wt BM-DCs (A, C, and D) and tlr2/9−/− BM-DCs (B) were infected with HSV-1 (3 MOI, 6 hours), treated with Pam3Csk4 (0.1 µg/ml) or ODN1826 (1 µM) for 1 hour (A), or pre-treated as described with DPI (3.7 µM) (C) or LNAC (6.4 mM) (D) prior to HSV-1 infection. Whole-cell extracts were harvested and LC3 II and GAPDH protein levels were measured by western blotting. LC3 II was normalized to GAPDH and shown as bar charts. Western blots representing panel A are shown. A.U., arbitrary unit.
Reactive oxygen species (ROS) are linked to induction of autophagy and are generated during HSV-1 infection (38,39). In order to evaluate whether ROS are involved in HSV-1-activated autophagy in myeloid cells, we treated BM-DCs with the antioxidants diphenyleneiodonium chloride (DPI) (Fig 3C) or N-acetyl-L-cysteine (LNAC) prior to HSV-1 infection and then analyzed LC3 II levels (Fig 4D). We have recently demonstrated that HSV induces ROS production in macrophages and this is abrogated by LNAC treatment (40). LC3 II formation was not influenced by pre-treatment with either DPI or LNAC as compared to cells that received vehicle as pre-treatment. Collectively, these data show that HSV-1-stimulated autophagy is independent of TLR2 and TLR9 and proceeds through a mechanism independent or ROS.
FIGURE 4.
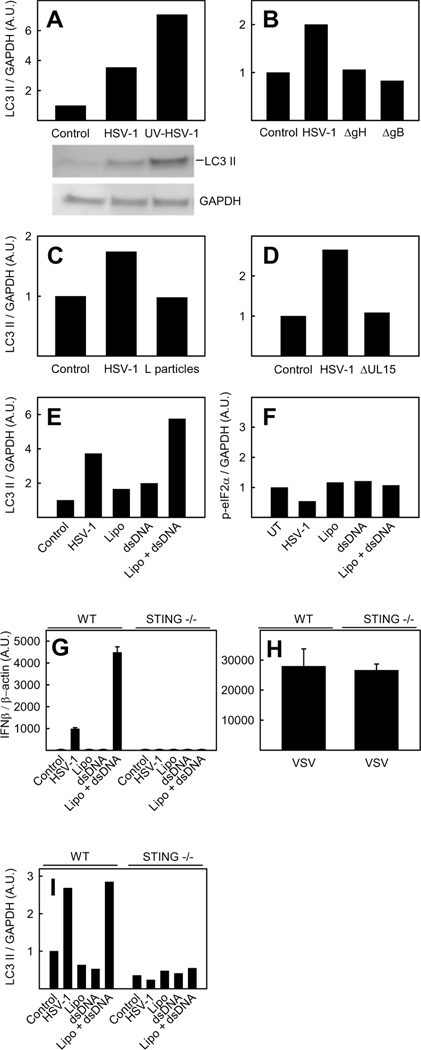
Role for viral replication, entry, viral genome, and STING in HSV-1-induced autophagy. Wt BM-DCs (A–I) and STING deficient BM-DCs (G–I) were infected with HSV-1 (A–G, and I), UV-inactivated HSV-1 (A), entry deficient ΔgB or ΔgH HSV-1 (B), HSV-1 L-particles (C), ΔUL15 HSV-1 (D), VSV (1.6 MOI) (H) or transfected with HSV 60mer DNA (E–G, and I). A MOI of 3 was used for infectious HSV-1 and for ΔgB and ΔgH HSV-1, HSV-1 L-particles, and ΔUL15 HSV-1 viral material corresponding to MOI 3 for the infectious virus was used. Whole-cell extracts were harvested 6 hours post treatment and LC3 II (A–E and I), eIF2α (F) and GAPDH (A–F, and I) protein levels were measured by western blotting or total RNA was extracted 6 h post infection (G–H) and expression of IFN-β mRNA was detected by RT-PCR and normalized to β-actin mRNA and shown as means of dublicates ± S.D. LC3 II (A–E) and p-eIF2α (F) were normalized to GAPDH and shown as bar charts. Western blots representing panel A are shown. A.U., arbitrary unit.
HSV-1-induced autophagy is independent of viral replication but dependent on viral entry, the presence of cytosolic viral DNA, and STING
To examine the existence of a potential novel autophagy-inducing pathway in non-permissive cells, we looked at the dependence on different viral components on HSV-1-induced autophagy in BM-DCs. First we examined the role of HSV-1 replication, which results in the production of double-stranded RNA (41). As seen in Fig 4A, UV-inactivated HSV-1 was able to induce a level of LC3 II even higher than seen in cells infected with untreated virus. Thus, replicative products are apparently not necessary for autophagy induction. Next, we investigated whether viral entry or binding to the cell membrane is necessary for HSV-1-stimulated autophagy. BM-DCs infected with the entry-incompetent glycoprotein (g)H- or gB-deficient HSV-1 (ΔgH and ΔgB), both capable of binding to cells, were unable to induce autophagy, thus demonstrating that viral entry is vital in activation of autophagy (Fig 4B) (42).
To determine if the autophagy-inducing factor of HSV-1 is located in the nucleocapsid and/or genome, we infected BM-DCs with HSV-1 L-particles, which are composed mainly of envelope and tegument proteins, lacking genome and nucleocapsid (43). L-particles were unable to stimulate LC3 II formation, indicating that the autophagy-inducing component is located in the nucleocapsid or the genome (Fig 4C). To further identify the viral component responsible for autophagy induction, we infected BM-DCs with the HSV-1 UL15 mutant (ΔUL15), which retains the nucleocapsid but largely fails to package viral DNA into the virion (44). Infection of BM-DCs with this virus mutant did not activate autophagy, indicating that the HSV-1 genome could be the autophagy-inducing component (Fig 4D). This was supported by the observation that transfection of a 60-base pair dsDNA oligonucleotide derived from the HSV-1 genome (HSV-1 60mer), activated LC3 II formation as seen in Fig 4E. Induction of autophagy by the 60mer oligonucleotide appears to take place in an eIF2α independent way, since we did not observe an increase in the phosphorylation of this subunit, supporting the existence of a novel PKR/eIF2α-independent alpha-herpesvirus-driven activation of autophagy (Fig 4F).
To examine whether STING played any role in HSV-1 DNA activated autophagy we first determined the dependency on this protein in the induction of IFN-β mRNA in STING deficient BM-DCs during HSV-1 infection or transfection of HSV-1 60mer DNA. As seen in Fig 4G, the induction of IFN-β was completely abrogated in cells lacking STING. Unlike HSV-1, the RNA virus VSV induced IFN-β mRNA in a completely STING-independent manner (Fig 4H). Next we looked at the ability of STING deficient BM-DCs to induce LC3 II formation. As seen in Fig 4I, LC3 II was not formed following either HSV-1 infection or HSV-1 60mer DNA transfection in cells lacking STING. Together these results demonstrate that HSV-1-stimulated autophagy in BM-DCs takes place in a replication-independent but entry- and viral genome-dependent manner. In addition, we found that an HSV-1 60mer transfected into the cytosol activates autophagy, and that activation of autophagy by either transfected DNA or HSV-1 takes place in a STING dependent mechanism. Cumulatively, these data support a role for viral DNA in the cytosol as a novel mediator of autophagy during HSV-1 infection, and also provides evidence for STING being essential for this process.
The HSV genome is released from the nucleocapsid to the cytosol during infection
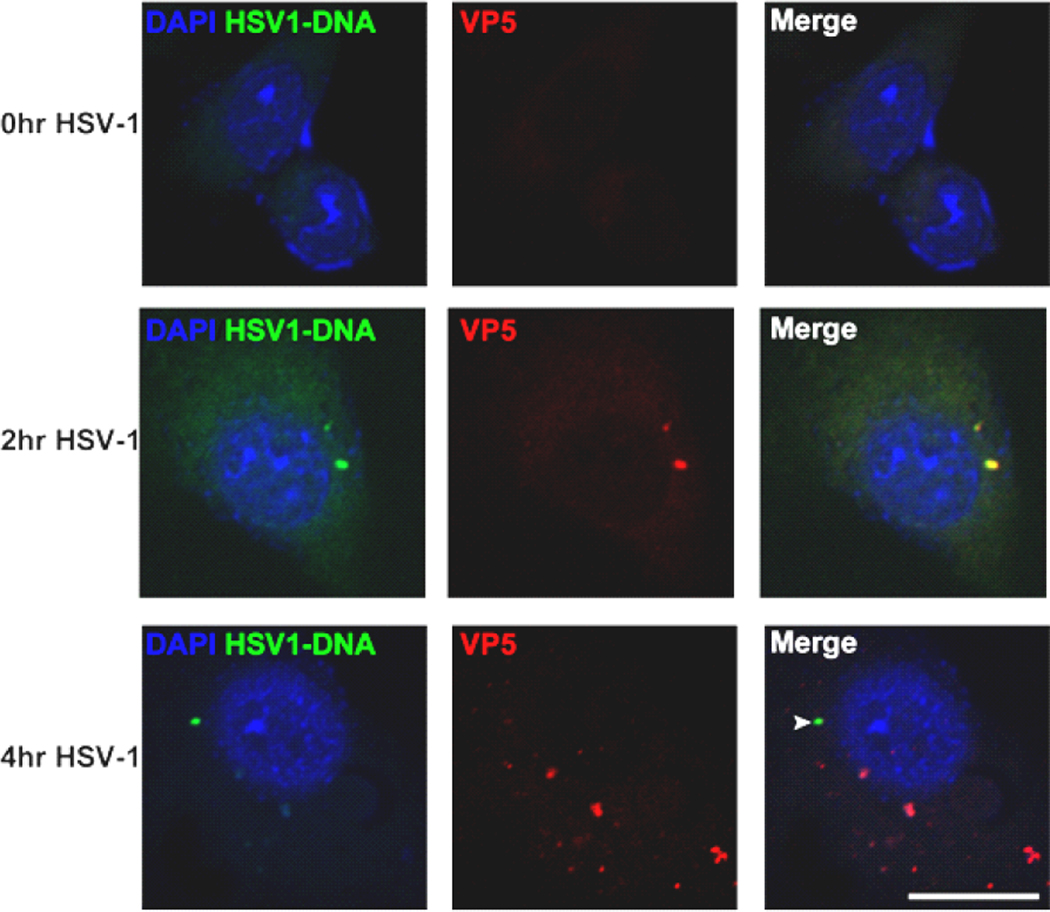
Recent studies have demonstrated the presence of HSV-1 DNA in the cytosol of macrophages (32). To examine whether HSV-1 DNA can also be observed in the cytoplasm of DCs, we used a specifically labeled oligonucleotide complementary to HSV-1 DNA. As seen in Fig 5, viral DNA and nucleocapsid protein VP5 co-localize at 2 h p.i, whereas at 4 h p.i. viral DNA is separated from VP5. This finding suggests that HSV-1 DNA is released from the viral capsid between 2 and 4 h p.i. and is present in the cytosol in an accessible form. Interestingly, there seems to be a temporal overlap between the release of HSV-1 genome and the onset of autophagy.
FIGURE 5.
Translocation of the HSV genome from nucleocapsid to cytosol during infection. Confocal microscopy of BM-DCs grown on coverslips and infected with HSV-1 (5 MOI). Cells were fixed at indicated time points and stained with antibody to viral capsid protein VP5 (red), DAPI (blue) for visualization of nuclei, and fluorescent in situ hybridization for HSV genome (green). Scale bar, 10 µm.
Inhibition of autophagy leads to decreased IFN-β induction during HSV-1 infection
Autophagy has been linked to detection of viral pathogen-associated molecular patterns (PAMPs) by PRRs resulting in type I IFN induction (12). Given the STING-dependent nature of the autophagy response, we wanted to investigate whether autophagy was essential for type I IFN production during infection with alpha-herpesviruses, BM-DCs were treated with 3-MA, a commonly used inhibitor of autophagosome formation, and IFN-β mRNA was monitored following infection. As seen in Fig 6A and B mRNA levels for IFN-β decreased in 3-MA-treated HSV-1- and PrV-, but not VSV-infected cells as compared to control cells. This difference in the ability of 3-MA to inhibit virus-induced IFN-β induction could be a consequence of different PRRs involved in the recognition of the investigated viruses. As was seen in Fig 4G and H, HSV-1 induces IFN-β in a STING dependent manner, whereas VSV drives this response through different pathways. VSV has been shown to be detected by a non-endosomal pathway involving MAVS, an adaptor protein in the cytosolic antiviral RIG-like receptor (RLR) pathway (45,46). This was supported by our observation in bone-marrow derived macrophages (BMMs), in which MAVS was essential for induction of IFN-β in response to VSV (Fig 6C). To confirm the data with 3-MA we used BM-DCs generated from Vav-iCre; Atg7Flox/Flox (hereafter called Vav-ATG7−/−) mice which do not express the essential autophagy gene ATG7 in the haematopoetic system were first examined for their ability to form LC3 II in response to HSV-1 infection. As seen in Fig 6D, basal and HSV-induced LC3 II formation was observed only in wt cells expressing ATG7 (Vav-iCre−; Atg7Flox/Flox). Next we examined the dependency on ATG7 for induction of IFN-β mRNA in response to HSV-1. Interestingly, BM-DCs deficient in ATG7 exhibited a reduced ability to express IFN-β compared to wt cells (Fig 6E). As seen in Fig 6F the induction of TNF-α mRNA was also reduced in cells lacking ATG7. Collectively, these results strongly suggest a role for autophagy (or autophagy-related proteins) in STING-dependent induction of IFN-β expression in response to viral DNA recognition.
FIGURE 6.
The role of autophagy in induction of IFN-β during HSV-1 infection. A, BM-DCs were pretreated for 20 min with 3-MA (10 mM) or left untreated and infected with HSV-1 (3 MOI), PrV (3 MOI), or VSV (1.6 MOI) (B). C, wt and MAVS−/− BMMs were infected with VSV (1.6 MOI) and wild type (Vav-iCre−; Atg7Flox/Flox) and Vav-ATG7−/− (Vav-iCre; Atg7Flox/Flox) BM-DCs were infected with HSV-1 (D–F). A–C and E–F, total RNA was extracted 6 h post infection. Expression of IFN-β and TNF-α mRNA was detected by RT-PCR and normalized to β-actin mRNA. The data are shown as means of dublicates ± S.D.. D, whole-cell extracts were harvested 6 hours post treatment and LC3 II and GAPDH (A–F, and I) protein levels were measured by western blotting. A.U., arbitrary unit.
Discussion
Previous studies have shown that HSV-1 induces autophagy in fibroblasts and neurons in a PKR/eIF2α dependent manner, and that this induction is suppressed by ICP34.5 mediating dephosphorylation of p-eIF2α and inhibition of Beclin 1 (7,16,19,21). Recently, this universal dependency on eIF2α in HSV-1-activated autophagy has been disputed by English and colleagues, showing that uninhibited autophagy is induced in a macrophage cell line followed HSV-1 infection in an apparently eIF2α-independent manner (9). This observation led us to investigate whether alpha-herpesviruses activate autophagy in non-permissive cells in an alternative way, which the virus is unable to block. In this study, we demonstrate that HSV-1 indeed does activate autophagy in myeloid cells, represented by BM-DCs and RAW264.7 cells, as measured by fluorescence microscopy of LC3 foci and western blot of LC3 II. We present data supporting that HSV-1-mediated stimulation of autophagy is independent of the classical PKR/eIF2α pathway but dependent on the presence of viral genome in the cytosol and STING, a vital component of the cytosolic DNA sensor system. Hence, our results indicate the existence of a novel autophagy-inducing mechanism mediated by cytosolic viral DNA.
Autophagosomes formed during HSV-1 infection of macrophages display two distinct morphologies. In addition to the conventional and well characterized double-membrane structure a second type of autophagosomes with four-layered membranes has been observed, originating from a coiling of the inner and outer nuclear membrane, but otherwise exhibiting normal macroautophagy “behavior” (9). In our study we did not observe LC3 foci in obvious proximity of the nuclear membrane, but this has to be definitively determined by electron microscopy, in which the number of membranes can be visualized.
In addition to HSV-1, PrV was also found to increase LC3 II formation, suggesting activation of autophagy in myeloid cells as a general phenomenon followed alpha-herpesvirus infection. The observed HSV-1 mediated autophagy induction was independent of eIF2α and ICP34.5 since both wt and ΔICP34.5 HSV-1 led to equal levels of LC3 II, starting at 2–4 h p.i., in spite of eIF2α being dephosphorylated following infection with the wt virus and strongly phosphorylated following infection with the ICP34.5 mutant. Strengthening the hypothesis of eIF2α-independent induction of autophagy followed HSV-1 infection, we also observed the formation of LC3 II to be independent of PKR. This independence of PKR was confirmed by the efficiency of UV-inactivated HSV-1 to stimulate autophagy. UV-treatment obstructs replication of HSV-1, a process essential for PKR activation (47).
As previously outlined, ICP34.5 not only blocks autophagy by dephosphorylating eIF2α, but also by inhibiting Beclin 1 (16). Beclin-1 works both in the early steps of autophagy initiation, upstream of LC3 II formation, and downstream of LC3 II formation in the maturation of the autophagosome (48). Some viruses inhibit maturation of the autophagosomes, thereby avoiding degradation of the sequestered material (49). We do not believe our findings are due to virus-mediated inhibition of autophagosomal turnover, since the process, including the STING dependence, can be mimicked by synthetic DNA in the absence of viral material
Oxidative stress caused by ROS is linked to both HSV-1 infection and induction of autophagy (38,39). Moreover, ROS has recently been demonstrated to play important roles in signaling downstream of RNA- and DNA-sensing PRRs (40,50). We inhibited ROS by DPI, an inhibitor of flavin-containing proteins essential in many ROS generating pathways, and the general antioxidant LNAC. Treatment with these inhibitors did not affect LC3 II formation, suggesting the ROS pathway not to be vital in DNA stimulated autophagy during HSV-1 infection in BM-DCs.
In this study aimed to identify the viral component(s) responsible for HSV-1-mediated autophagy. HSV-1 binding, especially of gD, has previously been found to activate intracellular signaling, and thus might be involved (51). Using entry-defective HSV-1 lacking gB or gH, we found that attachment of the virus in itself did not activate LC3 II formation. Our data also showed that HSV-1-induced autophagy is independent of viral replication, suggesting that de-novo-produced product or intermediate, such as dsRNA, is responsible for the process. Using the genome deficient L-particles and ΔUL15 HSV-1 we found that presence of viral genomic DNA is essential for induction of autophagy. The dependence on viral DNA was supported by the finding that transfection of synthetic dsDNA into DCs increased LC3 lipidation. Further supporting the notion that HSV-1 can induce autophagy in a PKR/eIF2α-independent manner, we observed no increase in p-eIF2α following DNA transfection. Importantly, using confocal microscopy we also observed HSV-1 DNA in the cytosol separated from the viral capsid protein VP5 at 4 h p.i. The process of viral DNA release seems to take place from 2 h p.i, since at this time point HSV-1 DNA and capsid were observed to co-localize. Interestingly, we were not able to observe LC3 II formation in STING−/− BM-DCs following either HSV-1 infection or transfection of dsDNA. HSV is known to be sensed by a range of cytosolic DNA receptors resulting in type I IFN induction (32,52,53,54,55). All cytosolic DNA sensors are believed to signal though STING since this protein is essential for cytoplasmic DNA activation of the IRF3 pathway, which is vital for the type I IFN response (56). In addition to type I IFN induction, cytosolic DNA also activates the inflammasome, resulting in IL-1β and IL-18 production (57–59). How STING is involved in autophagy still has to be elucidated, but given its function as a transmembrane protein involved in membrane dynamics during HSV-1 infection (56), it appears likely that STING is involved in integration of DNA sensing with membrane reorganization. Also, the involvement of STING in interaction between the autophagy and IFN pathways needs further clarification. In this respect it is interesting that in addition to STING, TBK1 is also involved in both autophagy and IFN responses (5). Other adaptor molecules of the PRRs, including myeloid differentiation factor 88 and toll/interleukin-1 receptor-domain-containing adaptor inducing IFN-β, involved in TLR signaling, have been demonstrated to positively regulate Beclin 1 and thereby promote autophagy (35).
With our present findings, and together with a recent publication by Preston and associates (60), autophagy can now be added to the list of cellular innate immunological processes activated by cytosolic DNA via a mechanism dependent on STING. The cellular DNA sensor(s) mediating autophagy remain(s) to be identified.
The data presented on a role for autophagy in the STING-dependent IFN response to alpha-herpesvirus infection in myeloid cells included a dual approach using both chemical inhibition and gene deletion. Treatment with 3-MA resulted in reduced IFN-β mRNA expression after infection with HSV-1 and PrV, but not VSV. In addition, ATG7-deficient DCs also exhibited reduced IFN-β and TNF-α expression during HSV-1 infection. These data, together with a previous report showing that HSV-1 induced IFN-α expression is reduced in plasmacytoid DCs unable to initiate autophagy(12), strongly indicate an essential role for autophagy or specific proteins of the autophagy machinery in activation of the cytokine response to HSV-1 infection. Since both 3-MA treatment and ATG7-deficiency results in reduced HSV-induced IFN-β expression, these authors favor the idea that the actual autophagocytic process is essential for STING-dependent IFN responses after DNA recognition. For instance, it is possible that DNA recognition leads to assembly of a signaling complex on autophagosomes, and that STING plays a role in the membrane dynamics required to bring this complex together at this location.
It has recently been reported by Preston and associates that transfection of human cytomegalovirus DNA into human foreskin fibroblasts induces autophagy (60). Using HSV-1 and PrV, we here demonstrate that alpha-herpesviruses also induces autophagy through their DNA, and further demonstrate that capsid-free viral genomic DNA is present in the cytoplasm after infection with a kinetics correlating with activation of autophagy. Importantly, we demonstrate that STING is essential for induction of autophagy by HSV. Together with the work by Preston and colleagues this report establishes cytoplasmic DNA as a potent inducer of autophagy during herpesvirus infections.
Acknowledments
The technical assistance of Kirsten Stadel Petersen is greatly appreciated. We thank Professor Soren Mogensen for critical reading of the manuscript.
Glossary
- PRR
pattern recognition receptor
- BM-DC
bone marrow-derived dendritic cell
- BMM
bone marrow-derived macrophage
- MEF
mouse embryonic fibroblast
- LC3
microtubule-associated protein 1 light chain 3
- PKR
dsRNA dependent protein kinase
- eIF2α
eukaryotic initiation factor 2α
- VSV
vesicular stomatitis virus
- PrV
Pseudorabies virus
- PAMP
pathogen associated molecular patterns
- ROS
reactive oxygen species
- DPI
diphenyleneiodonium chloride
- LNAC
N-acetyl-L-cysteine
- MOI
multiplicity of infection
- wt
wild type
Footnotes
This work was supported by research grants from The Danish Medical Research Council (grant no 09-072636), The Novo Nordisk Foundation, The Velux Foundation, The Lundbeck Foundation (grant # R17-A1526, and R34-3855), Elvira og Rasmus Riisforts almenvelgørende Fond, Aarhus University Research Foundation, Kathrine og Vigo Skovgaards Fond and NIH (AI092230 to B.H). S.B.R. is recipient of a PhD fellowship from the Faculty of Health Sciences, AU, DK, and K.A.H. was funded by a Marie Curie Incoming International Fellowship.
References
- 1.Hoyer-Hansen M, Jaattela M. Autophagy: an emerging target for cancer therapy. Autophagy. 2008;4:574–580. doi: 10.4161/auto.5921. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol.Rev. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orvedahl A, MacPherson S, Sumpter R, Jr, Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host.Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat.Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 6.Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J.Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 7.Talloczy Z, Virgin HW, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 8.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Munz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 9.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippe R, Desjardins M. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat.Immunol. 2009;10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, Mizushima N, Grinstein S, Iwasaki A. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–239. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leib DA, Alexander DE, Cox D, Yin J, Ferguson TA. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J.Virol. 2009;83:12164–12171. doi: 10.1128/JVI.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 13.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, Okuda K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc.Natl.Acad.Sci.U.S.A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, Akira S. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc.Natl.Acad.Sci.U.S.A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paludan SR, Bowie AG, Horan KA, Fitzgerald KA. Recognition of herpesviruses by the innate immune system. Nat.Rev.Immunol. 2011;11:143–154. doi: 10.1038/nri2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host.Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talloczy Z, Jiang W, Virgin HW, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc.Natl.Acad.Sci.U.S.A. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramaiah KV, Davies MV, Chen JJ, Kaufman RJ. Expression of mutant eukaryotic initiation factor 2 alpha subunit (eIF-2 alpha) reduces inhibition of guanine nucleotide exchange activity of eIF-2B mediated by eIF-2 alpha phosphorylation. Mol.Cell Biol. 1994;14:4546–4553. doi: 10.1128/mcb.14.7.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He B, Gross M, Roizman B. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc.Natl.Acad.Sci.U.S.A. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cassady KA, Gross M. The herpes simplex virus type 1 U(S)11 protein interacts with protein kinase R in infected cells and requires a 30-amino-acid sequence adjacent to a kinase substrate domain. J.Virol. 2002;76:2029–2035. doi: 10.1128/jvi.76.5.2029-2035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters GA, Khoo D, Mohr I, Sen GC. Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J.Virol. 2002;76:11054–11064. doi: 10.1128/JVI.76.21.11054-11064.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, Komatsu M, Simon AK. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc.Natl.Acad.Sci.U.S.A. 2010;107:832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect.Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikkelsen SS, Jensen SB, Chiliveru S, Melchjorsen J, Julkunen I, Gaestel M, Arthur JS, Flavell RA, Ghosh S, Paludan SR. RIG-I-mediated activation of p38 MAPK is essential for viral induction of interferon and activation of dendritic cells: dependence on TRAF2 and TAK1. J.Biol.Chem. 2009;284:10774–10782. doi: 10.1074/jbc.M807272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maggi LB, Jr, Heitmeier MR, Scheuner D, Kaufman RJ, Buller RM, Corbett JA. Potential role of PKR in double-stranded RNA-induced macrophage activation. EMBO J. 2000;19:3630–3638. doi: 10.1093/emboj/19.14.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng G, Yang K, He B. Dephosphorylation of eIF-2alpha mediated by the gamma(1)34.5 protein of herpes simplex virus type 1 is required for viral response to interferon but is not sufficient for efficient viral replication. J.Virol. 2003;77:10154–10161. doi: 10.1128/JVI.77.18.10154-10161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaplan AS, Vatter AE. A comparison of herpes simplex and pseudorabies viruses. Virology. 1959;7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 30.Malmgaard L, Melchjorsen J, Bowie AG, Mogensen SC, Paludan SR. Viral activation of macrophages through TLR-dependent and -independent pathways. J.Immunol. 2004;173:6890–6898. doi: 10.4049/jimmunol.173.11.6890. [DOI] [PubMed] [Google Scholar]
- 31.Roberts AP, Abaitua F, O'Hare P, McNab D, Rixon FJ, Pasdeloup D. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J.Virol. 2009;83:105–116. doi: 10.1128/JVI.01032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat.Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA, Jr, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van dK, Vaquero IEC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J.Biol.Chem. 2008;283:33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aravalli RN, Hu S, Rowen TN, Palmquist JM, Lokensgard JR. Cutting edge: TLR2-mediated proinflammatory cytokine and chemokine production by microglial cells in response to herpes simplex virus. J.Immunol. 2005;175:4189–4193. doi: 10.4049/jimmunol.175.7.4189. [DOI] [PubMed] [Google Scholar]
- 37.Krug A, Luker GD, Barchet W, Leib DA, Akira S, Colonna M. Herpes simplex virus type 1 activates murine natural interferon-producing cells through toll-like receptor 9. Blood. 2004;103:1433–1437. doi: 10.1182/blood-2003-08-2674. [DOI] [PubMed] [Google Scholar]
- 38.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kavouras JH, Prandovszky E, Valyi-Nagy K, Kovacs SK, Tiwari V, Kovacs M, Shukla D, Valyi-Nagy T. Herpes simplex virus type 1 infection induces oxidative stress and the release of bioactive lipid peroxidation by-products in mouse P19N neural cell cultures. J.Neurovirol. 2007;13:416–425. doi: 10.1080/13550280701460573. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez-Dosal R, Horan KA, Rahbek SH, Ichijo H, Chen ZJ, Mieyal JJ, Hartmann R, Paludan SR. HSV infection induces production of ROS, which potentiate signaling from pattern recognition receptors: role for S-glutathionylation of TRAF3 and 6. PLoS Pathog. 2011;7:e1002250. doi: 10.1371/journal.ppat.1002250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J.Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heldwein EE, Krummenacher C. Entry of herpesviruses into mammalian cells. Cell Mol.Life Sci. 2008;65:1653–1668. doi: 10.1007/s00018-008-7570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szilagyi JF, Cunningham C. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J.Gen.Virol. 1991;72(Pt 3):661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 44.Baines JD, Cunningham C, Nalwanga D, Davison A. The U(L)15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the U(L)15 gene product. J.Virol. 1997;71:2666–2673. doi: 10.1128/jvi.71.4.2666-2673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 46.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 47.Chou J, Roizman B. The gamma 1(34.5) gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programed cell death in neuronal cells. Proc.Natl.Acad.Sci.U.S.A. 1992;89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mehrpour M, Esclatine A, Beau I, Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Research. 2010;20:748–762. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- 49.Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soucy-Faulkner A, Mukawera E, Fink K, Martel A, Jouan L, Nzengue Y, Lamarre D, Vande VC, Grandvaux N. Requirement of NOX2 and reactive oxygen species for efficient RIG-I-mediated antiviral response through regulation of MAVS expression. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000930. e1000930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacLeod IJ, Minson T. Binding of herpes simplex virus type-1 virions leads to the induction of intracellular signalling in the absence of virus entry. PLoS One. 2010;5:e9560. doi: 10.1371/journal.pone.0009560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 53.Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J, Liu YJ. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc.Natl.Acad.Sci.U.S.A. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat.Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat.Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 59.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McFarlane S, Aitken J, Sutherland JS, Nicholl MJ, Preston VG, Preston CM. Early induction of autophagy in human fibroblasts after infection with human cytomegalovirus or herpes simplex virus type 1. J.Virol. 2011 doi: 10.1128/JVI.02435-10. [DOI] [PMC free article] [PubMed] [Google Scholar]